Abstract
The known genomic islands of Pseudomonas aeruginosa clone C strains are integrated into tRNALys (pKLC102) or tRNAGly (PAGI-2 and PAGI-3) genes and differ from their core genomes by distinctive tetranucleotide usage patterns. pKLC102 and the related island PAPI-1 from P. aeruginosa PA14 were spontaneously mobilized from their host chromosomes at frequencies of 10% and 0.3%, making pKLC102 the most mobile genomic island known with a copy number of 30 episomal circular pKLC102 molecules per cell. The incidence of islands of the pKLC102/PAGI-2 type was investigated in 71 unrelated P. aeruginosa strains from diverse habitats and geographic origins. pKLC102- and PAGI-2-like islands were identified in 50 and 31 strains, respectively, and 15 and 10 subtypes were differentiated by hybridization on pKLC102 and PAGI-2 macroarrays. The diversity of PAGI-2-type islands was mainly caused by one large block of strain-specific genes, whereas the diversity of pKLC102-type islands was primarily generated by subtype-specific combination of gene cassettes. Chromosomal loss of PAGI-2 could be documented in sequential P. aeruginosa isolates from individuals with cystic fibrosis. PAGI-2 was present in most tested Cupriavidus metallidurans and Cupriavidus campinensis isolates from polluted environments, demonstrating the spread of PAGI-2 across habitats and species barriers. The pKLC102/PAGI-2 family is prevalent in numerous beta- and gammaproteobacteria and is characterized by high asymmetry of the cDNA strands. This evolutionarily ancient family of genomic islands retained its oligonucleotide signature during horizontal spread within and among taxa.
The genome of a bacterium consists of a core that is common to all strains of a taxon and an accessory part that varies within and among clones of a taxon. The accessory genome represents the flexible gene pool that frequently undergoes acquisition and loss of genetic information and hence plays an important role for the adaptive evolution of bacteria (10). The flexible gene pool is made up of elements such as bacteriophages, plasmids, insertion elements, transposons, conjugative transposons, integrons, and genomic islands.
Genomic islands are chromosomal regions that are typically flanked by direct repeats and inserted at the 3′ end of a tRNA gene. They contain transposase or integrase genes that are required for chromosomal integration and excision and other mobility-related genes. Genomic islands are clone or strain specific and are never found in all clones of a taxon. Most islands are easily differentiated from the core genome by their atypical G+C contents and atypical oligonucleotide compositions, with steep gradients at their boundaries (37, 38). First identified in pathogenic bacteria (pathogenicity islands), genomic islands have since been detected in numerous nonpathogenic species. Genomic islands may confer fitness traits, increase metabolic versatility or adaptability, or promote bacterium-host interaction in terms of symbiosis, commensalism, or virulence (10).
The ubiquitous and metabolically versatile Pseudomonas aeruginosa is an important opportunistic pathogen for humans, plants, and animals (34). Several large genomic islands have been detected in strains from human infections and aquatic habitats. All known large genomic islands of P. aeruginosa but one (28) are integrated into tRNA genes. Two different types were identified, the islands PAGI-2/PAGI-3 (25) and pKLC102 (21)/PAPI-1 (16), respectively. PAGI-2 and PAGI-3 were sequenced in strains C and SG17M of the major clone C (41), an isolate from the lungs of a patient with cystic fibrosis and an isolate from a river. PAGI-2 and PAGI-3 integrate into tRNAGly genes adjacent to the PAO homolog PA2820. In both islands, the first open reading frame (ORF) adjacent to the tRNAGly gene encodes a bacteriophage P4-related multidomain integrase. PAGI-2 and PAGI-3 have a modular bipartite structure. The first part (the cargo region) adjacent to the tRNA gene consists of strain-specific ORFs encoding metabolic functions and transporters, the majority of which have homologs of known function in other eubacteria. The second part (the conserved part) is made up of a syntenic set of ORFs, the majority of which are either classified as conserved hypotheticals or related to DNA replication or mobility genes. Forty-seven of these ORFs are arranged in the same order in both islands with amino acid identities of 35 to 88%.
The other known large genomic islands are integrated into one of the two identical tRNALys genes adjacent to PAO1 homologs PA0976 and PA4541. The sequenced islands that integrated adjacent to PA4541 are the pathogenicity island PAPI-1 of strain PA14 (16) and the mobile genetic element pKLC102 of the clone C strain SG17M (21). The 104-kb pKLC102 and the 108-kb PAPI-1 share a phage module that confers integrase, the att element, and a syntenic set of conserved genes, similar to those detected in PAGI-2 and PAGI-3 (21). The other tRNALys gene adjacent to PA0976 is targeted by genomic islands of various sizes (4 to 81 kb in six sequenced strains) and with various gene contents (16, 21, 24). These islands encode the type III secretion effector protein ExoU, a potent cytotoxic lipase (43), in exoU-positive strains (24). The sequence analysis suggests that the exoU-containing genomic islands probably evolved from an ancestral plasmid similar to pKLC102. Subsequent integrations of insertion elements, deletions, and rearrangements may then have led to the contemporary diversity of the islands (24).
The integration sites for all of these large genomic islands are located in the three hypervariable regions of the P. aeruginosa chromosome (17, 39). Since the PAO gene contig of these regions spans genomic segments of various sizes in other clones (17), we hypothesized that genomic islands account for their pronounced plasticity. We were curious to know whether and to what extent the sequenced pKLC102 and PAGI-2 are prototypes for these suspected genomic islands. PAGI-2 and pKLC102 share a set of 36 homologous genes, 15 of which have been identified in numerous genomic islands of other proteobacteria (29). In this study, the presence of homologs of all ORFs of pKLC102 and PAGI-2 was investigated in a panel of 71 genetically unrelated strains from diverse habitats and geographic origins (30) to assess the abundance and conservation of these types of genomic islands in P. aeruginosa.
Genomic islands are typically stably integrated into the host chromosome. The reversible integration and excision of genomic islands has so far been documented for only a few cases, such as the clc element of Pseudomonas putida strain RR21 (14); pathogenicity islands of Vibrio cholerae (32), Shigella flexneri (42), and Yersinia pseudotuberculosis (27, 33); integrative and conjugative elements (ICEs) of Escherichia coli strain ECOR31 (44) and of Vibrio cholerae (7, 8); and the SaPIbov2 pathogenicity island of Staphylococcus aureus (51); the last two are not integrated into a tRNA gene. Among the P. aeruginosa islands, pKLC102 is known to coexist in episomal and chromosome-integrated forms in clone C strains (21, 41), but no information was available about the chromosomal stability of the other three sequenced large genomic islands. Hence, the relative numbers of integrated and episomal forms were determined for PAGI-2, PAGI-3, pKLC102, and PAPI-1 during growth in vitro. In parallel, the oligonucleotide usage (OU) patterns of the four genomic islands were analyzed to unravel their genomic signatures and any commonalities with each other and their P. aeruginosa host chromosome. In particular, pKLC102 proved to behave like a foreign selfish element, consistent with its exceptionally high mobility.
MATERIALS AND METHODS
Oligonucleotide usage statistics.
Overlapping oligonucleotide words of a certain length, lw, were counted in the sequence of Lseq nucleotides by shifting the window in steps of 1 nucleotide. The total word number (Wtotal) is Lseq − lw in a linear sequence or Wtotal = Lseq in a circular sequence. Since Lseq ≫ lw, Wtotal ≅ Lseq in all cases. For a given word length lw, an Nw of 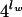 different words is possible for a sequence of four letters, A, T, G, and C. The observed counts of words (Co) were compared with the expected counts of words (Ce). Assuming the same distribution frequency for all words of a common length lw irrespective of their compositions and sequences, Ce matches the standard count number Cn0:
different words is possible for a sequence of four letters, A, T, G, and C. The observed counts of words (Co) were compared with the expected counts of words (Ce). Assuming the same distribution frequency for all words of a common length lw irrespective of their compositions and sequences, Ce matches the standard count number Cn0:
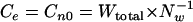 |
(1) |
Correspondingly, if we normalize OU by mononucleotide content using a zero-order Markov method (1), Ce becomes
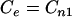 |
The deviation Δw of observed from expected counts is given by
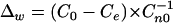 |
(2) |
In the present work, we used the following format for abbreviations of the different types of patterns: type_lwmer. Types are called “n0” if they are not normalized by mononucleotide frequency or “n1” if they are normalized by the zero-order Markov method. For example, the nonnormalized tetranucleotide usage pattern is an n0_4mer type, and the normalized tetranucleotide usage pattern is an n1_4mer type.
Variances OUV of word deviations were determined as follows:
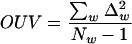 |
(3) |
For the comparison of sequences by OU patterns of the same type, the words in each sequence were ranked by Δw values according to equation 2. Rank numbers instead of word counts were used to simplify pattern comparison.
The distance D between two patterns was calculated as the sum of absolute distances between ranks of identical words in patterns i and j as follows:
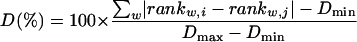 |
(4) |
whereby
 |
(5) |
Dmax is the maximal distance that is theoretically possible between two patterns of lw-long words (equation 5). Dmin is the minimal distance between two patterns. The minimal distance is zero for two independent sequences but has a positive value for the two complementary strands of the same DNA sequence, because the OU patterns designed for both strands of the same DNA molecule cannot be identical. The pattern skew (PS) describes this distance between opposite strands of the same DNA and is a measure of OU symmetry. The minimal theoretical distance between two patterns of opposite strands is realized if the words and their reverse complements are distributed with similar frequencies in the sequence, and it is
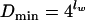 |
(6a) |
if lw is an odd number but
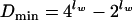 |
(6b) |
if lw is an even number, because palindromes, which occur in both strands with the same frequency, exist only in words with an even number of nucleotides, and the total number of all possible palindromes is 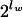 .
.
The computational program for determining OU patterns and their comparative analysis and storage in a database was written in Python 2.2 [http://www.python.org/] (38).
Strains.
Seventy-one P. aeruginosa strains from diverse origins and unrelated SpeI genotypes (Table 1) (30) were selected from an in-house strain collection. Moreover, the sequenced reference strains PAO1 and PA14 were included. Multilocus genotyping was performed in informative single-nucleotide polymorphisms (SNPs) of the loci oriC, oprL, alkB2, gltA, oprI, ampC, fliC, exoS, and exoU as described previously (5, 30). The 16 binary SNP genotypes of the 71 strains (Table 1) were represented by a four-digit hexadecimal code (see Table S1 in the supplemental material). The 16 SNPs were divided into four groups of 4 SNPs each, and the 16 possible combinations in each group were differentiated by 16 characters (0 to 9 and A to F). Sequential P. aeruginosa isolates were collected from the airways of 36 individuals with cystic fibrosis in half-year intervals after the onset of airway colonization over a period of up to 21 years. Strains were screened for the presence of PAGI-2 by PCR with specific primers for the gene C10 (25). Cupriavidus strains were supplied by M. Mergeay, Mol, Belgium (Table 1). Unless otherwise stated, strains were grown in liquid LB medium or on LB agar plates.
TABLE 1.
Description of strains
| Strain no. | Name | Place (yr) of isolation | Source | SNP genotypea |
|---|---|---|---|---|
| Pseudomonas aeruginosa collection | ||||
| 1 | ATCC 10145 | Prague, Czech Republic (≤1960) | Unknown | 46BA |
| 2 | ATCC 14886 | Osaka, Japan (≤1958) | Soil | EC2A (26; 55) |
| 3 | ATCC 15522 | United States (≤1967) | Soil | 481A |
| 4 | ATCC 15691 | Melbourne, Australia (1952) | Burn wound | 7D9A (13) |
| 5 | ATCC 21472 | Japan (≤1973) | Oil field soil | 3412 (6) |
| 6 | ATCC 21776 | Japan (≤1974) | Soil | 3412 (5) |
| 7 | ATCC 33348 | Bonn, Germany (≤1957) | Human infection | 2C1A (42; 54) |
| 8 | ATCC 33356 | Heidelberg, Germany (≤1955) | Human feces | CD9E |
| 9 | ATCC 33364 | (≤1978) | Human infection | E42A |
| 10 | ATCC 33818 | Unknown | Agaricus bisporus | 6CA2 |
| 11 | ATCC 33988 | Ponca City, OK | Fuel tank | 6C22 |
| 12 | 63741 | Hannover, Germany (1990) | Burn wound | 3C52 (33; 62) |
| 13 | A 5670 | Heidelberg, Germany (1992) | Wound | 7D9A (4) |
| 14 | A 5803 | Heidelberg, Germany (1992) | Trachea | F429 |
| 15 | AL 5846 | Heidelberg, Germany (1992) | Wound | D429 (39; 56) |
| 16 | 2733/92 | Copenhagen, Denmark (1992) | CFb patient | 3C2A |
| 17 | 2813 A/92 | Copenhagen, Denmark (1992) | CF patient | 4012 (37) |
| 18 | BST1 | Hannover, Germany (1985) | CF patient | E469 |
| 19 | KB1 | Sarstedt, Germany (1985) | CF patient | 059A |
| 20 | SS1 | Lueneburg, Germany (1985) | CF patient | 6D92 (60; 65) |
| 21 | MF6 | Bremen, Germany (1987) | CF patient | AC9A |
| 22 | PD1 | Hannover, Germany (1985) | CF patient | E59A |
| 23 | RN4 | Oldenburg, Germany (1986) | CF patient | D421 (41; 44; 51) |
| 24 | RP1 | Hannover, Germany (1985) | CF patient | 0C2E (38; 64) |
| 25 | Va 24437 | Halle, Germany (1992) | CF patient | 3C51 |
| 26 | Va 26232 | Halle, Germany (1992) | CF patient | EC2A (2; 55) |
| 27 | Va 27081 | Halle, Germany (1992) | CF patient | 081E |
| 28 | Va 27260 | Halle, Germany (1992) | CF patient | 239A |
| 29 | DM | Hamburg, Germany (1984) | CF patient | E84A |
| 30 | Zw 30 | Innsbruck, Austria (1997) | CF patient | B420 |
| 31 | Zw 31 | Innsbruck, Austria (1997) | CF patient | AC2E |
| 32 | Zw 41 | Verona, Italy (1997) | CF patient | 0192 |
| 33 | Zw 43 | Genova, Italy (1997) | CF patient | 3C52 (12; 62) |
| 34 | Zw 49 | Verona, Italy (1997) | CF patient | A5AA |
| 35 | Zw 54 | Milan, Italy (1997) | CF patient | 6C12 |
| 36 | Zw 64 | Lund, Sweden (1997) | CF patient | 279A |
| 37 | Zw 77 | London, Great Britain (1997) | CF patient | 4012 (17) |
| 38 | Zw 79 | Galway, Ireland (1997) | CF patient | 0C2E (24; 64) |
| 39 | Zw 81 | London, Great Britain (1997) | CF patient | D429 (15; 56) |
| 40 | Zw 83 | London, Great Britain (1997) | CF patient | 6E12 (46) |
| 41 | Zw 85 | Aberdeen, Great Britain (1997) | CF patient | D421 (23; 44; 51) |
| 42 | Zw 88 | London, Great Britain (1997) | CF patient | 2C1A (7; 54) |
| 43 | Zw 92 | Marseille, France (1997) | CF patient | EC22 |
| 44 | Zw 98 | The Hague, The Netherlands (1997) | CF patient | D421 (23; 41; 51) |
| 45 | Zw 102 | Leuven, Belgium (1997) | CF patient | 2E12 |
| 46 | Zw 113 | Rotterdam, Netherlands (1997) | CF patient | 6E12 (40) |
| 47 | Zw 117 | Vienna, Austria (1997) | CF patient | 0812 |
| 48 | Zw 119 | Poznań, Poland (1997) | CF patient | F469 |
| 49 | SG1 (= C) | Bueckeburg, Germany (1986) | CF patient | C40A (50) |
| 50 | SG31 (= SG17M) | Muelheim, Germany (1993) | River | C40A (49) |
| 51 | PT2 | Muelheim, Germany (1992) | Water | D421 (23; 41; 44) |
| 52 | PT6 | Muelheim, Germany (1992) | Water | 2992 |
| 53 | PT12 | Muelheim, Germany (1992) | Water | F419 |
| 54 | PT20 | Muelheim, Germany (1992) | Water | 2C1A (7; 42) |
| 55 | PT22 | Muelheim, Germany (1992) | Water | EC2A (2; 26) |
| 56 | PT36 | Muelheim, Germany (1992) | Water | D429 (15; 39) |
| 57 | 641HD11 | Muelheim, Germany (1992) | Water | 249A |
| 58 | Gr 2052 | Athens, Greece (1995) | Clinic | 2C92 (59) |
| 59 | Gr 2057 | Athens, Greece (1995) | Clinic | 2C92 (58) |
| 60 | Gr 2248 | Athens, Greece (1995) | Clinic | 6D92 (20; 65) |
| 61 | PAO-DSM 1707 | Melbourne, Australia (<1955) | Burn wound | 0002 |
| 62 | 892 | Hannover, Germany (1983) | CF patient | 3C52 (12; 33) |
| 63 | PAK | Japan (≤1960) | Unknown | 55AA |
| 64 | HJ2 | Cologne, Germany (1990) | CF patient | 0C2E (24; 38) |
| 65 | G7 | Stade, Germany (1986) | CF patient | 6D92 (20; 60) |
| 66 | H2 | Unknown | Clinic (catheter) | 241A |
| 67 | K9 | Husum, Germany (1985) | CF patient | 1BAE |
| 68 | DSM 288 | Goettingen, Germany (1990) | Hygiene institute | 0B92 (71) |
| 69 | DSM 939 | United States (≤1981) | Animal room water bottle | 049A |
| 70 | DSM 1128 | United States (1980) | Ear infection | EC38 |
| 71 | DSM 1253 | Stanford, CA (≤1949) | Burn wound | 0B92 (68) |
| Reference strains | ||||
| P. aeruginosa PA14 | United States (≤1995) | Burn wound | D421 | |
| P. aeruginosa TB | Hannover, Germany (1983) | CF patient | 3C52 | |
| P. putida KT2440 | Minoh City, Japan (1961) | TOL plasmid cured derivative of soil isolate mt-2 | ||
| Cupriavidus strains | ||||
| C. metallidurans CH34 | Liège, Belgium (1976) | Decantation tank, zinc factory | ||
| C. metallidurans CH42 | Liège, Belgium (1976) | Zinc factory | ||
| C. metallidurans CH79 | Liège, Belgium (1976) | Zinc factory | ||
| C. metallidurans KT01 | Goettingen, Germany (≤1987) | Wastewater | ||
| C. metallidurans KT02 | Goettingen, Germany (1984) | Sewage treatment plant | ||
| C. metallidurans KT21 | Goettingen, Germany (≤1987) | Sewage treatment plant | ||
| C. campinensis AE2700 | Leadville, CO (≤2002) | Unknown | ||
| C. campinensis AE2701 | Leadville, CO (≤2002) | Unknown |
SNP genotype defined by 13 SNPs and three additional markers, given as a code of four hexadecimal digits (for a description see table S1 in the supplemental material); the numbers in parentheses indicate the numbers of strains with identical genotypes.
CF, cystic fibrosis.
DNA preparation.
DNA manipulations followed standard procedures (2). High-molecular-weight chromosomal DNA of P. aeruginosa was prepared following the protocol of Goldberg and Ohman (15). Small-scale isolations of plasmid and cosmid DNAs were performed by using QIAprep spin miniprep kits (QIAGEN), while larger amounts of cosmid DNA were purified by using QIAtip100 columns (QIAGEN) following the instructions of the supplier.
Combinatorial PCR.
PCR was performed with PA14-, PAPI-1-, SG17M-, pKLC102-, C-, PAGI-2-, or PAGI-3-derived target-specific primer sequences (see Table S2 in the supplemental material) and 50 ng P. aeruginosa DNA in a 50-μl reaction mixture (5 μl 10× reaction buffer [Eurogentec], 3.3 μl 25 mM MgCl2, 1 μl dimethyl sulfoxide, 10 μl primer solution [5 μM each], 3 μl deoxynucleoside triphosphates [2 mM each], 1 U Goldstar DNA Polymerase [Eurogentec]). For PCR kinetics, aliquots of 5 μl were withdrawn at the indicated cycles, separated by electrophoresis, and stained with ethidium bromide. The relative amounts Ni and Nj of the template DNA sequences i and j in the reaction mixture were determined from the titration for the first reaction cycle n when the PCR products became visible by ethidium bromide fluorescence during the late exponential phase of PCR according to the following equation:
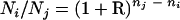 |
(7) |
Thus, the efficiency R of the thermocycler used during the exponential phase of PCR was determined as follows: R = 0.78 ± 0.02 for PCR products of 100 to 800 bp in length within the interval of reaction cycles 10 < n < 35 (6, 18).
Southern hybridization analysis.
To visualize the copy numbers of PAGI-2- and pKLC102-type islands in P. aeruginosa strains, XhoI- or NcoI-restricted genomic DNA was separated by agarose gel electrophoresis, blotted onto Hybond N+ membranes (Amersham), hybridized with digoxigenin (DIG)-labeled PCR-generated probes, and detected by chemiluminescent immunoreactive signals by applying standard procedures (40). According to BlastN analysis, the primer sequences were specific for PAGI-2 or pKLC102 and showed no homology to the PAO1 genomic sequence (49).
Macroarrays. (i) Design.
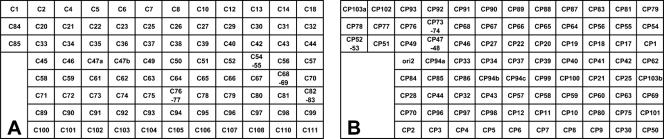
PCR products generated with PAGI-2- or pKLC102-derived primer sequences were spotted onto nylon membranes. The scheme is shown in Fig. 1. For the PAGI-2 macroarrays, 91 PCR products were distributed onto the membrane representing 93 of the 111 predicted ORFs (Fig. 1A). ORF C47 was represented by two different products (“C47a” and “C47b”); the adjacent genes C54 and C55, C76 and C77, and C82 and C83 were each represented by a single ORF-spanning PCR product.
FIG. 1.
Schematic diagram of the positions of ORF-derived PCR products on the PAGI-2 (A) and pKLC102 (B) macroarrays. (A) PAGI-2. ORF C47 is represented twice (C47a and C47b) by different PCR products. (B) pKLC102. ORF CP103 is represented twice (CP103a and CP103b) and CP94 three times (CP94a, CP94b, and CP94c) by different PCR products. Five (A) or 10 (B) positive or negative control dots were spotted in the lower left corner.
Eighty-five ORFs were represented in the pKLC102 macroarray (Fig. 1B), among which ORFs CP94 and CP103 were represented by three (“CP94a,” “CP94b,” “CP94c”) and two (“CP103a” and “CP103b”) PCR products, respectively. Three PCR products spanned two ORFs each (CP47 and CP48, CP52 and CP53, and CP73 and CP74). One spot (“ori2”) contained a part of oriV (21). Control PCR products were spotted in the left lower corner. In case of the PAGI-2 array, the five dots contained partial sequences of (from top to bottom) the P. aeruginosa genes gltA, fliC (type A), and fliC (type B) (positive controls) and of an intergenic sequence of Pseudomonas putida KT2440 and of the human ob gene (negative controls). In addition to these five control dots, the pKLC102 macroarray contained, in the second lane from the left, the five controls “ori1” of pKLC102, PA0977, and PA0981 of P. aeruginosa PAO1 and two PAGI-2 homologs of P. aeruginosa TB.
(ii) Production of macroarrays.
Probe sequences of 208 bp to 805 bp were generated by four PCRs with cosmids encoding pKLC102 (21) or PAGI-2 (25) sequences as templates. The contig CP39 to CP41 was amplified from P. aeruginosa C genomic DNA. The primer sequences are listed in Table S2 in the supplemental material. All PCRs were performed with 40 to 200 ng cosmid DNA or 100 to 200 ng genomic DNA in a final volume of 100 μl (10 μl 10× buffer [500 mM Tris-HCl, 160 mM NaNH4SO4, 0.1% {vol/vol} Tween 20, pH 8.8], 2 μl 50 mM MgCl2, 6 μl each of 5 μM primer A and B stock solutions, 2 μl dimethyl sulfoxide, 6 μl 8 mM deoxynucleoside triphosphates [2 mM each nucleotide], 2 U Taq DNA polymerase [InViTek]). After denaturation for 300 s at 96°C, 35 cycles were run (annealing for 45 s at 60°C or 58°C, elongation for 45 to 90 s at 72°C, and denaturation for 120 s at 94°C). According to agarose gel electrophoresis and subsequent ethidium bromide staining, more than 80% of all PCR products were at least 99.9% pure and all other PCR products were at least 98% pure. Macroarray copies were produced in parallel from the same stock of pooled PCR products to ensure that the corresponding ORFs were represented by identical amounts of DNA on each membrane. Hence, for each of the 96 PCR products, an aliquot of 50 μl of pooled PCR product, 85 μl Tris-EDTA buffer, and 15 μl 3 M NaOH was dispensed in a well of a 96-well plate, denatured for 30 min at 65°C, and chilled on ice. After the addition of 100 μl 3 M ammonium acetate, aliquots of 100 μl each were transferred by a minifold-dot-vacuum-blot apparatus (Schleicher & Schüll) onto a Hybond N+ nylon membrane soaked in 1 M ammonium acetate. The membrane was dried, and the DNA was immobilized by irradiation with UV light.
(iii) Hybridization of macroarrays.
Membranes were incubated for 2 to 16 h at 68°C with hybridization buffer (0.5 M sodium phosphate, 7% sodium dodecyl sulfate, 1 mM EDTA, 0.5% blocking reagent [Roche], pH 7.2), hybridized for 16 to 24 h at 68°C in the same buffer with DIG-labeled genomic DNA, and then washed twice for 30 to 45 min each at 68°C in washing buffer (40 mM sodium phosphate, 1% sodium dodecyl sulfate, 1 mM EDTA, pH 7.2). Detection of DIG-labeled fragments by anti-DIG conjugate antibody, enzymatic cleavage of CDP-Star, and exposure to X-ray films were performed as described previously (40).
(iv) Evaluation of macroarray hybridization signals.
Signals were classified as strong, weak, or negative according to the signal intensity of the hybridization of labeled PCR products of known sequence onto restricted cosmid DNA. Strong hybridization signals were obtained for homologs with 85% sequence identity or more. Control hybridizations of PAGI-2 onto the pKLC102 macroarray gave negative signals for all pKLC102-derived gene fragments of the array, whereas the reciprocal hybridization of pKLC102 onto the PAGI-2 array revealed weak signals for four of the 34 homologs. The nucleotide sequence identities of the PCR-amplified fragments with their homologous genes were 72%, 76%, 74%, and 63% for C49, C65, C71, and C108, respectively. The E values of the corresponding BlastN comparisons were 1E-80, 2E-115, 2E-127, and 2E-55. Importantly, the weak homolog of C108 in pKLC102 carried a 28-bp stretch of identical sequence, which may explain the occurrence of the weak cross-hybridization signal despite the lower overall homology. In general, however, applying the stringent hybridization conditions, a minimal sequence identity of 75% between the membrane-bound PCR product and the DIG-labeled genomic sample was estimated to be the threshold for generating hybridization signals.
(v) Parsimony analysis.
Parsimony analysis was performed with the program “PARS” from the software package “PHYLIP 3.66” (http://evolution.genetics.washington.edu/phylip.html). Signals obtained with the positive controls PAGI-2 and pKLC102 were defined as the standard normalized to “1” for all island ORFs on the macroarrays. In the cases of PAGI-2 subtypes, signals of C1 (integrase gene), C84 and C85 (transposon genes), and C68/C69 were excluded from the analysis because of possible cross-hybridization of homologs or occasional false-negative signals (C68/C69). Similarly, the ORFs CP84, CP85, CP86, and CP103 of pKLC102 were excluded because homologs are found elsewhere in the genome. The purified datasets of all strains were then either combined or separately evaluated by parsimony analysis with PAGI-2 and/or pKLC102 as a reference, respectively.
RESULTS AND DISCUSSION
Local tetranucleotide signature.
The local tetranucleotide usage was calculated for the four genomic islands pKLC102, PAPI-1, PAGI-2, and PAGI-3 (Fig. 2). Values for a 5-kb sliding window were compared with the global tetranucleotide usage of the whole P. aeruginosa PAO1 chromosome. The variance of tetranucleotide frequency, OUV, is the difference between the empirical frequency and the null hypothesis of an equal frequency of all 256 tetranucleotides (52). OUV is primarily shaped by the local G+C content in P. aeruginosa (52), and hence, we calculated OUV:n1_4mer normalized for mononucleotide frequencies (37). These OUV:n1_4mer values reflect the species-specific selection of tetranucleotides normalized for the high G+C content of P. aeruginosa and are an appropriate measure of the oligonucleotide signature of the genome. The local OUV:n1_4mer values of all four islands were consistently below the median OUV value of 0.37 of the P. aeruginosa PAO1 chromosome. In other words, the selection of tetranucleotides was less biased in the islands than in the P. aeruginosa core genome and hence might facilitate the horizontal spread of the islands to bacterial species with other oligonucleotide signatures.
FIG. 2.
Tetranucleotide usage of the four P. aeruginosa genomic islands pKLC102, PAPI-1, PAGI-2, and PAGI-3. Local OU patterns were analyzed in 5-kb sliding windows with steps of 0.5 kb. Curves of the distance D:n0_4mer, pattern skew PS:n0_4mer, and oligonucleotide variance OUV:n1_4mer are specified by color code: blue for D, green for PS and brown for OUV. Protein-coding genes are shown by red bars. The abscissa separates genes by their direction of transcription. The tetranucleotide usage of the genomic islands was significantly different from that of the whole chromosome. The median (inner quartile) values of local tetranucleotide patterns in the whole P. aeruginosa PAO1 chromosome were 13.9 (12.3 to 16.0) for D:n0_4mer, 21.4 (17.9 to 25.6) for PS:n0_4mer, and 0.37 (0.32 to 0.43) for OUV:n1_4mer.
The parameter “distance,” D, compares the rank order of tetranucleotide frequencies in two patterns (37), i.e., in this case, the rank order in a 5-kb window compared to that of the whole genome (see Materials and Methods). Local D values were similar to that of the P. aeruginosa core genome throughout the whole genomic island PAGI-2 (Fig. 2). The other three islands, however, showed high peak values in several regions. In the case of PAGI-3, almost all genes in the strain-specific cargo regions (see Table S3 in the supplemental material) (25), but none of the genes that are conserved among members of this family of genomic islands (39), harbored an atypical oligonucleotide composition. Peaks of D values were flanked by various small transposable elements, highlighting the complex architecture of PAGI-3 (see Table S3 in the supplemental material) (25). In pKLC102, the loci with atypical local oligonucleotide compositions were predominantly associated with genes that are necessary for conjugation and integration, such as those for sex pili, relaxase, and integrase (Table 2) (21). In PAPI-1, these loci were either flanked on one side by a direct or inverted repeat or were part of type IV pilus biogenesis machinery (16). In summary, regions with an atypical oligonucleotide composition encode repeats and/or elements of genetic mobility.
TABLE 2.
ORFs of the phage and plasmid modules of the genomic island pKLC102
| ORF no. or feature | Gene name | ORF/protein length (bp/amino acids) | Putative product | PAGI-2 homolog | Homolog producta | GenBank accession no.a | E valuea |
|---|---|---|---|---|---|---|---|
| CP1 | soj | 885/294 | Chromosome partitioning-related protein | C108 | Chromosome partitioning-related protein PA14_58910 (P. aeruginosa UCBPP-PA14) | YP_792889 | 4E-142 |
| CP9 | dnaB | 1,278/425 | Replicative DNA helicase | Replicative DNA helicase Paer2_01005575 (P. aeruginosa 2192) | ZP_00970992 | 0 | |
| CP16 | 255/84 | DNA binding protein | Hypothetical protein PA14_59060 (P. aeruginosa UCBPP-PA14) | YP_792903 | 1E-23 | ||
| CP17b | 1,734/577 | ParB-like nuclease | C107 | Hypothetical protein PA14_59070 (P. aeruginosa UCBPP-PA14) | YP_792904 | 0 | |
| CP18 | 756/251 | Conserved hypothetical protein | C106 | Hypothetical protein PaerP_01000019 (P. aeruginosa PA7) | ZP_01297689 | 2E-118 | |
| oriVc | 1,647 | ||||||
| CP19 | 729/242 | Conserved hypothetical protein | C104 | Hypothetical protein PA14_59130 (P. aeruginosa UCBPP-PA14) | YP_792909 | 2E-122 | |
| CP20b | 549/182 | Conserved hypothetical protein | C103 | Hypothetical protein PA14_59140 (P. aeruginosa UCBPP-PA14) | YP_792910 | 2E-71 | |
| CP22 | ssb | 489/162 | Single-stranded DNA binding protein | C102 | Single-stranded DNA binding protein PaerC_01005119 (P. aeruginosa C3719) | ZP_00965569 | 2E-83 |
| CP27 | topA | 1,920/639 | Topoisomerase I | C101 | DNA topoisomerase I (P. aeruginosa CF005) | AAR01278 | 0 |
| CP33-CP42 | pilLNOPQRSUVM | 10,643 | Sex pilus biogenesis cluster | Type IVB pilus proteins (P. aeruginosa UCBPP-PA14) | CP000438 | 0 | |
| CP56d | 2,256/751 | Helicase | C71 | DNA/RNA helicase Paer2_01005538 (P. aeruginosa 2192) | ZP_00971322 | 0 | |
| CP67e | 2,232/743 | TraG-/TraD-like conjugation protein | C65 | Hypothetical protein PaerP_01000052 (P. aeruginosa PA7) | ZP_01297722 | 0 | |
| CP102 | 1,920/639 | TraI-like conjugative relaxase | C36 | Hypothetical protein EXA2 (P. aeruginosa 6077) | ABD94612 | 0 | |
| CP103a | xerC | 1,284/427 | Phage-like integrase | −f | Putative integrase EXA1a (P. aeruginosa 6077) | ABD94670 | 0 |
Closest homolog according to PSI- and PHI-BLAST searches; copies of pKLC102 were not considered.
Homologs of CP17 and CP20 in the clc element are involved in the regulation of the expression of the phage P4-type integrase (45, 46).
No oriV-like structure in PAGI-2.
The ORF contig CP46 to CP56 is highly conserved in pKLC102- and PAGI-2-type islands.
The ORF contig CP64 to CP68 is highly conserved in pKLC102- and PAGI-2-type islands.
The parameter PS describes this distance D between opposite strands of the same DNA and is a measure of oligonucleotide symmetry (37, 38). Comparatively low local PS values, such as the 21% calculated for 5-kb sliding windows in the P. aeruginosa PAO1 chromosome, are typical of bacterial chromosomes that are characterized by strand symmetry and intrastrand parity of complementary oligonucleotides (37). The profiles of local PS values roughly followed those of local D values in all four genomic islands, but more importantly, the absolute values were within or above the upper outer quartile of local PS in the host chromosome. With the exception of one small peak, the local PS was scattered between 20% and 30% throughout PAGI-2. Of the four islands, PAGI-2 had the PS values most similar to those of its host chromosome. In contrast, higher basal values of about 30% and numerous peaks with anomalously high local PS were typical of the other three islands. The maximal values were close to or above the value of 60% of a random sequence, implying that in these peak regions no strand symmetry exists. In other words, oligonucleotide frequencies on the two strands were only weakly correlated in all four islands and were completely lost in the peak regions of pKLC102 (three segments), PAPI-1 (two segments), and PAGI-3 (three segments).
In summary, the local tetranucleotide signatures of all four islands were distinct from that of the P. aeruginosa chromosome. PAGI-2 is homogeneous in its tetranucleotide composition throughout the island, but pKLC102, PAPI-1, and PAGI-3 each contain regions of highly atypical tetranucleotide composition.
Chromosomal stability of island integration.
The atypical oligonucleotide signature, particularly the pronounced strand asymmetry, prompted us to investigate whether the islands could be spontaneously excised from their host chromosomes. All four islands are endowed with genetic elements of mobility. They harbor phage modules (Table 2) that encode chromosome-partitioning proteins (soj) at one terminus and integrases of the bacteriophage P4 subfamily (PAGI-2, PAGI-3 [25]) or a phage tyrosine integrase (pKLC102 [21] or PAPI-1 [16]) at the other end. PAPI-1 and pKLC102, moreover, include numerous ORFs that are related to plasmid-encoded replication and recombination functions.
Combinatorial PCR that spans the integration sites of the islands was applied to detect excised circularized islands and island-free chromosomes compared to integrated genomic islands. Overnight growing cultures were diluted with fresh liquid LB medium, and samples were then taken from the early-exponential to the late-stationary phase of growth. The relative copy number of circularized PAPI-1 was estimated to be 2% of that of PA14 chromosomes. About 0.3 to 1% of PA14 chromosomes did not carry an integrated PAPI-1 island. A copy number of 30 circular pKLC102 molecules per SG17M host chromosome was estimated from semiquantitative PCR kinetics (Fig. 3). During growth, the percentage of pKLC102-free chromosomes increased from about 2 to 3% in early exponential phase to approximately 10% in stationary phase (Fig. 3). In contrast, no circular forms of PAGI-2 or PAGI-3 were detected by combinatorial PCR. Hence, the spontaneous excision rates, if they occur, are below the sensitivity threshold (1 × 10−7) of the assay. Consistent with this finding, no strain C or strain SG17M chromosomes were identified by PCR that had cured PAGI-2 or PAGI-3, respectively.
FIG. 3.
Combinatorial PCR analysis of integrated and episomal versions of genomic islands PAPI-1 in strain P. aeruginosa PA14 and pKLC102 in P. aeruginosa SG17M. An aliquot from an exponentially growing culture was inoculated into 100 ml fresh medium adjusted to an optical density at 578 nm (OD578) of 0.2. Samples were then taken from the growing culture (from left to right) at OD578s of 0.9, 1.3, 2.0, 2.9, and 4.0 and after 24 h (left) or at OD578s of 0.9, 1.3, 2.0, and 4.0 and after 24 h (right). Bacteria were growing aerobically in 250-ml flasks in liquid LB medium at 37°C at a mixing frequency of 250 rpm. Chromosome-integrated islands were detected by PCR products spanning the 5′ tRNA (il) or the 3′ tRNA (ir) integration sites by utilizing PA14- and PAPI-1- or SG17M- and pKLC102-derived primer sequences. Circularized episomal forms (ce) were identified by PCR products spanning the breakpoints in PAPI-1 or pKLC102. PA14 or SG17M chromosomes (fa) devoid of PAPI-1 or pKLC102 were detected by PCR products spanning the tRNALys gene adjacent to the PAO1 homolog PA4541. PCR kinetics were performed with 50 ng P. aeruginosa DNA in a 50-μl reaction mixture. Aliquots of 5 μl were withdrawn at the indicated cycles, separated by electrophoresis, and stained with ethidium bromide.
If we assume that PA14 and SG17M cells will grow in rich medium at statistically indistinguishable rates irrespective of the presence or absence of the genomic island in their chromosome, the spontaneous excision rates can be estimated from the semiquantitative PCR kinetics (Fig. 3) to be at least 3 × 10−3 for PAPI-1 in strain PA14 and at least 10−1 for pKLC102 in strain SG17M. The latter estimate relies on steady-state values and hence is probably too low, because pKLC102, like its relative pKLK106, can reversibly integrate into and be excised from its tRNALys site (20).
The precise excision of enterobacterial pathogenicity islands has been reported to occur spontaneously at a frequency of 10−5 to 10−4 (27, 32, 42, 44), although mutations, deletions, and genome rearrangements are likely to be responsible for the inability of most genomic islands to achieve precise excision and mobilization. In the cases of pKLC102 and PAPI-1, the frequencies of spontaneous excision from the host chromosome are 1 or even 3 orders of magnitude higher. pKLC102 and PAPI-1 harbor the phage module with the xerC integrase gene, some plasmid-related genes, a type IV pilus biogenesis gene cassette, and a syntenic set of conserved ORFs, similar to those detected in PAGI-2 and PAGI-3 (Table 2). These features probably allow the islands to be excised exactly from the chromosome and to form a circular extrachromosomal intermediate of sufficient stability. The lower copy number of PAPI-1 indicates that circular forms were only present in a few percent of cells and probably modulate the phenotype of the PA14 community only to a minor extent. The opposite conclusion applies to pKLC102. Circular forms were in 10-fold excess of chromosomal forms, demonstrating that circular pKLC102 replicates in its host cell. Moreover, a substantial number of SG17M chromosomes became devoid of pKLC102 during growth to higher cell densities. These data verify the previous assignment of pKLC102 as a plasmid (20, 21, 41). The functional plasmid module of pKLC102 is apparently responsible for the highest mobility of a genomic island that, to our knowledge, has ever been reported. As a hybrid of phage and plasmid origin (Table 2), pKLC102 may be considered an intermediate between a mobile genetic element and a genomic island.
Epidemiology of PAGI-2- and pKLC102-like genomic islands in P. aeruginosa.
PAGI-2 and pKLC102 share a syntenic set of ORFs (21), homologs of which have been detected in more than 30 genomic islands of other beta- and gammaproteobacteria (29). The presence of these island types in numerous taxa suggests that they form a family with a deep evolutionary origin (29). However, since no epidemiological data have yet been reported, the roles of PAGI-2 and pKLC102 in the contemporary P. aeruginosa population are unknown. Therefore, we investigated the abundance and diversity of PAGI-2- and pKLC102-like genomic islands in 71 strains of unrelated SpeI genotypes (Table 1) (30). The panel included isolates from diverse habitats and geographic origins and was a representative sample of present-day P. aeruginosa clones. Note that 36 of the 71 strains share their SNP genotypes with at least one other strain in the panel (seven pairs, six trios, and one quadruple; the hexadecimal genotypes of strains are listed in Table 1). This finding implies that differences in the accessory genome frequently give rise to macrorestriction fragment patterns that are classified as distinct P. aeruginosa genotypes by accepted criteria (40), although the SNP genotypes of the core genomes are identical.
Macroarrays of PAGI-2 and pKLC102 ORFs (Fig. 1) were hybridized with the strains' DNAs under high stringency to suppress equivocal cross-hybridization signals of homologous genes (see Materials and Methods). The hybridization analyses were calibrated with samples and probes of known sequence so that a sequence identity of at least 75% was required for a positive signal. An identity of 85% or more between the two sequences yielded strong hybridization signals. Tables 3 and 4 show the results of macroarray hybridizations of strains with positive hybridization signals.
TABLE 3.
PAGI-2 macroarray hybridization patterns of island-positive strains
| PAGI-2 ORF | Hybridization of straina:
|
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 7 | 9 | 14 | 15 | 16 | 21 | 22 | 23 | 24 | 25 | 26 | 29 | 33 | 35 | 45 | 46 | 48 | 49 | 50 | 52 | 53 | 54 | 55 | 56 | 60 | 62 | 63 | 64 | 67 | 70 | |
| C1 | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | ? | x | ? | x | x | ? | x | x | x | x | x | |
| C2 | x | x | ? | x | x | ||||||||||||||||||||||||||
| C4 | x | x | ? | ? | x | ? | x | ? | ? | x | x | x | x | x | ? | x | |||||||||||||||
| C5 | x | x | x | x | x | ||||||||||||||||||||||||||
| C6 | x | x | x | x | x | ||||||||||||||||||||||||||
| C7 | x | x | x | x | x | ||||||||||||||||||||||||||
| C8 | x | x | x | x | x | ||||||||||||||||||||||||||
| C10 | x | x | x | x | x | ||||||||||||||||||||||||||
| C12 | x | x | x | x | x | ||||||||||||||||||||||||||
| C13 | x | x | x | x | x | ||||||||||||||||||||||||||
| C14 | x | x | x | x | x | ||||||||||||||||||||||||||
| C18 | x | x | x | x | x | ||||||||||||||||||||||||||
| C20 | x | x | x | x | x | ||||||||||||||||||||||||||
| C21 | x | x | x | x | x | ||||||||||||||||||||||||||
| C22 | x | x | x | x | x | ||||||||||||||||||||||||||
| C23 | x | x | x | x | x | ||||||||||||||||||||||||||
| C25 | x | x | x | x | x | ||||||||||||||||||||||||||
| C26 | x | x | x | x | x | ||||||||||||||||||||||||||
| C27 | x | x | x | x | x | ||||||||||||||||||||||||||
| C29 | x | x | x | x | x | ||||||||||||||||||||||||||
| C30 | x | x | x | x | x | ||||||||||||||||||||||||||
| C31 | x | x | x | x | x | ||||||||||||||||||||||||||
| C32 | x | x | x | x | x | ||||||||||||||||||||||||||
| C33 | x | x | x | x | x | ||||||||||||||||||||||||||
| C34 | x | x | x | x | x | ||||||||||||||||||||||||||
| C35 | x | x | x | x | x | ||||||||||||||||||||||||||
| C36 | x | ? | x | ? | ? | x | ? | x | x | ? | x | ? | ? | x | ? | x | ? | x | x | ? | ? | ? | ? | x | |||||||
| C37 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | ? | x | x | x | x | x | |||
| C38 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| C39 | x | x | x | x | x | x | x | ? | x | x | ? | x | x | ? | x | x | x | x | ? | ? | x | x | x | x | ? | x | ? | x | x | x | |
| C40 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C42 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x |
| C43 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x |
| C44 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C45 | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | |
| C46 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C47 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C49 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C50 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x |
| C51 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C52 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C54 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C55 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C56 | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||
| C57 | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | |||||||
| C58 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||
| C59 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||
| C61 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||
| C62 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||
| C63 | x | x | x | x | x | x | x | ? | x | x | x | x | x | ? | ? | x | ? | ? | x | x | ? | x | x | x | |||||||
| C64 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x |
| C65 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C66 | ? | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | ? | x | ? | x | x | |
| C67 | x | x | x | x | ? | x | x | x | x | x | x | ? | x | ? | x | x | x | x | x | x | x | x | ? | x | x | ? | x | ? | x | x | |
| C70 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C71 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C72 | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | ? | x | x | x | x | x | |
| C73 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C74 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C75 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C76 | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| C77 | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| C78 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | ? | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | |
| C79 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C80 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | ||||
| C81 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C82 | x | x | ? | x | x | x | x | ? | ? | x | x | x | x | x | x | x | x | ? | ? | x | x | ||||||||||
| C83 | x | x | ? | x | x | x | x | ? | ? | x | x | x | x | x | x | x | x | ? | ? | x | x | ||||||||||
| C84 | x | x | x | ? | x | x | x | ? | ? | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x |
| C85 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | ? | ? | x | x |
| C89 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C90 | x | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C91 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C92 | x | x | ? | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | ? | x | ? | x | x | |||||||
| C93 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C94 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C95 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | ? | x | x |
| C96 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| C97 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | ||
| C98 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | x | x | x | x | ? | x | x | ? | x | x | ||
| C99 | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | ? | x | x | ? | x | x | x | x | ? | x | x | x | ? | x | x | |
| C100 | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||
| C101 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C102 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C103 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C104 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C105 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C106 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C107 | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| C108 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| C110 | x | x | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | ? | x | x | x | ||||
| C111 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | ? | x | ? | x | x | |||
x, strong hybridization signal; ?, weak hybridization signal.
TABLE 4.
pKLC102 macroarray hybridization patterns of island-positive strains
| pKLC-ORF | Hybridization of straina:
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 9 | 10 | 12 | 13 | 14 | 15 | 16 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 28 | 29 | 31 | 33 | 35 | 36 | 37 | 39 | 41 | 44 | 45 | 46 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 58 | 59 | 60 | 62 | 65 | 66 | 68 | 69 | 70 | 71 | |
| CP1 | x | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| CP2 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP3 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP4 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||
| CP5 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP6 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP7 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||
| CP8 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP9 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| CP10 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||
| CP11 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| CP12 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| CP17 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP18 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||
| oriV | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||
| CP19 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP20 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP21 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||
| CP22 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP25 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||
| CP27 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| CP28 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||
| CP30 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP32 | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||||||||||||
| CP33 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||
| CP34 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||
| CP37 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||
| CP39 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||
| CP40 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| CP41 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP42 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||
| CP43 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP44 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||
| CP46 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP47 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP48 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP49 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP50 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||
| CP51 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||
| CP52 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||
| CP53 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||
| CP54 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| CP55 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP56 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP57 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||
| CP58 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||||
| CP59 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||
| CP60 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||
| CP62 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||
| CP63 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||
| CP64 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP65 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP66 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP67 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP68 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP69 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| CP70 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||
| CP73 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP74 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP75 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP76 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP77 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP78 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||
| CP79 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||
| CP80 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP81 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP83 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
| CP84 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||
| CP85 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||
| CP86 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||
| CP87 | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||||||||||||||||||||||||
| CP88 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP89 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP90 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP91 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |
| CP92 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||
| CP93 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||
| CP94 | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||||||||
| CP96 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||||||||||||||||||||||
| CP97 | x | x | x | x | x | |||||||||||||||||||||||||||||||||||||||||||||
| CP98 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||||||||||||||||||||||
| CP99 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||
| CP100 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||
| CP101 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||||||
| CP102 | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x |
| CP103 | x | ? | ? | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ? | ? | x | ? | x | x | x | ? | x | x | x | x | ? | x | x | x | x | x | x | x | x | x | x | x | ||||
x, strong hybridization signals; ?, weak hybridization signals.
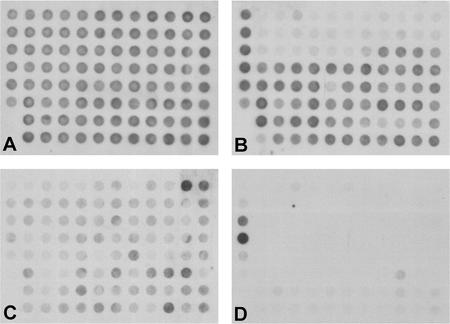
PAGI-2 type islands were detected in 31 of the 71 strains (44%). Twelve strains were harboring one island, 11 strains two islands, 7 strains three islands, and 1 strain four islands. The identified islands were grouped into 10 subtypes according to their hybridization patterns (see Figure S1 in the supplemental material). Typical examples are shown in the upper panel of Fig. 4. Two environmental isolates from aquatic habitats in the Rhine-Ruhr area (Germany) and an ear infection isolate from the United States were carrying PAGI-2 (Fig. 4B). The pattern of Fig. 4C was typical of 13 strains. The strain C-specific cargo genes encoding metabolic functions stretching from C2 to C35 were absent (see Table S3 in the supplemental material), but the region of conserved hypotheticals was present in the genomic DNA. Subtypes differed in the variable hybridization signals of ORFs C76, C77, C82, C83, and C92. Ten strains like that shown in Fig. 4D also lacked the gene contig C56 to C63, which is characterized by a consistent low G+C content of 59% (25). Figures 4E and F show singular cases: the combination of strain C cargo genes with the lack of C56 to C63 (Fig. 4E) and the strain with the fewest hybridization signals.
FIG. 4.
Examples of PAGI-2 (upper two rows) and pKLC102 (lower two rows) subtype macroarray hybridization patterns. The PAGI-2 macroarrays show (A) strain PAO (DSM1707) (negative control), (B) strain C (positive control), (C) strain 7 (subtype G1b), (D) strain 3 (subtype G2a), (E) strain 54 (subtype G2c), and (F) strain 63 (subtype G4). The pKLC102 macroarrays show (G) strain PAO (DSM1707) (negative control), (H) strain SG17M (positive control), (I) strain 6 (subtype K1c), (J) strain 10 (subtype K3c), (K) strain 36 (subtype K3d), and (L) strain 53 (subtype K4).
pKLC102-type islands were identified in 50 of the 71 strains (70%). Fifteen subtypes were differentiated by hybridization pattern (see Fig. S1b in the supplemental material), eight of which were represented by a single strain. Nine clinical and environmental isolates, including an oil field isolate from Japan, harbored pKLC102 (Fig. 4H). Strong oriV-reacting signals were observed in 10 subtypes, suggesting that these 26 strains may also harbor mobile genomic islands, as strain SG17M does. The most common subtype, K1d (see the supplemental material), was shared by 14 strains. It lacked homologs for eight pKLC102 ORFs, including the major putative virulence factor chvB CP94 (21). Weak hybridization signals indicated that the sequences of oriV and 14 other ORFs should substantially deviate from the pKLC102 blueprint. Combinatorial PCR of DNAs from subtype K1d strains revealed extrachromosomal circular forms in yields similar to those obtained with strain SG17M (data not shown), suggesting that the most abundant subtype can also replicate in its host cell.
Figure 5 summarizes the hybridization results. The signal patterns of the PAGI-2 macroarray were in accordance with the known bipartite structure of individual cargo and syntenic homologs in the sequenced islands PAGI-2 and PAGI-3 (25). The “cargo” genes C2 to C35, which have homologs with known functions in other eubacteria (see Table S3 in the supplemental material), were detected only in PAGI-2 and a close derivative thereof (subtype G2c). PAGI-2 subtypes vary in their attributes encoded by the accessory clusters of “cargo” genes. The commonalities of PAGI-2-type islands are 68 to 77 homologs that include genes related to replication or genetic mobility or that are conserved hypotheticals with unknown functions. Thirty-six of these ORFs have homologs in pKLC102. pKLC102-type islands were more diverse than PAGI-2 types in their combinations of gene cassettes, in accordance with their nested arrangements of island- and subtype-specific ORFs (21), but they apparently carried fewer strain-specific cargo genes. The backbone of more than 50% of the ORFs, including the 36 PAGI-2 homologs, was found to be highly conserved among all pKLC102-type islands. For 90% of the pKLC102 ORFs, homologous sequences were identified in the majority of islands. Only the contig CP94 to CP101 and ORF CP32 were missing in most strains (Table 4). The least abundant ORF, CP32, had the lowest G+C content (41.6%) of all ORFs in pKLC102 and served as the integration site for an integron in strain C (21), which led to large genome rearrangements in sequential isolates from individuals with cystic fibrosis (22). In summary, the diversity of PAGI-2 islands is mainly caused by the insertion of one large block of strain-specific cargo genes, whereas the diversity of pKLC102 islands is primarily generated by subtype-specific combinations of gene cassettes.
FIG. 5.
Summary of macroarray hybridization data for 31 PAGI-2-type-positive (A) and 50 pKLC102-type-positive (B) P. aeruginosa strains. The shading indicates the percentages of island-positive strains with a hybridization signal for the respective ORF. Black, ≥96% of strains positive; dark gray, 90 to 95% positive; light gray, 50 to 89% positive; white, <50% positive.
Figure 6 visualizes the outcome of the parsimony analysis of the relatedness of strains classified by their PAGI-2 and pKLC102 hybridization patterns. The broad diversity introduced by subtype-typical ORFs is highlighted by the multiple nodes in the dendrogram. Importantly, no segregation of PAGI-2 and pKLC102 subtypes was noted. In other words, no restrictions in the combination of subtypes of the two classes were observed in the 26 strains that harbor both PAGI-2- and pKLC102-type islands in their chromosomes. All nodes were occupied by a single strain, implying that microevolution in the large genomic islands contributed substantially to the interclonal diversity of our P. aeruginosa strain panel.
FIG. 6.
Relatedness of macroarray hybridization patterns of 55 PAGI-2- and/or pKLC102-positive P. aeruginosa strains. The unrooted tree is based on the parsimony analysis (“PHYLIP 3.66”) of the hybridization data.
Spread and loss of PAGI-2.
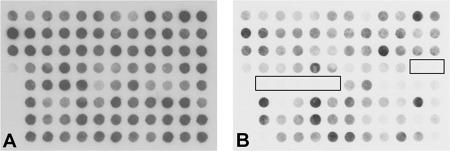
No spontaneous excision of PAGI-2 from its strain C chromosome was demonstrated during growth in vitro (see above), but nevertheless, we still suspected that PAGI-2-type islands are mobilized from their host chromosomes in vivo and can spread to other strains—at least at low frequency—because the closely related clc element of Pseudomonas putida RR21, which shares 85 to 100% nucleotide sequence identity in the conserved region with PAGI-2 (14), was found to be capable of self-transfer to other beta- and gammaproteobacteria (35, 36). Therefore, we searched our P. aeruginosa strain collection of sequential airway isolates from 36 individuals with cystic fibrosis for excision events of PAGI-2-type islands. Typical PAGI-2 genes were detected in 50 isolates collected from six chronically colonized patients. Two index cases of loss of PAGI-2 were identified (Fig. 7). One patient was chronically cocolonized with PAGI-2-positive and PAGI-2-negative clone C strains (Fig. 7A and B), where the first PAGI-2-negative strain (Fig. 7B) was isolated from the patient's airways 2 years after the acquisition of P. aeruginosa clone C. At least another PAGI-2 subtype was retained in the PAGI-2-negative clone C strain (7B). Note that the hybridization pattern shown in Fig. 7A represents the sequenced PAGI-2 of the strain C genome (25). In other words, the sequenced PAGI-2 was spontaneously excised from its host chromosome in the cystic fibrosis lung. The other case was another P. aeruginosa clone C carrier who had lost PAGI-2 in her last clone C-positive culture 17 years after the acquisition of clone C (Fig. 7C and D) and subsequently became superinfected with two other P. aeruginosa clones.
FIG. 7.
Loss of PAGI-2-type islands in sequential P. aeruginosa airway isolates from patients with cystic fibrosis. (Upper row) PAGI-2 macroarray hybridization patterns of clone C strains SG1 (A) and SG3 (B), indicating the loss of PAGI-2 in the later isolate SG3 while another PAGI-2 subtype was retained. SG1 (strain C) was isolated from the patient's first P. aeruginosa-positive sputum specimen; SG3 is the sixth isolate, collected 2 years later. (Lower row) PAGI-2 macroarray hybridization patterns of clone C strains NN18 (C) and NN86 (D), indicating the loss of a PAGI-2-type island(s) in strain NN86, which was isolated from the patient′s last clone C-positive culture 17 years after the acquisition of clone C.
The cystic fibrosis lung is an atypical and evolutionarily very recent niche for P. aeruginosa, which inhabits numerous aquatic, as well as animal and plant, host-associated environments, but to date, sequential isolates from cystic fibrosis patients are the only source that is available for longitudinal studies on the microevolution of the P. aeruginosa genome in the time frame of years or decades (20, 47, 50). For all other habitats of P. aeruginosa, the natural history and spread of a genomic island can be deduced only from cross-sectional sequence comparisons of well-characterized strains from a documented source. The PAGI-2-type clc element, for example, is almost 100% identical over the whole length of a chromosomal region in the betaproteobacterium Burkholderia xenovorans LB400 (14, 25). Similarly, a contig with close to 100% nucleotide sequence identity to PAGI-2 was identified in the betaproteobacterial Cupriavidus (formerly Ralstonia) metallidurans CH34 genome (25). To test whether this finding is typical for Cupriavidus, a collection of six C. metallidurans strains from Belgium and Germany and two Cupriavidus campinensis strains from the United States were tested for the presence of PAGI-2 by macroarray hybridization (Fig. 8). Five isolates from sewage and heavy metal ion-polluted environments, including the two strains from North America, harbored complete copies of PAGI-2 (Fig. 8A). C. metallidurans strain CH79 was carrying a PAGI-2 subtype with a complete set of cargo genes. These results demonstrate the transcontinental spread of PAGI-2-type islands into diverse clinical and environmental habitats (Table 1) and phylogenetically unrelated betaproteobacteria (genera Cupriavidus and Burkholderia) and gammaproteobacteria (genus Pseudomonas).
FIG. 8.
PAGI-2 macroarray hybridization patterns of Cupriavidus strains C. campinensis AE2701 (A) and C. metallidurans CH79 (B). The boxes highlight absent hybridization signals.
Diversity of pKLC102 and PAGI-2 type islands in proteobacteria.
More than 30 contiguous sequences in the databases were found to exhibit significant sequence similarity to at least 15 ORFs of the syntenic set of conserved hypotheticals in pKLC102 and PAGI-2. For proteobacteria other than P. aeruginosa, the boundaries of the genomic islands could be precisely defined for 17 strains by oligonucleotide signatures and tRNA integration sites. The similarity of these genomic islands was evaluated by the distance D:n0_4mer of their selection of tetranucleotides (Fig. 9). Three branches could be distinguished. Haemophilus pathogenicity islands clustered with that of Neisseria gonorrhoeae, and enterobacterial pathogenicity islands clustered with islands of the phytopathogen Erwinia carotovora and of the entomopathogen Photorhabdus luminescens. PAGI-2 and pKLC102 belonged to the most clearly separated group, characterized by a rather homogeneous profile of tetranucleotide selection (Fig. 9). The beta- and gammaproteobacterial host strains of this group are endowed with broad metabolic versatility, particularly their capability to degrade complex aliphatic and aromatic hydrocarbons, and as a corollary, they share highly similar metabolic or fitness islands that in the extreme can be identical in sequence, as has been seen for PAGI-2 in Cupriavidus and P. aeruginosa (this work) or for the clc element in P. putida RR21 and Burkholderia xenovorans LB400 (14). The host strains of the islands belong to taxa that, prior to the introduction of the rRNA gene-based taxonomy, had been classified as pseudomonads based on lifestyle and metabolic features (48). Consistent with this abandoned chemotaxonomic classification, the genomic islands of the “Pseudomonas” group encode a wide range of metabolic features and resistance determinants. The similar oligonucleotide usage of the islands probably facilitates the spread of this gene pool among strains that previously had been lumped together as “pseudomonads” due to their metabolic versatility.
FIG. 9.
Similarity of pKLC102-type genomic islands in proteobacteria based on the distance of oligonucleotide usage. The distance D:n0_4mer of tetranucleotides was calculated for each genomic island. The matrix of D values obtained was sorted for the degree of evolutionary relationship between the genomic islands by the Fitch-Margoliash criterion, assuming a constant molecular clock, and by the least-squares methods using the KITSCH program of the PHYLIP library (11). The bar indicates branch length 5. Branch lengths are not drawn exactly to scale. Short branches are exaggerated in length so that they are more visible.
The similar profiles of the tetranucleotide usage of pKLC102-type islands of phylogenetically distinct hosts suggest that the horizontal transfer of pKLC102 ancestors between “pseudomonads” happened in the past, but the data provide no clue as to why pKLC102 remained so mobile and was not irreversibly captured by its host chromosomes. pKLC102 did not lose the phage and plasmid modules, like the exoU-encoding islands that became irreversibly fixed in the chromosome by secondary events, such as rearrangements and the acquisition and loss of genetic elements (24). Inspection of the pattern skew of oligonucleotide composition (Fig. 10) provided an argument as to why the genomic islands of the pKLC102/PAGI-2 family are endowed with exceptionally high mobility. Figure 10 displays the n0_4mer PSs of 22 pKLC102-type genomic islands and their host chromosomes. The pattern skew was only a few percent for most chromosomes, in accordance with previous analyses showing that bacterial chromosomes are characterized by strand symmetry and intrastrand parity of complementary oligonucleotides (37). Oligonucleotides and their reverse complements share physicochemical properties, such as base-stacking energy, propeller twist angle, bendability, and position preference (3, 4), and occur with similar frequencies in bacterial chromosomes, in accordance with Chargaff's second parity rule (9). In contrast, no such correlation was observed for a random sequence (PS ≈ 50%). The 22 studied genomic islands of the pKLC102 family were intermediate between chromosome and random sequences. The n0_4mer PS values of 18 islands were above the 95% confidence intervals of the n0_4mer PS values of 155 completely sequenced bacterial chromosomes and 316 plasmids (37) (Fig. 10). PAGI-2 belonged to the four islands with PS values within the confidence interval. The PS of PAGI-3 was within the inner quartiles of the PS of the pKLC102 family. PAPI-1 and pKLC102, together with four other islands from Erwinia carotovora SCRI1043, Photorhabdus luminescens TT01, Pseudomonas fluorescens Pf-5, and Yersinia enterocolitica 8081, exhibited the highest PSs of ≥26%. Thus, the tetranucleotide frequency of the complementary strands was least correlated in these six members of the pKLC102 family. Stably integrated genomic islands have an atypical oligonucleotide composition compared to that of the core genome, but strand symmetry is locally maintained (38). The conjugative islands of the pKLC102/PAGI-2 family, particularly the six islands with the highest PSs, however, do not adhere to this rule and not only locally (Fig. 2), but also globally, perturb strand symmetry. Typically, the pattern skew values of the two replisomes that replicate bidirectionally from the origin and meet in the terminal region are approximately the same (37) and hence compensate for each other to values close to zero for the whole chromosome (Fig. 10). These data mean that both replisomes carry the same burden of physicochemical constraints exerted by strand asymmetry (37). The integration of a pKLC102 element perturbs this subtle balance. As long as the PS of the foreign genetic element remains high, spontaneous excision from the chromosome will occur. In summary, the high pattern skew, together with functional phage and/or plasmid modules, may account for the high mobility of the islands of the pKLC102/PAGI-2 family, suggesting that they behave like selfish parasitic DNA that is prone to horizontal spread within and among taxa.
FIG. 10.
Pattern skew of pKLC102-type genomic islands (squares) and their corresponding chromosomes (triangles). Pattern skew values (n0_4mer PS) are plotted against the logarithmic scale of sequence lengths. The gray-shaded area depicts the 95% confidence intervals of variation of n0_4mer PS values in 155 completely sequenced bacterial chromosomes and 316 plasmids (37). The PS values of n0_4mer patterns of bacterial chromosomes are typically in the range of 1 to 8%. Outliers in the investigated panel are Haemophilus ducreyi 35000HP, with a PS value of about 9%, and Xylella fastidiosa 9a5c, with an extreme value of 24.3%. The n0_4mer PS values of pKLC102-type genomic islands exceed the 95% confidence interval. The genomic islands, identified by their host strains, with the name of the island given in brackets if available, were as follows: 1, Azoarcus sp. strain EbN1; 2, Erwinia carotovora subsp. atroseptica SCRI1043; 3, Haemophilus ducreyi 3500HP; 4, Haemophilus influenzae 86-028NP (ICEHin-like); 5, Haemophilus somnus 129PT; 6, Methylobium petroleophilum PM1; 7, Photorhabdus luminescens TT01; 8, P. aeruginosa C (pKLC102); 9, P. aeruginosa C (PAGI-2); 10, P. aeruginosa PA14 (PAPI-1); 11, P. aeruginosa SG17M (PAGI-3); 12, Pseudomonas fluorescens Pf-5; 13, Pseudomonas syringae pv. syringae B728a; 14, Salmonella enterica subsp. enterica serovar Typhi CT18 (SPI-7); 15, Xylella fastidiosa 9a5c; 16, Yersinia enterocolitica 8081; 17, H. influenzae 1056.b (ICEHin 1056); 18, Neisseria gonorrhoeae MS11 (GGI); 19, Nitrosomonas eutropha C71; 20, Pseudomonas putida RR21 (clc-transposon); 21, P. syringae pv. phaseolicola 1302A (PPHG-1); 22, Yersinia pseudotuberculosis 32777 (YAPI).
The role of pKLC102/PAGI-2-type islands in bacterial evolution.
The gene repertoire of a bacterial cell consists of genes that have been transmitted vertically over long periods of time and of genes that were acquired or generated at various points in the lineage, including some very recently (26). Horizontal gene transfer provides most of the diversity in the genomic repertoire, but the majority of these horizontally acquired genes that persist in genomes are transmitted strictly vertically (26). Hence, despite substantial horizontal gene transfer, the phylogenetic relationships between taxa are robust, as indicated by the congruence of gene trees based on rRNA gene sequences, gene contents, or average amino acid identities of shared genes (23).
The genomic islands of the pKLC102/PAGI-2 family are a major exception to this rule. The islands spread across barriers of taxa while retaining the identity of their oligonucleotide signature. Family members were identified at high frequency in the global P. aeruginosa population but were also widespread among other beta- and gammaproteobacteria. Identical islands were detected in phylogenetically distinct clades and isolates from diverse habitats and geographic origins. High strand asymmetry and phage and/or plasmid modules make up the signature of this evolutionarily ancient island family.
pKLC102/PAGI-2 family members share a syntenic set of homologs. Accessory gene clusters are nestled in this core and encode island-specific features. The sequence diversity of these family-typical core genes is higher than that of vertically transmitted orthologs, probably because divergent evolutionary forces act on sequences during horizontal and vertical transmission. Genes that are irreversibly captured by the host chromosome minimize strand asymmetry and become subject to purifying selection, like the genes of the core genome (13, 19), whereas the self-transfer of genomic islands into phylogenetically distinct host chromosomes counterselects strand symmetry, loss of the island-typical oligonucleotide signature, and loss of sequence diversity. Thus, ongoing horizontal transfer maintains a higher sequence diversity of a genetic element than its irreversible incorporation into a host genome.
The functions of most genes of the conserved module of the pKLC102/PAGI-2 family are still unknown, although at least a subset should be involved in the excision, transfer, integration, or stabilization of the island (45, 46). Moreover, mutagenesis studies in PAPI-1 demonstrated that genes of the conserved core are involved in the animal and plant virulence of strain PA14 (16). Future research should unravel in more detail to what extent the syntenic gene set is not only essential for the maintenance of the genomic islands of the pKLC102/PAGI-2 family, but also affects the phenotype of its host strain, as has been demonstrated for individual cargo genes (12, 14, 31).
Supplementary Material
Acknowledgments
We cordially thank Max Mergeay, Laboratories for Microbiology and Radiobiology, SCK.CEN, Mol, Belgium, for the supply of Cupriavidus strains.
J.K., D.W., and O.R. were members of the International Research Training Group Pseudomonas: Pathogenicity and Biotechnology (IRTG 653 of the Deutsche Forschungsgemeinschaft). Financial support by the priority program Ecology of Bacterial Pathogens: Molecular and Evolutionary Aspects and by the Collaborative Research Program SFB 587 (project A9) of the Deutsche Forschungsgemeinschaft is gratefully acknowledged.
Footnotes
Published ahead of print on 28 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almagor, H. 1983. A Markov analysis of DNA sequences. J. Theor. Biol. 104:633-645. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl, ed. 1994. Current protocols in molecular biology. Wiley, New York, NY.
- 3.Baisnee, P. F., S. Hampson, and P. Baldi. 2002. Why are complementary DNA strands symmetric? Bioinformatics 18:1021-1033. [DOI] [PubMed] [Google Scholar]
- 4.Baldi, P., and P. F. Baisnee. 2000. Sequence analysis by additive scales: DNA structure for sequences and repeats of all lengths. Bioinformatics 16:865-889. [DOI] [PubMed] [Google Scholar]
- 5.Bragonzi, A., L. Wiehlmann, J. Klockgether, N. Cramer, D. Worlitzsch, G. Döring, and B. Tümmler. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261-3269. [DOI] [PubMed] [Google Scholar]
- 6.Bremer, S., T. Hoof, M. Wilke, R. Busche, B. Scholte, J. R. Riordan, G. Maass, and B. Tümmler. 1992. Quantitative expression patterns of multidrug-resistance P-glycoprotein (MDR1) and differentially spliced cystic-fibrosis transmembrane-conductance regulator mRNA transcripts in human epithelia. Eur. J. Biochem. 206:137-149. [DOI] [PubMed] [Google Scholar]
- 7.Burrus, V., J. Marrero, and M. K. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173-183. [DOI] [PubMed] [Google Scholar]
- 8.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chargaff, E. 1951. Structure and function of nucleic acids as cell constituents. Fed. Proc. 10:344-360. [PubMed] [Google Scholar]
- 10.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1986. Distance methods: reply to Farris. Cladistics 2:130-143. [DOI] [PubMed] [Google Scholar]
- 12.Frantz, B., and A. M. Chakrabarty. 1987. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc. Natl. Acad. Sci. USA 84:4460-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, R., J. W. Drake, and A. L. Hughes. 2004. Genome-wide patterns of nucleotide substitution reveal stringent functional constraints on the protein sequences of thermophiles. Genetics 167:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillard, M., T. Vallaeys, F. J. Vorholter, M. Minoia, C. Werlen, V. Sentchilo, A. Pühler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg, J. B., and D. E. Ohman. 1984. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J. Bacteriol. 158:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, J., R. L. Baldini, E. Deziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman, and L. G. Rahme. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuer, T., C. Bürger, G. Maass, and B. Tümmler. 1998. Cloning of prokaryotic genomes in yeast artificial chromosomes: application to the population genetics of Pseudomonas aeruginosa. Electrophoresis 19:486-494. [DOI] [PubMed] [Google Scholar]
- 18.Hoof, T., J. R. Riordan, and B. Tümmler. 1991. Quantitation of mRNA by the kinetic polymerase chain reaction assay: a tool for monitoring P-glycoprotein gene expression. Anal. Biochem. 196:161-169. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, A. 2004. Evidence for abundant slightly deleterious polymorphisms in bacterial populations. Genetics 169:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiewitz, C., K. Larbig, J. Klockgether, C. Weinel, and B. Tümmler. 2000. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNALys gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology 146:2365-2373. [DOI] [PubMed] [Google Scholar]
- 21.Klockgether, J., O. Reva, K. Larbig, and B. Tümmler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kresse, A. U., S. D. Dinesh, K. Larbig, and U. Römling. 2003. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 47:145-158. [DOI] [PubMed] [Google Scholar]
- 23.Konstantinidis, K. T., and J. M. Tiedje. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187:6258-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulasekara, B. R., H. D. Kulasekara, M. C. Wolfgang, L. Stevens, D. W. Frank, and S. Lory. 2006. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188:4037-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tümmler. 2002. Gene islands integrated into tRNA Gly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerat, E., V. Daubin, H. Ochman, and N. A. Moran. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 28.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morales, G., L. Wiehlmann, P. Gudowius, C. van Delden, B. Tümmler, J. L. Martinez, and F. Rojo. 2004. Structure of Pseudomonas aeruginosa populations analyzed by single nucleotide polymorphism and pulsed-field gel electrophoresis genotyping. J. Bacteriol. 186:4228-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, T. A., C. Werlen, J. Spain, and J. R. van der Meer. 2003. Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ. Microbiol. 5:163-173. [DOI] [PubMed] [Google Scholar]
- 32.Rajanna, C., J. Wang, D. Zhang, Z. Xu, A. Ali, Y. M. Hou, and D. K. Karaolis. 2003. The vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J. Bacteriol. 185:6893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakin, A., C. Noelting, P. Schropp, and J. Heesemann. 2001. Integrative module of the high-pathogenicity island of Yersinia. Mol. Microbiol. 39:407-415. [DOI] [PubMed] [Google Scholar]
- 34.Ramos, J. L., ed. 2004. Pseudomonas, vol. 1 to 3. Kluwer Academic, New York, NY.
- 35.Ravatn, R., S. Studer, D. Springael, A. J. B. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravatn, R., S. Studer, A. J. B. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reva, O. N., and B. Tümmler. 2004. Global features of sequences of bacterial chromosomes, plasmids and phages revealed by analysis of oligonucleotide usage patterns. BMC Bioinformatics 5:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reva,O. N., and B. Tümmler. 2005. Differentiation of regions with atypical oligonucleotide composition in bacterial genomes. BMC Bioinformatics 6:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Römling, U., J. Greipel, and B. Tümmler. 1995. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol. Microbiol. 17:323-332. [DOI] [PubMed] [Google Scholar]
- 40.Römling, U., T. Heuer, and B. Tümmler. 1994. Bacterial genome analysis by pulsed field gel electrophoresis techniques. Adv. Electrophoresis 7:353-406. [Google Scholar]
- 41.Römling, U., K. D. Schmidt, and B. Tümmler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 42.Sakellaris, H., S. N. Luck, K. Al-Hasani, K. Rajakumar, S. A. Turner, and B. Adler. 2004. Regulated site-specific recombination of the she pathogenicity island of Shigella flexneri. Mol. Microbiol. 52:1329-1336. [DOI] [PubMed] [Google Scholar]
- 43.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert, S., S. Dufke, J. Sorsa, and J. Heesemann. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51:837-848. [DOI] [PubMed] [Google Scholar]
- 45.Sentchilo, V., R. Ravatn, C. Werlen, A. J. Zehnder, and J. R. van der Meer. 2003. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J. Bacteriol. 185:4530-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sentchilo, V., A. J. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 47.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 49.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 50.Tümmler, B. 2006. Clonal variations in Pseudomonas aeruginosa, p. 35-68. In J.-L. Ramos and R. C. Levesque (ed.), Pseudomonas, vol. 4, p. 35-68. Springer, Heidelberg, Germany. [Google Scholar]
- 51.Ubeda, C., M. A. Tormo, C. Cucarella, P. Trotonda, T. J. Foster, I. Lasa, and J. R. Penades. 2003. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol. 49:193-210. [DOI] [PubMed] [Google Scholar]
- 52.Weinel, C., K. E. Nelson, and B. Tümmler. 2002. Global features of the Pseudomonas putida KT2440 genome sequence. Environ. Microbiol. 4:809-818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.