Abstract
Thermobifida fusca is a moderately thermophilic soil bacterium that belongs to Actinobacteria. It is a major degrader of plant cell walls and has been used as a model organism for the study of secreted, thermostable cellulases. The complete genome sequence showed that T. fusca has a single circular chromosome of 3,642,249 bp predicted to encode 3,117 proteins and 65 RNA species with a coding density of 85%. Genome analysis revealed the existence of 29 putative glycoside hydrolases in addition to the previously identified cellulases and xylanases. The glycosyl hydrolases include enzymes predicted to exhibit mainly dextran/starch- and xylan-degrading functions. T. fusca possesses two protein secretion systems: the sec general secretion system and the twin-arginine translocation system. Several of the secreted cellulases have sequence signatures indicating their secretion may be mediated by the twin-arginine translocation system. T. fusca has extensive transport systems for import of carbohydrates coupled to transcriptional regulators controlling the expression of the transporters and glycosylhydrolases. In addition to providing an overview of the physiology of a soil actinomycete, this study presents insights on the transcriptional regulation and secretion of cellulases which may facilitate the industrial exploitation of these systems.
Thermobifida fusca (formerly known as Thermomonospora fusca) is an aerobic, moderately thermophilic, filamentous soil bacterium that is a major degrader of plant cell walls in heated organic materials, such as compost heaps, rotting hay, manure piles, or mushroom growth medium (1). It produces spores that can be allergenic and has been associated with a condition called farmers lung (50). Its extracellular glycoside hydrolases (cellulases and xylanases) have been studied extensively because of their thermostability, broad pH range (4 to 10), and high activity. It appears to degrade all major plant cell wall polymers except lignin and pectin and can grow on most simple sugars and carboxylic acids. It belongs to the phylum of Actinobacteria and was first isolated from decaying wood (2).
T. fusca has been the source organism for isolating and studying multiple secreted cellulases and other carbohydrate-degrading enzymes (12, 15). Using classical biochemical methods, six different cellulases have been identified: four endocellulase genes (7, 12, 15) and two exocellulases (18, 53). In addition, an intracellular β-glucosidase that degrades cellobiose to glucose (46), an extracellular xyloglucanase (17), two secreted xylanases, and a GH family 81 β-1,3-glucanase (4, 11, 16, 22, 31) have been cloned and characterized. Secreted cellulases have great biotechnological promise for utilization in the degradation of agricultural products and waste to produce sugars that can be subsequently converted to ethanol. Several complete genomic sequences of the phylum Actinobacteria are currently available. The availability of these complete genomic sequences of the phylum Actinobacteria (29) enables sequence comparisons, which can provide valuable information for the biotechnological application of these microbes.
MATERIALS AND METHODS
Genome sequencing and assembly.
The complete genome of T. fusca was sequenced at the Joint Genome Institute using a combination of 3-kb and fosmid (40-kb) libraries. Library construction, sequencing, finishing, and automated annotation steps were performed as described at the JGI web page (http://www.jgi.doe.gov/sequencing/index.html). Predicted coding sequences (CDSs) were manually analyzed and evaluated using an Integrated Microbial Genomes (IMG) annotation pipeline (http://img.jgi.doe.gov).
Genome analysis.
Comparative analysis of T. fusca with related organisms was performed using a set of tools available in IMG. Unique and orthologous T. fusca genes were identified by using BLASTp (cutoff scores of E < 10−2 and 20% identity and reciprocal hits with cutoff scores of E < 10−5 and 30% identity, respectively). Signal peptides were identified using the SignalP 3.0 (3) and TMHMM (25) at default values. Whole-genome comparisons were performed using MUMmer (27).
Nucleotide sequence accession numbers.
The sequence data described here have been deposited in GenBank (CP000088).
RESULTS AND DISCUSSION
Genome features and comparative Actinobacteria genomics.
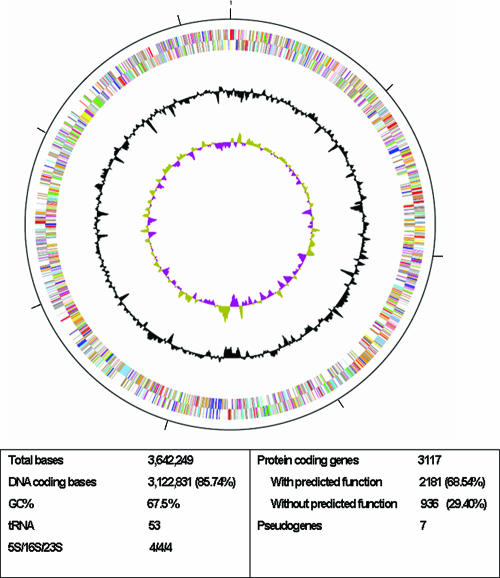
The T. fusca genome consists of a single circular chromosome with 3,642,249 bp. The GC content is 67.5%, and there are 3,117 predicted CDSs in the genome. The overall genome statistics are listed in Fig. 1. Among the predicted genes, 68% have been assigned a function. Twenty-six percent (830 genes) display sequence similarity to other organisms in the database with no known function, and 106 genes (3.3%) appear to be unique in T. fusca. There are four rrn loci arranged in 5S-23S-16S operons.
FIG. 1.
Circular representation of the genome of T. fusca. From outside to inside, the first two circles represent the COG assignment for predicted coding sequences on the plus and minus strands, respectively. Colors indicate the following: dark gray, hypothetical proteins; light gray, conserved hypothetical and unknown function; brown, general function prediction; red, replication and repair; green, energy metabolism; blue, carbon and carbohydrate metabolism; cyan, lipid metabolism; magenta, transcription; yellow, translation; orange, amino acid metabolism; pink, metabolism of cofactors and vitamins; light red, purine and pyrimidine metabolism; lavender, signal transduction; sky blue, cellular processes. The two innermost circles represent the percent G+C content and G+C skew values, respectively.
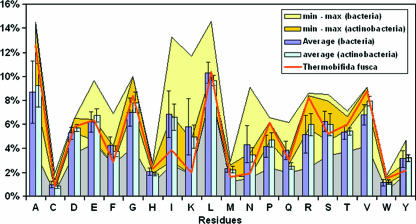
According to Suhre and Claverie (47), thermophilic proteomes exhibit an increased content of charged (Asp, Glu, Lys, and Arg) versus polar (Asn, Gln, Ser, and Thr) residues. This trend is a common feature for organisms with optimal growth temperatures higher than 55°C. T. fusca exhibits the same trend with the striking exception of Lys residues, which appear to be close to the minimum for bacteria in IMG. The reverse trend is observed for Ala, which is elevated in T. fusca while most thermophiles have fewer Ala residues (Fig. 2).
FIG. 2.
Amino acid utilization in T. fusca, based on a comparison of the percent content of amino acid residues in all sequenced Bacteria, Actinobacteria, and T. fusca.
T. fusca has 412 (∼13%) unique genes when compared to the 32 Actinobacteria genomes present in IMG. From these 412 genes only 83 CDSs have InterPro hits, and the rest are hypothetical proteins with no functional hits. Comparisons between representatives of the five major Actinobacteria genera, T. fusca, Streptomyces coelicolor, Mycobacterium tuberculosis, Nocardia farcinica, and Corynebacterium diphtheriae, indicate that there are 1,101 genes (∼30%) genes shared between these five organisms. T. fusca has 660 unique genes (20%) compared to the above five genomes. Overall, comparisons between these five genomes both in terms of gene similarity (Table 1) and synteny (Fig. 3) indicate that T. fusca is most closely related to S. coelicolor and N. farcinica.
TABLE 1.
Global genomic comparisons among five representatives of Actinobacteriaa
| Comparison organism(s) | No. of genes with (or without) a homolog ina:
|
||||
|---|---|---|---|---|---|
| T. fusca | S. coelicolor | M. tuberculosis H37Rv | C. diphtheriae NCTC 13129 | N. farcinica IFM 10152 | |
| Comparisons for unique genes | |||||
| T. fusca | (838) | (1,436) | (1,840) | (1,077) | |
| S. coelicolor | (4,310) | (5,038) | (6,125) | (4,007) | |
| M. tuberculosis H37Rv | (2,184) | (1,764) | (2,548) | (1,413) | |
| C. diphtheriae | (1,191) | (1,058) | (1,076) | (921) | |
| N. farcinica IFM 10152 | (3,196) | (2,495) | (2,894) | (4,109) | |
| T. fusca + S. coelicolor | (673) | (1,055) | (409) | ||
| T. fusca + M. tuberculosis | (75) | (555) | (85) | ||
| T. fusca + C. diphtheriae | (53) | (151) | (51) | ||
| T. fusca + N. farcinica | (170) | (444) | (814) | ||
| Comparisons for common genes | |||||
| T. fusca | 2,279 | 1,681 | 1,277 | 2,040 | |
| S. coelicolor | 3,905 | 3,177 | 2,090 | 4,208 | |
| M. tuberculosis | 1,815 | 2,235 | 1,451 | 2,586 | |
| C. diphtheriae | 1,129 | 1,262 | 1,244 | 1,399 | |
| N. farcinica | 2,740 | 3,441 | 3,042 | 1,827 | |
Results for comparisons that identified unique genes (without homologs in the organisms being compared) are indicated in parentheses. Results for comparisons that identified common genes (with homologs in the organisms being compared) are shown without parentheses. The calculations were based on a maximum E-value of 10−3 and a minimum identity of 30%.
FIG. 3.
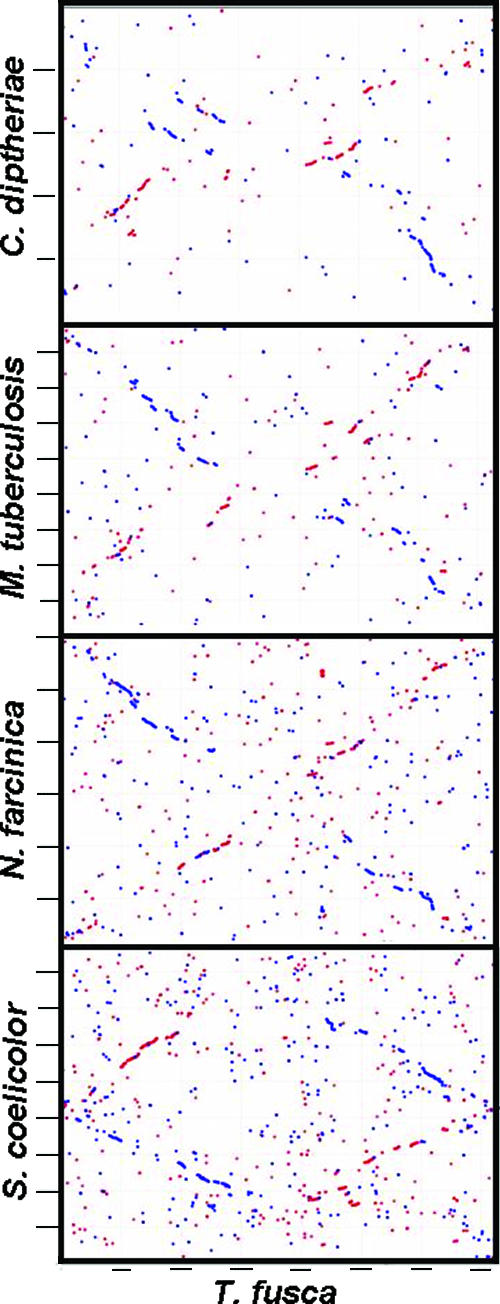
Synteny plots between T. fusca (horizontal axis) and five representative genomes of Actinobacteria: Streptomyces coelicolor, Nocardia farcinica, Mycobacterium tuberculosis, and Corynebacterium diphtheriae.
Cellulases and glycoside hydrolases.
The genome of T. fusca encodes a total of 45 hydrolytic enzymes predicted to act on oligo- and/or polysaccharides as identified by the CAZy ModO database (http://afmb.cnrs-mrs.fr/CAZY/) (Table 2). These enzymes include 36 glycoside hydrolases, 9 carbohydrate esterases, and 2 polysaccharide lyases. The glycoside hydrolases are distributed in 22 families (GH families), with the GH_13 family being the most abundant with six individual members represented in the genome. Computational analysis of signal peptides predicts 16 glycoside hydrolases are secreted. The majority of these secreted proteins (13 out of 16) have a signal sequence resembling the twin-arginine translocation (TAT) signal in their N terminus. Analysis of the secretion systems will be presented in a subsequent section.
TABLE 2.
Summary of glycoside hydrolases and CBM-containing proteins in T. fusca
| Locus tag | Family | Description | Binding module | Signal peptide? | Potential TAT signal? | Reference(s) |
|---|---|---|---|---|---|---|
| Tfu0082 | CE_1 | Acetyl xylan esterase | Yes | Yes, RRIPARVWVALTAVLGLGAALLGTTALAPRAEAA | ||
| Tfu2990 | CE_3 | Acetyl xylan esterase | Cellulose, CBM2 | Yes | No | |
| Tfu1621 | CE_4 | Acetyl xylan esterase/chitin deacetylase | LBP | Yes | No | |
| Tfu2788 | CE_4 | Acetyl xylan esterase/chitin deacetylase | Cellulose, CBM2 | Yes | Yes, PRRSPLRKRLLVALCALGLAFTSAATAHAQV | |
| Tfu2789 | CE_4 | Acetyl xylan esterase/chitin deacetylase | Yes | No | ||
| Tfu2473 | CE_9 | 6P-NAG deacetylase/6P-NAGal deacetylase | No | |||
| Tfu0084 | CE_14 | GlcNAc-Pl deacetylase | No | |||
| Tfu0449 | CE_14 | GlcNAc-Pl deacetylase | No | |||
| Tfu0486 | CE_14 | GlcNAc-Pl deacetylase | No | |||
| Tfu0937 | GH_1 | β-Glucosidase/cellobiase | No | 46 | ||
| Tfu1629 | GH_1 | Glucosidase/galactosidase | Yes | No | ||
| Tfu0915 | GH_2 | Galactosidase | No | |||
| Tfu1607 | GH_3 | Xylosidase/acetylhexosaminidase | β-1,3-Glucan, CBM6 | No | ||
| Tfu2486 | GH_3 | Xylosidase/acetylhexosaminidase | Yes | |||
| Tfu2768 | GH_4 | Glucuronidase/galactosidase/glucosidase | No | |||
| Tfu0900 | GH_5 | β-Mannanase (EC 3.2.1.78) | Cellulose, CBM2 | Yes | Yes, RKRLAVAAATVLALLASVFALTQPANAAT | 14 |
| Tfu0901 | GH_5 | Endocellulase (E5) | Cellulose, CBM2 | Yes | Yes, RKGGPPVAVAVTAALALLIALLSPGVAQAAG | 28 |
| Tfu2712 | GH_5 | Cellulase | Cellulose, CBM3 | Yes | Yes, RRLRAGAAAIAIGASALIPLTSSPAAASG | 38 |
| Tfu0620 | GH_6 | β-1,4-Exocellulase (E3) | Cellulose, CBM2 | Yes | Yes, RRSWMRRGLAAASG | 28, 53 |
| Tfu1074 | GH_6 | β-1,4-Endoglucanase (E2) | Cellulose, CBM2 | Yes | No | 28 |
| Tfu1627 | GH_9 | β-1,4-Endoglucanase (E1) | Cellulose/amorphous/xylan, CBM2, CBM4 | Yes | Yes, RRPRSRSPLVAL | 19 |
| Tfu2176 | GH_9 | Cellulase (E4) | Cellulose, CBM2, CBM3 | Yes | Yes, PRRRGRHSRARRF | 19, 28 |
| Tfu2791 | GH_10 | Xylanase | Yes | No | 22 | |
| Tfu2923 | GH_10 | Xylanase | Cellulose, CBM2 | Yes | Yes, RHRPSRRARRSLSLLLTSALTAAGLLVTAAPAQAES | 4 |
| Tfu1213 | GH_11 | Endo 1,4-β-d-xylanase | Cellulose, CBM2 | Yes | Yes, RRRFRPRLLIGKAFAAALVAVVTMIPSTAAHAAV | 16 |
| Tfu0582 | GH_13 | Amylase/pullulanase | No | |||
| Tfu0584 | GH_13 | Amylase | No | |||
| Tfu0585 | GH_13 | Amylase/pullulanase | No | |||
| Tfu0833 | GH_13 | Amylase/pullulanase | No | |||
| Tfu0985 | GH_13 | Amylase/pullulanase | Starch, CBM20 | Yes | Yes, RRSLAALLAALLGCATSLVALTVAASPAHAAP | |
| Tfu1891 | GH_13 | Amylase/pullulanase | No | |||
| Tfu0046 | GH_15 | Amylase/dextranase | No | |||
| Tfu1345 | GH_15 | Amylase/dextranase | No | |||
| Tfu0580 | GH_18 | Chitinase | No | |||
| Tfu0868 | GH_18 | Exochitinase | Yes | Yes, RRRTFAPTWVVLLVAAGVVALC | ||
| Tfu0480 | GH_23 | Lysozyme | No | |||
| Tfu2594 | GH_23 | Lysozyme | No | |||
| Tfu1613 | GH_31 | Amylase/isomaltase | No | |||
| Tfu1615 | GH_42 | Galactosidase | No | |||
| Tfu1616 | GH_43 | Xylanase/galactan galactosidase | Yes | |||
| Tfu1959 | GH_48 | β-1,4-Exocellulase (E6-celF) | Cellulose, CBM2 | Yes | Yes, RRWRTLASGALAAALAAAVLSPGVAHAAV | 18 |
| Tfu3044 | GH_65 | Trehalase/maltose phosphorylase | No | |||
| Tfu1612 | GH_74 | Xyloglucanase, endo-β-1,4-glucanase | Cellulose, CBM2 | Yes | Yes, RRRGIIARALTCIAAAATVAAVG | 17 |
| Tfu2205 | GH_77 | Amylomaltase or 4-glucanotransferase | No | |||
| Tfu2130 | GH_81 | Glucanase | Yes | Yes, RRRWRRATTSAATAALLCGALLTFPSAPAAA | 31 | |
| Tfu1614 | GH_95 | Fucosidase | No | |||
| Tfu0153 | PL_1 | Pectate-lyase (EC 4.2.2.2) | Yes | Yes, MRRAATLGVALALPLTLAAPSSALAQP | ||
| Tfu2168 | PL_1 | Pectate-lyase (EC 4.2.2.2) | Yes | Yes, VGRSITRRLASTLATAAVVTAGLTLPVSPASAQT | ||
| Tfu0644 | Cellulose, CBM2 | No | ||||
| Tfu1268 | Chitin, CBM33 | Yes | ||||
| Tfu1643 | Xylan/mannose/ galactose, CBM13 | No | No | |||
| Tfu1665 | Cellulose/chitin, CBM2, CBM33 | Yes | No | |||
| Tfu2009 | Rhamnogalacturonan lyase | Cellulose, CBM2 | Yes | Yes, RRRPVRFGAALAAFVLGATGAAALPSPAHAAA |
Fourteen enzymes have been isolated and studied (including six enzymes with various cellulolytic activities and cellulases E1 to E6, as well as a β-mannanase and an endo-xylanase) from T. fusca (Table 2). In addition to these enzymes, 28 more putative glycosyl hydrolases and enzymes potentially involved in plant cell wall degradation were identified in the genome (mainly with dextran/starch- and xylan-degrading functions). Two enzymes were identified to be similar to endochitinase and exochitinase (Tfu0580 and Tfu0868, respectively). The absence of N-acetyl-hexosaminidase implies that chitin degradation proceeds either with an alternative pathway or by gradual degradation of the substrate by an exochitinolytic enzyme. Deacetylases of chitin, xylan, and N-acetyl-glucosamine were identified, suggesting that the organism can utilize these substrates for energy production. T. fusca contains several enzymes involved in the degradation of xylan: endo-β-1,4-xylanase (Tfu1213, Tfu2791, and Tfu2923), xylosidase (Tfu1613), α-l-arabinofuranosidase (Tfu1616), and several CDSs with putative acetylxylan esterase activity. In addition to the above-mentioned carbohydrases, T. fusca possesses two CDSs with significant similarity to pectate lyases, Tfu0153 and Tfu2168 (Table 2). These two genes encode secreted proteins that possess a signal peptide that may be interacting with the TAT system. Pectate lyases (EC 4.2.2.2) catalyze the eliminative cleavage of de-esterified pectin. However, T. fusca does not appear to possess a pectin methylesterase or a pectin acetylesterase gene. These observations agree with experimental evidence showing that T. fusca cannot grow on pectin but does grow on its de-esterifed derivative, polygalacturonic acid (pectate), which is the major pectin polymer in the cell walls of most plants. Besides pectate lysases, T. fusca has a gene (Tfu2009) predicted to be a rhamnogalacturonan lyase, which may be involved in the degradation of rhamnogalacturonan, a constituent of pectin and pectate.
Besides the glycohydrolases mentioned above, the T. fusca genome encodes a set of four CDSs that possess a CBM domain without any identifiable glycohydrolase catalytic domain. Tfu0644 is predicted to have one predicted transmembrane helix, exposing the CBM2 domain to the extracellular face of the membrane. Tfu1268 is a secreted protein containing a CBM33 chitin binding domain. Tfu1665 is a secreted protein that contains two CBMs: a chitin binding domain at its N terminus (CBM33) and a C-terminal cellulose binding module (CBM2). We hypothesize that these proteins probably participate in plant cell wall degradation by providing scaffold services to hydrolytic enzymes and facilitating their action. Tfu1643 is an intracellular protein with a ricin-type lectin domain which presumably binds and interacts with carbohydrates.
Transcription and translation.
There are 29 large and 17 small ribosomal subunit proteins for the assembly of the ribosome, and the typical prokaryotic translation initiation factors (IF1, IF2, and IF3) and elongation factors (Tu and G) are present. Fifty-three CDSs code for tRNAs covering all 20 amino acids.
The transcriptional regulation of six T. fusca cellulase gnes (Tfu0620, Tfu0901, Tfu1074, Tfu1627, Tfu1959, and Tfu2176) is mediated by the CelR gene product (Tfu0938), which binds to a 14-bp inverted repeat (5′-TGGGAGCGCTCCCA) in their 5′-upstream regions (44, 45). CelR acts as a repressor, and physiological concentrations of cellobiose (the major end product of cellulases) cause dissociation of the CelR-DNA complex. The 14-bp inverted repeat mentioned above is found in an additional five locations in the T. fusca genome.
It is found upstream of Tfu0934 which, together with Tfu935 and Tfu936, constitutes a disaccharide ABC transport cassette. The chromosomal location of this cassette is immediately upstream of the intracellular β-glucosidase bglC (Tfu0937) (46) and the CelR-coding gene (Tfu0938). Spiridonov et al. characterized the operon Tfu935-Tfu938 and postulated that imperfect palindromes located upstream of Tfu935 may represent putative CelR binding sites. However, the existence of the perfect palindrome 5′-upstream of Tfu934 suggests that the set of Tfu0934-Tfu0938 may be transcriptionally coregulated by CelR. There are four additional genes with perfect CelR binding sites at their 5′ region: Tfu1135, Tfu1508, Tfu1665, and Tfu2844. Tfu1135 is a unique protein in T. fusca with no apparent homologs or similar genes in other organisms. Tfu1508 is a membrane protein of the major facilitator family, and it may be involved in the transport of the cellulose hydrolysis products into the cell. Tfu1665 is a secreted protein that contains three carbohydrate binding domains: an N-terminal chitin binding domain, a C-terminal cellulose binding domain (CBM_2 class), and a fibronectin type III binding region located in the middle of the protein. This structure suggests that Tfu1665 may act as a scaffold protein mediating the action of other carbohydrate-hydrolyzing enzymes. It does not contain a TAT signal, and presumably it is secreted via the sec pathway. Finally, Tfu2844 is an oxidoreductase of unknown specificity.
Besides the above-mentioned genes that contain perfect CelR binding palindromes, an additional secreted xylanase (Tfu2923) has an imperfect palindrome in its upstream region (5′-TGGGAGCGCTCCCG). Interestingly, this imperfect palindrome is also located next to the perfect one at the regulatory regions upstream of Tfu0621 and Tfu1959, suggesting that it may also participate in the regulation of the expression of these cellulases. The same perfect palindrome is found in 10 locations in the genome of S. coelicolor and four locations in the genome of Streptomyces avermitilis. In the case of S. coelicolor the palindrome is located upstream of six secreted glycosylhydrolases (SCO0554, SCO0643, SCO0765, SCO1187, SCO6546, and SCO7637), a protein that contains a cellulose binding domain (SCO5786), and one lac-type transcriptional regulator (SCO2794). In the case of S. avermitilis, the palindrome is located upstream of two secreted glycosylhydrolases (SAV555 and SAV1854) as well as the cellobiose transporter (SAV5256, SAV5255, and SAV5254) and a lac-type transcriptional regulator (SAV5257).
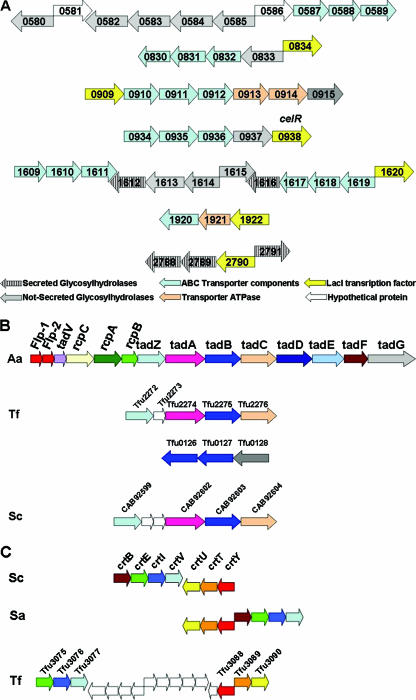
There are six additional genes (Tfu2790, Tfu1620, Tfu1710, Tfu0834, Tfu1922, and Tfu0909) that have significant similarity to CelR and belong to the lactose repressor family (lac), and they might be involved in carbohydrate-induced transcriptional regulation. These genes may respond to different agonists than CelR. CelR is probably not a universal transcriptional regulator, since its levels are diminished when T. fusca grows on glucose or xylan. All of the above lac-type CDSs except Tfu1710 are localized on the chromosome adjacent to saccharide transporters and glycoside hydrolases (Fig. 4A). Based on this observation, we hypothesize that they regulate the expression of the respective genomic regions.
FIG. 4.
Organization of T. fusca chromosomal regions. Each arrow represents one gene. Locus tags and putative gene names are indicated. Tf, T. fusca; Aa, A. actinomycetemcomitans; Sc, S. coelicolor; Sa, S. avermitilis. A. Chromosomal clustering of T. fusca secreted and intracellular glycoside hydrolases with carbohydrate transporters and lac-type transcription factors. B. Comparison of the tight adherence locus of A. actinomycetemcomitans to the chromosomal regions of T. fusca and S. coelicolor that contain homologous genes. C. Comparison of the S. coelicolor cluster encoding genes predicted to participate in carotene biosynthesis to the locus of homologous genes in S. avermitilis and T. fusca.
Protein transport, secretion, adherence, and motility.
T. fusca has a complete set of proteins comprising a sec system for general protein secretion. It has homologs of the secA (Tfu2490 and Tfu0761), secY (Tfu2626), secE (Tfu2660), secG (Tfu2014), and yajC (Tfu2092) secretion components. A signal peptidase (Tfu0667) is also present. Most organisms have one copy of the SecA protein family, and recent evidence suggests that the second copy has distinct functions (5). In Actinobacteria a second copy of secA exists in Bifidobacterium longum DJO10A, all corynebacteria and mycobacteria sequenced to date, and S. avermitilis. T. fusca has all the components of the sec-independent twin-arginine protein translocation (TAT) system: Tfu1768 (tatC), Tfu1769 (tatA/E), Tfu0381(tatB), and Tfu0398 (tatD). The distinguishing feature of the TAT system is its ability to translocate fully folded proteins, and it may have significant implications for the secretion of functional cellulases. The genome does not contain any homologs of the type I and type III secretion systems. However, CDSs with similarity to components of the type II secretion system have been identified. Previous work demonstrated that the secretion of a Cel5 cellulase by Erwinia chrysanthemi was carried out by a type II protein secretion system (6). On the contrary, Faury et al. reported that the TAT export system mediates the secretion of an active xylanase from Streptomyces lividans (9). Moreover, recent work demonstrated that the TAT system is the major route for protein export in S. coelicolor (52). Therefore, we investigated whether the T. fusca genome encodes components of the type II secretion apparatus.
Type II secretion systems generally consist of 12 proteins and share many components with the type IV pilus biogenesis machinery (35). Examination of the genome revealed the existence of CDSs similar to type II and IV secretion components distributed between two operons. The first operon contained three CDSs with homologies to pilus assembly protein TadB (Tfu0126 and Tfu0127) and the cytoplasmic ATPase of the PulE type (Tfu0128). The second operon, located between Tfu2271 and Tfu2276, contains proteins with homologies to the pilus assembly proteins CpaB (Tfu2271), TadG (Tfu2273), and GSPII_F (Tfu2275 and Tfu2276). It also contains two predicted ATPases, Tfu2274, which contains a pulE-like domain (Pfam GSPII_E), and Tfu2272, which is also an ATPase containing a receiver domain at its amino-terminal region. However, we were unable to identify any homologs of the secretin (T2SD), which is the component forming channels for protein efflux, in T. fusca and other Actinobacteria. On the contrary, the gene similarities and structures of the respective chromosomal regions resemble the tight adherence (tad) locus (Fig. 4B) identified in the gram-negative bacterium Actinobacillus actinomycetemcomitans (20, 37, 42). The tad locus has been identified as an important mechanism for the production of pili that mediate nonspecific adherence to surfaces and formation of biofilms (21). However, both T. fusca and S. coelicolor lack homologs of either RcpA (a secretin family protein) or prepilins. We hypothesize that these particular functions have been replaced by nonorthologous genes in T. fusca. Overall, based on the above observations, we propose that T. fusca lacks a type II secretion system, and the above-mentioned CDSs constitute a system for nonspecific adherence to surfaces analogous to the one characterized in Actinobacillus actinomycetemcomitans (36, 37). Consequently, we hypothesize that the secretion of cellulases in T. fusca is mediated by the TAT translocation system and the general sec secretion system.
Transport systems.
Approximately 6% (180 CDSs) of the T. fusca genome encodes genes for transport. T. fusca has the core phosphotransferase system components E1 (Tfu2489) and Hpr (Tfu2487), as do all published Actinobacteria except Tropheryma (reduced genome) and mycobacteria. However, unlike other Actinobacteria, it has no identifiable homologs of PTS transporters. Therefore, we hypothesize that T. fusca does not have an active PTS transport system, and the E1 and Hpr components function in signaling pathways. This is a major difference with the carbohydrate transport system of S. coelicolor, which utilizes the PTS system to transport fructose and N-acetylglucosamine (33, 34).
T. fusca has an extensive set of transporters for carbohydrate uptake. Eight ABC disaccharide transporter cassettes were identified in the genome, and the majority of them (five) are localized on the chromosome next to glycoside hydrolases (Fig. 4A). The transport system (Tfu0934-Tfu0936) that clusters with the CelR transcription factor (see above) exhibits high similarity to the cellobiose/cellotriose transport system of Streptomyces reticuli (40). This system lacks an ATPase component in both organisms. However, T. fusca harbors a homolog of the MsiK (multiple sugar import) protein, Tfu1763. MsiK has been characterized in Streptomyces, and it was shown to assist both in cellobiose and maltose transport (41). The T. fusca maltose transport system consists of three CDSs, Tfu0830, Tfu0831, and Tfu0832. It is clustered on the chromosome with Tfu0833, which is an intracellular 1,4-α-d-glucan glucanohydrolase, probably involved in the hydrolysis of maltose. The adjacent CDS (Tfu0834) is a lacI-type transcriptional regulation and may be involved in the transcription of the transporter-glycohydrolase operon. The chromosomal region between Tfu1609 and Tfu1620 (12 CDSs) contains two transport systems, a set of two secreted and three intracellular glycosyl hydrolases, as well as a lac-type transcriptional regulator (Fig. 4A). Tfu1617, Tfu1618, and Tfu1619 are components of the xylobiose transporter based on their high similarity to the recently characterized xylobiose transporter from Streptomyces thermoviolaceus (49). Tfu1612 is the secreted xyloglucanase possessing a potential TAT targeting signal peptide characterized by Irwin et al. (17). Tfu1616 is a GH_43 family protein encoding an uncharacterized secreted glycoside hydrolase activity, whereas Tfu1613, Tfu1614, and Tfu1615 are intracellular glycoside hydrolases. Tfu1609, Tfu1610, and Tfu1611 are components of a putative disaccharide transport system with unknown specificity. Tfu1620 is a lac-type transcriptional regulator, and we postulate that it regulates the expression of the gene set Tfu1609-Tfu1620.
The genome of T. fusca also has an ortholog (Tfu1668) of the GlcP sugar permease characterized in S. coelicolor as a major glucose uptake system (51). Overall, our analysis identified the presence of cellobiose/cellotriose, maltose and xylobiose ABC transport systems, and a permease of the major facilitator superfamily for glucose uptake.
There are four putative amino acid ABC transport systems (Tfu0304-Tfu0306, Tfu0910-Tfu0914, Tfu1182-Tfu1187, and Tfu2928-Tfu2930).
Genes encoding a siderophore transport system were identified (Tfu0656, Tfu0657, and Tfu1491-Tfu1494) as well as a siderophore biosynthesis cluster (Tfu1865-Tfu1867). In addition, there is a ferrous iron transport protein (Tfu0820). There are four additional heavy metal transport systems (Tfu0857-Tfu0858, Tfu0931-Tfu0932, Tfu1661-Tfu1663, and Tfu2312-Tfu2313).
DNA repair and replication and stress.
Compared to many other representatives of Actinobacteria, especially some pathogenic species, T. fusca has reduced DNA repair capabilities and fewer mechanisms of resistance to reactive oxygen species (Table 1). T. fusca has Mn-dependent and heme catalases (Tfu0886 and Tfu1649, respectively) and an Mn-dependent superoxide dismutase (Tfu0957), while other enzymes present in N. farcinica, Streptomyces spp., and Mycobacterium spp., such as catalase/peroxidase and Cu-Zn superoxide dismutase, are missing. Similarly, only a homolog of the organic hydroperoxide resistance protein Ohr (Tfu0697) is present (8), while peroxiredoxin reductase AhpD and peroxiredoxin AhpC are absent.
T. fusca is capable of repairing UV-induced cyclobutane pyrimidine dimers due to the presence of type I deoxyribopyrimidine photolyase (Tfu0534). Deaminated bases are removed via a base excision mechanism (uracil-DNA glycosylase Tfu1341 and G/U-mismatch DNA glycosylase Tfu1918), while oxidized bases are removed by endonuclease III (Tfu0118), formamidopyrimidine-DNA glycosylase (Tfu0652), and A/G-specific adenine glycosylase (Tfu2875), and the resulting abasic sites are processed by the DNA (apurinic or apyrimidinic site) lyase activities of some these enzymes or by endonuclease IV (Tfu1956) and endonuclease V (Tfu1400). The repertoire of pathways for repair of alkylated bases is limited in T. fusca to base excision by DNA-3-methyladenine glycosylase I (Tfu0498) and alkylation reversal via DNA-N1-methyladenine dioxygenase (Tfu1427) (23); no homologs of other methyladenine glycosylases or a suicidal protein-cysteine S-methyltransferase AidA were found. Genes coding for the subunits of excinuclease ABC, an enzyme responsible for nucleotide excision repair, are also present in the genome (Tfu2024, Tfu1196, and Tfu2021). Similar to other Actinobacteria, the genes coding for a methyl-directed mismatch repair system (MutSLH proteins) are absent. T. fusca has only one error-prone DNA polymerase responsible for translesion synthesis (Tfu1096). Unlike many other representatives of Actinobacteria, no homologs of proteins responsible for double-strand break repair by nonhomologous end joining (3) were found in the genome of T. fusca (13).
One of the most striking features of T. fusca is the genetic organization of the recA gene (Tfu0803), which has two inteins (see the New England BioLabs intein database [http://www.neb.com/neb/inteins.html]) inserted at both the RecA-a and RecA-b sites (4). While intein-harboring recA genes appear to be quite common among both pathogenic and free-living mycobacteria (39), none of the genes sequenced so far has two intein insertions. Since it has been demonstrated that the intein-free RecA protein from Mycobacterium smegmatis and single intein-containing RecA protein from Mycobacterium tuberculosis display different activities in DNA strand exchange (10), RecA from T. fusca represents an interesting model to further clarify the mechanistic basis and the factors that contribute to the extent of DNA strand transfer in various Actinobacteria. Other features of recombinational DNA repair include the absence of a RecBCD complex or its AddAB equivalog with DNA helicase and exonuclease activities. Some of the essential components of an alternative RecF recombination pathway operating in recBC mutants in E. coli (43) are also missing; these include helicase RecQ and 5′-3′-exonuclease RecJ. However, despite the apparent absence of any helicase or exonuclease activity typically associated with DNA recombination, other components of a recombination apparatus (24), such as RecF, RecO, RecR, and RecN proteins (Tfu0004, Tfu0852, Tfu0044, and Tfu2032, respectively) and enzymes responsible for Holliday junction resolution (Tfu2093, Tfu2094, Tfu2095, and Tfu0646), are present in the genome. Further analysis of the phylogenetic distribution of the genes coding for various components of recombination machinery in Actinobacteria reveals no correlation between the genetic structure of the recA gene and the presence of the proteins involved in RecBCD or RecF recombination pathways.
Central metabolism.
The T. fusca genome encodes all the enzymes necessary to carry out glycolytic degradation of monosaccharides. Additionally, it has the Entner-Douodoroff pathway for glucose utilization.
All the enzymes of the tricarboxylic acid cycle are present. The presence of the glyoxylate cycle bypass from isocitrate to malate is indicative of its ability to grow on additional carbon sources such as acetate. The existence of homologs of glycerol kinase (Tfu0787), glycerol-3-phosphate dehydrogenase (Tfu0631), and triose phosphate isomerase (Tfu2015) indicates its ability to convert glycerol to glyceraldehyde-3-phosphate, an intermediate of glycolysis. Therefore, glycerol can also serve as a carbon source. It has the gluconeogenic enzymes pyruvate carboxylase (Tfu2554), phosphoenolpyruvate carboxykinase (Tfu0083), and fructose-1,6-bisphosphatase (Tfu0464) indicative of gluconeogenesis.
It has the machinery for the de novo biosynthesis of all amino acids except asparagine. However, it possesses a class II aspartyl-tRNA synthetase (Tfu2086), indicative of its ability to synthesize Asn-tRNA through transamidation of Asp-tRNA (32).
All the enzymes necessary for the biosynthesis of purines and pyrimidines have been identified. T. fusca has the enzymes for NAD, coenzyme A, riboflavin, pyridoxal, folate, cobalamin, and menaquinone biosynthesis. Two pathways have been described for the biosynthesis of quinolinate, an intermediate in NAD biosynthesis (26). The tryptophan-to-quinolinate pathway is present in T. fusca as well as in both Streptomyces strains sequenced to date, but it is absent in the rest of the Actinobacteria. T. fusca also utilizes the aspartate-to-quinolinate pathway, which is common to all Actinobacteria. T. fusca does not have a biotin pathway biosynthesis, since homologs of BioB, BioF, and BioD have not been identified. However, it possesses a transporter (Tfu2314) for scavenging biotin from the environment. The biosynthetic pathway of thiamine is also incomplete.
Lipid and cell wall metabolism.
T. fusca is able to synthesize and metabolize saturated and unsaturated fatty acids, as well as other major lipid classes, such as phospholipids, glycolipids, and isoprenoids. T. fusca appears to have all the enzymes necessary for fatty acid biosynthesis. It utilizes exclusively a type II fatty acid synthesis system in common with Streptomyces and in contrast to Mycobacterium, Corynebacterium, and Nocardia, which utilize a type I fatty acid synthase in addition to the type II system. It generates unsaturated fatty acids by a fatty acid desaturase mechanism (Tfu0413). An interesting feature of the T. fusca genome lies in the existence of a PlsX homolog in the absence of its PlsY counterpart. PlsX and PlsY were recently shown to constitute a novel mechanism for phospholipid biosynthesis initiation through the formation of acylphosphates (30). T. fusca appears to lack both a PlsB as well as a PlsY homolog, raising the intriguing possibility that an as-yet-unidentified enzyme participates in the initiation of phospholipid biosynthesis.
Two secreted triacylglycerol lipases can be identified in the genome (Tfu0882 and Tfu0883). It has the enzymes for β-oxidation of both odd and even carbon number fatty acids. In contrast to Mycobacteria and Streptomyces, it does not have homologs of diacylglycerol acyltransferases and, therefore, it probably lacks the machinery for endocellular triacylglycerol accumulation. Hence, these observations suggest that T. fusca can hydrolyze extracellular lipids, take up the resulting fatty acids, and utilize them subsequently as a carbon source. T. fusca has all the genes for the biosynthesis of phosphatidylglycerol, phosphatidylserine, phosphatidylethanolamine, and phosphatidylinositol. Phosphatidylinositol is further decorated with mannosyl groups, as evidenced by the existence of a phosphatidylinositol mannosyltransferase homolog (Tfu2101). It utilizes a phospholipase d-type of cardiolipin synthase (Tfu2817). This type of enzyme catalyzes the synthesis of cardiolipin from two molecules of phosphatidylglycerol. In contrast, Mycobacterium and Nocardia appear to possess a “eukaryotic”-type reaction via a CDP-alcohol phosphatidyltransferase, which catalyzes cardiolipin formation from CDP-diacylglycerol and phosphatidylglycerol.
Additionally, T. fusca and both S. coelicolor and S. avermitilis have two novel classes of CDP-alcohol phosphatidyltransferases (Tfu2439 and Tfu2359), indicative of their ability to synthesize as-yet-unidentified phospholipids. The most attractive hypothesis is that these enzymes catalyze the formation of novel structures of phosphatidyl sugars.
It possesses a complete nonmevalonate pathway for isoprenoid biosynthesis. The existence of genes with homology to phytoene synthase (Tfu3076), phytoene dehydrogenase (Tfu3075), and lycopene cyclase (Tfu3088) is indicative of a putative carotenogenesis process (Fig. 4C). A recent study established the ability of S. coelicolor to carry out light-induced synthesis of carotenoids (48).
All the genes (murC, murD, murE, and murF) that encode enzymes for the conversion of d-glutamate to UDP-MurNac pentapeptide are present, as are other enzymes in the peptidoglycan synthetic pathway, such as GlmU, MurA, and MurB.
Conclusions.
The genome sequence of T. fusca provides the means for a detailed analysis of the cellular mechanisms controlling the expression and secretion of plant cell wall-degrading enzymes by this soil bacterium. T. fusca utilizes a variety of enzymes attacking cellulose, xylan, and pectin, major components of plant cell walls. Detailed genomic analysis provides evidence for the utilization of the TAT secretion system for the export of these enzymes to the extracellular space, the existence of multiple transcription factors regulating the expression of glycosylhydrolases, and oligo/polysaccharide transport systems. These observations open further research directions for understanding the mechanisms of plant cell wall hydrolysis and utilization by soil actinomycetes.
Acknowledgments
This study was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program and by the University of California, Lawrence Berkeley National Laboratory under contract DE-AC02-05CH11231 and Lawrence Livermore National Laboratory under contract DE-AC02-06NA25396.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Bachmann, S. L., and A. J. McCarthy. 1991. Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl. Environ. Microbiol. 57:2121-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellamy, W. D. 1977. Cellulose and lignocellulose digestion by thermophilic actinomycetes for single cell protein production. Dev. Ind. Microbiol. 18:249-254. [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, J., J. J. Coque, J. Velasco, and J. F. Martin. 1997. Cloning, expression in Streptomyces lividans and biochemical characterization of a thermostable endo-beta-1,4-xylanase of Thermomonospora alba ULJB1 with cellulose-binding ability. Appl. Microbiol. Biotechnol. 48:208-217. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453-464. [DOI] [PubMed] [Google Scholar]
- 6.Chapon, V., M. Czjzek, M. El Hassouni, B. Py, M. Juy, and F. Barras. 2001. Type II protein secretion in gram-negative pathogenic bacteria: the study of the structure/secretion relationships of the cellulase Cel5 (formerly EGZ) from Erwinia chrysanthemi. J. Mol. Biol. 310:1055-1066. [DOI] [PubMed] [Google Scholar]
- 7.Collmer, A., and D. B. Wilson. 1983. Cloning and expression of a Thermomonospora YX endocellulase gene in E. coli. Biotechnology 1:594-601. [Google Scholar]
- 8.Cussiol, J. R., S. V. Alves, M. A. de Oliveira, and L. E. Netto. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 278:11570-11578. [DOI] [PubMed] [Google Scholar]
- 9.Faury, D., S. Saidane, H. Li, and R. Morosoli. 2004. Secretion of active xylanase C from Streptomyces lividans is exclusively mediated by the Tat protein export system. Biochim. Biophys. Acta 1699:155-162. [DOI] [PubMed] [Google Scholar]
- 10.Ganesh, N., and K. Muniyappa. 2003. Characterization of DNA strand transfer promoted by Mycobacterium smegmatis RecA reveals functional diversity with Mycobacterium tuberculosis RecA. Biochemistry 42:7216-7225. [DOI] [PubMed] [Google Scholar]
- 11.Ghangas, G. S., Y. J. Hu, and D. B. Wilson. 1989. Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. J. Bacteriol. 171:2963-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghangas, G. S., and D. B. Wilson. 1988. Cloning of the Thermomonospora fusca endoglucanase E2 gene in Streptomyces lividans: affinity purification and functional domains of the cloned gene product. Appl. Environ. Microbiol. 54:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hefferin, M. L., and A. E. Tomkinson. 2005. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amsterdam) 4:639-648. [DOI] [PubMed] [Google Scholar]
- 14.Hilge, M., S. M. Gloor, W. Rypniewski, O. Sauer, T. D. Heightman, W. Zimmermann, K. Winterhalter, and K. Piontek. 1998. High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca-substrate specificity in glycosyl hydrolase family 5. Structure 6:1433-1444. [DOI] [PubMed] [Google Scholar]
- 15.Hu, Y. J., and D. B. Wilson. 1988. Cloning of Thermomonospora fusca genes coding for beta-1-4 endoglucanases E1, E2 and E5. Gene 71:331-337. [DOI] [PubMed] [Google Scholar]
- 16.Irwin, D., E. D. Jung, and D. B. Wilson. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irwin, D. C., M. Cheng, B. Xiang, J. K. Rose, and D. B. Wilson. 2003. Cloning, expression and characterization of a family-74 xyloglucanase from Thermobifida fusca. Eur. J. Biochem. 270:3083-3091. [DOI] [PubMed] [Google Scholar]
- 18.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 19.Jung, E. D., G. Lao, D. Irwin, B. K. Barr, A. Benjamin, and D. B. Wilson. 1993. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl. Environ. Microbiol. 59:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9:429-437. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. H., D. Irwin, and D. B. Wilson. 2004. Purification and characterization of Thermobifida fusca xylanase 10B. Can. J. Microbiol. 50:835-843. [DOI] [PubMed] [Google Scholar]
- 23.Koivisto, P., T. Duncan, T. Lindahl, and B. Sedgwick. 2003. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J. Biol. Chem. 278:44348-44354. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 25.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 26.Kurnasov, O., V. Goral, K. Colabroy, S. Gerdes, S. Anantha, A. Osterman, and T. P. Begley. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10:1195-1204. [DOI] [PubMed] [Google Scholar]
- 27.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lao, G., G. S. Ghangas, E. D. Jung, and D. B. Wilson. 1991. DNA sequences of three β-1,4-endoglucanase genes from Thermomonospora fusca. J. Bacteriol. 173:3397-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liolios, K., N. Tavernarakis, P. Hugenholtz, and N. C. Kyrpides. 2006. The Genomes On Line Database (GOLD) v. 2: a monitor of genome projects worldwide. Nucleic Acids Res. 34:D332-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, Y. J., Y. M. Zhang, K. D. Grimes, J. Qi, R. E. Lee, and C. O. Rock. 2006. Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Mol. Cell 23:765-772. [DOI] [PubMed] [Google Scholar]
- 31.McGrath, C. E., and D. B. Wilson. 2006. Characterization of a Thermobifida fusca β-1,3-glucanase (Lam81A) with a potential role in plant biomass degradation. Biochemistry 45:14094-14100. [DOI] [PubMed] [Google Scholar]
- 32.Min, B., J. T. Pelaschier, D. E. Graham, D. Tumbula-Hansen, and D. Soll. 2002. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. USA 99:2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nothaft, H., D. Dresel, A. Willimek, K. Mahr, M. Niederweis, and F. Titgemeyer. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 185:7019-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 36.Perez, B. A., P. J. Planet, S. C. Kachlany, M. Tomich, D. H. Fine, and D. H. Figurski. 2006. Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation. J. Bacteriol. 188:6361-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planet, P. J., S. C. Kachlany, D. H. Fine, R. DeSalle, and D. H. Figurski. 2003. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34:193-198. [DOI] [PubMed] [Google Scholar]
- 38.Posta, K., E. Beki, D. B. Wilson, J. Kukolya, and L. Hornok. 2004. Cloning, characterization and phylogenetic relationships of cel5B, a new endoglucanase encoding gene from Thermobifida fusca. J. Basic Microbiol. 44:383-399. [DOI] [PubMed] [Google Scholar]
- 39.Saves, I., M. A. Laneelle, M. Daffe, and J. M. Masson. 2000. Inteins invading mycobacterial RecA proteins. FEBS Lett. 480:221-225. [DOI] [PubMed] [Google Scholar]
- 40.Schlosser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlosser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J. Bacteriol. 179:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiner, H. C., K. Sinatra, J. B. Kaplan, D. Furgang, S. C. Kachlany, P. J. Planet, B. A. Perez, D. H. Figurski, and D. H. Fine. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 100:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, G. R. 1989. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell 58:807-809. [DOI] [PubMed] [Google Scholar]
- 44.Spiridonov, N. A., and D. B. Wilson. 2000. A celR mutation affecting transcription of cellulase genes in Thermobifida fusca. J. Bacteriol. 182:252-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiridonov, N. A., and D. B. Wilson. 1999. Characterization and cloning of celR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J. Biol. Chem. 274:13127-13132. [DOI] [PubMed] [Google Scholar]
- 46.Spiridonov, N. A., and D. B. Wilson. 2001. Cloning and biochemical characterization of BglC, a beta-glucosidase from the cellulolytic actinomycete Thermobifida fusca. Curr. Microbiol. 42:295-301. [DOI] [PubMed] [Google Scholar]
- 47.Suhre, K., and J. M. Claverie. 2003. Genomic correlates of hyperthermostability, an update. J. Biol. Chem. 278:17198-17202. [DOI] [PubMed] [Google Scholar]
- 48.Takano, H., S. Obitsu, T. Beppu, and K. Ueda. 2005. Light-induced carotenogenesis in Streptomyces coelicolor A3(2): identification of an extracytoplasmic function sigma factor that directs photo-dependent transcription of the carotenoid biosynthesis gene cluster. J. Bacteriol. 187:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsujibo, H., M. Kosaka, S. Ikenishi, T. Sato, K. Miyamoto, and Y. Inamori. 2004. Molecular characterization of a high-affinity xylobiose transporter of Streptomyces thermoviolaceus OPC-520 and its transcriptional regulation. J. Bacteriol. 186:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van den Bogart, H. G., G. Van den Ende, P. C. Van Loon, and L. J. Van Griensven. 1993. Mushroom worker's lung: serologic reactions to thermophilic actinomycetes present in the air of compost tunnels. Mycopathologia 122:21-28. [DOI] [PubMed] [Google Scholar]
- 51.van Wezel, G. P., K. Mahr, M. Konig, B. A. Traag, E. F. Pimentel-Schmitt, A. Willimek, and F. Titgemeyer. 2005. GlcP constitutes the major glucose uptake system of Streptomyces coelicolor A3(2). Mol. Microbiol. 55:624-636. [DOI] [PubMed] [Google Scholar]
- 52.Widdick, D. A., K. Dilks, G. Chandra, A. Bottrill, M. Naldrett, M. Pohlschroder, and T. Palmer. 2006. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 103:17927-17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, S., G. Lao, and D. B. Wilson. 1995. Characterization of a Thermomonospora fusca exocellulase. Biochemistry 34:3386-3395. [DOI] [PubMed] [Google Scholar]