Abstract
The enzyme diversity of the cellulolytic system produced by Clostridium cellulolyticum grown on crystalline cellulose as a sole carbon and energy source was explored by two-dimensional electrophoresis. The cellulolytic system of C. cellulolyticum is composed of at least 30 dockerin-containing proteins (designated cellulosomal proteins) and 30 noncellulosomal components. Most of the known cellulosomal proteins, including CipC, Cel48F, Cel8C, Cel9G, Cel9E, Man5K, Cel9M, and Cel5A, were identified by using two-dimensional Western blot analysis with specific antibodies, whereas Cel5N, Cel9J, and Cel44O were identified by using N-terminal sequencing. Unknown enzymes having carboxymethyl cellulase or xylanase activities were detected by zymogram analysis of two-dimensional gels. Some of these enzymes were identified by N-terminal sequencing as homologs of proteins listed in the NCBI database. Using Trap-Dock PCR and DNA walking, seven genes encoding new dockerin-containing proteins were cloned and sequenced. Some of these genes are clustered. Enzymes encoded by these genes belong to glycoside hydrolase families GH2, GH9, GH10, GH26, GH27, and GH59. Except for members of family GH9, which contains only cellulases, the new modular glycoside hydrolases discovered in this work could be involved in the degradation of different hemicellulosic substrates, such as xylan or galactomannan.
Cellulose, a long polymer of β-1,4-glucose, is the major component of the plant cell wall (39). Cellulolytic bacteria and fungi secrete many different types of cellulases to catalyze efficient degradation of this recalcitrant substrate. Many cellulolytic, anaerobic microorganisms secrete multienzyme complexes, called cellulosomes (2, 9, 41). The large number and the diversity of enzymes secreted by these microorganisms reflect the complex chemical composition of the polysaccharides surrounding the cellulose fibrils in the plant cell wall. Cellulosomal enzymes are active against numerous substrates, such as crystalline cellulose, and the backbone or side chains of xylans, mannans, and pectins, and the enzymes display various modes of action (endo-, exo-, or processive substrate degradation) (2, 9, 41). Most of the cellulosomal enzymes cleave glycosidic bonds by hydrolysis, but a few of them utilize a beta-elimination mechanism (37, 41). Cell wall-degrading enzymes are classified into three distinct groups: glycoside hydrolases, polysaccharide lyases, and carbohydrate esterases (CAZY database [http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html]) (6). Each cellulosomal enzyme contains, in addition to its catalytic domain, a noncatalytic domain termed a dockerin which is able to interact with the repetitive homologous cohesin domains of a noncatalytic scaffolding protein (2, 9, 41). A carbohydrate binding module (CBM) promotes the binding of the scaffolding protein and therefore the binding of the cellulosome to cellulose.
In addition to cellulosomes, saprophytic clostridia secrete noncellulosomal cellulases and hemicellulases (2, 8, 26, 41). These enzymes, which lack a dockerin domain, are not incorporated into the multienzyme complexes. They bind to the substrate by specific CBMs and constitute a complementary enzymatic system for the hydrolysis of plant cell wall polysaccharides. Both free (noncellulosomal) and cellulosomal enzymes are necessary for effective plant cell wall degradation, and the two enzymatic systems probably act synergistically (8, 26).
At this time, the sequence of the complete genome of the mesophilic organism Clostridium cellulolyticum is not available. However, 12 genes encoding C. cellulolyticum cellulosomal components have been found in a large 26-kb cluster, cipC-cel48F-cel8C-cel9G-cel9E-orfX-cel9H-cel9J-man5K-cel9M-rgl11Y-cel5N (29, 30, 31, 35). The first gene of the cluster, cipC, encodes the scaffolding protein CipC, composed of a family 3 CBM (CAZY database [http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html] CBM classification), two X2 domains having unknown functions, and eight cohesins (35). Each cohesin is able to interact with a dockerin domain. Eight genes encode cellulases, which belong to families GH48, GH8, GH9, and GH5. The man5K and rgl11Y genes encode a mannanase and a rhamnogalacturonan lyase, respectively (37, 38). In the gene cluster, orfX encodes a membrane-associated protein having an unknown function and harboring a cohesin domain (35). In addition to this cluster of genes, two isolated genes encoding GH5 cellulases (cel5A and cel5D) and a two-gene cluster (cel5I-cel44O) have been isolated (29). Cel5I, encoded by the cel5I gene and lacking dockerin, is a noncellulosomal enzyme. Thus, 13 genes encoding dockerin-containing proteins are known to date, while only eight cohesins are present in CipC. The number of enzymes potentially able to bind to the scaffolding protein CipC is higher than the number of cohesins in CipC, indicating that there is heterogeneity in the composition of the cellulosomes produced. Moreover, about 12 dockerin-containing proteins were detected by one-dimensional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis of the cellulolytic system produced by cellulose-grown cells of a cipC insertional mutant of C. cellulolyticum designated cipCMut1 (31). None of the enzymes encoded by the cel gene cluster was found in this strain, indicating that numerous dockerin-containing proteins are encoded by genes not yet isolated.
The aim of the present study was to investigate the enzyme composition and diversity of the cellulolytic system of C. cellulolyticum grown on crystalline cellulose. For this purpose, the cellulolytic system produced by C. cellulolyticum was analyzed by two-dimensional (2D) PAGE. The proteins containing a dockerin domain were detected using a biotin-labeled cohesin-containing protein. Of the 30 dockerin-containing proteins detected, most of the known proteins, namely, Cel48F, Cel8C, Cel9G, Cel9E, Man5K, Cel9M, Cel5A, Cel5N, Cel9J, and Cel44O, were located on the 2D gel. In addition, the cellulolytic system of C. cellulolyticum contains 30 noncellulosomal proteins that lack a dockerin domain. Genes encoding new dockerin-containing proteins, detected by 2D PAGE, were cloned and sequenced by a suitable PCR method, designated Trap-Dock PCR, followed by DNA walking.
MATERIALS AND METHODS
Bacterial strains.
C. cellulolyticum ATCC 35319 was used as the source of genomic DNA. For production of cellulosomes and noncellulosomal enzymes, C. cellulolyticum was grown anaerobically for 6 days at 32°C on basal medium (H10) (16) supplemented with MN300 cellulose (7.5 g/liter; Serva).
Preparation of the cellulolytic system of C. cellulolyticum.
C. cellulolyticum was grown on crystalline cellulose as described above. A 6-day culture was filtered through a 3-μm-pore-size glass filter (GF/D glass microfiber filter; Whatman). Residual cellulose fibrils retained by the filter, on which cellulosomal and some noncellulosomal enzymes (free enzymes) were bound, were washed successively with 50 mM phosphate buffer (pH 7) and 12.5 mM phosphate buffer (pH 7). A fraction, called the Fc fraction, containing the cellulolytic system (cellulosomal and noncellulosomal enzymes) was eluted from the residual cellulose with water as described by Perret et al. (38). The Fc fraction was concentrated by ultrafiltration with an Amicon cell (membrane molecular mass cutoff, 10 kDa).
2D PAGE, Western blotting, and detection of dockerin-containing proteins.
Two 7-cm-long ReadyStrip linear immobilized pH gradient (IPG) strips (pH 4 to 7 and pH 3.9 to 5.1; Bio-Rad, Richmond, CA) were used. Seventy-five or 150 μg of the Fc fraction was incubated for 1 h at room temperature in 125 μl (final volume) of denaturing solution containing 8 M urea, 9 M thiourea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), traces of bromophenol blue, 2.8 mg/ml dithiothreitol, and 2% (vol/vol) IPG buffer (pH 4 to 7 or pH 3.9 to 5.1). After this incubation, the denaturing solution containing the sample was used to passively hydrate the IPG strips. The IPG strips were then placed on a Multiphor II electrophoresis unit (Amersham Pharmacia Biotech) for isoelectric focusing (IEF). For the second dimension, the IPG strips were equilibrated for 20 min in 10 ml of equilibration solution (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% [vol/vol] glycerol, 2% [wt/vol] SDS, traces of bromophenol blue, 10 mg/ml dithiothreitol). The IPG strips were then subjected to a second electrophoresis on an SDS-polyacrylamide gel containing 8% (vol/vol) or 10% (vol/vol) polyacrylamide (Prosieve 50 gel solution for protein analysis; TEBU). Electrophoresis was performed using a Hoefer mini VE system (Amersham Pharmacia Biotech). After fixation (10% [vol/vol] acetic acid, 25% [vol/vol] isopropanol), proteins were visualized by silver or Coomassie blue staining (21, 34). A protein silver-staining kit (Amersham Bioscience) was used.
Proteins were blotted onto nitrocellulose (Ba83; Schleicher & Schuell) for Western blotting with specific antibodies (31, 36) and for detection of dockerin-containing protein with biotinylated miniCipC (36).
N-terminal sequencing.
After 2D PAGE, proteins were transferred onto a polyvinylidene difluoride membrane (Schleicher & Schuell), incubated with fixation buffer, stained with Ponceau Red, cut out, and sequenced by the Edman method by using an Applied Biosystems 470A sequencer (Plateforme Protéome, Institut de Biologie Structurale et Microbiologie [IBSM], CNRS, Marseille, France).
Zymograms.
Zymograms with xylan (oat spelt xylan; Sigma) and carboxymethyl cellulose (CMC) were obtained by incorporating 0.1% (wt/vol) of each substrate into the polyacrylamide gels used for second-dimension electrophoresis (3). After electrophoresis, the gels were washed twice 30 min with 100 mM potassium phosphate buffer (pH 7) containing 25% (vol/vol) isopropanol and twice for 15 min with the same buffer without isopropanol. The gels were then incubated for 90 min at 37°C in 100 mM potassium phosphate buffer (pH 7). The enzymatic activities were visualized after 30 min of staining with Congo red (0.5%, wt/vol) and destaining with 1 M NaCl.
Trap-Dock PCR.
Genomic DNA from C. cellulolyticum (150 ng) was used as a template to amplify genes encoding P90, P50, and P41a. Three oligonucleotide pairs were used (oligonucleotides dock-In and p90dir, oligonucleotides dock-In and p50dir, and oligonucleotides dock-In and p41dir) at final concentrations of 10 μM for primer dock-In and 2 μM for the other primers (Table 1). The degeneracy of each primer was decreased by use of inosine for substituting 4-base wobbles instead of using all 4-base substitutions. Taq DNA polymerase (Promega), which allowed synthesis of DNA with an inosine-containing template, was used for amplification. Compared to standard PCR conditions, lower annealing and elongation temperatures were used (40°C and 65°C, respectively) and higher PCR cycle numbers (40 to 45 cycles) were required. Each amplified DNA fragment was directly cloned into the pGEM-T vector (Promega) to obtain the recombinant plasmids designated pGp90, pGp50, pGp41/1200, and pGp41/400. The recombinant vector pGp41/1200 contains a 1,200-bp fragment amplified with primers dock-In and p41dir, whereas pGp41/400 contains a 400-bp fragment amplified with the same oligonucleotides. Each insert was sequenced (dye terminator method) by GATC Biotech (Germany).
TABLE 1.
Primer sequences
| Primer | Sequencea |
|---|---|
| dock-In | ITCDATIGCITCDATRTTNCCITCRTTRTTNACITCICC |
| p41dir | GCNACNCCIACIGGIAARIGNITNAARGAYGTNCA |
| p41d1 | CTAACAATTATAAGGTTCATGGA |
| p41r1 | AAACTATTTTGTGAAGGTTCTAA |
| p41d2 | ACTAATTCGACTGAACTTCAAA |
| p41r2 | CACGTACATCCATCTCAGTTAT |
| pGH9d1 | GACAACGGTAAGCTTCTTTGGGGAGAGG |
| pGH9d2 | TTGAATACAAAAATGGCGTAGGTTCC |
| pGH9r2 | ACTCTGCCGGCATTATCTGTGTACCC |
| pGH9r3 | GATACTGCATCAAAGTCCGGCG |
| pGH9d4 | CAGGCAGGTCGTGATGAAAGTTAC |
| pGH9r4 | CTTACATCATCCCAGCAGTGCG |
| pGH9d5 | TGCCGGAAGCGAAACATTTTG |
| pGH9r5 | ATGTGTTTAAGAGTAGGACGTTG |
| p90dir | GGNACIGAITAIAAITAIGCIAARITNITNCAITA |
| p90d1 | GGCAGAATAAGAGTGTGCTTGACCC |
| p90r1 | TCCAGGTGTATTGGTTATTTTCGCC |
| p90d2 | TGTGTTTGTGACAACGGCGTTCTAG |
| p90r2 | GATGACTCCAGTCATCAGTCGGATTCCA |
| pMand1 | AACATACCCTTTCCCGTCAGACG |
| pManr1 | TGTAAGATGCCATCCGTACCAAGG |
| pMand2 | GCTGTACAGACTGCTATTCG |
| pManr2 | CTGAAAAGTACTGGAACACCT |
| p50dir | TNAAYAAYICNITNGGNITNACNCCICCIATGGG |
| p50d1 | GGATGCAAAGACATTTGCTTCATGGG |
| p50r1 | CAAGTTCCCATTGGAATCTCTTGCAGG |
| p50d2 | GACCAGCGGAGGCAATATTGAGATTAG |
| p50r2 | TATGAGCCCGTAAAACTGCCTTTGTCAG |
| p50d5 | CGTATACTTGTTCCGGCTGGG |
| p50r5 | GAGGCAATTGTACCCGCACC |
| p50d12 | CACTAAGTCCGACCCGACAG |
| p50r12 | GGTATGGCCTTGAATCAGGG |
| p59d6 | GATCTGGCAAGCGGCTATTAC |
| p59r6 | TGATAGAGATGTTATGCCATGCA |
| p59d7 | ATGTGGATTTCGGCGATGG |
| p59r7 | CCACATCCTCTCCGCCTTC |
In degenerate primers, I = inosine, Y = C or T, N = A, G, C, or T, R = A or G, and D = G, A, or T.
Chromosomal DNA walking.
5′ and 3′ DNA regions of each gene were amplified by inverse PCR (42) (Expand High Fidelity; Roche Applied Science) using total chromosomal DNA of C. cellulolyticum digested with several restriction enzymes (EcoRI, AccI, PvuII, PstI, SspI, HaeIII, HaeII, and HindIII) and circularized. Primers listed in Table 1 were used to amplify DNA sequences flanking the xyn10A gene (p41d1, p41r1, p41d2, and p41r2), to amplify DNA sequences flanking the cel9Q gene (pGH9d1, pGH9d2, pGH9r2, pGH9r3, pGH9d4, pGH9r4, pGH9d5, and pGH9r5), to amplify DNA sequences flanking the cel9P gene (p90d1, p90r1, p90d2, p90r2, pMand1, pManr1, pMand2, and pManr2), and to amplify DNA sequences flanking the gal27A gene (p50d1, p50r1, p50d2, p50r2, p50d5, p50r5, p50d6, p50r6, p50d7, p50r7, p50r12 p50d12, p59r6, p59d6, p59r7, and p59d7).
Amino acid sequence analysis.
Protein sequences were compared to the amino acid sequences in the NCBI sequence databases using the BLAST program. The statistical significance of matches with database-registered proteins was calculated for each new protein using two different programs, protein-protein BLAST (BlastP) and a search for short, nearly exact matches.
Protein domain compositions were analyzed by using ProDom (protein domain analysis; http://protein.toulouse.inra.fr/prodom/current/html/home.php), a database of protein families and domains.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under the following accession numbers: DQ778331 for a 2.286-kb DNA fragment containing xyn10A, DQ778332 for a 3.012-kb DNA fragment containing cel9Q, DQ778333 for a 8.777-kb DNA fragment containing genes encoding CBM-containing proteins (orf3, gal27A, gal59A, and orf4), and DQ778334 for a 5.411-kb DNA fragment containing man26A and cel9P.
RESULTS
Two-dimensional electrophoresis profile of the cellulolytic system of C. cellulolyticum.
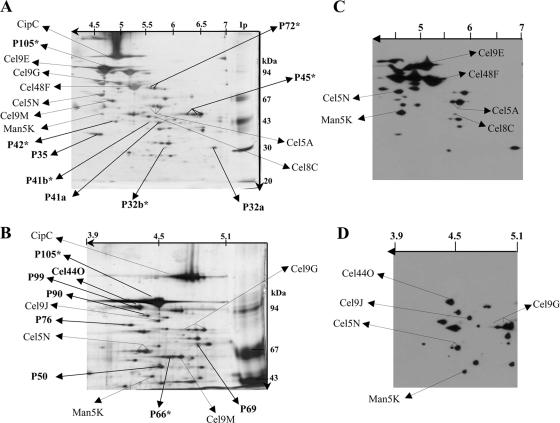
The components of the cellulolytic system of C. cellulolyticum (Fc fraction), prepared as described in Materials and Methods, were separated by two 2D PAGE (Fig. 1A and B). About 60 proteins were silver stained, which was significantly more than the 15 proteins detected by a previous one-dimensional SDS electrophoresis analysis of the Fc fraction stained with Coomassie blue (15).
FIG. 1.
Composition of the cellulolytic system of C. cellulolyticum and localization of the dockerin-containing proteins. (A and B) Proteins of the Fc fraction were separated on two-dimensional gels (12% polyacrylamide with IEF at pH 4 to 7 [A] or 8% polyacrylamide with IEF at pH 3.9 to 5.1 [B]) and silver stained. The gel images are oriented with the IEF dimension horizontal and the SDS-PAGE dimension vertical. (C and D) After blotting, proteins separated by 2D electrophoresis on 12% polyacrylamide with IEF at pH 4 to 7 (C) and proteins separated by 2D electrophoresis on 8% polyacrylamide with IEF at pH 3.9 to 5.1 (D) were incubated with biotinylated miniCipC. The positions of known proteins, including CipC, Cel48F, Cel8C, Cel9G, Cel9E, Man5K, Cel9M, Cel5A, Cel5N, Cel9J, and Cel44O are indicated by arrows, as are the positions of some unknown proteins. Each unknown protein, indicated by bold type, is designated “P” followed by a number referring to the molecular mass of the protein. The N-terminal sequences of these proteins and their enzymatic activities as determined by zymogram assays are shown on Table 3. P32b, P41b, P42, P45, P66, P72, and P105 (in bold type with an asterisk) did not interact with biotinylated miniCipC and presumably lack dockerin.
Eight of the 13 known cellulosomal proteins (CipC, Cel48F, Cel8C, Cel9G, Cel9E, Man5K, Cel9M, and Cel5A) were identified by 2D Western blot analysis using specific antibodies. As expected, the scaffolding protein CipC appears to be one of the most abundant proteins of the system, together with the dockerin-containing cellulases Cel48F and Cel9E (Fig. 1A and B). N-terminal sequencing performed for CipC, Cel48F, Cel9E, Cel9M, Cel9G, Cel5A, and Man5K confirmed the results obtained by 2D Western blotting (Table 2). Antibodies against Cel5A and against Cel9G recognized two and three proteins with slightly different isoelectric points and the same apparent molecular mass. These proteins, having the same amino-terminal sequence, appeared to be isoforms of the cellulases Cel5A and Cel9G.
TABLE 2.
Known proteins belonging to the cellulolytic system of C. cellulolyticum detected on two-dimensional gels
| Protein | N-terminal sequencea | Theoretical isoelectric point | Theoretical molecular mass (Da) |
|---|---|---|---|
| CipC | AGTGWSVQF | 4.96 | 155,725 |
| Cel48F | ASSPANKVYQ | 5.15 | 77,620 |
| Cel8C | ADQIPFPYDA | 5.47 | 47,199 |
| Cel9G | AGTYNYGEAL | 5.04 | 76,109 |
| Cel9E | FALVGAGDLI | 5.16 | 94,165 |
| Man5K | ATTYKLGDVD | 4.63 | 45,071 |
| Cel9M | AGTHDYSTAL | 4.84 | 54,504 |
| Cel5A | YDASLIPNLQ | 5.44 | 50,710 |
| Cel5N | AVDTNNDDWL | 4.61 | 56,641 |
| Cel44O | ADXSXAINV | 4.60 | 90,898 |
| Cel9J | GSTTPPFNYA | 4.86 | 81,194 |
Except for Cel8C, the N-terminal sequences were determined experimentally. The extremely close migration zones of Cel44O and P99 made individual N-terminal-extremity sequencing very difficult, and in the N-terminal sequence of Cel44O, X indicates an amino acid that was not clearly identified.
Since antibodies raised against the five remaining known cellulosomal proteins (Cel5N, Cel9J, Cel9H, Cel5D, and Cel44O) were not available, the putative migration zones of these proteins were determined on the basis of their theoretical isoelectric points and molecular masses (Table 2), and N-terminal sequencing was performed. This procedure resulted in localization of two Cel5N isoforms, Cel9J, and Cel44O (Fig. 1A and B).
P105, a major protein in terms of abundance, which was previously detected by Maamar et al. (31) and had the N-terminal sequence ASGSYEFEDQATVLKDLGIWQGDTT, was easily detected on 2D gels (Fig. 1A and B). Its N terminus did not exhibit any similarity with known proteins involved in plant cell wall degradation.
Dockerin-containing proteins were detected with biotinylated miniCipC (36). About 30 distinguishable proteins possess a dockerin domain, including the two major cellulases (Cel9E and Cel48F), as well as Cel8C, Cel5A, Man5K, and Cel9G (Fig. 1C and D). Surprisingly, Cel9M, a cellulosomal protein, did not interact with the biotinylated miniCipC in the experimental conditions used (Fig. 1D). This result indicates that the number of cellulosomal proteins is probably underestimated by the method used. So far, 13 genes encoding C. cellulolyticum cellulosomal cellulases have been isolated and sequenced. Of the 30 dockerin-containing proteins revealed by 2D PAGE of the cellulolytic system of C. cellulolyticum, 10 were already known. Thus, at least 20 cellulosomal proteins produced by C. cellulolyticum grown on crystalline cellulose remain unknown.
In addition, the cellulolytic system of C. cellulolyticum contains about 30 proteins that lack a dockerin domain.
N-terminal sequencing of new Fc fraction components.
The N-terminal sequences of 11 new components of the C. cellulolyticum cellulolytic system were determined by Edman sequencing of proteins separated by 2D electrophoresis (Table 3 and Fig. 1A and B). For some of these components (P90, P50, P42, P35, and P32a) a BLAST search (BLAST for short nearly exact matches) against the sequences deposited in the NCBI database revealed identities with carbohydrate-active enzyme modules (Table 3). The N terminus of dockerin-containing protein P90 exhibits 90% identity with the N terminus of the catalytic domain of Cel9T from Clostridium thermocellum (28), and P50 exhibits 83% identity with the N terminus of the catalytic module of the Aga27A α-galactosidase from Clostridium josui (22). The level of identity between the P35 N-terminal sequence and the dockerin domain of the EngY cellulase from Clostridium cellulovorans (9) was 81%. Although P42 was not detected with biotinylated miniCipC, its amino-terminal sequence exhibits similarity to the dockerin domain of a putative cellulosomal enzyme detected by sequencing of the genome of C. thermocellum (Table 3). Finally, the BLAST search carried out using P32 revealed 88% identity with the N terminus of Xyn11D, a xylanase from the rumen protozoan Polyplastron multivesiculatum (7).
TABLE 3.
New components of the Fc fraction of C. cellulolyticum
| Proteina | N-terminal sequence | Identity with another protein or domain
|
Zymogram activityc | |
|---|---|---|---|---|
| %b | Protein or domain (accession no.) | |||
| P99 | AAAIXVSIDTI | NS | CMCase | |
| P90 (= Cel9P) | GTEYNYAKLLQYXQ | 90 | Cel9T, C. thermocellum (ABO44407.2) | ND |
| P76 | ATDSTXPGFXVE | NS | ND | |
| P72d | MTTNKKLEA | NS | ND | |
| P66d | AEPGVA | NS | CMCase | |
| P69 | Not sequenced | NS | CMCase | |
| P50 | LNNSLGLTPXMG | 83 | Aga27A, C. josui (ABO25362.1) | ND |
| P45d | XKLVPAHGGKSLV | ND | ||
| P42d | LEPPCLYGDXNGD | 69 | Dockerin domain of a 537-amino acid protein (CtheDRAFT 0895 ZP_00510083 from C. thermocellum) | ND |
| P41a (= Xyn10A) | ATPTGKRLKEVQ | NS | Xylanase | |
| P41bd | Not sequenced | NS | Xylanase | |
| P32a | ATXITENKTGTI | 88 | Xyn11D, P. multivesiculatum (AJ516958) | Xylanase |
| P32bd | Not sequenced | NS | Xylanase | |
| P35 | EASSADNEVVGSYTLFGDLNND | 81 | Eng9Y dockerin domain, C. cellulovorans (AAG59608) | ND |
Each protein was designated “P” followed by a number indicating the molecular mass. P90 and P41a correspond to Cel9P and Xyn10A, respectively. The genes encoding these two proteins were cloned by Trap-Dock PCR and sequenced in this study (for details see Fig. 2 and the text). The proteins listed share CMCase or xylanase activity detected by zymogram analysis after 2D electrophoresis or have been subjected to Edman sequencing.
NS, no significant identity found.
ND, not detected.
Protein that did not interact with the biotinylated miniCipC.
Zymogram pattern of the Fc fraction.
Proteins of the cellulolytic system (Fc fraction) of C. cellulolyticum, separated by 2D PAGE, were examined for the ability to hydrolyze two different substrates, xylan and CMC, incorporated into the gels. Carboxymethyl cellulase (CMCase) activity was detected for several dockerin-containing proteins (Cel44O and/or P99, P69, Cel5N, Cel5A, Cel9M, and Cel8C) and for one noncellulosomal protein (P66) (Table 3). CMC is typically hydrolyzed by endocellulases, such as Cel5A, Cel9M, and Cel8C (41). Cel5N, a noncharacterized member of the GH5 family encoded by the last gene of the 26-kb cel gene cluster, appears to be highly active against this substrate. Family GH5 includes enzymes active against cellulose (such as Cel5A), glucan, or mannan (such as Man5K) (12, 38). Cel5N, which is able to hydrolyze CMC, is probably a new GH5 cellulase. Cel44O and P99 have identical isoelectric points and similar molecular masses, and thus the CMCase activity detected in their migration zone can be attributed to Cel44O or P99 or both.
To determine the distribution of xylanase activities in the Fc fraction, we performed a zymogram analysis with xylan. Two major proteins with xylanase activities were detected at about 41 kDa (data not shown). One of these proteins, designated P41a, contained a dockerin module, and the other protein, designated P41b, was unable to interact with biotinylated miniCipC (Fig. 1A and C) and probably lacked dockerin. The N terminus of P41a is shown in Table 3. Two proteins with minor activity with xylan were found at a molecular mass of 32 kDa: P32a, a dockerin-containing protein, and P32b, a protein lacking dockerin (Table 3). P32a could belong to family GH11, as indicated by the NCBI BLAST search results (Table 3). The endocellulase Cel5A also showed weak activity with xylan, as previously reported (12).
Cloning of genes encoding new dockerin-containing proteins.
A consensus sequence was derived from alignment of the known C. cellulolyticum dockerin sequences. In this consensus sequence, GDVNNDGNIDAID, residues at positions 1, 2, 6, 10, 11, and 13 (underlined) are invariant. A reverse inosine-containing degenerate oligonucleotide designated dock-In (5′-ITCDATIGCITCDATRTTNCCITCRTTRTTNACITCICC-3′) was designed by back-translation of the consensus amino acid sequence (Table 1). This primer is putatively able to anneal to all DNA regions encoding a dockerin module.
The N-terminal amino acid sequence of a dockerin-containing protein detected by 2D PAGE could be used to design a direct inosine-containing degenerate oligonucleotide. This direct oligonucleotide along with the reverse oligonucleotide dock-In could be used to amplify by PCR the corresponding gene in genomic DNA. In this study, genes encoding P90, P50, and P41a were chosen as target genes.
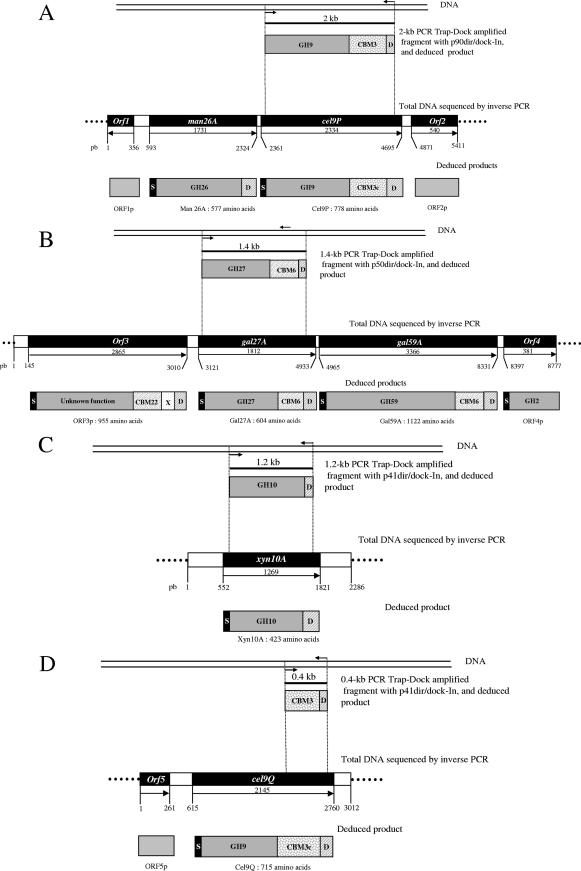
Trap-Dock PCR performed using oligonucleotides p90dir and dock-In and oligonucleotides p50dir and dock-In allowed amplification of two fragments, which were 2 and 1.4 kb long, respectively (Fig. 2A and B). Two PCR products, which were 1.2 and 0.4 kb long, were obtained using primers p41dir and dock-In (Fig. 2C and D). The PCR products were cloned into the pGEM-T Easy vector and then sequenced. BlastP and ProDom analyses with the NCBI protein sequence database were performed for each deduced amino acid sequence. For the 2-kb fragment, the deduced amino acid sequence revealed a modular cellulase containing an N-terminal catalytic domain belonging to family GH9, associated with a family 3c CBM (Fig. 2A). The translated sequence of the 1.4-kb DNA fragment consisted of two distinct domains, a family GH27 catalytic domain and a CBM6 domain (Fig. 2B). The sequences of the 1.2- and 0.4-kb PCR products, obtained with oligonucleotides p41dir and dock-In, revealed two distinct new open reading frames (ORFs) (Fig. 2C and D). The first ORF encoded a GH10 protein (Fig. 2C). This result is in good agreement with the catalytic activity detected with a zymogram, since all known enzymes of this family are xylanases. The 0.4-kb PCR product, obtained by less specific annealing of the p41dir primer, encoded a small peptide consisting of a CBM3c domain (Fig. 2D). This type of CBM has been found to be associated only with family GH9 catalytic domains (14, 32). The peptide deduced from translation of the 0.4-kb Trap-Dock PCR product could probably be part of a new modular family 9 cellulase (Fig. 2D).
FIG. 2.
Isolation and characterization of new genes encoding dockerin-containing proteins. Genes encoding Cel9P (P90) (A), Gal27A (B), and Xyn10A (P41a) (C) were amplified from the total genomic DNA of C. cellulolyticum by Trap-Dock PCR using convergent inosine-containing degenerate primers. A nonspecific fragment (length, 0.4 kb) was amplified with oligonucleotides p41dir and dock-In (D). Chromosomal DNA walking was performed to clone DNA flanking regions of each gene. S, signal sequence; GH, glycoside hydrolase; D, dockerin domain; X, domain with unknown function. For glycoside hydrolases and CBMs, the number indicates the family.
DNA walking and nucleotide sequence analysis.
For each sequence obtained, DNA regions located upstream and downstream were amplified by inverse PCR. The primers used are listed in Table 1.
A 5,411-bp DNA fragment from cel9P was sequenced (Fig. 2A). cel9P encodes a 778-amino-acid protein. The putative N terminus of the mature form of Cel9P, GTEYNYAKLLQYSQ, is the same as the N terminus determined for P90 (Table 3). The catalytic domain of Cel9P is 69% identical to Cel9T from C. thermocellum (28). In addition, Cel9P contains a CBM3c domain. GH9 cellulases can have different modular arrangements. The catalytic domain can be isolated, as it is in Cel9T from C. thermocellum, fused to a CBM3c domain, or fused to a CBM4 domain and an immunoglobulin-like domain. So far, three GH9 cellulases containing a CBM3c domain have been identified in C. cellulolyticum: Cel9G (accession number M87018), Cel9H (accession number AF316823), and Cel9J (accession number AF316823). These enzymes exhibit 33, 28, and 38% identity with Cel9P, respectively. The 1,731-bp ORF located upstream of cel9P encodes a cellulosomal GH26 mannanase, designated Man26A (Fig. 2A). Man26A from C. cellulolyticum exhibits a high level of identity (72%) to Man26B from C. thermocellum (accession number AB044406; Cthe DRAFT 2304 ZP_00504676) (27). Upstream of man26A, orf1 encodes a protein exhibiting homology with a PrsA foldase protein precursor from Clostridium acetobutylicum (accession number Q97E99) (24). Downstream of cel9P, orf2 encodes a transposase exhibiting 53% identity with a putative transposase from Thermoanaerobacter ethanolicus ATCC 33223 (Teth39DRAFT 1803 ZP_00779197).
An 8,777-bp fragment from the initial 1.4-kb fragment amplified by Trap-Dock PCR with primers p50dir and dock-In was sequenced by successive inverse PCRs (Fig. 2B). Three ORFs were found in this DNA fragment. One of these ORFs encodes a 604-amino-acid protein designated Gal27A, which exhibits 60% identity with the Aga27A α-galactosidase from C. josui (accession number ABO25362) (22). Aga27A is a three-domain cellulosomal enzyme consisting of a signal sequence, a GH27 catalytic domain, and a C-terminal dockerin domain. Gal27A possesses an additional CBM6 domain (Fig. 2B). Surprisingly, the N-terminal sequence of the mature form of Gal27A (AWDNGLAKTPPMG) was found to be different from the N-terminal sequence of P50. Nevertheless, the N-terminal sequence of P50 (LNNSLGLTPXMG) is, without any ambiguity, the N-terminal sequence of a GH27 α-galactosidase (22). Thus, two different GH27 α-galactosidases are potentially produced and incorporated into the cellulosomes of C. cellulolyticum. A 3,366-bp gene was found 32 bp downstream of gal27A and was designated gal59A. This gene appears to encode a modular enzyme consisting of a GH59 catalytic domain followed by a CBM6 domain and a dockerin domain. An incomplete gene (orf4) encoding a putative GH2 enzyme was found 66 bp downstream of gal59A (Fig. 2B). A 2,865-bp gene (orf3) encoding a 955-amino-acid protein was identified upstream of gal27A. NCBI BLASTP analyses of the putative catalytic domain of this protein (amino acids 23 to 697) did not reveal any similarity to protein sequences deposited in the NCBI database. The sequence from amino acid 698 to amino acid 842 consists of a CBM22 followed by a small domain with an unknown function (X) and a dockerin domain. The CBM22 exhibits 38% identity with the N-terminal CBM of the same family of XynD, a GH10 xylanase from C. thermocellum (accession number AJ585345) (45).
The gene encoding P41a and the flanking regions were sequenced by inverse PCR. This 1,269-bp gene encodes a 423-amino-acid GH10 xylanase (Fig. 2C). The putative N-terminal sequence of the mature enzyme, renamed Xyn10A (ATPTGKRLKDVQSR), is in good agreement with the results of the Edman sequencing performed for P41a (Table 3). Xyn10A exhibited striking homology with xylanases from different thermophilic species, such as C. thermocellum, Thermotoga sp., Thermobacillus xylanilyticus, and Thermobifida alba. For instance, Xyn10A from C. cellulolyticum is 47% identical to XynA from the hyperthermophile Thermotoga maritima strain FjSS3-B.1 (accession number U33060) (40), 51% identical to XylB from T. maritima strain MSB8 (accession number AY340789) (43), 47% identical to XylA from T. alba (accession number Z81013) (4), and 42% identical to the catalytic domain of the bifunctional enzyme Xyn10Z from C. thermocellum (accession number M22624) (5, 17). No new ORF was found in the flanking regions.
Chromosomal DNA walking around the nonspecific 0.4-kb PCR product (obtained with primers p41dir and dock-In) allowed isolation of a 2,145-bp gene, cel9Q, encoding a GH9 domain fused to a CBM3c domain (Fig. 2D). This 715-residue protein has an N-terminal signal sequence and a C-terminal dockerin domain. Cel9Q is 71% identical to Cel9N from C. thermocellum (accession number AJ275974) (46) and 70% identical to a hypothetical GH9 cellulase from the same bacterium (ChteDRAFT 0785 ZP_00510325) (44). Cel9Q exhibits 44, 40, 31, and 32% identity with Cel9G, Cel9H, Cel9J, and Cel9P from C. cellulolyticum, respectively. An ORF (orf5) encoding a putative ADP-ribose pyrophosphatase was found upstream of cel9Q, whereas no new ORF was found in the 252-bp sequence downstream.
DISCUSSION
An exceptionally high number of cellulosomal proteins is produced by the thermophilic bacterium C. thermocellum. Indeed, analysis of the recently sequenced genome of this Clostridium revealed the presence of 71 different dockerin-containing proteins (44). These proteins are cellulases, xylanases, chitinases, mannanases, lichenases, carbohydrate esterases, pectinases, putative proteases, and protease inhibitors. Thirteen of them contain domains with unknown functions. They belong to various glycoside hydrolase families (GH2, GH5, GH8, GH9, GH10, GH11, GH16, GH18, GH26, GH28, GH30, GH39, GH53, GH43, GH81, GH44, GH48, GH54, and GH74), polysaccharide lyase families (PL1, PL9, PL10, and PL11), and carbohydrate esterase families (CE1, CE2, CE3, CE4, and CE12). Thirty-three proteins contain a CBM, and at least 10 are bifunctional (i.e., have two different catalytic domains) (44).
In the absence of the genome sequence of C. cellulolyticum, the aim of the present work was to identify genes and gene products that define the diversity of the cellulolytic system of this Clostridium species. This study provided proteome information (2D PAGE and zymogram analysis), as well as information concerning identification of the genes encoding specific enzymes (Trap-Dock PCR) that contribute to degradation of vegetal biomass.
Sixty proteins were detected by two-dimensional analysis of the C. cellulolyticum cellulolytic system. Thirty of these proteins lack dockerin and are not incorporated into multienzyme complexes. They are probably targeted to their cognate substrates by specific CBMs. The real impact of the free enzymes in plant cell wall degradation by C. cellulolyticum is still unknown. At least 30 proteins are cellulosomal. Zymogram analyses with CMC and xylan as substrates allowed us to identify new cellulosomal and noncellulosomal CMCases, as well as new cellulosomal and noncellulosomal xylanases. C. cellulolyticum is able to grow on xylan, and it has been shown previously that two xylanases (31 and 54 kDa) are incorporated into cellulosomes purified from a xylan-grown culture (33). The 31-kDa protein detected by Mohand-Oussaid et al. (33) may correspond to P32a. However, no xylanase activity was found to be associated with a 54-kDa dockerin-containing protein. Xyn10A is the first xylanase clearly identified in C. cellulolyticum. Its corresponding gene was cloned and sequenced (accession number DQ778331). Cellulosome-producing bacteria generally have xylanases belonging to families GH10 and GH11. As suggested from its N-terminal sequence, P32b could be a GH11 xylanase. In addition to these xylanases, other xylanases could be detected in C. cellulolyticum grown on xylan. Indeed, in C. cellulovorans, production of xylanases and hemicellulolytic enzymes depends on the growth substrates (18, 19, 25).
Seven new genes encoding dockerin-containing proteins were isolated by Trap-Dock PCR and inverse PCR. Two of these genes, cel9P and cel9Q, encode modular GH9 cellulases (accession numbers DQ778334 and DQ778332, respectively). In these proteins, the catalytic domain is attached to a CBM3c domain, as it is in Cel9G, Cel9J, and Cel9H isolated from the same clostridia. To date, this type of CBM has been found to be associated only with a GH9 catalytic domain. Usually, the role of a CBM is to target the enzyme to its specific substrate. CBM3c, together with the catalytic domain, allows the binding of one cellulose chain to the active site. CBM3c and GH9 catalytic domains are structurally and functionally connected. In 2005, Fierobe et al. (13) clearly demonstrated that cellulosomal GH48 and GH9-CBM3c enzymes play a crucial role in the degradation of cellulose by cellulosomes. Indeed, incorporation into a mini-cellulosome of the processive Cel48F and of Cel9G allowed the workers to obtain the highest level of hydrolyzing activity (11, 13). In addition, together with the scaffolding protein CipC, Cel48F is one of the most prominent proteins, and we hypothesize that at least one Cel48F molecule is bound per scaffolding protein in cellulosomes. This hypothesis is supported by the recently published results of Han et al. (18). These workers found that the composition of subpopulations of cellulosomes from C. cellulovorans, purified from different carbon substrates, such as cellulose, xylan, and pectin, indicated that the three major subunits, scaffolding protein CbpA, endoglucanase EngE (GH9), and cellobiohydrolase ExgS (GH48), were present in all the purified cellulosomal populations. Strangely, GH9-CBM3c enzymes, which are essential for effective hydrolysis of the crystalline cellulose, are not massively produced by C. cellulolyticum or by the other known cellulosome-producing clostridia. The poor production of individual GH9-CBM3c enzymes could be compensated for by the redundancy of the genes encoding proteins having the same domain arrangements. This hypothesis may be supported by the recent analysis of the genome of C. thermocellum, in which seven genes encoding cellulases containing a catalytic GH9 domain linked to a CBM3c were found (44).
In the present work, the genes encoding hemicellulases Man26A, Gal27A, and Xyn10A were sequenced. Man26A (accession number DQ778334), like its C. thermocellum homologue (accession number AB044406), is likely to be a mannan backbone-hydrolyzing enzyme (27). An alpha-galactosidase, such as Aga27A (22) from C. josui, hydrolyzes α-galactosidic linkages located at the nonreducing ends of various galactose-containing substrates, such as galactomannan. In the cellulosome of C. cellulolyticum, Man26A and Gal27A (accession number DQ778333) could act synergistically to degrade galactomannan. According to its N-terminal sequence, P50 is a GH27 α-galactosidase that is different from Gal27A. These two galactosidases may be produced in different growth conditions. Synthesis of GH27 and Gal27A on different growth substrates, such as crystalline cellulose, xylan, galactomannan, and plant cell wall material, should be studied.
The presence of a gene encoding a modular GH59 enzyme is surprising. Indeed, GH59 enzymes are found in eukaryotic organisms, such as Homo sapiens, Mus musculus (mouse), Canis familiaris (dog), and Rattus norvegicus (rat), and the GH59 family comprises enzymes whose only known activity is galactocerebrosidase (6). Galactocerebrosidases catalyze the following reaction: d-galactosyl-N-acylsphingosine + H2O → d-galactosyl + N-acylsphingosine. They are responsible for the lysosomal catabolism of some galactolipids (10, 23). The only bacterial member of the GH59 family is a protein designated AcbD from Actinoplanes sp. strain SE50 (accession number Y18523) (20). This enzyme is involved in the biosynthesis of the α-glucosidase inhibitor belonging to the acarbose α-glucosidase inhibitor family. Acarbose (C25H45N3O16) is a linear tetrasaccharide of α-d-glucose residues, which is modified at the nonreducing end, is able to inhibit α-glucosidases, and is used in the treatment of diabetes patients. Although the presence of a dockerin module and a CBM6 domain clearly indicates a role in the plant cell wall degradation process, the precise role of Gal59A in C. cellulolyticum remains mysterious. The gal59A gene is in a cluster containing at least four genes. This cluster groups together genes encoding CBM-containing enzymes probably involved in hemicellulose degradation.
About 10 genes encoding proteins with a dockerin domain associated with modules having unknown functions have been found in the C. thermocellum genome (44). In C. cellulolyticum, orf3, a gene encoding a protein with a module having an unknown function linked to a CBM22 and a dockerin domain, has been found. The catalytic domain of ORF3p might constitute a new carbohydrate-active enzyme family. Indeed, a BLASTP search against the NCBI protein database did not reveal any homology with proteins or modules involved in plant cell wall degradation. CBM22 generally has a xylan-binding function (1, 6). They are associated with xylanolytic domains, which often belong to family GH10, or are present in bifunctional enzymes, such as XynY from C. thermocellum, which has the following modular organization: CBM22-GH10-CBM22-D-CE1 (accession number X83269) (5).
The results of the present work clearly show the diversity of enzymes that can be incorporated into cellulosomes. The hemicellulolytic system of C. cellulolyticum appears to be extremely rich. All the genes isolated in this study encode cellulosomal proteins. In the work reported here, crystalline cellulose was used as the reference substrate for the growth of C. cellulolyticum. Analysis of the compositions of the cellulolytic systems of C. cellulolyticum grown on different substrates (xylan, mannan, or straw) may reveal regulation of the synthesis of the different components.
Acknowledgments
We are very grateful to Régine Lebrun (Plateforme Proteome, IBSM, Marseille, France) for N-terminal sequencing. We thank Alain Dolla (CNRS Marseille UPR 9036, IBSM) for technical assistance and advice concerning 2D PAGE. We thank Laurence Casalot and Denise Henrissat for correcting the English.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Ali, E., R. Araki, G. Zhao, M. Sakka, S. Karita, T. Kimura, and K. Sakka. 2005. Functions of family-22 carbohydrate-binding modules in Clostridium josui Xyn10A. Biosci. Biotechnol. Biochem. 69:2389-2394. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., J. P. Bélaïch, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 3.Béguin, P. 1983. Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Anal. Biochem. 131:333-336. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, J., J. J. Coque, J. Velasco, and J. F. Martin. 1997. Cloning, expression in Streptomyces lividans and biochemical characterization of a thermostable endo-beta-1,4-xylanase of Thermomonospora alba ULJB1 with cellulose-binding ability. Appl. Microbiol. Biotechnol. 48:208-217. [DOI] [PubMed] [Google Scholar]
- 5.Blum, D. L., I. A. Kataeva, X. L. Li, and L. G. Ljungdahl. 2000. Feruloyl esterase activity of the Clostridium thermocellum cellulosome can be attributed to previously unknown domains of XynY and XynZ. J. Bacteriol. 182:1346-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 7.Devillard, E., C. J. Newbold, K. P. Scott, E. Forano, R. J. Wallace, J. P. Jouany, and H. J. Flint. 1999. A xylanase produced by the rumen anaerobic protozoan Polyplastron multivesiculatum shows close sequence similarity to family 11 xylanases from gram-positive bacteria. FEMS Microbiol. Lett. 181:145-152. [DOI] [PubMed] [Google Scholar]
- 8.Doi, R. H., J. S. Park, C. C. Liu, L. M. Malburg, Y. Tamaru, A. Ichiishi, and A. Ibrahim. 1998. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 9.Doi, R. H., and Y. Tamaru. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 10.Dolcetta, D., L. Perani, M. I. Givogri, F. Galbiati, A. Orlacchio, S. Martino, M. G. Roncarolo, and E. Bongarzone. 2004. Analysis of galactocerebrosidase activity in the mouse brain by a new histological staining method. J. Neurosci. Res. 77:462-464. [DOI] [PubMed] [Google Scholar]
- 11.Fierobe, H. P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Bélaïch, R. Lamed, Y. Shoham, and J. P. Bélaïch. 2002. Degradation of cellulose substrate by cellulose chimeras. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 12.Fierobe, H. P., C. Gaudin, A. Bélaïch, M. Loutfi, E. Faure, C. Bagnara, D. Baty, and J. P. Bélaïch. 1991. Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173:7956-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fierobe, H. P., F. Mingardon, A. Mechaly, A. Bélaïch, M. T. Rincon, S. Pagès, R. Lamed, C. Tardif, J. P. Bélaïch, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 14.Gal, L., C. Gaudin, A. Bélaïch, S. Pagès, C. Tardif, and J. P. Bélaïch. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal, L., S. Pagès, C. Gaudin, A. Bélaïch, C. Reverbel-Leroy, C. Tardif, and J. P. Bélaïch. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giallo, J., C. Gaudin, J. P. Bélaïch, E. Petitdemange, and F. Caillet-Mangin. 1983. Metabolism of glucose and cellobiose by cellulolytic mesophilic Clostridium sp. strain H10. Appl. Environ. Microbiol. 45:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grepinet, O., M. C. Chebrou, and P. Béguin. 1998. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J. Bacteriol. 170:4582-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, S. O., H. Y. Cho, H. Yukawa, M. Inui, and R. H. Doi. 2004. Regulation of expression of cellulosomes and noncellulosomal (hemi)cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources. J. Bacteriol. 186:4218-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2005. Effect of carbon source on the cellulosomal subpopulations of Clostridium cellulovorans. Microbiology 151:1491-1497. [DOI] [PubMed] [Google Scholar]
- 20.Hemker, A., A. Stratmann, K. Goeke, W. Schröder, J. Lenz, W. Piepersberg, and H. Pape. 2001. Identification, cloning, expression, and characterization of the extracellular acarbose-modifying glycosyltransferase, AcbD, from Actinoplanes sp. strain SE50. J. Bacteriol. 183:4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heukeshoven, J., and R. Dernick. 1985. Simplified method for silver staining of proteins in polyacrylamide gels and mechanism of silver staining. Electrophoresis 6:103-112. [Google Scholar]
- 22.Jindou, S., S. Karita, E. Fujino, T. Fujino, H. Hayashi, T. Kimaru, K. Sakka, and K. Ohmiya. 2002. Alpha-galactosidase Aga27A, an enzymatic component of the Clostridium josui cellulosome. J. Bacteriol. 184:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo, Y., D. A. Wenger, V. Gallo, and I. D. Duncan. 2005. Galactocerebrosidase-deficient oligodendrocytes maintain stable central myelin by exogenous replacement of the missing enzyme in mice. Proc. Natl. Acad. Sci. USA 102:18670-18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontinen, V. P., and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8:727-737. [DOI] [PubMed] [Google Scholar]
- 25.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurokawa, J., E. Hemjinda, T. Arai, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Sequence of the Clostridium thermocellum mannanase gene man26B and characterization of the translated product. Biosci. Biotechnol. Biochem. 65:548-554. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa, J., E. Hemjinda, T. Arai, T. Kimura, K. Sakka, and K. Ohmiya. 2002. Clostridium thermocellum cellulase CelT, a family 9 endoglucanase without an Ig-like domain or family 3c carbohydrate-binding module. Appl. Microbiol. Biotechnol. 59:455-461. [DOI] [PubMed] [Google Scholar]
- 29.Maamar, H. 2003. Ph.D. thesis. Université de Provence, Marseille, France.
- 30.Maamar, H., L. Abdou, C. Boileau, O. Valette, and C. Tardif. 2006. Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum. J. Bacteriol. 188:2614-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maamar, H., O. Valette, H. P. Fierobe, A. Bélaïch, J. P. Bélaïch, and C. Tardif. 2004. Cellulolysis is severely affected in Clostridium cellulolyticum strain cipCMut1. Mol. Microbiol. 51:589-598. [DOI] [PubMed] [Google Scholar]
- 32.Mandelman, D., A. Bélaïch, J. P. Bélaïch, N. Aghajari, H. Driguez, and R. Haser. 2003. X-ray crystal structure of the multidomain endoglucanase Cel9G from Clostridium cellulolyticum complexed with natural and synthetic cello-oligosaccharides. J. Bacteriol. 185:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohand-Oussaid, O., S. Payot, E. Guedon, E. Gelhaye, A. Youyou, and H. Petitdemange. 1999. The extracellular xylan degradative system in Clostridium cellulolyticum cultivated on xylan: evidence for cell-free cellulosome production. J. Bacteriol. 181:4035-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 35.Pagès, S., A. Bélaïch, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Bélaïch. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagès, S., A. Bélaïch, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J. P. Bélaïch. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagès, S., O. Valette, L. Abdou, A. Bélaïch, and J. P. Bélaïch. 2003. A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. J. Bacteriol. 185:4727-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perret, S., A. Bélaïch, H. P. Fierobe, J. P. Bélaïch, and C. Tardif. 2004. Towards designer cellulosomes in clostridia: mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. J. Bacteriol. 186:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter, W. D. 2002. Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5:536-542. [DOI] [PubMed] [Google Scholar]
- 40.Saul, D. J., L. C. Williams, R. A. Reeves, M. D. Gibbs, and P. L. Bergquist. 1995. Sequence and expression of a xylanase gene from the hyperthermophile Thermotoga sp. strain FjSS3-B.1 and characterization of the recombinant enzyme and its activity on kraft pulp. Appl. Environ. Microbiol. 61:4110-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tardif, C., A. Bélaïch, H. P. Fierobe, S. Pagès, P. de Philip, and J. P. Bélaïch. 2006. Clostridium cellulolyticum: cellulosomes and cellulolysis. In I. Kataeva (ed.), Cellulosome. Nova Sciences Publishers, Inc., Hauppauge, NY.
- 42.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhengqiang, J., A. Kobayashi, M. M. Ahsan, L. Lite, M. Kitaoka, and K. Hayashi. 2001. Characterization of a thermostable family 10 endo-xylanase (XynB) from Thermotoga maritima that cleaves p-nitrophenyl-beta-d-xyloside. J. Biosci. Bioeng. 92:423-428. [DOI] [PubMed] [Google Scholar]
- 44.Zverlov, V. V., J. Kellermann, and W. H. Schwarz. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646-3653. [DOI] [PubMed] [Google Scholar]
- 45.Zverlov, V. V., N. Schantz, P. Schmitt-Kopplin, and W. H. Schwarz. 2005. Two new major subunits in the cellulosome of Clostridium thermocellum: xyloglucanase Xgh74A and endoxylanase Xyn10D. Microbiology 151:3395-3401. [DOI] [PubMed] [Google Scholar]
- 46.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2003. Two new cellulosome components encoded downstream of celI in the genome of Clostridium thermocellum: the nonprocessive endoglucanase CelN and the possibly structural protein CseP. Microbiology 149:515-524. [DOI] [PubMed] [Google Scholar]