Abstract
Vibrio harveyi hemolysin, an important virulence determinant in fish pathogenesis, was further characterized, and the enzyme was identified as a phospholipase B by gas chromatography. Site-directed mutagenesis revealed that a specific residue, Ser153, was critical for its enzymatic activity and for its virulence in fish.
Vibrio harveyi is a serious pathogen for many marine vertebrates and invertebrates (3, 4, 8). Extracellular products and, in particular, hemolysin have been considered to be important for virulence (4, 23). Hemolysin is arguably the most widely distributed exotoxin among pathogenic vibrios, and there is much epidemiological and experimental evidence to suggest involvement in disease pathogenesis (8, 19, 24). Although generally referred to as hemolysins for their capacity to lyse erythrocytes, these toxins correspond to two different modes of action, involving either a pore-forming protein or a phospholipase enzyme (17). Hemolysis may result from the phospholipase enzyme activity demonstrated by some species of bacteria, including phospholipase C of Pseudomonas aeruginosa and phospholipase D in Photobacterium damselae subspecies damselae. However, most other hemolysins act as pore-forming proteins (8, 9, 15). Hemolysin with both of these activities has also been reported (14).
Zhang et al. (25) cloned and sequenced two closely related hemolysin genes from a highly pathogenic isolate, V. harveyi VIB 645. Both of the genes (designated vhhA and vhhB) had an open reading frame of 1,254 nucleotides, which is the same size as that of the tlh gene of Vibrio parahaemolyticus (13). Furthermore, nucleotide sequence alignment revealed that the identities of vhhA and vhhB to tlh were 77.5% and 77.2%, respectively (25). Thermolabile hemolysin (TLH) of V. parahaemolyticus, also named lecithin-dependent hemolysin (LDH), which had phospholipase A2/lysophospholipase activity (20), represents a major type of hemolysin in vibrios (24). In previous work, we obtained purified recombinant V. harveyi hemolysin (VHH) protein and showed that it was cytotoxic to flounder gill cells in tissue culture, had strong pathogenicity to flounder when injected intraperitoneally, and had phospholipase activity on egg yolk agar (26). In this study, we compared the characteristics and activities of VHH and the null-mutant protein of VHH obtained via site-directed mutagenesis. Also, the type of phospholipase activity of VHH was determined by gas chromatography (GC). Moreover, three-dimensional models were used to illustrate the properties of VHH. These studies were part of a fundamental effort to demonstrate the attack mechanism of the hemolysin on the eukaryotic cell. Another purpose was to obtain a nonvirulent protein usable as a component of a potential vaccine against V. harveyi.
The amino acid sequence of VHH (translated from the nucleotide sequence of the vhhA gene [NCBI accession number AF293430]) was submitted to a protein database (PROSITE; http://www.expasy.org/prosite) to identify the motifs and active site. The data obtained revealed that VHH might belong to a recently discovered lipase/esterase family, i.e., the GDSL protein family, on the basis of the presence of the conserved active Gly-Asp-Ser-Leu motif (1, 21). To confirm this possibility, the amino acid sequence of VHH was aligned with the sequences of known members of this enzyme family from different bacteria, including Escherichia coli, Pseudomonas putida, Aeromonas hydrophila, Vibrio mimicus, V. parahaemolyticus, and Xenorhabdus luminescens (1, 6, 7, 12, 16, 18, 20, 22). The results indicated that VHH should be considered a GDSL enzyme, based mainly on the sharing of identical invariant residues and similar conserved sequence motifs with the proteins mentioned above (data not shown). Thus, an important residue of the GDSL motif, i.e., Ser153 in VHH, was chosen to be replaced by a glycine, a neutral residue. The introduced glycine was selected because as a general consideration, a neutral residue could not have any impact on a catalytic reaction, regardless of its contribution to protein conformation.
A previously constructed expression vector, pET-24d(+), plus the complete open reading frame of the vhhA gene, i.e., pET-24d(+)-vhhA (26), was used as a template for site-directed PCR-based mutagenesis using a Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutagenesis primers were designed to change AGC (encoding a serine residue) to GGC (encoding a glycine residue). The synthetic oligonucleotides which served as primers were as follows: 5′-GCA TTG GGT GAC GGC TTG TCT GAT ACA GGC A-3′ (forward) and 5′-TGC CTG TAT CAG ACA AGC CGT CAC CCA ATG C-3′ (reverse) (the boldface letters indicate the changed nucleotides). The mutation was performed according to the manufacturer's specifications. The mutagenic vhhA gene in the pET-24d(+) vector from a single colony was sequenced, and the result suggested that the mutation was introduced into the expected site and that no second-site mutations were present (data not shown). Then, the plasmid of the same clone was transformed into E. coli BL21 (Novagen, Madison, WI) to obtain the mutant recombinant protein.
The mutant protein was overexpressed by isopropyl-β-d-thiogalactopyranoside (IPTG) induction and purified by metal chelating affinity chromatography, as previously described (26). The concentration of the purified protein was determined to be 424 μg/ml by the Bradford method (5).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis was performed on a Mini-PROTEAN 3 cell apparatus (Bio-Rad) using a 12% (wt/vol) polyacrylamide gel according to Laemmli (10). Approximately 1 μg of mutant VHH protein was subjected to the electrophoresis. The same dose of purified VHH protein obtained during a previous work was used as a control. The result indicated that the expressed recombinant protein had the same molecular weight as the native VHH protein and was purified (data not shown).
Western blotting was performed to check the immunoreactivity of the mutant VHH protein. Electrophoresis was performed as described above, and then the proteins were transferred to a polyvinylidene fluoride membrane (Amersham Biosciences, England) by use of a semidry transfer unit (Hoefer TE70; Amersham Biosciences). Then, the membrane was treated using a Western blotting kit (Boster, Wuhan, China), according to the manufacturer's instructions. An anti-VHH antibody was prepared as previously described, and the dilution for immunoblotting was 1:1,000 (26). The result revealed that the mutant VHH protein retained an immunoreactivity against antiserum similar to that of VHH, because the two types of proteins were subjected to electrophoresis at the same dose and their reactions with the antiserum showed similar levels of sensitivity by Western blotting (data not shown).
Fifty microliters of VHH (50 μg/ml) and 50 μl of mutant VHH protein (50 μg/ml) were added to Oxford cups, which were placed on LB agar supplemented with 1% (vol/vol) egg yolk (for phospholipase activity assay) or 3% (vol/vol) fish blood cells (for hemolytic activity assay). After incubation at 37°C overnight, the presence of an opalescent zone (for phospholipase assay) or a brown zone (for hemolytic assay) around the well was considered to be evidence of a positive reaction. The results were as follows. On the egg yolk emulsion plate, the purified VHH formed an inner cloudy zone and an outer distinct opalescent zone with a relative transparent ring between them. This suggested the potential multiple functions of VHH in hydrolyzing different substrates or at least different bonds of a single substrate, as well as the potential substrate preferences of this protein. These possibilities were supported to a certain extent by the results of the GC analysis. In contrast, the purified mutant protein at the same dose as the VHH showed no activity (Fig. 1a). On fish blood agar, the purified VHH formed a clear brown zone, suggesting strong hemolysis. However, the mutant protein at the same dose as the VHH showed absolutely no activity (Fig. 1b).
FIG. 1.
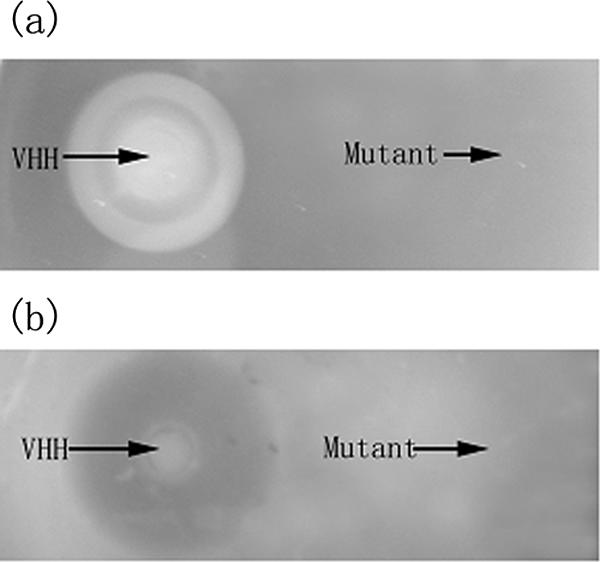
Results of phospholipase activity (a) and hemolytic activity (b) tests. Mutant VHH protein lost all the enzymatic activities.
According to our previous work (26), the 50% lethal dose of the purified VHH protein was 1.2 μg protein g−1 fish when a flounder was injected intraperitoneally. Moreover, purified VHH killed all experimental animals when the dose was 6.1 μg protein g−1 fish. Because the enzymatic assay indicated that the mutant VHH protein lost all its hemolytic activity, a higher dose, i.e., 8.0 μg protein g−1 fish, was used to investigate its virulence in fish. Thus, turbot (Scophthalmus maximus) with an average weight of 5.2 g from quarantined stocks that were recognized as disease-free (2) were used as models to assess pathogenicity as described below. A group of 10 turbot were infected by intraperitoneal injection with 0.1-ml volumes of undiluted mutant VHH protein preparations (amounting to ∼8.0 μg protein g−1 of fish). The positive controls were injected with the same dose of VHH, whereas the negative controls were injected with 0.1-ml volumes of Tris-buffered saline. The infected animals were maintained for up to a week in covered polypropylene tanks supplied with aerated seawater (50% of the volume was changed daily) at ∼20°C. Dead and moribund fish and the survivors at the end of the experiment were examined pathologically (2). The results were as follows. All of the fish in the positive control group injected with purified VHH died within 12 h, whereas the fish of the experimental group injected with purified mutant protein and the negative control group survived for at least a week and showed no disease symptoms. Most infected fish of the positive control group revealed hemorrhaging in the peritoneal cavity and darkening of the body (i.e., melanosis), which were also the major symptoms exhibited by the fish infected by V. harveyi (23). The results indicated that the mutant VHH protein lost its pathogenicity to fish. These results together with those of the Western blotting suggested that the mutant VHH protein might be considered as a component of a potential fish vaccine against V. harveyi, for it was nonvirulent to fish and had an immunoreactivity similar to that of the pathogenic VHH.
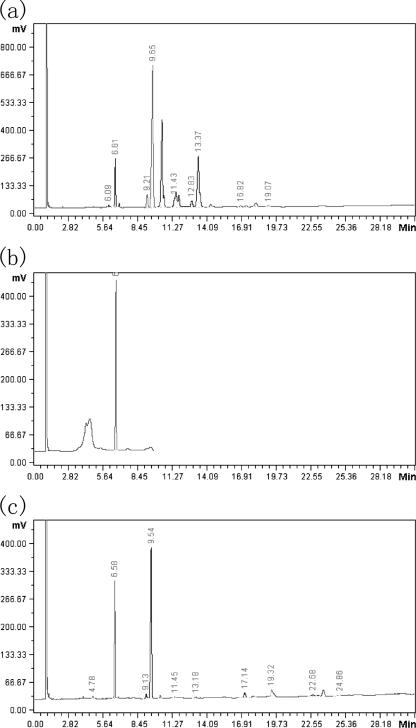
The phospholipase activity suggested that the VHH might be a multiple-function enzyme. To investigate this point, the type of phospholipase activity of VHH was identified by gas chromatography. In general, the type of phospholipase was determined according to the hydrolyzed bond of the lecithin substrate of the enzyme. The particular free fatty acid released by the enzyme would correspond to particular bond of the substrate. Thereby, the enzyme type could be determined by identification of the free fatty acids present in the enzyme-substrate reaction medium. Hence, a special compound, a phosphatidylcholine (lecithin) bearing two different fatty acid chains, l-α-phosphatidyl-choline β-palmitoyl γ-oleoyl (PCPO; Sigma), was used as the substrate for VHH in the present study (11). Pure oleic acid and palmitic acid served as controls. The substrate was dissolved in 10× reaction buffer (1.0 M Tris-HCl [pH 8.0], 1.0% [vol/vol] Triton X-100) to a final concentration of 5 mmol/liter. Then, 100 μl of the substrate suspension was incubated with 900 μl of enzyme solution at 37°C for 2 h. The free fatty acids present in the reaction medium were extracted with 200 μl of solvent (2:1, light petroleum-ether). After evaporation of the organic phase, the sample as well as pure oleic acid and palmitic acid was methylated with 3 ml of boron trifluoride (14% [wt/vol] in methanol) for 30 min. The methyl esters of the sample and controls were then subjected to GC analysis, which was performed using an HP-5890II apparatus (Hewlett Packard, Palo Alto, CA), equipped with a flame ionization detector. A capillary Carbowax 20 M column (0.25-mm inner diameter by 30 m; film thickness, 0.25 μm; Ohio Valley Speciality Co.) was used for GC with nitrogen as the carrier gas (1 ml/min). The column was operated isothermically at 190°C, whereas the injection port and detector were set at 250°C. The fatty acids released by VHH from the substrate were identified by comparison of the retention times between the peaks of the controls and the sample (Fig. 2). The results showed that both the oleic acid and the palmitic acid were released from PCPO by VHH, suggesting that this enzyme was a phospholipase B or a lysophospholipase.
FIG. 2.
GC analysis of the hydrolytic type of the phospholipase activity of VHH. (a) Control a. The highest peak corresponds to the control oleic acid methyl ester. (b) Control b. The highest peak corresponds to the control palmitic acid methyl ester. (c) Sample. According to the retention time of the peaks in panels a and b, the left peak corresponds to the released palmitic acid and the right one corresponds to the released oleic acid.
Upton and Buckley (21) first classified GDSL enzymes by the characteristic motif Gly-Asp-Ser-Leu situated near the N terminus. These enzymes have five consensus sequences (I to V) and four invariant important catalytic residues, namely Ser, Gly, Asn, and His in blocks I, II, III, and V, respectively, which led to a new designation of these enzymes as the SGNH-hydrolase family. The serine residue in block I, which was also located at the active GDSL motif, was usually considered to be an important member of the active site. Hence, to obtain a null-mutant protein of VHH by introducing one or more mutations, the corresponding residue of VHH, i.e., Ser153, was chosen for the first mutagenesis operation. However, it was interesting that a single residue change led to the loss of all of VHH's hemolytic and phospholipase activities and its pathogenicity to fish. To explain this, we turned to the steric structure of the active site and action mode of GDSL enzymes.
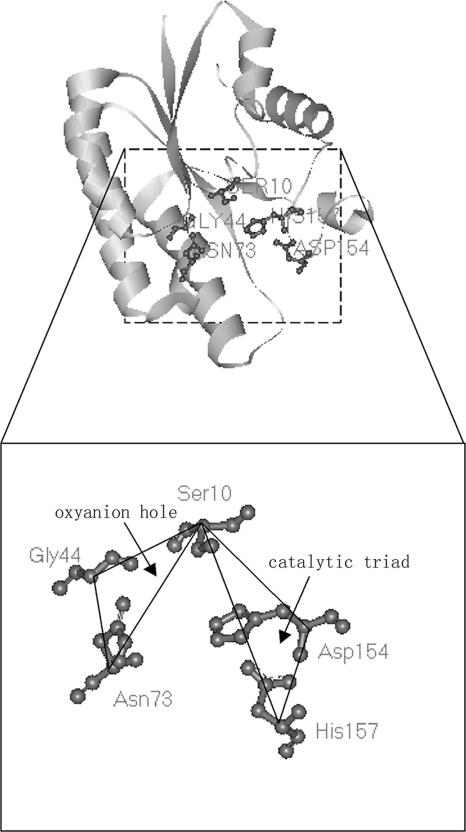
Based on (i) the description by Akoh et al. (1) of the hypothesis of the catalytic mechanism of SGNH-hydrolases which had substantial crystallographic evidence, (ii) the action mode of the catalytic triad in the serine proteases which shares a mechanism of action similar to that of the GDSL enzymes, and (iii) the relative positions of the conserved active residues of a representative GDSL enzyme, thioesterase I from E. coli, revealed by the three-dimensional model constructed with published coordinate data from the Protein Data Bank (Fig. 3), we concluded that the active site of GDSL enzymes consisted of two subgroups of residues, each containing three conserved members, irrespective of the residues of the substrate-binding site. One subgroup involved the Ser residue in block I, the Gly residue in block II, and Asn in block III. They constituted the so-called “oxyanion hole.” The other subgroup included the Ser in block I and the His in block V, as well as another conserved residue, Asp, also in block V and the third amino acid preceding His. These residue subgroups composed the catalytic triad, which had the same residues in the serine proteases. The members of each subgroup formed a triangle in the steric structure. Moreover, two triangles shared a common vertex, i.e., the active Ser residue in block I (Fig. 3). Each of the five residues described above played a key role in the function of the enzyme. Furthermore, it was proposed that the Ser in block I served as the nucleophile. The hydroxyl group was deprotonated by the His residue in block V to make the Ser more nucleophilic, which resulted in the formation of a tetrahedral oxyanion in a transition state. The protonated His was stabilized by the third member of the catalytic triad, the Asp in block V. As to the oxyanion hole, the amide protons of the Ser in block I and the Gly in block II, together with the Hδ of the Asn in block III, served as the proton donors of the oxyanion hole that stabilized the tetrahedral intermediate.
FIG. 3.
Key residues of the active site in GDSL enzymes. A three-dimensional model of a representative GDSL enzyme, thioesterase I from E. coli, was generated with structural information available in the Protein Data Bank to illustrate the basic residues of the catalytic triad and oxyanion hole in an SGNH hydrolase. Ser10, Gly44, Asn73, Asp154, and His157 in this protein corresponded to the residues Ser, Gly, Asn, and His in blocks I, II, III, and V of a general GDSL enzyme, respectively. The alpha carbon atoms of these residues are linked with dark lines to show their relative positions. The figure was drawn using a WebLab Viewer (Accelrys, Cambridge, United Kingdom).
On the basis of the active-site features and action mode of the GDSL enzymes, we found that the active serine of the GDSL motif is the center residue of the active site and acts as the main “performer” in the catalytic reaction, with the assistance of other key residues. Hence, it was understandable that the replacement of Ser153 in VHH resulted in the loss of all the functions examined.
In conclusion, V. harveyi hemolysin was demonstrated to be a lysophospholipase or a phospholipase B. A particular single base change introduced by the substitution of a glycine for Ser153 resulted in the loss of the hemolytic and phospholipase activities of VHH as well as its pathogenicity to fish. Since the mutant protein was purified and proved to have an immunoreactivity similar to that of VHH, this mutant protein might be considered as a component of a potential vaccine against V. harveyi. This study of TLH-like hemolysin is the first to provide substantial experimental data to support the hypothesis on the mechanism of action of GDSL enzymes. Moreover, the fact that disruption of the active site of phospholipase activity of VHH resulted in the loss of hemolytic activity and pathogenicity to fish suggests that the phospholipase activity of VHH probably plays a major role in hemolysis.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (30371119), the program New Century Excellent Talents in University (NCET-04-0645), and a grant from the Major State Basic Research Development Program of China (973 Program; grant 2006CB101803)
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Akoh, C. C., G.-C. Lee, Y.-C. Liaw, T.-H. Huang, and J.-F. Shaw. 2004. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43:534-552. [DOI] [PubMed] [Google Scholar]
- 2.Austin, B., and D. A. Austin. 1989. Methods for the microbiological examination of fish and shellfish. Ellis Horwood, Chichester, United Kingdom.
- 3.Austin, B., and D. A. Austin. 1999. Bacterial pathogens: disease of farmed and wild fish. Springer-Praxis, Godalming, United Kingdom.
- 4.Austin, B., and X.-H. Zhang. 2006. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43:119-124. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cho, H., and J. E. Cronan, Jr. 1993. Escherichia coli thioesterase I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J. Biol. Chem. 268:9238-9245. [PubMed] [Google Scholar]
- 7.Essar, D. W., L. Eberly, and I. P. Crawford. 1990. Evolutionary differences in chromosomal locations of four early genes of the tryptophan pathway in fluorescent pseudomonads: DNA sequences and characterization of Pseudomonas putida trpE and trpGDC. J. Bacteriol. 172:867-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida, T., and T. Honda. 1997. Hemolysins produced by vibrios. J. Toxicol. Toxin Rev. 16:215-227. [Google Scholar]
- 9.Kreger, A. S., A. W. Bernheimer, L. A. Etkin, and L. W. Daniel. 1987. Phospholipase D activity of Vibrio damsela cytolysin and its interaction with sheep erythrocytes. Infect. Immun. 55:3209-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lee, J.-H., S.-H. Ahn, S.-H. Kim, Y.-H. Choi, K.-J. Park, and I.-S. Kong. 2002. Characterization of Vibrio mimicus phospholipase A (PhlA) and cytotoxicity on fish cell. Biochem. Biophys. Res. Commun. 298:269-276. [DOI] [PubMed] [Google Scholar]
- 12.Lo, Y.-C., S.-C. Lin, J.-F. Shaw, and Y.-C. Liaw. 2003. Crystal structure of Escherichia coli thioesterase I/protease I/lysophospholipase L1: consensus sequence blocks constitute the catalytic center of SGNH-hydrolases through a conserved hydrogen bond network. J. Mol. Biol. 330:539-551. [DOI] [PubMed] [Google Scholar]
- 13.Nishibuchi, M., and J. B. Kaper. 1985. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 162:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal, S., A. Datta, G. B. Nair, and B. Guhathakurta. 1998. Use of monoclonal antibodies to identify phospholipase C as the enterotoxic factor of the bifunctional hemolysin-phospholipase C molecule of Vibrio cholerae O139. Infect. Immun. 66:3974-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker, M. W., and S. C. Feil. 2005. Pore-forming protein toxins: from structure to function. Prog. Mol. Biol. Biophys. 88:91-142. [DOI] [PubMed] [Google Scholar]
- 16.Robertson, D. L., S. Hilton, K. R. Wong, A. Koepke, and J. T. Buckley. 1994. Influence of active site and tyrosine modification on the secretion and activity of the Aeromonas hydrophila lipase/acyltransferase. J. Biol. Chem. 269:2146-2150. [PubMed] [Google Scholar]
- 17.Rowe, G. E., and R. A. Welch. 1994. Assays of haemolytic toxins. Methods Enzymol. 235:657-669. [DOI] [PubMed] [Google Scholar]
- 18.Shaw, J.-F., R.-C. Chang, K.-H. Chuang, Y.-T. Yen, Y.-J. Wang, and F.-G. Wang. 1994. Nucleotide sequence of a novel arylesterase gene from Vibro mimicus and characterization of the enzyme expressed in Escherichia coli. Biochem. J. 298:675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinoda, S. 1999. Protein toxins produced by pathogenic vibrios. J. Nat. Toxins 8:259-269. [PubMed] [Google Scholar]
- 20.Shinoda, S., H. Matsuoka, T. Tsuchie, S. Miyoshi, S. Yamamoto, H.Taniguchi, and Y. Mizuguchi. 1991. Purification and characterization of a lecithin-dependent haemolysin from Escherichia coli transformed by a Vibrio parahaemolyticus gene. J. Gen. Microbiol. 137:2705-2711. [DOI] [PubMed] [Google Scholar]
- 21.Upton, C., and J. T. Buckley. 1995. A new family of lipolytic enzymes? Trends Biochem. Sci. 20:178-179. [DOI] [PubMed] [Google Scholar]
- 22.Wang, H., and B. C. Dowds. 1993. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J. Bacteriol. 175:1665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, X.-H., and B. Austin. 2000. Pathogenicity of Vibrio harveyi to salmonids. J. Fish Dis. 23:93-102. [Google Scholar]
- 24.Zhang, X.-H., and B. Austin. 2005. Haemolysins in Vibrio species. J. Appl. Microbiol. 98:1011-1019. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X.-H., P. G. Meaden, and B. Austin. 2001. Duplication of hemolysin genes in a virulent isolate of Vibrio harveyi. Appl. Environ. Microbiol. 67:3161-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong, Y.-B., X.-H. Zhang, J.-X. Chen, Z.-H. Chi, B.-G. Sun, Y. Li, and B. Austin. 2006. Overexpression, purification, characterization, and pathogenicity of Vibrio harveyi hemolysin VHH. Infect. Immun. 74:6001-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]