Abstract
Vibrio cholerae, the causative agent of cholera, can undergo phenotypic variation generating rugose and smooth variants. The rugose variant forms corrugated colonies and well-developed biofilms and exhibits increased levels of resistance to several environmental stresses. Many of these phenotypes are mediated in part by increased expression of the vps genes, which are organized into vps-I and vps-II coding regions, separated by an intergenic region. In this study, we generated in-frame deletions of the five genes located in the vps intergenic region, termed rbmB to -F (rugosity and biofilm structure modulators B to F) in the rugose genetic background, and characterized the mutants for rugose colony development and biofilm formation. Deletion of rbmB, which encodes a protein with low sequence similarity to polysaccharide hydrolases, resulted in an increase in colony corrugation and accumulation of exopolysaccharides relative to the rugose variant. RbmC and its homolog Bap1 are predicted to encode proteins with carbohydrate-binding domains. The colonies of the rbmC bap1 double deletion mutant and bap1 single deletion mutant exhibited a decrease in colony corrugation. Furthermore, the rbmC bap1 double deletion mutant was unable to form biofilms at the air-liquid interface after 2 days, while the biofilms formed on solid surfaces detached readily. Although the colony morphology of rbmDEF mutants was similar to that of the rugose variant, their biofilm structure and cell aggregation phenotypes were different than those of the rugose variant. Taken together, these results indicate that vps intergenic region genes encode proteins that are involved in biofilm matrix production and maintenance of biofilm structure and stability.
Vibrio cholerae, the etiologic agent of the diarrheal disease cholera (34), is found naturally in environmental aquatic habitats both as a free-living organism in the water column and in a biofilm state attached to different surfaces, including mucilaginous surfaces of phytoplankton, chitinous surfaces of zooplankton, and abiotic surfaces (19). It has been proposed that this facultative human pathogen uses biofilm formation and phenotypic variation as survival strategies (63, 64, 71).
Biofilms are surface-attached microbial communities composed of microorganisms and the extrapolymeric substances they produce (15, 49, 51). Biofilm formation begins with the transport and attachment of bacteria to surfaces. After initial attachment, colonization of a surface is mediated by the movement and growth of attached bacteria, followed by the formation of microcolonies, which are often surrounded by extrapolymeric substances. Further growth of bacteria and continued production of exopolysaccharides lead to the development of mature biofilms characterized by pillars and mushroom-like structures. It has been shown that the development of these structures depends on several factors, including biomass growth rate, twitching motility, signaling molecules, and exopolysaccharide production (15, 49, 51). Cells in the mature biofilms can return to the planktonic stage through detachment, thus completing the cycle of biofilm development (49, 63). Detachment of mature biofilms may occur as a result of nutritional and oxygen availability or the presence of other environmental stresses (11, 22, 49, 56, 59). Compared to surface attachment and mature biofilm development, the molecular mechanisms of the detachment process are largely unknown. Nonetheless, polysaccharide lyases have been reported to be involved in biofilm detachment and cell dispersal in several biofilm-forming microorganisms (4, 11, 17, 35, 50).
In V. cholerae, the sequence of events leading to the initial attachment and subsequent formation of mature biofilms on surfaces under laboratory conditions, as well as the genes required for these steps, has been reported (63). The mannose-sensitive hemagglutinin type IV pilus and flagellum facilitate attachment to abiotic surfaces. Under laboratory conditions, the mannose-sensitive hemagglutinin type IV pilus is also required for efficient colonization of biotic surfaces, such as the chitinous exoskeleton of zooplankton (14) and cellulose fibers (62). Chitin-regulated pilus and toxin-coregulated pilus also facilitate attachment to chitinous surfaces (44, 53). Furthermore, Vibrio polysaccharide (VPS), which is part of the extracellular matrix, is required for the development of mature biofilms (63, 71).
V. cholerae can undergo phenotypic variation in response to environmental stresses, resulting in rugose and smooth colonial variants (48, 60, 64, 71). Compared to the smooth variant, the rugose variant forms corrugated colonies and well-developed biofilms and exhibits increased levels of resistance to osmotic and oxidative stresses. Most of these rugose-associated phenotypes are due in part to increased production of VPS (48, 54, 60, 71), which is mediated by proteins encoded by the vps genes (vpsA to -K, VC0917 to -27 [vps-I cluster]; vpsL to -Q, VC0934 to -9 [vps-II cluster]) (Fig. 1). Mutations in any of the vps genes yield a smooth colonial morphology and reduced capacity to form biofilms (71). The vps genes are organized into regions encoding vps-I (11.5 kb) and vps-II (6.6 kb) on the large chromosome, separated by an 8.3-kb intergenic region (Fig. 1). We previously reported that rbmA (VC0928), the first gene located in the vps intergenic region, is required for the full development of corrugated rugose colony morphology as well as pillar and mushroom-like structures of biofilms (20). We hypothesized that the genes located in the vps intergenic region may have a role in rugose colony development and biofilm formation. We generated null mutants of these genes and undertook a complete phenotypic characterization. In this study, we show that genes located downstream of rbmA, termed rbmB, rbmC, rbmD, rbmE, and rbmF (rugosity and biofilm structure modulators B to F; VC0929 to -33) encode proteins that modulate rugose colony development and biofilm formation in V. cholerae. In addition, we also show that bap1 (biofilm-associated protein 1; VC1888) (47) (also called vcp [vps-coregulated protein] [30]), which has peptide sequence similarity to that of rbmC, encodes a protein that is also involved in rugose colony development and biofilm formation in V. cholerae.
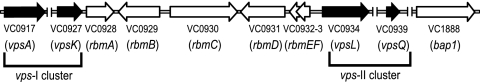
FIG. 1.
Genetic organization of the vps intergenic region. The genetic organization of rbmABCDEF and bap1 (open arrows) on the V. cholerae chromosome is depicted with locus annotations under each open reading frame. The vps-I and vps-II gene clusters, as well as the first and last genes of each cluster (vpsA and vpsK and vpsL and vpsQ) are marked and labeled. Unlinked chromosomal DNA regions are indicated (∥). Illustration is not to scale.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All V. cholerae and Escherichia coli strains were routinely grown aerobically, in Luria-Bertani (LB) medium, at 30°C and 37°C, respectively, unless otherwise noted. Agar medium contained 1.5% (wt/vol) granulated agar (Difco), except for motility studies, where the agar concentration was 0.3% (wt/vol). Concentrations of antibiotics used were as follows: ampicillin (100 μg/ml), rifampin (100 μg/ml), and gentamicin (30 μg/ml). Pellicle formation experiments were carried out in glass culture tubes (18 by 150 mm) containing 5 ml of LB medium or LB medium supplemented with ampicillin and were inoculated with 200-fold-diluted overnight-grown cultures. The tubes were incubated at 30°C under static conditions for 2 days. Cultures grown for colony morphology observations were also incubated at 30°C for 2 days.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 (φ80lacΔM15) | Promega |
| CC118 (λ pir) | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λ pir | 26 |
| S17-1 (λ pir) | Tpr SmrrecA thi pro rK− mK+RP4:2-Tc:MuKm Tn7 λ pir | 16 |
| V. cholerae strains | ||
| FY_Vc_2 | O1 El Tor A1552, rugose variant, Rifr | 69 |
| FY_Vc_1040 | RΔrbmB, FY_Vc_2 ΔrbmB, Rifr | This study |
| FY_Vc_686 | RΔrbmC, FY_Vc_2 ΔrbmC, Rifr | This study |
| FY_Vc_690 | RΔrbmD, FY_Vc_2 ΔrbmD, Rifr | This study |
| FY_Vc_742 | RΔrbmEF, FY_Vc_2 ΔrbmEF, Rifr | This study |
| FY_Vc_1367 | RΔbap1, FY_Vc_2 Δbap1, Rifr | This study |
| FY_Vc_1400 | RΔrbmC Δbap1, FY_Vc_1367 ΔrbmC, Rifr | This study |
| FY_Vc_222 | FY_Vc_2 mTn7-GFP, Gmr | 20 |
| FY_Vc_1318 | FY_Vc_1040 mTn7-GFP, Gmr | This study |
| FY_Vc_1321 | FY_Vc_686 mTn7-GFP, Gmr | This study |
| FY_Vc_1387 | FY_Vc_690 mTn7-GFP, Gmr | This study |
| FY_Vc_1324 | FY_Vc_742 mTn7-GFP, Gmr | This study |
| FY_Vc_1392 | FY_Vc_1367 mTn7-GFP, Gmr | This study |
| FY_Vc_1395 | FY_Vc_1400 mTn7-GFP, Gmr | This study |
| Plasmids | ||
| pGP704-sacB28 | pGP704 derivative, mob/oriT sacB, Apr | G. Schoolnik |
| pFY-271 | pGP704-sacB28::ΔrbmB, Apr | This study |
| pFY-272 | pGP704-sacB28::ΔrbmC, Apr | This study |
| pFY-273 | pGP704-sacB28::ΔrbmD, Apr | This study |
| pFY-274 | pGP704-sacB28::ΔrbmEF, Apr | This study |
| pFY-330 | pGP704-sacB28::Δbap1, Apr | This study |
| pACYC177 | Multicopy cloning vector, Apr | New England Biolabs |
| pFY-361 | prbmB, pACYC177::rbmB, Apr | This study |
| pFY-362 | prbmC, pACYC177::rbmC, Apr | This study |
| pFY-363 | prbmD, pACYC177::rbmD, Apr | This study |
| pFY-364 | prbmEF, pACYC177::rbmEF operon, Apr | This study |
| pFY-366 | pbap1, pACYC177::bap1, Apr | This study |
| pBAD/Myc-His B | Arabinose-inducible expression vector with C-terminal Myc epitope and six His codons, Apr | Invitrogen |
| pFY-417 | prbmB-myc, pBAD/Myc-His B::rbmB coding region, Apr | This study |
| pFY-420 | prbmC-myc, pBAD/Myc-His B::rbmC coding region, Apr | This study |
| pFY-429 | pbap1-myc, pBAD/Myc-His B::bap1 coding region, Apr | This study |
| pvpsA::lacZ | pRS415 vpsA promoter, Apr | 13 |
| pvpsL::lacZ | pRS415 vpsL promoter, Apr | 13 |
| pMCM11 | pGP704::mTn7-GFP, Gmr Apr | G. Schoolnik |
| pUX-BF13 | oriR6K helper plasmid, mob/oriT, provides Tn7 transposition function in trans, Apr | 6 |
Recombinant DNA techniques.
DNA manipulations were carried out by standard molecular techniques (55). Restriction enzymes and DNA modification enzymes were obtained from New England Biolabs. PCRs were carried out using primers purchased from Operon Technologies and the High-Fidelity PCR kit (Roche). Primers used in this study are listed in the Table 2.
TABLE 2.
Primers used in this study
| Primer type and name | Nucleotide sequence |
|---|---|
| Deletion primers | |
| rbmB_del_A | GATCTCTAGAGTGAGCGAGTGGAACAGTCA |
| rbmB_del_B | GAACGCATTTCTCGTTTCTTTCGAACATGTCA |
| rbmB_del_C | AAGAAACGAGAAATGCGTTCGAATCATCCTC |
| rbmB_del_D | GATCCCATGGGACCGACAGCAAGTGCAATA |
| rbmC_del_A | GATCTCTAGAAGCTGGGCTAAACAGAACAAG |
| rbmC_del_B | TAATCGCCACCAGGCGGCTGATAATACACC |
| rbmC_del_C | CAGCCGCCTGGTGGCGATTAAATTCACGATG |
| rbmC_del_D | GATCCCATGGGGCAGTTTAATGGCGATCAT |
| rbmD_del_A | GATCTCTAGAGAAGGATGGCTCTAGCGTGT |
| rbmD_del_B | CAATGAAATTGGCAGTTTAATGGCGATCAT |
| rbmD_del_C | TTAAACTGCCAATTTCATTGAGCGACCATGC |
| rbmD_del_D | GATCGAGCTCGGACCCAACCGAGATTATCA |
| rbmEF_del_A | GATCTCTAGACAACTTTGCATCCAAGACCA |
| rbmEF_del_B | TTCATTTGATCCGGGCTTAACCAATACCTT |
| rbmEF_del_C | TTAAGCCCGGATCAAATGAAGCGGTAGTGTTT |
| rbmEF_del_D | GATCGAGCTCGAAACCAATTGAAAACAACCAA |
| bap1_del_A | CTATGAGCTCGCTACCTCCCGTCTGTTTCA |
| bap1_del_B | GCGGAACGCGCTGTTTCATGGCTTGACCTTC |
| bap1_del_C | CATGAAACAGCGCGTTCCGCTGAAGTAAAGA |
| bap1_del_D | CTAGTCTAGAAGCTTCGGCTTTAACCCAAT |
| Complementation primers | |
| rbmB_com F | GATCCCCGGGGTGAGCGAGTGGAACAGTCA |
| rbmB_com R | GATCCTCGAGGACCGACAGCAAGTGCAATA |
| rbmC_com F | GATCCCCGGGCTAGAAAATGCTTCTTGAAA |
| rbmC_com R | GATCCTCGAGTTGCTCTTCATCTCCTG |
| rbmD_com F | GATCCCCGGGCGACTCATCGCTTTACT |
| rbmD_com R | GATCCTCGAGCCCAGATAACTGACTTG |
| rbmEF_com F | GATCAAGCTTCAACTTTGCATCCAAGACCA |
| rbmEF_com R | GATCCTCGAGGAAACCAATTGAAAACAACCAA |
| bap1_com F | GATCCCCGGGGACTCGATCGTGAGTTT |
| bap1_com R | GATCAAGCTTGGTTTATTCCTTTATCTTTTTA |
| Myc-tagging primers | |
| rbmB_myc F | GATCCTCGAGGGTGCTGTTATACTTAAATC |
| rbmB_myc R | GATCGAATTCCATCTTTAATAAAGTGCTGTATA |
| rbmC_myc F | GATCCTCGAGAATGACGTCTCACTATATTG |
| rbmC_myc R | GATCGAATTCCGTAGACAAACTGGAAGC |
| bap1_myc F | GATCGAATTCATGAAACAGACAAAAACGTT |
| bap1_myc R | GATCTCTAGACGCTTCAGCGGAACGC |
Generation of in-frame rbmBCDEF and bap1 deletion mutants and complemented strains.
Deletion mutants of rbmBCDEF and bap1 were generated in the rugose genetic background according to the protocol previously published (20, 42). Briefly, 5′ regions of the genes were amplified by PCR with deletion primers del_A and del_B, while 3′ regions of the genes were amplified by PCR with deletion primers del_C and del_D. The two PCR products generated were then joined via splicing by overlap extension PCR (28, 29, 41) with del_A and del_D primers and cloned into a pGP704-sacB28 suicide plasmid, resulting in deletion plasmids pFY-271, pFY-272, pFY-273, pFY-274, and pFY-330 for rbmB, rbmC, rbmD, rbmEF, and bap1, respectively. The deletion plasmids were maintained in E. coli CC118 (λ pir). Biparental matings were carried out with the V. cholerae rugose variant and the conjugative strain E. coli S17-1 (λ pir) harboring various deletion plasmids. Ampicillin- and rifampin-resistant transconjugants, resulting from single homologous recombinations, were selected and subjected to sucrose-based selection, similar to that described by Fullner and Mekalanos (21). Ampicillin-sensitive and sucrose-resistant V. cholerae deletion strains, which had undergone double homologous recombinations, were selected. A double deletion mutant of rbmC and bap1 was generated by deleting bap1 in a rugose rbmC null mutant using the same method described above. The deletion mutants were verified by PCR and designated RΔrbmB, RΔrbmC, RΔrbmD, RΔrbmEF, RΔbap1, and RΔrbmC Δbap1.
Complementation plasmids prbmB, prbmC, prbmD, prbmEF, and pbap1 were constructed by cloning PCR-amplified rugose variant chromosomal fragments of rbmB, rbmC, rbmD, rbmEF, and bap1 (including the upstream and downstream regulatory elements) into the vector pACYC177 (New England Biolabs), using respective forward and reverse complementation primers. Ampicillin-resistant strains were selected after electroporation of various complementation plasmids into respective deletion strains. Complementation strains were verified via PCR.
Generation of Myc-tagged RbmB, RbmC, and Bap1.
The coding regions of rbmB, rbmC, and bap1 (including the start codon but excluding the stop codon) were amplified with respective forward and reverse Myc-tagging primers and directionally cloned into the vector pBAD/Myc-His B (Invitrogen), in frame with the upstream start codon as well as downstream Myc epitope and six-histidine codons. The resulting C-terminal Myc-tagged expression plasmids prbmB-myc, prbmC-myc, and pbap1-myc were electroporated into the rugose variant and various deletion mutants. Ampicillin-resistant strains harboring the Myc-tagged expression plasmids were selected and verified via PCR. V. cholerae deletion strains carrying the various Myc-tagged expression plasmids were used in cellular fractionation experiments. The prbmB-myc and pbap1-myc plasmids were also used in overexpression experiments in the presence of 0.1% (wt/vol) arabinose, except for colony morphology determinations of the rugose variant harboring prbmB-myc or the vector, for which the arabinose concentration was 0.01% (wt/vol).
Generation of GFP-tagged strains.
V. cholerae deletion strains and rugose variant were tagged with green fluorescent protein (GFP) according to the procedure previously described (20). Briefly, triparental matings were carried out with donor E. coli S17-1 (λ pir) carrying pMCM11, helper E. coli S17-1 (λ pir) harboring pUX-BF13, and various V. cholerae strains. Transconjugants were selected on thiosulfate-citrate-bile salts-sucrose (Difco) agar medium containing gentamicin at 30°C. GFP-tagged V. cholerae strains were verified by PCR and used in flow cell and cell aggregation experiments.
β-Galactosidase assays.
β-Galactosidase assays were carried out in MultiScreen-HA 96-well microtiter plates fitted onto a MultiScreen filtration system (Millipore) using a previously published procedure (20) which is similar to that described by Miller (46). Exponential cultures were harvested when optical densities at 600 nm (OD600) reached 0.3 to 0.5. The assays were repeated with four different biological replicates and at least eight technical replicates.
Quantitative biofilm assays.
Biofilm formation assays were carried out in polyvinyl chloride microtiter plates with 100 μl of 200-fold-diluted overnight-grown cultures incubated at 30°C for 8 h, according to the method previously described (69). The assays were repeated with three different biological replicates and at least eight technical replicates.
Cell aggregation experiments, flow cell experiments, and CLSM.
Cell aggregation experiments were carried out using hanging drop slides (Fisher Scientific) with 10-fold-diluted overnight-grown cultures of GFP-tagged V. cholerae strains. Flow cell experiments were carried out according to the procedure previously described (42). Briefly, overnight-grown cultures of GFP-tagged V. cholerae strains were diluted to an OD600 of 0.1, and 350-μl aliquots of the diluted cultures were inoculated by injection into the flow cell chambers. Before inoculation, the chambers were sterilized and equilibrated with 0.5% (vol/vol) hypochlorite, followed by sterile MilliQ water and 2% LB (0.2 g/liter tryptone, 0.1 g/liter yeast extract, 9 g/liter NaCl) or 2% LB supplemented with ampicillin at a flow rate of 4.5 ml/h. After inoculation, the chambers were allowed to stand inverted, with no flow, for 1 h. Flow was resumed at a rate of 4.5 ml/h with chambers standing upright. Flow cell experiments were carried out at room temperature. Overexpression of rbmB, for the flow cell studies, was achieved by growing the bacterial strains in 2% LB supplemented with ampicillin for 24 h, followed by changing the growth medium to 2% LB supplemented with ampicillin and 0.1% (wt/vol) arabinose. Overexpression of bap1 was achieved by growing the bacterial strains in 2% LB supplemented with ampicillin and 0.1% arabinose after inoculation. Confocal laser scanning microscopy (CLSM) images of the biofilms and cell aggregates were captured with a LSM 5 PASCAL system (Zeiss) at 488-nm excitation and 543-nm emission wavelengths. Three-dimensional images of the biofilms were reconstructed using Imaris software (Bitplane) and quantified using COMSTAT (27). Flow cell and cell aggregate experiments were carried out with at least two different biological replicates.
VPS extractions and analyses.
VPS extractions and analyses were carried out according to procedures similar to those previously described (18). Briefly, overnight-grown cultures (100 μl) were spread on petri plates (100 by 15 mm) containing LB agar medium and incubated overnight at 30°C. Bacteria from these plates were harvested by scraping and suspended in 10 ml of phosphate-buffered saline (PBS; 20 mM sodium phosphate [pH 7.3], 100 mM NaCl). Optical densities of the samples were determined after vortexing, and the samples were diluted to the same OD with PBS. Equal volumes of the samples were transferred to 25-ml Erlenmeyer flasks and shaken on a rotary shaker at 4°C for 5 h. The VPS suspensions were separated from the cells and debris by repeated centrifugation at 20,000 × g for 30 min, followed by 13,000 × g for 30 min. Crude VPS was precipitated with 3 volumes of ethanol at 4°C overnight, followed by centrifugation at 20,000 × g for 30 min. The pellets were washed with 70% ethanol and suspended in 3 ml of nuclease buffer (40 mM Tris-Cl [pH 8.0], 10 mM MgCl2, 2 mM CaCl2). DNase I (New England Biolabs) and RNase A (QIAGEN) were added to the VPS suspensions at final concentrations of 0.7 units/ml and 5 μg/ml, respectively, followed by incubation at 37°C shaking for 3 h. Proteinase K (Merck) was then added at a final concentration of 70 μg/ml, and the suspensions were further incubated with shaking overnight at 37°C. VPS suspensions were aliquoted into 2-ml centrifuge tubes, and phenol-chloroform extractions (equal volumes) were carried out three times, followed by precipitation with 3 volumes of ethanol, washing with 70% ethanol, and suspension in 250 μl of water. Equal amounts of VPS samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The upper (5% polyacrylamide) stacking portion of the SDS-PAGE gel was 6.5 cm long, while the lower (12% polyacrylamide) portion of the gel was 2 cm long. The gels were stained with Stains-All (U.S. Biochemical) according to a previously published method (39). VPS analyses were repeated with two different biological replicates.
Cellular fractionations.
Overnight-grown cultures of Myc-tagged V. cholerae strains were diluted 200-fold and inoculated into fresh LB medium containing ampicillin. Cells in the exponential growth phase (OD600 of 0.3 to 0.4) were induced with 0.1% (wt/vol) arabinose for 1 h. Cells were harvested by centrifugation at 15,000 × g for 15 min, washed with PBS, and resuspended in 10 mM Tris-Cl (pH 8.0). Cell lysis was carried out with sonication (10 10-s pulses), and cell debris was separated from whole-cell lysate by repeated centrifugation at 15,000 × g for 15 min. Culture supernatants were collected by repeated centrifugation at 15,000 × g for 15 min, followed by deoxycholic acid sodium salt monohydrate-trichloroacetic acid (Na-DOC/TCA) precipitation (10). Briefly, Na-DOC was added to culture supernatants to obtain a final concentration of 0.02% (wt/vol), followed by incubation on ice for 30 min. TCA (Sigma) was then added to obtain a final concentration of 10% (vol/vol), followed by incubation on ice for 2 h. Proteins in the culture supernatants were pelleted by centrifugation at 15,000 × g for 30 min. Pellets were washed with ice-cold acetone and resuspended in 10 mM Tris-Cl (pH 8.0). Protease inhibitors aprotinin, leupeptin, and pepstatin (Roche) were used at 2 μg/ml, 5 μg/ml, and 0.7 μg/ml, respectively. Protein concentrations were estimated using a Coomassie Plus protein assay kit (Pierce) with bovine serum albumin as standard. Protein samples were loaded at 40 μg and 5 μg for immunoblot analyses of Myc-tagged proteins (RbmC-Myc and Bap1-Myc) and OmpU, respectively.
Immunoblot analyses.
Immunoblot analyses were carried out according to a procedure previously described (20). Rabbit polyclonal antisera against V. cholerae OmpU (provided by K. Klose) was used at a dilution of 1:100,000, while mouse monoclonal antibody against the Myc epitope (Santa Cruz Biotechnology) was used at a dilution of 1:1,000. Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies (Santa Cruz Biotechnology) were used at a dilution of 1:2,500. Immunoblots were developed with the SuperSignal West Pico chemiluminescent kit (Pierce). Immunoblot analyses were carried out with at least two different biological replicates. Immunoblot analysis of the outer membrane protein OmpU was carried out as a control on the cellular fractions.
RESULTS
RbmB affects rugose colony morphology and biofilm structure formation.
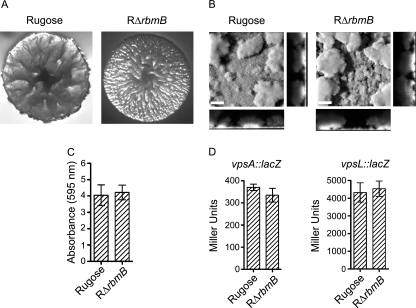
rbmB (VC0929) is the second gene located in the vps intergenic region (Fig. 1) and is annotated as a hypothetical protein in The Institute for Genomic Research (TIGR) Comprehensive Microbial Resource (CMR) database. Quick BlastP (NCBI BlastP 2.2.13) searches revealed that RbmB has low peptide sequence similarity to Alteromonas fortis ι-carrageenase CgiA (E-value, 1e-05) (7) and Bacillus circulans α-1,3-glucanase Agl (E-value, 4e-06) (68), both of which are polysaccharide hydrolases. The similarities of peptide sequences of RbmB and CgiA, calculated using the European Molecular Biology Open Source Software Suite (EMBOSS) alignment programs, revealed that RbmB and CgiA exhibit 30.1% similarity over the entire length of the peptide sequences. Similarly, RbmB and Agl exhibit 13.4% similarity over the entire length of the peptide sequences. These findings suggested that rbmB could encode a protein that might function as a polysaccharide hydrolase. To determine the role of rbmB in colony morphology development and biofilm formation, an in-frame deletion mutant of rbmB was generated in the rugose genetic background (RΔrbmB). The deletion mutant exhibited an increased colony corrugation compared to the rugose variant (Fig. 2A), and this phenotype was partially complemented (reduced corrugation) by introducing a wild-type copy of rbmB in a multicopy plasmid to the RΔrbmB strain (data not shown). Because RΔrbmB exhibited altered colony morphology, we hypothesized that biofilm formation by the mutant may also be altered. To test this hypothesis, we compared the structures of biofilms formed by these two strains (Fig. 2B) using flow cell systems and CLSM, and we also analyzed total biofilm formation using the crystal violet staining assay (Fig. 2C). Although we did not observe significant differences in the biofilm-forming capacities of the two strains, biofilm structures formed by the RΔrbmB mutant were subtly different from those formed by the rugose variant. COMSTAT analyses of the CLSM images of the biofilm structures formed after 24 h postinoculation revealed that the mutant formed thicker biofilms than those of the rugose variant (Table 3). The mutant also formed biofilms that were more heterogeneous in nature, as RΔrbmB biofilms had a higher roughness coefficient compared to the rugose variant. Total biomass and substrate coverage of the biofilms formed by RΔrbmB and rugose variant were similar. These results indicate that RbmB affects colony rugosity and subtly affects biofilm structures. We also compared motility, pellicle formation, growth profiles, and cell aggregations of RΔrbmB and the rugose variant, but we did not observe significant differences (data not shown).
FIG. 2.
RbmB modulates rugose colony development and biofilm formation. (A) Colony morphologies of the rugose variant and RΔrbmB. (B) CLSM images of the top-down views (large panels) and orthogonal views (side panels) of biofilms formed by the rugose variant and RΔrbmB in a flow cell system after 24 h of incubation. Bars, 40 μm. (C) Quantitative analysis of biofilm formation by the rugose variant and RΔrbmB after 8 h of growth at 30°C under static conditions. Error bars represent standard deviations. (D) Expression of the vpsA::lacZ and vpsL::lacZ fusion genes in the rugose variant and RΔrbmB grown at 30°C to the mid-exponential growth phase. Error bars represent standard deviations.
TABLE 3.
COMSTAT quantitative analysis of biofilm structures formed by V. cholerae rugose variant and deletion mutants of rbmB-F and bap1a
| Strain | Total biomass (μm3/μm2) | Thickness (μm)
|
Substrate coverage | Roughness coefficient | |
|---|---|---|---|---|---|
| Avg | Maximum | ||||
| Rugose | 15.6 (3.3) | 17.5 (4.9) | 56.7 (15.1) | 1.0 (0.01) | 0.5 (0.1) |
| RΔrbmB | 15.7 (3.3) | 18.2 (5.4) | 65.6 (12.5) | 1.0 (0.02) | 0.7 (0.1) |
| RΔrbmC | 13.7 (1.8) | 15.0 (2.0) | 52.8 (2.1) | 1.0 (0.01) | 0.6 (0.1) |
| RΔbap1 | 16.9 (2.9) | 19.2 (3.7) | 54.3 (9.7) | 0.9 (0.1) | 0.5 (0.04) |
| RΔrbmC Δbap1 | 0.4 (0.3) | 0.5 (0.3) | 20.9 (5.8) | 0.1 (0.04) | 1.9 (0.1) |
| RΔrbmD | 15.7 (2.3) | 20.4 (3.4) | 58.3 (11.1) | 1.0 (0.02) | 0.4 (0.1) |
| RΔrbmEF | 16.1 (1.0) | 17.3 (1.2) | 53.0 (1.9) | 1.0 (0.001) | 0.3 (0.04) |
Values presented are means of data from at least five z-series image stacks taken at 24 h. The numbers in parentheses indicate standard deviations.
RbmB affects VPS accumulation and may be a polysaccharide lyase.
Because VPS has been linked to rugosity and biofilm formation in V. cholerae (71), we investigated whether the increased corrugation of the colony morphology and altered biofilm structures in RΔrbmB are due to increased amounts of VPS. To this end, VPS was extracted from RΔrbmB and the rugose variant, which were normalized to the same cell density, and the extracted VPS samples were analyzed on a 5% SDS-PAGE gel and stained with Stains-All. VPS accumulation by the deletion mutant was noticeably higher compared to the rugose variant as shown in Fig. 3. To determine if this increase was due to an increase in expression of the vps genes in the deletion mutant, we monitored the expression of vpsA and vpsL using vpsA::lacZ and vpsL::lacZ fusion plasmids (13) in β-galactosidase assays. As shown in Fig. 2D, there were no significant differences in vpsA and vpsL expression levels between the rugose variant and RΔrbmB. Both the rugose variant and RΔrbmB harboring the vector showed negligible background β-galactosidase activities (data not shown). Taken together, the results demonstrate that vps gene expression levels were not altered in RΔrbmB and that the more-corrugated colony morphology, subtle altered biofilm structures, and increased VPS accumulation observed in the deletion mutant are due to the loss of RbmB function.
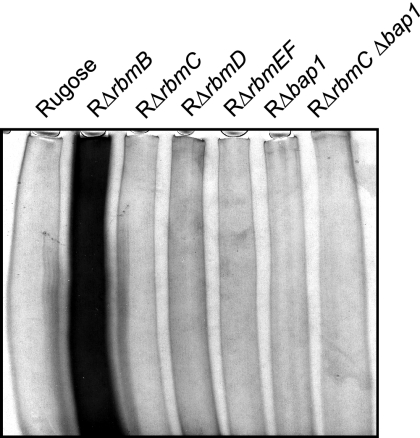
FIG. 3.
SDS-PAGE analysis of VPS accumulation. Equal volumes (50 μl) of VPS samples were analyzed from the rugose variant, RΔrbmB, RΔrbmC, RΔrbmD, RΔrbmEF, RΔbap1, and RΔrbmC Δbap1. Cultures were normalized to the same cell density. The gel was stained with Stains-All, and the 5% polyacrylamide stacking portion of the gel is shown.
Computational analyses using the Simple Modular Architecture Research tool (SMART) revealed that RbmB contains six domains composed of parallel β-helix repeats (PbH1; T116-S147, S181-G210, T234-Y256, Y257-P279, W289-S310, and I321-S342), which are often found in enzymes with polysaccharide substrates (25, 32, 33, 45). ModBase software (for three-dimensional protein models calculated by comparative modeling) also predicted a right-handed parallel β-helix protein fold for RbmB, with an E-value of 6e-19 to rhamnogalacturonase A from Aspergillus aculeatus (52). Two putative N-acetylglucosamine-binding sites and seven putative mannose-binding sites were also predicted in RbmB by ModBase. These predictions are consistent with the possible role of RbmB as a polysaccharide lyase in breaking down VPS, which is composed of glucose, galactose, N-acetylglucosamine, and mannose (71), in the extracellular matrix of V. cholerae biofilms.
Polysaccharide lyases reported to date are (or have been predicted to be) extracellular or periplasmic (1, 4, 17, 35, 50, 65). The TIGR-CMR database indicated the presence of a putative 31-amino-acid-long signal peptide in RbmB, but SignalP 3.0 (for signal peptide cleavage site prediction) did not have a strong prediction for a cleavage site (probability of cleavage between S31 and E32 is 0.319 by the SignalP-NN method and 0.023 by the SignalP-HMM method). We attempted to determine the localization of RbmB with immunoblot analyses using Myc- and His-tagged RbmB. Due to the presence of several contaminating signals around the predicted sizes of the tagged RbmB (45.7 kDa without the predicted signal peptide; 49.9 kDa with the signal peptide) using both anti-Myc and anti-His antibodies, we were unable to determine its cellular localization. We are currently generating anti-RbmB antibodies to determine the localization of this putative polysaccharide lyase.
Overexpression of rbmB affects colony morphology and biofilm structures.
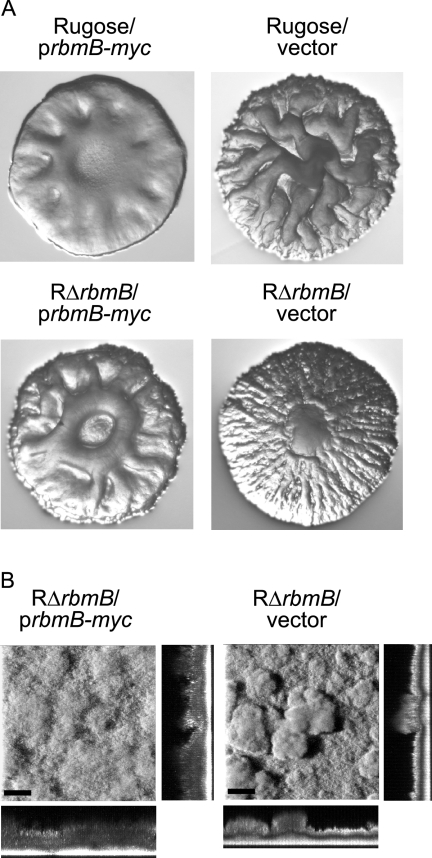
Since the lack of functional RbmB alters colony corrugation and biofilm structures, we hypothesized that increased production of RbmB may also alter these phenotypes. Indeed, overexpression of rbmB from an arabinose-inducible promoter in both the rugose variant and RΔrbmB mutant resulted in decreased colony corrugation, while the strains carrying the vector remained corrugated (Fig. 4A). We also determined that overexpression of RbmB in mature biofilms leads to changes in biofilm structures where the biofilms become less compact and more homogeneous compared to the biofilms formed by the same strain carrying the vector only (Fig. 4B). These findings further suggest that RbmB may be degrading the VPS, which is required for the development of colony corrugation and biofilm formation, by acting as a polysaccharide lyase.
FIG. 4.
Overexpression of rbmB affects colony morphology and biofilm structures. (A) Colony morphologies of the rugose variant and RΔrbmB harboring the overexpression plasmid prbmB-myc or the vector pBAD/Myc-His B grown in the presence of arabinose. (B) CLSM images of the top-down views (large panels) and orthogonal views (side panels) of biofilms formed by RΔrbmB harboring prbmB-myc or the vector in a flow cell system after 24 h postinduction. Mature biofilms were allowed to form in the flow cell chambers in the absence of arabinose for 24 h, followed by overexpression of rbmB with the addition of 0.1% (wt/vol) arabinose to the growth medium. Bars, 40 μm.
RbmC and Bap1 modulate rugose colony morphology.
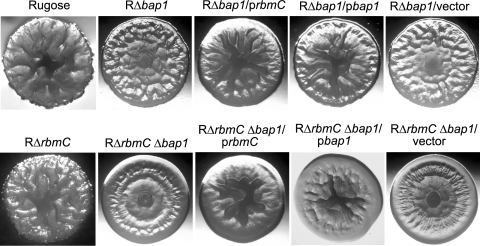
The third gene in the vps intergenic region is rbmC (VC0930) (Fig. 1), which is predicted to encode a protein exhibiting 46.7% similarity to Bap1 (VC1888). It has been reported that a transposon insertion into VC0930, in an El Tor biotype C6706 rugose variant, converts the rugose colony morphology to smooth (2). Furthermore, Bap1 is required for biofilm formation in the classical biotype O395 strain (30), and expression levels of both rbmC and bap1 are coregulated with the vps genes in an El Tor biotype A1552 strain (70). To investigate the functions of RbmC and Bap1 in rugosity, we generated single and double deletions of rbmC and bap1 in the rugose genetic background (RΔrbmC, RΔbap1, and RΔrbmC Δbap1). Although colony morphology of RΔrbmC (Fig. 5) was not noticeably different from that of the rugose variant, RΔbap1 and double deletion RΔrbmC Δbap1 formed colonies that were more flat and less wrinkled than the rugose variant. An increase in smoothness was also obvious when comparing RΔbap1 and the double mutant RΔrbmC Δbap1, indicating that RbmC and Bap1 have an additive effect on development of colony corrugation. RΔbap1 and RΔrbmC Δbap1 carrying the complementation plasmids prbmC and pbap1 exhibited increased corrugation in the center of the colony (partial complementation), suggesting that RbmC and Bap1 may have similar functions. Colony morphologies of strains carrying the control vector pACYC177 were similar to those of the respective deletion mutants. Together, these results show that RbmC and Bap1 modulate the development of corrugated colonies.
FIG. 5.
RbmC and Bap1 are involved in rugose colony development. Colony morphologies are shown for the rugose variant, RΔrbmC, RΔbap1, RΔbap1 harboring prbmC, RΔbap1 harboring pbap1, RΔbap1 harboring the vector pACYC177, RΔrbmC Δbap1, RΔrbmC Δbap1 harboring prbmC, RΔrbmC Δbap1 harboring pbap1, and RΔrbmC Δbap1 harboring the vector pACYC177.
RbmC and Bap1 are secreted proteins and are critical for pellicle and biofilm formation.
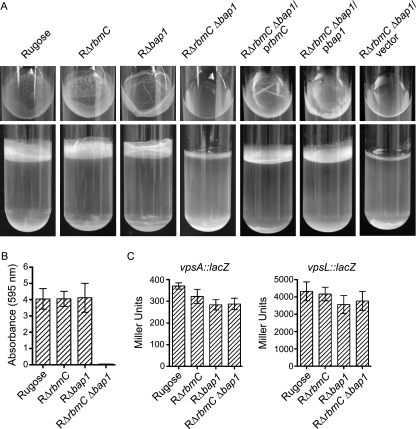
We next investigated whether the pellicle (biofilm formed at the air-liquid interface) and biofilm formation on solid surfaces by the mutants were altered. After 2 days of growth at 30°C under static conditions, the pellicle formed by RΔrbmC (Fig. 6A) was not significantly different from that formed by the rugose variant. On the other hand, RΔbap1 formed a pellicle with an altered structure (top panel), while the double deletion mutant RΔrbmC Δbap1 was incapable of forming pellicles. The defect in the pellicle formation phenotype of the RΔrbmC Δbap1 mutant could be complemented by introducing the wild-type copy of either rbmC or bap1 on a multicopy plasmid, again suggesting that RbmC and Bap1 may have a similar function. RΔrbmC Δbap1 harboring the control vector pACYC177 remained impaired in pellicle formation. Although total biofilm formation by the two single deletion mutants (RΔrbmC and RΔbap1) was not significantly different compared to the rugose variant, the double deletion mutant (RΔrbmC Δbap1) was unable to form biofilms (Fig. 6B). Using vpsA::lacZ and vpsL::lacZ fusion plasmids, we determined that vps gene expression was not significantly altered in these deletion mutants (Fig. 6C), indicating that the observed defect in biofilm formation was not due to decreased vps transcription.
FIG. 6.
RbmC and Bap1 are involved in pellicle and biofilm formation. (A) Pellicle formation by the rugose variant, RΔrbmC, RΔbap1, RΔrbmC Δbap1, RΔrbmC Δbap1 harboring prbmC, RΔrbmC Δbap1 harboring pbap1, and RΔrbmC Δbap1 harboring the vector pACYC177 after 2 days of incubation at 30°C. Top and side views of the culture tubes are showed in the top and bottom panels, respectively. (B) Quantitative comparison of biofilm formation by the rugose variant, RΔrbmC, RΔbap1, and RΔrbmC Δbap1 after 8 h of growth at 30°C under static conditions. Error bars represent standard deviations. (C) Expression of the vpsA::lacZ and vpsL::lacZ fusion genes in the rugose variant, RΔrbmC, RΔbap1, and RΔrbmC Δbap1 grown at 30°C to the mid-exponential growth phase. Error bars represent standard deviations.
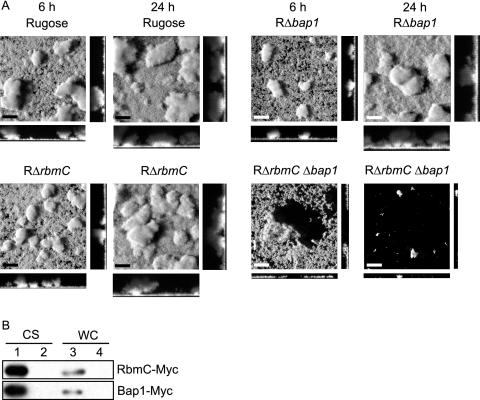
We further investigated biofilm structure formation in these deletion mutants using flow cell systems and CLSM (Fig. 7A) and analyzed the images with COMSTAT (Table 3). As shown in Fig. 7A, RΔrbmC Δbap1 formed a thin layer of biofilm, which detached after 6 h. Only a few microcolonies remained attached to the substratum after 24 h. This is reflected in the COMSTAT analyses, where the total biomass, biofilm thickness, and substrate coverage of the double deletion mutant were dramatically lower at 24 h postinoculation compared to those of the rugose variant. In contrast, RΔrbmC and RΔbap1 single mutants formed biofilm structures similar to those of the rugose variant at both 6 and 24 h, which were also reflected in the COMSTAT analyses of the images. We also extracted and analyzed VPS from the deletion mutants and the rugose variant but did not observe differences in the amount of VPS accumulated by the different strains (Fig. 3). It should be noted that growth profiles, cell aggregations, and motilities of the deletion mutants and the rugose variant were also similar (data not shown). Together, these results indicate that RbmC and Bap1 are critical for pellicle and biofilm formation and the altered phenotypes are due to the loss of RbmC and Bap1 functions.
FIG. 7.
RbmC and Bap1 are secreted proteins and are involved in maintaining biofilm architecture. (A) CLSM images of the top-down views (large panels) and orthogonal views (side panels) of biofilms formed by the rugose variant, RΔrbmC, RΔbap1, and RΔrbmC Δbap1 in a flow cell system after 6 and 24 h of incubation. Bars, 40 μm. (B) Immunoblot analysis of proteins in the CS and WC fractions from arabinose-induced RΔrbmC (top) and RΔbap1 (bottom) harboring the overexpression plasmids prbmC-myc and pbap1-myc (lanes 1 and 3) as well as the vector pBAD/Myc-His B (lanes 2 and 4).
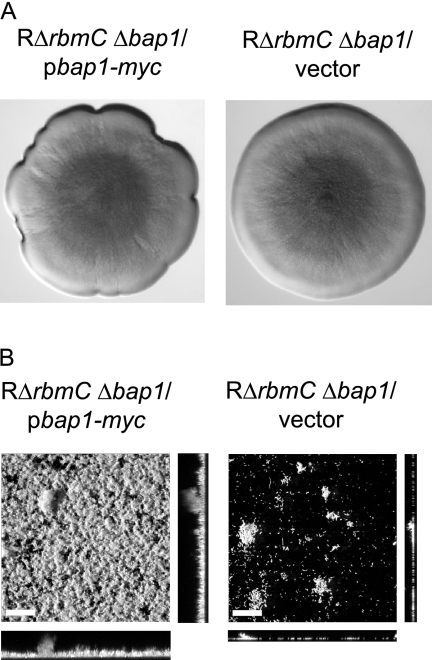
Since the RΔrbmC Δbap1 mutant forms colonies with dramatically decreased colony corrugation and is defective in forming mature biofilms, we hypothesized that increased production of Bap1 may alter colony morphology and biofilm phenotypes. We overexpressed Bap1 from an arabinose-inducible promoter and analyzed morphology of the colonies developed on LB agar medium supplemented with ampicillin and 0.1% arabinose. It should be noted that growth on arabinose reduces colony corrugation of the rugose variant (3), and the RΔrbmC Δbap1 mutant formed smooth-looking colonies on LB agar medium supplemented with 0.1% arabinose. Increased expression of bap1 in the double deletion mutant resulted in only a slight increase in colony roughness at the edges of the colony compared to the same strain carrying the vector (Fig. 8A). In contrast, the biofilm-forming capacity of the RΔrbmC Δbap1 mutant harboring the overexpression plasmid pbap1-myc was dramatically increased compared to the RΔrbmC Δbap1 mutant carrying the vector (Fig. 8B). These results suggest that increased Bap1 expression leads to a modest change in colony morphology and results in the formation of more-structured biofilms.
FIG. 8.
Overexpression of bap1 affects colony morphology and biofilm structure. (A) Colony morphologies of RΔrbmC Δbap1 harboring the overexpression plasmid pbap1-myc or the vector pBAD/Myc-His B. Strains were grown in the presence of 0.1% (wt/vol) arabinose. (B) CLSM images of the top-down views (large panels) and orthogonal views (side panels) of biofilms formed by RΔrbmC Δbap1 harboring pbap1-myc or the vector in a flow cell system after 24 h of growth in the presence of 0.1% (wt/vol) arabinose. Bars, 40 μm.
Computational analyses revealed that both RbmC and Bap1 contain FG-GAP and carbohydrate-binding domains as well as three putative N-acetylglucosamine-binding sites. The InterPro database indicated that both RbmC and Bap1 contain one possible calcium-binding site, two and one FG-GAP domains, respectively, and one galactose mutarotase-like domain. The FG-GAP domains are found in the extracellular N-terminal region of the integrin α-chain, which has been reported to be involved in ligand recognition and binding with proteins in the extracellular matrix or surface proteins on other cells (5, 43). The galactose mutarotase-like domains, on the other hand, are described by the Structural Classification of Proteins database as probable carbohydrate-binding domains in enzymes acting on sugars. The ModBase software predicted both RbmC and Bap1 with a protein fold of 1jv2A (E values of 1e-12 and 5e-14, respectively), a human integrin αVβ3 (66). Three putative N-acetylglucosamine-binding sites were also predicted in both RbmC and Bap1 by the ModBase software. Further detailed analysis using the SAM (sequence alignment and modeling) software (36, 37) revealed that D220-A499 and T640-G821 of RbmC are homologous to the β-propeller FG-GAP domains of the integrin α-chain and that Q500-T644 and G821-Y957 are homologous to sugar-binding lectin domains. These computational predictions are consistent with the possible roles of RbmC and Bap1 in maintaining rugosity of colony morphology and stabilizing pellicle and biofilm structures.
SignalP predicted that both RbmC and Bap1 are secreted proteins. RbmC is predicted to contain a signal peptide with cleavage between A22 and T23 (probabilities of 0.806 and 0.870 for the two SignalP methods), while Bap1 is predicted to contain a signal peptide with cleavage between A26 and S27 (probabilities of 0.780 and 0.930). To determine whether RbmC and Bap1 are secreted, we carried out immunoblot analyses using anti-Myc antibodies on proteins in TCA-precipitated culture supernatant (CS) and whole-cell lysate (WC) fractions from cultures of RΔrbmC harboring prbmC-myc and RΔbap1 harboring pbap1-myc. Immunoblots of such analyses (Fig. 7B) showed the presence of immunoreactive polypeptides corresponding to the expected sizes for RbmC-Myc (105 kDa) and Bap1-Myc (75.4 kDa) (without the predicted signal peptides) in the CS fractions of the deletion strains harboring their respective overexpression plasmids (lane 1). Such signals were absent in the CS fractions (lane 2) and WC fractions (lane 4) from strains carrying the vector pBAD/Myc-His B. Only weak bands corresponding to the sizes of RbmC-Myc and Bap1-Myc were detected in the WC fractions from strains harboring prbmC-myc or pbap1-myc (lane 3). These results indicate that RbmC and Bap1 are predominantly secreted.
RbmD, RbmE, and RbmF are involved in biofilm structure formation and cell aggregation.
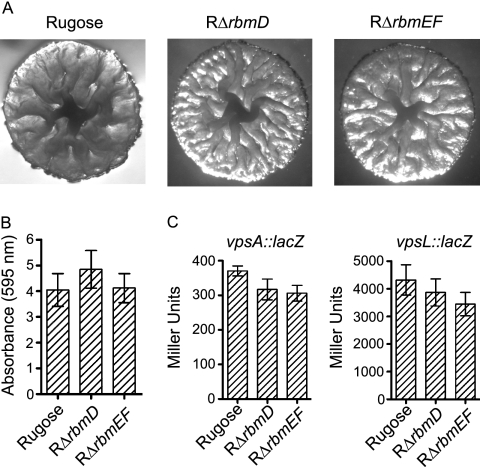
The last three genes in the vps intergenic region are the 1,317-bp rbmD (VC0931), 288-bp rbmE (VC0932), and 405-bp rbmF (VC0933) (Fig. 1). Since RbmA to -C are capable of modulating colony morphology and biofilm formation, we hypothesized that RbmD to -F may also play similar roles. To investigate this hypothesis, we generated in-frame deletions of rbmD and rbmEF in the rugose genetic background (RΔrbmD and RΔrbmEF). rbmE and rbmF were deleted together, as they shared 88-bp overlapping coding sequences. RΔrbmD and RΔrbmEF exhibited similar colony morphology as the rugose variant (Fig. 9A). Quantitative biofilm assays (Fig. 9B) and expression analyses of vpsA and vpsL (Fig. 9C) did not reveal significant differences between the deletion mutants and the rugose variant. Furthermore, we also did not observe significant differences in motility, pellicle formation, growth profiles (data not shown), and VPS accumulation when comparing RΔrbmD and RΔrbmEF to the rugose variant (Fig. 3).
FIG. 9.
Colony morphology and biofilm formation phenotypes of the rbmDEF deletion mutants and the rugose variant are similar. (A) Colony morphology of the rugose variant, RΔrbmD, and RΔrbmEF. (B) Quantitative comparison of biofilm formation by the rugose variant, RΔrbmD, and RΔrbmEF after 8 h of growth at 30°C under static conditions. Error bars represent standard deviations. (C) Expression of the vpsA::lacZ and vpsL::lacZ fusion genes in the rugose variant, RΔrbmD, and RΔrbmEF grown at 30°C to the mid-exponential growth phase. Error bars represent standard deviations.
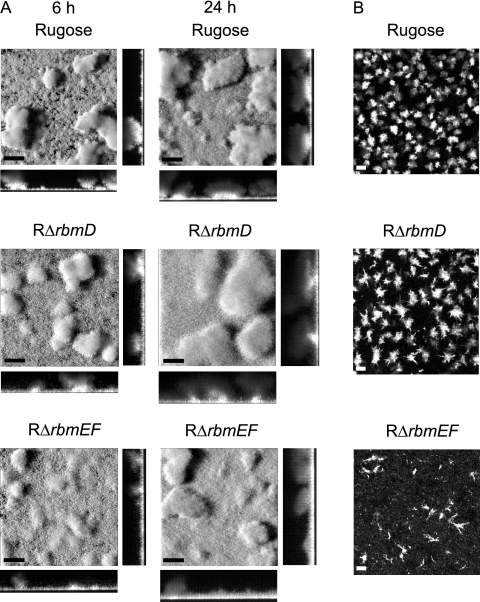
Analyses of RΔrbmD and RΔrbmEF biofilms formed using flow cell systems revealed that their biofilm structures were different from those of the rugose variant (Fig. 10A). The pillars of the biofilms formed by the deletion mutants appear to be less compact than those formed by the rugose variant at both 6 and 24 h. These differences were also reflected in the COMSTAT analyses (Table 3), in which the roughness coefficients of the deletion mutants were lower than for the rugose variant, indicating more homogeneous biofilm structures. On the other hand, total biomass, biofilm thickness, and substrate coverage were similar between the mutants and the rugose variant.
FIG. 10.
RbmD, RbmE, and RbmF are involved in biofilm structure formation and cell aggregation. (A) CLSM images of the top-down views (large panels) and orthogonal views (side panels) of biofilms formed by the rugose variant, RΔrbmD, and RΔrbmEF in a flow cell system after 6 and 24 h of incubation. (B) CLSM images of the top-down views of cell aggregates from overnight-grown cultures of the rugose variant, RΔrbmD, and RΔrbmEF. Bars, 40 μm.
The rugose variant forms aggregates when grown in suspension. We compared aggregation phenotypes of the RΔrbmD and RΔrbmEF mutants to that of the rugose variant. RΔrbmD formed cell aggregates that were larger and less compact, while RΔrbmEF formed string-like aggregates that were smaller in size compared to the cell aggregates formed by the rugose variant (Fig. 10B). Complementation of the altered cell aggregation phenotype was achieved with RΔrbmD and RΔrbmEF harboring prbmD and prbmEF, respectively (data not shown). Together, these results indicate that RbmD and RbmEF are involved in cell aggregation and biofilm structure formation and that the altered phenotypes are due to the loss of RbmD and RbmEF functions.
The TMHMM 2.0 program (for trans-membrane helices predictions) predicted 12 trans-membrane helices in RbmD, suggesting that RbmD is located on the membrane. RbmD is also predicted to contain a 30-amino-acid signal peptide (probabilities of cleavage between A30 and W31 are 0.390 and 0.666 for the two SignalP methods). Although RbmD is annotated as a conserved hypothetical protein in the TIGR-CMR database, Quick BlastP searches and EMBOSS alignment programs revealed that RbmD has peptide sequence similarity to O-antigen polymerases from Pseudoalteromonas atlantica (53.7% similarity) and Shewanella frigidimarina (54.1% similarity), with E-values of 5e-76 and 2e-70, respectively. The InterPro database (for integrated resource of protein families, domains, and functional sites) also indicated the presence of a Wzy_C domain in RbmD, which is found in O-antigen polymerases involved in the synthesis of the outer membrane lipopolysaccharide O-antigen (8). It is therefore possible that RbmD is involved in the biosynthesis of VPS in V. cholerae, consistent with the observations that RbmD is involved in wild-type biofilm structure formation and cell aggregation. In contrast, computational analyses of RbmE and RbmF, which are predicted to be organized in an operon, did not reveal significant domain structures. They are annotated as hypothetical proteins in the TIGR-CMR database, and Quick BlastP searches also did not retrieve protein matches with significant E-values. While RbmE does not contain a signal peptide and is not predicted to be secreted, SignalP predicted RbmF to contain a 22-amino-acid signal peptide (probabilities of cleavage between A22 and E23 are 0.890 and 0.999 for the two SignalP methods). The localization and involvement of RbmD to -F in VPS biosynthesis in V. cholerae remain to be investigated.
DISCUSSION
Phenotypic variation between smooth and rugose variants and the expression of rugose-associated phenotypes are believed to be linked to the survival of V. cholerae in aquatic environments. Many of the rugose-associated phenotypes, including increased capacity to form biofilms and increased biocide resistance, are mediated in part by the functions of the vps genes. We recently showed that rbmA, which is located between the two vps gene clusters, plays a crucial role in biofilm formation in V. cholerae. This finding, together with our observations that expression of the genes located in the vps intergenic region, as well as bap1 (VC1888) (Fig. 1), were increased in the rugose variant compared to the smooth variant (70), prompted us to investigate the function of the vps intergenic region in rugose-associated phenotypes (such as VPS production and biofilm formation). Other studies have also reported differential expression of genes in the vps intergenic region at different stages of growth, under different growth conditions, and between different biotypes of V. cholerae (9, 30, 47). Transcription of the genes located in the vps intergenic region and bap1 have been reported to be induced in planktonic wild-type cells when grown in the presence of mannose (47). Expression levels of rbmABC and bap1 have also been reported to be up-regulated in the presence of bile (30). Furthermore, transcriptional levels of rbmABC and bap1 were increased in the El Tor biotype compared to the classical biotype of V. cholerae (9).
In this study, we showed that rbmB, which may encode a polysaccharide lyase, is critical for development of rugose colony morphology and biofilm formation (Fig. 2 to 4). Consistent with its predicted role as a polysaccharide lyase, computational analyses revealed that RbmB contains six PbH1 domains with a right-handed parallel β-helix protein fold, commonly found in enzymes with polysaccharide substrates, including several bacterial pectate and pectin lyases (25, 32, 33), fungal and bacterial polygalacturonases and rhamnogalacturonases (32, 33), and ι-carrageenase from Alteromonas fortis (45). The presence of putative N-acetylglucosamine- and mannose-binding sites in RbmB further reinforces the notion that RbmB may function as a polysaccharide lyase in V. cholerae, breaking down the VPS that is composed of glucose, galactose, N-acetylglucosamine, and mannose (71). The exact ligand-binding sites, substrate specificity, and enzymatic properties of RbmB as well as its role in biofilm detachment in V. cholerae remain to be investigated.
Although there are a number of studies reporting the role(s) of polysaccharide lyases in biofilm formation and detachment processes, the underlying molecular mechanisms remain largely unknown. In Pseudomonas aeruginosa, alginate lyase has been reported to degrade alginate into shorter-length polymers, leading to cell sloughing and detachment of bacteria from mature biofilms (11, 24). The gene encoding the P. aeruginosa periplasmic alginate lyase, algL, is located within the alginate biosynthesis cluster (12, 57). Interestingly, alginate production and alginate lyase expression have been reported to be coregulated by the positive transcriptional regulator AlgB (57), and alginate lyase also plays a role in alginate biosynthesis in P. aeruginosa (1). A recent report also suggested the possible bifunctional roles of AlgL in a multiprotein secretion complex and in degrading free alginate polymers in the periplasm (31). Genes encoding alginate lyases in other Pseudomonas and Azotobacter species have also been reported to be located within their respective alginate biosynthesis gene clusters (65). It is therefore not surprising that the putative polysaccharide lyase-encoding gene, rbmB, is located within the two vps clusters in V. cholerae and its transcription is regulated by the vps positive regulator, VpsR (70). Several alginate lyases from a number of marine Vibrio species have also been studied, including V. halioticoli (58), V. harveyi AL-128 (40), V. alginolyticus ATCC 17749 (40), Vibrio sp. strain YWA (61), Vibrio sp. strain QY101 (23), and Vibrio sp. strain O2 (38). However, the biological roles of these alginate lyases in these Vibrio species remain to be investigated. Polysaccharide lyases in other microorganisms have also been reported to play a role in biofilm detachment and cell dispersal, including an endo-β-1,4-mannanase in Xanthomonas campestris (17), a β-N-acetylglucosaminidase in Actinobacillus actinomycetemcomitans (35), and an endo-polysaccharide hydrolase in Methanosarcina mazei (67).
RbmC and its homolog, Bap1 (30, 47), were previously annotated in the TIGR-CMR database as hemolysin-related proteins, and they also share peptide similarity to the hemolysin protein HlyA from V. cholerae. RbmC exhibits 28.2% sequence similarity to HlyA, while the sequence similarity between Bap1 and HlyA was found to be 20.7%. The facts that transcription of rbmC and bap1 are positively regulated by the positive regulator VpsR (70) and that rbmC and bap1 are coregulated together with the vps clusters (47, 70) led us to hypothesize that RbmC and Bap1 may have a role in colony morphology and biofilm formation rather than function as hemolysin proteins. Indeed, a single deletion of bap1 and double deletions of rbmC and bap1 resulted in strains with less-corrugated colony morphologies (Fig. 5) and altered pellicle formation (Fig. 6). The double deletion mutant was also impaired in forming mature biofilm structures (Fig. 7A). We also showed that RbmC and Bap1 are secreted proteins (Fig. 7B), consistent with their roles in maintaining rugosity of colony morphology and stabilizing pellicle and biofilm structures. RbmC and Bap1 are also predicted to contain FG-GAP and carbohydrate-binding domains as well as putative N-acetylglucosamine-binding sites. These computational predictions are consistent with the roles of RbmC and Bap1 in maintaining rugosity of colony morphology as well as stabilizing pellicle and biofilm structures (Fig. 5 to 8), possibly through binding to carbohydrates in the extracellular matrix of V. cholerae, suggesting that RbmC and Bap1 may act as lectins.
Ali et al. (2) reported that a transposon insertion into VC0930 in the chromosome of a V. cholerae O1 El Tor C6706 rugose variant converts rugose colony morphology to smooth. However, an in-frame deletion of rbmC generated in the O1 El Tor A1552 rugose variant in this study did not exhibit altered colony morphology compared to the rugose variant (Fig. 5), a finding likely to be due to the differences in the genetic make-up of these two strains. A study by Hung et al. (30) reported that a bap1 deletion mutant in V. cholerae classical strain O395 was defective in biofilm production. The bap1 deletion mutant generated in the V. cholerae O1 El Tor A1552 rugose variant in this study formed biofilms similar to those of the rugose variant (Fig. 7A). The differences could be attributed to the different biotypes used in the studies.
Deletion mutants of the last three genes in the vps intergenic region, rbmDEF, did not exhibit significant differences in colony morphology and total biofilm formation relative to the rugose variant (Fig. 9). However, the mutants were impaired in wild-type biofilm structure formation and cell aggregation (Fig. 10), suggesting that the proteins play a role in cell-cell or cell-matrix adhesion. Computational analyses predicted that the putative membrane-bound RbmD contains a Wzy_C domain found in O-antigen polymerase (8), suggesting a possible role of RbmD in VPS biosynthesis in V. cholerae. This possibility needs to be further investigated.
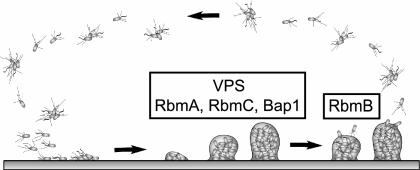
Biofilm formation is widely believed to be employed by many aquatic microorganisms, including V. cholerae, as a survival strategy in natural environments, and many studies have been carried out to understand the structural and regulatory components of this developmental process (15, 49, 51, 63, 71). However, the molecular bases of many areas in biofilm formation and detachment processes remain elusive. In V. cholerae, the rugose variant biofilm developmental cycle begins with the attachment of single planktonic or aggregated cells to surfaces, followed by the formation of microcolonies and mature biofilms (Fig. 11). It has been shown that the formation of mature biofilm structures requires the production of VPS (71) and that the secreted matrix protein RbmA is also involved in maintaining the biofilm architecture (20). Here, we report that other secreted matrix proteins, RbmC and Bap1, are also involved in stabilizing the biofilm matrix. Furthermore, we also identified a putative polysaccharide lyase, RbmB, that may play a role in degrading VPS in the extracellular matrix, leading to the dissemination of V. cholerae in natural aquatic environments.
FIG. 11.
Model of the biofilm developmental cycle in V. cholerae. Biofilm formation in the rugose variant begins with attachment of single or aggregated cells to surfaces. This is followed by the formation of microcolonies and mature biofilm structures, which require the presence of VPS as well as matrix proteins RbmA, RbmC, and Bap1, which are predicted to act as lectins. The biofilm developmental cycle is completed with detachment of bacteria from mature biofilms, which is likely to be mediated in part by RbmB, predicted to act as a VPS lyase.
Acknowledgments
This work was supported by grants from The Ellison Medical Foundation and NIH (AI055987) to F.H.Y.
We acknowledge Kevin Karplus for carrying out the bioinformatic analyses. We thank Karen Ottemann, Chad Saltikov, and members of the Yildiz laboratory for their suggestions.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Albrecht, M. T., and N. L. Schiller. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:3869-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, A., Z. H. Mahmud, J. G. Morris, Jr., S. Sozhamannan, and J. A. Johnson. 2000. Sequence analysis of TnphoA insertion sites in Vibrio cholerae mutants defective in rugose polysaccharide production. Infect. Immun. 68:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali, A., J. G. Morris, Jr., and J. A. Johnson. 2005. Sugars inhibit expression of the rugose phenotype of Vibrio cholerae. J. Clin. Microbiol. 43:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 5.Baneres, J. L., F. Roquet, A. Martin, and J. Parello. 2000. A minimized human integrin α5β1 that retains ligand recognition. J. Biol. Chem. 275:5888-5903. [DOI] [PubMed] [Google Scholar]
- 6.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 7.Barbeyron, T., G. Michel, P. Potin, B. Henrissat, and B. Kloareg. 2000. ι-Carrageenases constitute a novel family of glycoside hydrolases, unrelated to that of κ-carrageenases. J. Biol. Chem. 275:35499-35505. [DOI] [PubMed] [Google Scholar]
- 8.Bengoechea, J. A., E. Pinta, T. Salminen, C. Oertelt, O. Holst, J. Radziejewska-Lebrecht, Z. Piotrowska-Seget, R. Venho, and M. Skurnik. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 184:4277-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 74:3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollag, D. M., M. D. Rozycki, and S. J. Edelstein. 1996. Protein methods, 2nd ed., p. 84-85. Eiley-Lis, Inc., New York, N.Y.
- 11.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd, A., M. Ghosh, T. B. May, D. Shinabarger, R. Keogh, and A. M. Chakrabarty. 1993. Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene 131:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiavelli, D. A., J. W. Marsh, and R. K. Taylor. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 16.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 17.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enos-Berlage, J. L., and L. L. McCarter. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 182:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong, J. C., K. Karplus, G. K. Schoolnik, and F. H. Yildiz. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 188:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894-906. [DOI] [PubMed] [Google Scholar]
- 23.Han, F., Q. H. Gong, K. Song, J. B. Li, and W. G. Yu. 2004. Cloning, sequence analysis and expression of gene alyVI encoding alginate lyase from marine bacterium Vibrio sp. QY101. DNA Seq. 15:344-350. [DOI] [PubMed] [Google Scholar]
- 24.Hatch, R. A., and N. L. Schiller. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:974-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrissat, B., S. E. Heffron, M. D. Yoder, S. E. Lietzke, and F. Jurnak. 1995. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 107:963-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 28.Horton, R. M. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol. Biol. 67:141-149. [DOI] [PubMed] [Google Scholar]
- 29.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 30.Hung, D. T., J. Zhu, D. Sturtevant, and J. J. Mekalanos. 2006. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 59:193-201. [DOI] [PubMed] [Google Scholar]
- 31.Jain, S., and D. E. Ohman. 2005. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 73:6429-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins, J., O. Mayans, and R. Pickersgill. 1998. Structure and evolution of parallel β-helix proteins. J. Struct. Biol. 122:236-246. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins, J., and R. Pickersgill. 2001. The architecture of parallel β-helices and related folds. Prog. Biophys. Mol. Biol. 77:111-175. [DOI] [PubMed] [Google Scholar]
- 34.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karplus, K., R. Karchin, C. Barrett, S. Tu, M. Cline, M. Diekhans, L. Grate, J. Casper, and R. Hughey. 2001. What is the value added by human intervention in protein structure prediction? Proteins Struct. Funct. Genet. 45:86-91. [DOI] [PubMed] [Google Scholar]
- 37.Karplus, K., S. Katzman, G. Shackleford, M. Koeva, J. Draper, B. Barnes, M. Soriano, and R. Hughey. 2005. SAM-T04: what's new in protein-structure prediction for CASP6. Proteins Struct. Funct. Bioinformat. 61:135-142. [DOI] [PubMed] [Google Scholar]
- 38.Kawamoto, H., A. Horibe, Y. Miki, T. Kimura, K. Tanaka, T. Nakagawa, M. Kawamukai, and H. Matsuda. 2006. Cloning and sequencing analysis of alginate lyase genes from the marine bacterium Vibrio sp. O2. Mar. Biotechnol. 8:481-490. [DOI] [PubMed] [Google Scholar]
- 39.Kelley, J. T., and C. D. Parker. 1981. Identification and preliminary characterization of Vibrio cholerae outer membrane proteins. J. Bacteriol. 145:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamikado, M., C. H. Tseng, K. Yamaguchi, and T. Nakamura. 1992. Two types of bacterial alginate lyases. Appl. Environ. Microbiol. 58:2474-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre, B., P. Formstecher, and P. Lefebvre. 1995. Improvement of the gene splicing overlap (SOE) method. BioTechniques 19:186-188. [PubMed] [Google Scholar]
- 42.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 43.Loftus, J. C., J. W. Smith, and M. H. Ginsberg. 1994. Integrin-mediated cell adhesion: the extracellular face. J. Biol. Chem. 269:25235-25238. [PubMed] [Google Scholar]
- 44.Meibom, K. L., X. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 101:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michel, G., W. Helbert, R. Kahn, O. Dideberg, and B. Kloareg. 2003. The structural bases of the processive degradation of ι-carrageenan, a main cell wall polysaccharide of red algae. J. Mol. Biol. 334:421-433. [DOI] [PubMed] [Google Scholar]
- 46.Miller, J. H. 1972. Assay of β-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Moorthy, S., and P. I. Watnick. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174:1364-1368. [DOI] [PubMed] [Google Scholar]
- 49.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 50.Ott, C. M., D. F. Day, D. W. Koenig, and D. L. Pierson. 2001. The release of alginate lyase from growing Pseudomonas syringae pathovar phaseolicola. Curr. Microbiol. 42:78-81. [DOI] [PubMed] [Google Scholar]
- 51.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 52.Petersen, T. N., S. Kauppinen, and S. Larsen. 1997. The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix. Structure 5:533-544. [DOI] [PubMed] [Google Scholar]
- 53.Reguera, G., and R. Kolter. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187:3551-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice, E. W., C. H. Johnson, R. M. Clark, K. R. Fox, J. D. Reasoner, M. E. Dunnigan, P. Panigrahi, J. A. Johnson, and J. G. Morris, Jr. 1993. Vibrio cholerae O1 can assume a ‘rugose’ survival form that resist killing by chlorine, yet retains virulence. Int. J. Environ. Health Res. 3:89-98. [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiller, N. L., S. R. Monday, C. M. Boyd, N. T. Keen, and D. E. Ohman. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugimura, I. I., T. Sawabe, and Y. Ezura. 2000. Cloning and sequence analysis of Vibrio halioticoli genes encoding three types of polyguluronate lyase. Mar. Biotechnol. 2:65-73. [DOI] [PubMed] [Google Scholar]
- 59.Thormann, K. M., R. M. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Y. H., G. L. Yu, X. M. Wang, Z. H. Lv, X. Zhao, Z. H. Wu, and W. S. Ji. 2006. Purification and characterization of alginate lyase from marine Vibrio sp. YWA. Acta Biochim. Biophys. Sin. 38:633-638. [DOI] [PubMed] [Google Scholar]
- 62.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, P. B. 1938. The rugose variant of vibrios. J. Pathol. 46:1-6. [Google Scholar]
- 65.Wong, T. Y., L. A. Preston, and N. L. Schiller. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54:289-340. [DOI] [PubMed] [Google Scholar]
- 66.Xiong, J. P., T. Stehle, B. Diefenbach, R. Zhang, R. Dunker, D. L. Scott, A. Joachimiak, S. L. Goodman, and M. A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin αVβ3. Science 294:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xun, L. Y., R. A. Mah, and D. R. Boone. 1990. Isolation and characterization of disaggregatase from Methanosarcina mazei LYC. Appl. Environ. Microbiol. 56:3693-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yano, S., M. Wakayama, and T. Tachiki. 2006. Cloning and expression of an α-1,3-glucanase gene from Bacillus circulans KA-304: the enzyme participates in protoplast formation of Schizophyllum commune. Biosci. Biotechnol. Biochem. 70:1754-1763. [DOI] [PubMed] [Google Scholar]
- 69.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 71.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]