Abstract
Salmonella enterica serovar Typhimurium must successfully transition the broad fluctuations in oxygen concentrations encountered in the host. In Escherichia coli, FNR is one of the main regulatory proteins involved in O2 sensing. To assess the role of FNR in serovar Typhimurium, we constructed an isogenic fnr mutant in the virulent wild-type strain (ATCC 14028s) and compared their transcriptional profiles and pathogenicities in mice. Here, we report that, under anaerobic conditions, 311 genes (6.80% of the genome) are regulated directly or indirectly by FNR; of these, 87 genes (28%) are poorly characterized. Regulation by FNR in serovar Typhimurium is similar to, but distinct from, that in E. coli. Thus, genes/operons involved in aerobic metabolism, NO· detoxification, flagellar biosynthesis, motility, chemotaxis, and anaerobic carbon utilization are regulated by FNR in a fashion similar to that in E. coli. However, genes/operons existing in E. coli but regulated by FNR only in serovar Typhimurium include those coding for ethanolamine utilization, a universal stress protein, a ferritin-like protein, and a phosphotransacetylase. Interestingly, Salmonella-specific genes/operons regulated by FNR include numerous virulence genes within Salmonella pathogenicity island 1 (SPI-1), newly identified flagellar genes (mcpAC, cheV), and the virulence operon (srfABC). Furthermore, the role of FNR as a positive regulator of motility, flagellar biosynthesis, and pathogenesis was confirmed by showing that the mutant is nonmotile, lacks flagella, is attenuated in mice, and does not survive inside macrophages. The inability of the mutant to survive inside macrophages is likely due to its sensitivity to the reactive oxygen species generated by NADPH phagocyte oxidase.
Salmonella enterica serovar Typhimurium is a gram-negative facultative intracellular pathogen. Serovar Typhimurium infections usually result from ingestion of contaminated food or water. The organism generally targets and colonizes the intestinal epithelium of the host and causes gastroenteritis (i.e., salmonellosis) (22). During a Salmonella infection, the growth phase and growth conditions of the organism are important in attachment, invasion, and the regulation of many of the virulence genes (19, 40, 53, 73). Cells grown under limited oxygen concentrations are more invasive and adhere better to mammalian cells than do aerobically grown or stationary-phase cells (53). Salmonella invasion genes have been identified and localized (24, 46, 54, 63). During infection, serovar Typhimurium must adapt to changes in [O2] encountered in the gastrointestinal tracts of the host (33). Therefore, we hypothesized that the molecular mechanism(s) that controls adaptation to changing [O2] must play an important role in the virulence of serovar Typhimurium.
In Escherichia coli, transitions from aerobic to anaerobic environments, or vice versa, involve changes in a large number of genes (25, 29). However, upon sudden reappearance of oxygen, these cellular processes must be reversed in a precise and orderly fashion to ensure the safe transition to the oxygenated environment. This complex regulatory system has been extensively studied in E. coli, where the DNA-binding protein FNR (6, 27, 44), encoded by fnr, senses changes in [O2] and controls the expression of the different genes either alone or in cooperation with other regulators, e.g., ArcA (11, 15, 30, 32, 42, 74). Recently, three independent studies have examined the global changes in gene expression in E. coli strain MC4100 (70) and strain MG1655 (14, 43) in response to mutations in fnr. In spite of the relatedness of the two E. coli strains, the two studies examining FNR in the MG1655 strain were in closer agreement to each other than to that in the MC4100 strain. These variations were attributed to differences in the genetic backgrounds of the strains and the techniques used (14, 43).
In serovar Typhimurium, an fnr homolog, oxrA, was cloned and some of the genes regulated by OxrA were identified (76, 87). Previous studies examined the role of FNR (OxrA) in the invasiveness of serovar Typhimurium LT2 and concluded that it was not involved in the regulation of invasion (53). However, the LT2 strain used in that study (53) was later found to be avirulent due to an altered rpoS allele (77, 89). The present study is the first addressing the global regulatory role of FNR in serovar Typhimurium metabolism and virulence in a murine model of mucosal and acute infection. We used the well-characterized virulent strain of serovar Typhimurium (ATCC 14028s) growing anaerobically under conditions reported to avoid the effects of pH and catabolite repression on transcription (57). The results indicate that in serovar Typhimurium, as in E. coli, the FNR modulon encompasses the core metabolic and energy functions as well as motility. However, Salmonella-specific components of the FNR modulon were also identified, such as the eut operon (required for ethanolamine utilization) and many of the virulence genes in Salmonella pathogenicity island 1 (SPI-1), as well as the srfABC operon. The fnr mutant was shown to be nonmotile, lacking flagella, and attenuated in vivo.
MATERIALS AND METHODS
Bacterial strains.
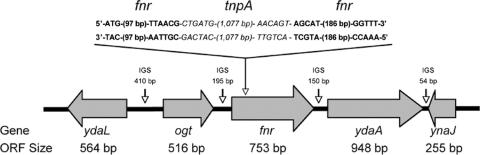
Wild-type (WT) serovar Typhimurium (ATCC 14028s) and its isogenic fnr mutant (NC 983) were used throughout. The mutant strain was constructed by transducing the fnr::Tn10 mutation from serovar Typhimurium [SL2986/TN 2958 (fnr::Tn10)] to strain 14028s using P22 phage (all from the culture collection of S. Libby). The transductants were plated on Evans blue-uranine agar, and the tetracycline marker was eliminated (8). The Tets and FNR− phenotypes were confirmed by the inability of NC983 (fnr mutant) to grow on media containing tetracycline (10 μg/ml) and by its inability to grow anaerobically on M9 minimal medium containing glycerol plus nitrate, respectively. Sequence analysis of fnr and neighboring genes (i.e., ogt and ydaA, respectively) in NC 893 showed that the remnant of Tn10 (tnpA) interrupts fnr between bp 106 and 107 and has no polar effect on ogt or ydaA (Fig. 1).
FIG. 1.
Location of the tnpA insertion (between bp 106 and 107) in the fnr gene. WT fnr sequences are in bold, and the sequences of the beginning and ending junctions of the tnpA insert are in italics. Arrows indicate the direction of transcription. IGS, intergenic spacer region. (Complete DNA sequences [i.e., ogt, tnpA/fnr junctions, and ydaA] are available at GenBank accession number AH015911.)
For complementation studies, a low-copy-number plasmid expressing fnr (pfnr) was constructed. The complete fnr sequence starting from the stop codon of ogtA (TGA [indicated in boldface type]) to 21 bp downstream of fnr (i.e., a 972 bp fragment) was amplified from WT strain 14028s with the following primers: fnr-Forward, 5′-ATATCCATGGTGAATATACAGGAAAAAGTGC-3′ (an NcoI site is underlined); fnr-Reverse, 5′-ATATATTCAGCTGCATCAATGGTTTAGCTGACG-3′ (a PvuII site is underlined). The PCR product was digested with NcoI and PvuII and ligated into the low-copy-number vector pACYC184 cut with NcoI and PvuII. Thus, in the new plasmid (pfnr) the Cmr gene in pACYC184 is replaced with the fnr gene. The plasmid (pfnr) was electroporated and maintained in E. coli DH5α. Transformants were confirmed for Tetr (15 μg/ml) and Cms (20 μg/ml) on Luria-Bertani (LB) plates, and the presence of the fnr gene was confirmed by restriction analysis using EcoRI and HindIII. The plasmid isolated from DH5α was used to complement the fnr mutant. Transformants were selected on LB plates containing tetracycline (15 μg/ml).
Growth conditions.
The WT and the fnr mutant were grown anaerobically at 37°C in MOPS (morpholinepropanesulfonic acid)-buffered (100 mM, pH 7.4) LB broth supplemented with 20 mM d-xylose (LB-MOPS-X). This medium was used in order to avoid the indirect effects of pH and catabolite repression (57). A Coy anaerobic chamber (Coy, Ann Arbor, MI) and anaerobic gas mixture (10% H2, 5% CO2, and 85% N2) were used. All solutions were preequilibrated for 48 h in the chamber. Cells from frozen stocks were used to inoculate LB-MOPS-X broth. Cultures were grown for 16 h and used to inoculate fresh anoxic media. The anaerobic growth kinetics of the mutant and the WT strains were similar, and the doubling times of the fnr mutant and the WT were 53.9 ± 1.2 and 45.4 ± 2.9 min, respectively.
RNA isolation.
Anaerobic cultures were used to inoculate three independent flasks each containing 150 ml of anoxic LB-MOPS-X. The three independent cultures were grown to an optical density at 600 nm (OD600) of 0.25 to 0.35, pooled, and treated with RNAlater (QIAGEN, Valencia, CA) to fix the cells and preserve the quality of the RNA. Total RNA was extracted with the RNeasy RNA extraction kit (QIAGEN), and the samples were treated with RNase-free DNase (Invitrogen, Carlsbad, CA). The absence of contaminating DNA and the quality of the RNA was confirmed by PCR amplification of known genes and by using agarose gel electrophoresis. Aliquots of the RNA samples were kept at −80°C for use in the microarray and quantitative real-time reverse transcription-PCR (qRT-PCR) studies.
Microarray studies.
Serovar Typhimurium microarray slides were prepared and used as previously described (66). The SuperScript Indirect cDNA labeling system (Invitrogen) was used to synthesize the cDNA for the hybridizations. Each experiment consisted of two hybridizations, on two slides, and was carried out in Corning Hybridization Chambers at 42°C overnight. Dye swapping was performed to avoid dye-associated effects on cDNA synthesis. The slides were washed at increasing stringencies (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS], 42°C; 0.1% SSC, 0.1% SDS, room temperature; 0.1% SSC, room temperature). Following hybridization, the microarrays were scanned for the Cy3 and Cy5 fluorescent signals with a ScanArray 4000 microarray scanner from GSI Lumonics (Watertown, MA). The intensity of every spot was codified as the sum of the intensities of all the pixels within a circle positioned over the spot itself and the background as the sum of the intensities of an identical number of pixels in the immediate surroundings of the circled spot.
Data analysis.
Cy3 and Cy5 values for each spot were normalized over the total intensity for each dye to account for differences in total intensity between the two scanned images. The consistency of the data obtained from the microarray analysis was evaluated by two methods: (i) a pair-wise comparison, calculated with a two-tailed Student's t test and analyzed by the MEAN and TTEST procedures of SAS-STAT statistical software (SAS Institute, Cary, NC) (the effective degrees of freedom for the t test were calculated as described previously [71]); and (ii) a regularized t test followed by a posterior probability of differential expression [PPDE (p)] method. These statistical analyses are implemented in the Cyber-T software package available online at the website of the Institute for Genomics and Bioinformatics of the University of California, Irvine (www.igb.uci.edu). The signal intensity at each spot from the FNR mutant and the WT were background subtracted, normalized, and used to calculate the ratio of gene expression between the two strains. All replicas were combined, and the median expression ratios and standard deviations were calculated for open reading frames (ORFs) showing ≥2.5-fold change.
qRT-PCR.
qRT-PCR (34) was used to validate the microarray data (64), where 19 genes were randomly chosen from the differentially expressed genes. This technique was also used to confirm the expression of a set of selected genes. qRT-PCRs were carried out with the QuantiTect SYBR green RT-PCR kit (QIAGEN) and an iCycler (Bio-Rad, Hercules, CA), and the data were analyzed by the Bio-Rad Optical System software, version 3.1, according to manufacturer specifications. To ensure accurate quantification of the mRNA levels, three amplifications for each gene were made with 1:5:25 dilutions of the total RNA. Measured mRNA levels were normalized to the mRNA levels of the housekeeping gene rpoD (σ70). Normalized values were used to calculate the ratios of the expression levels in the fnr mutant relative to the WT.
Logo graph and promoter analysis.
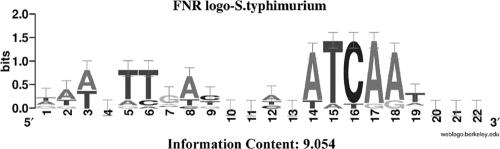
The information matrix for the generation of the FNR logo was produced by using the alignment of the E. coli FNR binding sequences, available at http://arep.med.harvard.edu/ecoli_matrices/ (68). The alignment of the FNR motifs from this website did not include the motifs present in the sodA and mutM promoters (31, 55); therefore, they were included in our analysis. To account for differences in nucleotide usage or slight variations in consensus sequences, a second alignment was built for serovar Typhimurium using the 5′ regions of the homologous genes originally used to build the E. coli information matrix. The alignment was used to prepare a new information matrix using the Patser software (version 3d), available at http://rsat.ub.ac.be/rsat/ (82). A graphical representation (Fig. 2) of the matrices through a logo graph was obtained with Weblogo software (version 2.8.1, 18 October 2004), available at http://weblogo.berkeley.edu/ (17).
FIG. 2.
Logo graph of the information matrix obtained from the consensus alignment of FNR motif sequences for serovar Typhimurium (derived from the corresponding FNR-regulated genes in E. coli). The total height of each column of characters represents the amount of information for that specific position, and the height of each character represents the frequency of each nucleotide.
Motility assay and electron microscopy.
The motilities of the WT, the fnr mutant, and the complemented mutant/pfnr were evaluated under anoxic conditions. Ten microliters of anaerobically grown (16 h) cells were spotted onto LB-MOPS-X agar (0.6% agar) plates and incubated at 37°C for 24 h. The diameter of the growth halo was used as a measure of motility. Scanning electron microscopy (SEM) was used to examine the morphology of the extracellular surfaces. WT and fnr cultures were grown anaerobically (OD600, 0.3 to 0.4) and centrifuged, and the pellets were resuspended in a fixative solution (3% glutaraldehyde in 0.1 M phosphate-buffered saline [PBS] [pH 7.4]) under anaerobic conditions. The fixed samples were rinsed in 0.1 M PBS buffer, postfixed with 1% osmium tetraoxide in 0.1 M PBS for 2 h, and rinsed with PBS, all at 4°C. An aliquot of each sample was filtered through a 0.1-μm filter. Each filter was dehydrated through a graded ethanol series (up to 100%), brought to room temperature, critical point dried with liquid CO2 (Tousimis Research, Rockville, MD), placed on stubs, and sputter coated with Au/Pd (Anatech Ltd., Denver, NC). Samples were viewed at 15 kV with a JEOL 5900LV SEM (JEOL USA, Peabody, MA). Transmission electron microscopy (TEM) and negative staining were used to visualize the flagella. WT and fnr cultures were grown anaerobically (OD600, 0.3 to 0.4), and a 20-μl aliquot of each sample was separately placed on a Formvar-carbon grid. The grids were washed with 0.1 M sodium acetate (pH 6.6), negatively stained with 2% phosphotungstic acid (PTA), and air dried for 5 min before being viewed at 80 kV with a JEOL JEM-100S TEM (JEOL USA, Peabody, MA).
Pathogenicity assays.
Immunocompetent 6- to 8-week-old C57BL/6 mice and their congenic iNOS−/− and pg91phox−/− immunodeficient mice (bred in the University of Colorado Health Science Center [UCHSC] animal facility according to Institutional Animal Care and Use Committee guidelines) were used in this study. Stationary-phase serovar Typhimurium (WT and fnr mutant) cultures grown aerobically in LB-MOPS-X broth were used, and the cells were diluted in PBS. For oral (p.o.) challenge, groups of 10 mice were gavaged with 5 × 106 or 5 × 107 CFU in 200 μl of PBS/mouse. For intraperitoneal (i.p.) challenge, groups of five mice were inoculated with 250 CFU in 500 μl of PBS/mouse. Mortality was scored over a 15- to 30-day period.
Macrophage assay.
Peritoneal macrophages were harvested from C57BL/6 mice and pg91phox−/− immunodeficient mice (bred in the UCHSC animal facility) 4 days after intraperitoneal inoculation with 1 mg/ml sodium periodate and used as previously described (18). Macrophages were challenged (multiplicity of infection of 2) for 25 min with the different test strains. Stationary-phase cultures grown aerobically in LB-MOPS-X broth were used as outlined above. Prior to infection, each strain was opsonized with 10% normal mouse serum for 20 min. After the challenge, extracellular bacteria were removed from the monolayers by washing with prewarmed RPMI medium (Cellgro, Herndon, VA) containing gentamicin (6 mg/ml) (Sigma), the Salmonella-infected macrophages were lysed at indicated time points, and the surviving bacteria were enumerated on LB agar plates. The results are expressed as percent survival relative to the number of viable intracellular bacteria recovered at time zero (i.e., after washing and removal of the extracellular bacteria, ∼25 min after infection).
Microarray data.
The microarray data are accessible via GEO accession number GSE3657 at http://www.ncbi.nlm.nih.gov/geo.
RESULTS
Transcriptome profiling.
Out of 4,579 genes, the two-tailed Student t test produced a set of 1,664 coding sequences showing significant differences (P < 0.05) between the fnr mutant and the WT. We restricted our analyses to include highly affected genes (i.e., having a ratio of ≥2.5-fold). Under this constraint, 311 genes were differentially expressed in the fnr mutant relative to the WT; of these, 189 genes were up-regulated and 122 genes were down-regulated by FNR (see Table S1 in the supplemental material). The 311 FNR-regulated genes were classified into clusters of orthologous groups (COGs) as defined at http://www.ncbi.nlm.nih.gov/COG (78, 79, 88) (Table 1). It should be noted that throughout the study we compared the levels of transcription in the fnr mutant to that in the WT strain. Thus, genes repressed by FNR possess values of >1, while genes activated by FNR have values of <1.
TABLE 1.
Classification of FNR-regulated genes according to COGs
| Functional gene groupa | No. of genes
|
|
|---|---|---|
| FNR activated | FNR repressed | |
| Cellular processes | 54 | 9 |
| Cell division and chromosome partitioning | 0 | 1 |
| Cell envelope and biogenesis, outer membrane | 4 | 3 |
| Cell motility and secretion | 30 | 0 |
| Posttranslational modification, protein turnover, chaperones | 8 | 2 |
| Inorganic ion transport and metabolism | 7 | 2 |
| Signal transduction mechanisms | 5 | 1 |
| Defense mechanisms | 1 | 0 |
| Information storage and processing | 9 | 5 |
| Translation, ribosomal structure, and biogenesis | 0 | 2 |
| Transcription | 6 | 2 |
| DNA replication, recombination, and repair | 3 | 1 |
| Intracell trafficking | 3 | 0 |
| Metabolism | 62 | 81 |
| Energy production and conversion | 18 | 30 |
| Amino acid transport and metabolism | 21 | 21 |
| Nucleotide transport and metabolism | 7 | 2 |
| Carbohydrate transport and metabolism | 10 | 23 |
| Coenzyme metabolism | 1 | 3 |
| Lipid metabolism | 2 | 2 |
| Secondary metabolite biosynthesis, transport, and catabolism | 3 | 0 |
| Unknown | 60 | 27 |
| General function prediction only | 12 | 5 |
| Function unknown | 10 | 9 |
| Poorly characterized | 38 | 13 |
| Total | 189 | 122 |
The differentially expressed genes were classified according to COGs as defined at http://www.ncbi.nlm.nih.gov/COG.
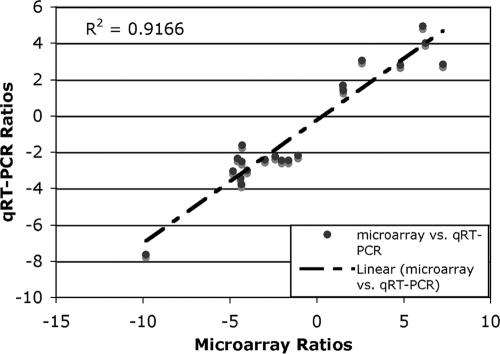
In order to globally validate the microarray data (64), we randomly selected 19 of the 311 differentially expressed genes for qRT-PCR. The measured levels of mRNA were normalized to the mRNA levels of the housekeeping gene rpoD. The specific primers used for qRT-PCR and the normalized mRNA levels are shown in Table 2. The microarray and qRT-PCR data were log2 transformed and plotted (Fig. 3). The correlation between the two sets of data was 0.94 (P < 0.05).
TABLE 2.
Validation of microarray data by qRT-PCR of randomly selected genes relative to the housekeeping gene rpoDa
| Locusb | Name | Primer sequencec | Fragment (bp) | Serovar Typhimurium gene functiond | Ratio of fnr mutant/WT
|
Log2 ratio
|
||
|---|---|---|---|---|---|---|---|---|
| qRT-PCRe | Microarrayf | qRT-PCRg | Microarrayh | |||||
| STM3217 | aer | 5′-CGTACAACATCTTAATCGTAGC-3′ | 163 | Aerotaxis sensor receptor; senses cellular redox state or proton motive force | 0.190 | 0.210 | −2.4 | −2.3 |
| 5′-TTCGTTCAGATCATTATTACCC-3′ | ||||||||
| STM1919 | cheM | 5′-GCCAATTTCAAAAATATGACG-3′ | 114 | Methyl-accepting chemotaxis protein II; aspartate sensor-receptor | 0.036 | 0.120 | −4.8 | −3.1 |
| 5′-GTCCAGAAACTGAATAAGTTCG-3′ | ||||||||
| STM0441 | cyoC | 5′-TATTTAGCTCCATTACCTACGG-3′ | 134 | Cytochrome o ubiquinol oxidase subunit III | 153.967 | 7.096 | 7.3 | 2.8 |
| 5′-GGAATTCATAGAGTTCCATCC-3′ | ||||||||
| STM1803 | dadA | 5′-TAACCTTTCGCTTTAATACTCC-3′ | 155 | d-Amino acid dehydrogenase subunit | 2.835 | 3.169 | 1.5 | 1.7 |
| 5′-GATATCAACAATGCCTTTAAGC-3′ | ||||||||
| STM0964 | dmsA | 5′-AGCGTCTTATCAAAGAGTATGG-3′ | 154 | Anaerobic dimethyl sulfoxide reductase, subunit A | 0.001 | 0.005 | −9.8 | −7.6 |
| 5′-TCACCGTAGTGATTAAGATAACC-3′ | ||||||||
| STM2892 | invJ | 5′-TTGCTATCGTCTAAAAATAGGC-3′ | 128 | Surface presentation of antigens; secretory proteins | 0.246 | 0.182 | −2.0 | −2.5 |
| 5′-TTGATATTATCGTCAGAGATTCC-3′ | ||||||||
| STM2324 | nuoF | 5′-GGATATCGAGACACTTGAGC-3′ | 163 | NADH-dehydrogenase I, chain F | 2.894 | 2.600 | 1.5 | 1.4 |
| 5′-GATTAAATGGGTATTACTGAA CG-3′ | ||||||||
| STM0650 | STM0650 | 5′-CAACAGCTTATTGATTTAGTGG-3′ | 130 | Putative hydrolase, C terminus | 0.476 | 0.219 | −1.1 | −2.2 |
| 5′-CTAACGATTTTTCTTCAATGG-3′ | ||||||||
| STM2787 | STM2787 | 5′-AAGCGAATACAGCTATGAACC-3′ | 144 | Tricarboxylic transport | 28.241 | 6.892 | 4.8 | 2.8 |
| 5′-ATTAGCTTTTGCAGAACATGG-3′ | ||||||||
| STM4463 | STM4463 | 5′-AAGGTATCAGCCAGTCTACG-3′ | 142 | Putative arginine repressor | 0.325 | 0.181 | −1.6 | −2.5 |
| 5′-CGTATGGATAAGGATAAATTCG-3′ | ||||||||
| STM4535 | STM4535 | 5′-TAAGCCAGCAGGTAGATACG-3′ | 139 | Putative PTS permease | 6.053 | 8.217 | 2.6 | 3.0 |
| 5′-CGACATAAAGAGATCGATAACC-3′ | ||||||||
| STM2464 | eutN | 5′-AGGACAAATCGTATGTACCG-3′ | 153 | Putative detox protein in ethanolamine utilization | 0.062 | 0.125 | −4.0 | −3.0 |
| 5′-ACCAGCAGTACCCACTCTCC-3′ | ||||||||
| STM2454 | eutR | 5′-GGTAAAAGAGCAGCATAAAGC-3′ | 118 | Putative regulator; ethanolamine operon (AraC/XylS family) | 0.043 | 0.195 | −4.6 | −2.4 |
| 5′-ATTATCACTCAAGACCTTACGC-3′ | ||||||||
| STM2470 | eutS | 5′-AATAAAGAACGCATTATTCAGG-3′ | 137 | Putative carboxysome structural protein; ethanol utilization | 0.049 | 0.073 | −4.3 | −3.8 |
| 5′-GTTAAAGTCATAATGCCAATCG-3′ | ||||||||
| STM1172 | flgM | 5′-AGCGACATTAATATGGAACG-3′ | 126 | Anti-FliA (anti-sigma) factor; also known as RflB protein | 0.050 | 0.174 | −4.3 | −2.5 |
| 5′-TTTACTCTGTAAGTAGCTCTGC-3′ | ||||||||
| STM3692 | lldP | 5′-TGATTAAACTCAAGCTGAAAGG-3′ | 189 | LctP transporter; l-lactate permease | 76.492 | 16.003 | 6.3 | 4.0 |
| 5′-CCGAAATTTTATAGACAAAGACC-3′ | ||||||||
| STM3693 | lldR | 5′-GAACAGAATATCGTGCAACC-3′ | 153 | Putative transcriptional regulator for lct operon (GntR family) | 68.378 | 30.597 | 6.1 | 4.9 |
| 5′-GAGTCTGATTTTCTCTTTGTCG-3′ | ||||||||
| STM1923 | motA | 5′-GGTTATCGGTACAGTTTTCG-3′ | 194 | Proton conductor component of motor; torque generator | 0.048 | 0.092 | −4.4 | −3.4 |
| 5′-TAGATTTTGTGTATTTCGAACG-3′ | ||||||||
| STM4277 | nrfa | 5′-GACTAACTCTCTGTCGAAAACC-3′ | 159 | Nitrite reductase; periplasmic cytochrome c552 | 0.051 | 0.324 | −4.3 | −1.6 |
| 5′-ATTTTATGGTCGGTGTAGAGC-3′ | ||||||||
STM3211 (rpoD) was used as the reference gene where no significant change in expression level was observed. The primer sequences used for rpoD were as follows: 5′-CGATGTCTCTGAAGAAGTGC-3′ (forward) and 5′-TTCAACCATCTCTTTCTTCG-3′ (reverse). The size of the fragment generated was 150 bp.
Location of the ORF in the serovar Typhimurium LT2 genome.
For each set, the first sequence is the forward primer, and the second sequence is the reverse primer.
Functional classification according to the KEGG (Kyoto Encyclopedia of Genes and Genomes) database.
Expression levels based on qRT-PCR. Values are ratios between the fnr mutant and the WT, where values of <1 indicate that FNR acts as an activator and values of >1 indicate that FNR acts as a repressor.
Expression levels based on microarray data. Values are ratios between the fnr mutant and the WT, where values of <1 indicate that FNR acts as an activator and values of >1 indicate that FNR acts as a repressor.
Expression levels based on qRT-PCR, comparing the fnr mutant with the WT. Values are signal-to-log2 ratios.
Expression levels based on microarray data, comparing the fnr mutant with the WT. Values are signal-to-log2 ratios.
FIG. 3.
Correlation between the microarray and qRT-PCR data for 19 selected genes. The ratios of changes in gene expression, from the microarray and qRT-PCR experiments, for the FNR mutant relative to the WT were log2 transformed and linearly correlated. The genes selected and the primers used in qRT-PCR are listed in Table 2.
To determine whether a binding site for FNR might be present in the region upstream of the candidate FNR-regulated genes, we searched the 5′ regions of these genes for the presence of a putative FNR-binding motif using a Salmonella logo graph (Fig. 2). One hundred ten out of the 189 genes activated by FNR (58%) and 59 out of the 122 genes repressed by FNR (48%) contained at least one putative FNR-binding site (see Table S1 in the supplemental material).
FNR as a repressor.
Transcription of the genes required for aerobic metabolism, energy generation, and nitric oxide detoxification was repressed by FNR. In particular, the genes coding for cytochrome c oxidase (cyoABCDE), cytochrome cd complex (cydAB), NADH-dehydrogenase (nuoBCEFJLN), succinyl-coenzyme A (CoA) metabolism (sucBCD), fumarases (fumB, stm0761, and stm0762), and the NO·-detoxifying flavohemoglobin (hmpA) were expressed at higher levels in the fnr mutant than in the WT (see Table S1 in the supplemental material). Also, genes required for l-lactate metabolism (lldPRD) and for the production of phosphoenolpyruvate (pykF), oxaloacetate (ppc), and acetoacetyl-CoA (yqeF) were expressed at higher levels in the mutant than in the WT (see Table S1 in the supplemental material).
FNR as an activator.
Several genes associated with anaerobic metabolism, flagellar biosynthesis, motility, chemotaxis, and Salmonella pathogenesis were activated by FNR. The genes constituting the dms operon, dmsABC (encoding the anaerobic dimethyl sulfoxide reductase), required for the use of dimethyl sulfoxide (DMSO) as an anaerobic electron acceptor (7), had the lowest expression levels (i.e., −200-, −62-, and −23-fold, respectively) in the fnr mutant relative to the WT (see Table S1 in the supplemental material). Two other operons coding for putative anaerobic DMSO reductases (STM4305 to STM4307 and STM2528 to STM2530) were also under positive control by FNR. The genes required for the conversion of pyruvate to phosphoenolpyruvate (pps), Ac-CoA (aceF), Ac-P (pta), and OAc (ackA), as well as those for the production of formate (tdcE, yfiD, focA) and d-lactate (ldhA), were expressed at lower levels in the fnr mutant than in the WT (see Table S1 in the supplemental material). In addition, the genes coding for a universal stress protein (ynaF), a ferritin-like protein (ftnB), an ATP-dependent helicase (hrpA), and aerotaxis/redox sensing (aer) were also positively regulated by FNR (see Table S1 in the supplemental material).
The genes for ethanolamine utilization (eut operon) (48, 67, 75) had lower transcript levels in the fnr mutant (Fig. 4B). Although the FNR-dependent genes for tetrathionate utilization (ttrABCSR), a major anaerobic electron acceptor, were not affected by the lack of FNR, this was not surprising since tetrathionate is also needed to induce expression (67).
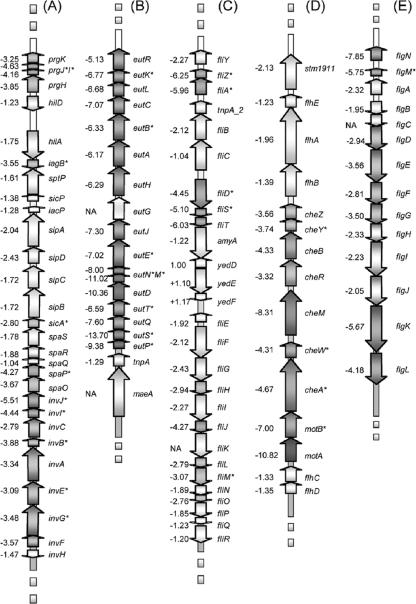
FIG. 4.
Scheme representing the structural organization of the major genes involved in virulence/SPI-1 (A), ethanolamine utilization (B), and flagellar biosynthesis and motility/swarming (C to E). The names of genes are listed to the right of the arrows, an asterisk next to the gene indicates the presence of at least one FNR motif in the 5′ region, and the numbers to the left of the arrows indicate the ratio of gene expression in the fnr mutant relative to that in the WT.
Several of the middle flagellar (class 2) genes (e.g., flgNMDEFGKL and fliZADSTHJLM) and late flagellar (class 3) genes (e.g., cheZYBRMWA, motBA, aer, trg, and tsr) had lower transcript levels in the fnr mutant than in the WT (Fig. 4C to E). There was no significant difference in the transcript levels of the early flagellar genes (class 1) flhD and flhC, whose gene products FlhD/FlhC are the master regulators of flagellar biosynthesis (Fig. 4D). In addition, many newly identified flagellar genes (23) (i.e., mcpA, mcpC, and cheV) had lower expression levels in the fnr mutant (see Table S1 in the supplemental material), while the expression of mcpB was not affected.
Several genes in SPI-1 (e.g., prgKJIH, iagB, sicA, spaPO, invJICBAEGF) had lower levels of expression in the fnr mutant than in the WT (Fig. 4A). This region contains genes coding for a type three secretion system and for proteins required for invasion and interaction with host cells. The data also showed that genes belonging to the other SPIs were unaffected by the lack of FNR. However, the virulence operon srfABC, which is located outside SPI-2 and regulated by a two-component regulatory system (SsrAB) located on SPI-2 (86, 90), was differentially regulated by FNR (see Table S1 in the supplemental material). The effects of FNR on a subset of the above-mentioned invasion and virulence genes were further confirmed by measuring the levels of mRNA in the fnr mutant and the WT strains by qRT-PCR (Table 3).
TABLE 3.
qRT-PCR of selected invasion and virulence genesa
| Locusb | Name | Primer sequencec | Fragment size (bp)d | Ratioe |
|---|---|---|---|---|
| STM2893 | invI | 5′-CTTCGCTATCAGGATGAGG-3′ | 161 | −9.27 |
| 5′-CGAACAATAGACTGCTTACG-3′ | ||||
| STM2874 | prgH | 5′-GGCTCGTCAGGTTTTAGC-3′ | 190 | −8.45 |
| 5′-CTTGCTCATCGTGTTTCG-3′ | ||||
| STM2871 | prgK | 5′-ATTCGCTGGTATCGTCTCC-3′ | 199 | −8.56 |
| 5′-GAACCTCGTTCATATACGG-3′ | ||||
| STM2886 | sicA | 5′-GATTACACCATGGGACTGG-3′ | 207 | −3.92 |
| 5′-CAGAGACTCATCTTCAGTACG-3′ | ||||
| STM1593 | srfA | 5′-AGGCGGCATTTAGTCAGG-3′ | 176 | −4.33 |
| 5′-GACAGGTAAGCTCCACAGC-3′ | ||||
| STM1594 | srfB | 5′-GGTACCAGAAATACAGATGG-3′ | 190 | −6.55 |
| 5′-GCCGATATCAATCGATGC-3′ |
STM3211 (rpoD) was used as the reference gene where no significant change in expression level was observed. The primer sequences used for rpoD were as follows: 5′-CGATGTCTCTGAAGAAGTGC-3′ (forward) and 5′-TTCAACCATCTCTTTCTTCG-3′ (reverse). The size of the fragment generated was 150 bp.
Location of the ORF in the serovar Typhimurium LT2 genome.
For each set, the first sequence is the forward primer, and the second sequence is the reverse primer.
Size of the amplified PCR product.
Ratio of transcription levels in the fnr mutant to those in the WT.
Effects of FNR on motility and flagella.
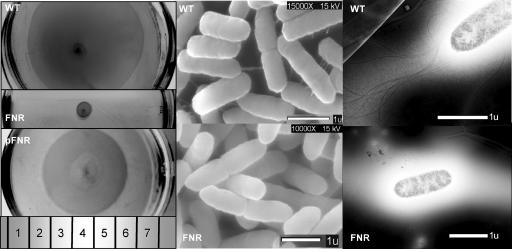
Expression of the flagellar biosynthesis, motility, and chemotaxis genes was lower in the fnr mutant than in the WT. Therefore, we compared the WT, fnr mutant, and the mutant cells harboring pfnr for motility in soft agar under anaerobic conditions (Fig. 5, left panel). The data indicate that the fnr mutant was nonmotile (Fig. 5, left panel; compare the middle and top sections) and that the lack of motility was complemented (∼75%) by the inclusion of pfnr (Fig. 5, left panel, bottom section). The <100% complementation by pfnr is probably due to extra copies of the global regulator FNR (32). We also compared the WT and the mutant for the presence of flagella by SEM (Fig. 5, center panel) and TEM (Fig. 5, right panel). Taken together, these data show that the fnr mutant is nonmotile due to the lack of flagella.
FIG. 5.
Comparison of the WT, the fnr mutant, and the mutant strain harboring pfnr for motility (left) and comparison of the WT and the mutant for the presence of surface appendages by SEM (center) and for the presence of flagella by negative staining and TEM (right). Cells were grown anaerobically in LB-MOPS-X media, and samples were prepared as described in Materials and Methods.
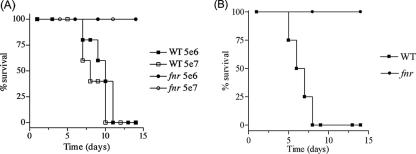
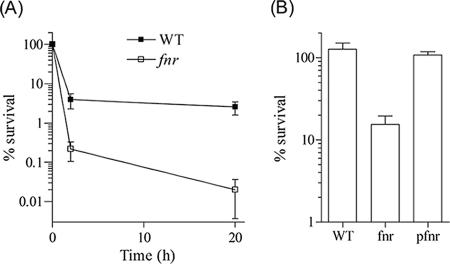
Effects of FNR on pathogenicity and killing by macrophages.
Because our data (Fig. 4 and 5 and Table S1 in the supplemental material) have shown that FNR positively regulates the expression of various loci, such as motility and SPI-1 genes that are important determinants for Salmonella pathogenesis (9, 85), we tested the virulence of fnr in a murine model of mucosal and acute infection. In immunocompetent C57BL/6 mice, the fnr mutant was completely attenuated over a 15-day period following an oral challenge with 5 × 106 or 5 × 107 CFU/mouse, while the WT strain killed all mice within 10 or 12 days, respectively (Fig. 6A). The mutant strain was also 100% attenuated when 250 CFU/mouse were inoculated i.p. (Fig. 6B). The different Salmonella strains were also tested for the ability to survive killing by macrophages (Fig. 7). Similar numbers of fnr mutant and WT cells were recovered from the macrophages 25 min after infection (designated as time zero postinfection) (data not shown). Data in Fig. 7A indicate that the lack of FNR resulted in a dramatic reduction in the ability of Salmonella to replicate in macrophages. Interestingly, most of the killing of the WT by macrophages took place during the first 2 h postinfection (i.e., the WT resisted further killing beyond 2 h), while the viability of the fnr mutant continued to decline by ∼1 log between 2 and 20 h postinfection (Fig. 7A and B). Data in Fig. 7B also show that this phenotype is complemented in fnr mutant cells harboring pfnr.
FIG. 6.
Comparison of the fnr mutant and the WT strain for virulence in 6- to 8-week-old C57BL/6 mice. (A) Groups of 10 mice were inoculated p.o. with 5 × 106 and 5 × 107 CFU/mouse. (B) Groups of five mice were challenged i.p. with 250 CFU/mouse, as described in Materials and Methods. Percent survival is the number of mice surviving relative to the number of mice challenged at time zero.
FIG. 7.
Comparison of the WT, the fnr mutant, and the mutant strain harboring pfnr for survival inside peritoneal macrophages from C57BL/6 mice. The macrophages were harvested and treated as described in Materials and Methods. (A) Comparison between the fnr mutant and the WT strain. The number of viable cells found inside the macrophages, at time zero, following the removal of extracellular bacteria by washing/gentamicin treatment is defined as 100% survival. (B) Comparison between the WT, the fnr mutant, and the pfnr-complemented mutant. The number of viable cells found inside macrophages at 20 h is expressed as percent survival relative to that found inside macrophages at 2 h.
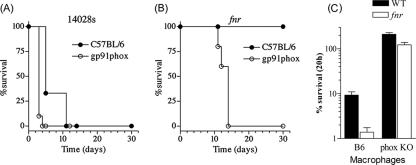
Congenic iNOS−/− mice (unable to make NO·) and pg91phox−/− mice (defective in oxidative burst oxidase) were used to examine the roles of reactive nitrogen and oxygen species (RNS and ROS), respectively, in resistance to an acute systemic infection with FNR-deficient or WT Salmonella. The fnr mutant was as attenuated in iNOS−/− mice as in congenic WT C57BL/6 controls (data not shown). In sharp contrast, the fnr mutant killed pg91phox−/− mice, albeit at a lower rate than the WT strain (Fig. 8A and B). Consistent with the in vivo data, the WT and the isogenic fnr mutant survived to similar extents in NADPH oxidase-deficient macrophages isolated from pg91phox−/− mice (Fig. 8C).
FIG. 8.
Virulence of the WT and the fnr mutant in C57BL/6 mice and congenic gp91phox−/− mice and survival of the bacteria inside peritoneal macrophages. The mice were challenged i.p. with 250 CFU/mouse, as described in Materials and Methods. (A) C57BL/6 and gp91phox−/− mice treated with the WT strain. (B) C57BL/6 and gp91phox−/− mice treated with the fnr mutant. (C) Survival of the WT and the fnr mutant inside macrophages from C57BL/6 and gp91phox−/− mice. The number of viable cells at 20 h is expressed as percent survival relative to that found inside the macrophages at time zero.
DISCUSSION
We have shown that FNR is a global anaerobic regulator in serovar Typhimurium ATCC 14028s, where it serves, directly or indirectly, as an activator or a repressor of at least 311 genes. In particular, we demonstrate that FNR is a regulator of virulence in serovar Typhimurium. The role of FNR in serovar Typhimurium has previously been examined (53, 76, 87), and a few comments are in order. Some of the genes identified as OxrA (FNR)-regulated (76, 87) were also identified in the present study (e.g., dmsA, pepT, dcuB). However, prior to the present report, none of the virulence genes (i.e., SPI-1 or srfABC) had been identified as a part of the Salmonella FNR modulon. Although several studies have shown that oxygen limitation induces adhesion and invasion in Salmonella (19, 40, 53, 73), it was concluded that “oxrA (fnr) is not involved in the regulation of Salmonella invasiveness” (53). However, in a recent study (69), fnr was shown to be essential for the virulence of serovar Typhimurium SL1344 in the enteritis mice model. Thus, the most likely explanation for the differences between our findings and those reported previously (53, 76, 87) relates to the strain of Salmonella employed. Herein, we used the virulent strain ATCC 14028s, while the previous studies (53, 76, 87) used serovar Typhimurium LT2, which was later found to be avirulent due to an altered rpoS allele (77, 89). In addition, the proteomes of the ATCC 14028 and the LT2 strains are significantly different (1).
FNR and metabolism.
The regulation of anaerobic growth and metabolism by FNR in E. coli is well characterized and has been used to explain similar processes in serovar Typhimurium. However, the similarity between the two genomes is only about 75 to 80% (62). Indeed, our data confirmed that several aspects of their metabolic regulation are similar (see Table S1 in the supplemental material). For example, the regulation of the dmsABC operon, encoding the three subunits of the anaerobic DMSO reductase (7), is similar in both organisms. Alignment of the region from −400 to +50 of dmsA in Salmonella and E. coli showed that the two sequences are almost identical. Also, an FNR motif was identified upstream from the starting ATG of dmsA. Two other operons, each encoding three subunits of putative anaerobic DMSO reductases, were identified and found to be regulated by FNR. Furthermore, the amino acid sequences coded by these putative operons are only slightly homologous to the dms operon, thus suggesting that their functions may be similar but distinct from each other (data not shown).
FNR was also found to repress many of the genes encoding enzymes involved in aerobic electron transport and oxidative phosphorylation (e.g., cyoABCDE, cydAB, and nuoBCEFJLN), as well as some TCA cycle enzymes (e.g., sucBCD, fumB, STM0761, and STM0762). The involvement of FNR, together with ArcA, in the anaerobic regulation of the TCA cycle has been observed (65).
The data have also revealed that FNR activates genes required for the anaerobic metabolism of pyruvate for the production of d-lactate, acetyl-CoA, acetate, and formate that may have a role in virulence (51) and at the same time represses genes required for the utilization of l-lactate and for the production oxaloacetate and pyruvate from phosphoenolpyruvate (see Table S1 in the supplemental material).
Ethanolamine, which is available in the host, can be used by enterobacteria as a source of carbon, nitrogen, and energy. The ability to use ethanolamine seems to correlate with virulence, as a knockout of eutB in Listeria monocytogenes reduces its ability to replicate intracellularly (41). Furthermore, in nonpathogenic E. coli, the eut operon is incomplete and its expression is not affected by FNR (14, 43, 70). Our data showed that all of the genes in the eut operon, including the positive regulator eutR, were activated by FNR (Fig. 4B). However, an FNR-binding motif was not found in the promoter of eutR, though several members of this operon have at least one FNR motif (Fig. 4B). This may suggest that FNR acts indirectly on the eutR promoter or that it activates genes downstream from eutR.
FNR and flagellar biosynthesis/motility/chemotaxis.
Our data indicated that FNR positively regulates the expression of genes involved in flagellar biosynthesis, motility, and chemotaxis (Fig. 4 and 5). The flagellar genes are organized in three classes that are expressed in a hierarchical fashion (12). Early genes (class 1) consist of the master regulator operon flhDC, coding for a transcriptional activator of the middle genes (class 2). The middle genes include fliA (encoding σ28, required for the expression of the late genes), fliM (encoding the anti-σ28), and the genes for the synthesis of the flagellar basal body. Late flagellar genes (class 3) consist of fliC (encoding flagellin) and the genes for chemotaxis (12). Our data are similar to those reported for E. coli MG1655 (14, 43) but different from those reported for E. coli MC4100 (70). This discrepancy is most likely due to the low transcript level of fnr in WT E. coli MC4100 (72).
We also found that FNR activates other genes related to motility/chemotaxis, such as yhjH and ycgR (see Table S1 in the supplemental material). In E. coli, yhjH and ycgR are members of the flagellar regulon and, in conjunction with H-NS, control the flagellar motor function (47). Furthermore, we found that the regulation of flagellar biosynthesis, motility, and chemotaxis by FNR is independent of the flagellar master operon flhDC. This finding is interesting but not surprising, because the regulation of this complex regulon is known to take place at multiple levels in response to different environmental signals (12).
FNR and pathogenesis.
Our data have shown that FNR positively regulates the expression of several loci involved in flagellar biosynthesis, chemotaxis, acetate metabolism, and SPI-1 invasion genes that are important in Salmonella pathogenesis (26, 50, 51, 59). FliZ and FliA have previously been shown to control the transcription of hilA and hilC, the regulators of a large number of SPI-1 genes (35, 60). In addition, hilA is activated by SirA in response to the accumulation of acetate and acetyl-P in the bacterial cytoplasm (51). Indeed, our data showed that FNR activates the transcription of fliZ and fliA (Fig. 4C) as well as the genes involved in the synthesis of acetate and acetyl-P (i.e., the eut operon, aceF, pta, and ackA) (Fig. 4B and Table S1 in the supplemental material). Certainly, further studies are needed to establish whether the effect of FNR on hilA and hilC is direct (i.e., via FliZ and FliA) or indirect (i.e., via fatty acid metabolites). In any case, the data revealed that the transcription of many SPI-1 genes (i.e., prgKJIH, iagB, sicA, spaPO, invJICBAEGF), as well as genes recently identified as having a role in pathogenesis (i.e., mcpA, mcpC, cheV, and srfAB) (23), was significantly reduced in the fnr mutant (Fig. 4A and Table S1 in the supplemental material). Furthermore, the positive regulation of SPI-1 genes by FNR, the global anaerobic activator, corroborates previous findings showing that oxygen limitation increases the expression of SPI-1 invasion genes (3, 39, 46) and that an fnr mutant is nonvirulent in the enteritis mice model (69).
SPI-1 is believed to be essential for serovar Typhimurium invasion of host cells but is not required for its survival inside macrophages. Thus, in the mouse typhoid model, SPI-1 mutants are attenuated when administered p.o. but are fully virulent when delivered intraperitoneally i.p. (24). However, our data showed that the fnr mutant is 100% attenuated in mice following either the p.o. or i.p. route of infection (Fig. 6) and is also highly attenuated in macrophages (Fig. 7). These results were unexpected, because survival inside macrophages usually requires the induction of SPI-2 virulence genes (13), which, according to our microarray data, were not affected by the lack of FNR. Intriguingly, recent findings indicate that SPI-1 genes are required for Salmonella persistence inside macrophages (10) and for persistent systemic infection in mice (52). These findings (10, 52) corroborate our results and may suggest a new role for FNR in coordinating the expression of virulence genes within and outside SPI-1 with those required for intracellular survival.
Another plausible explanation for the reduced survival of the fnr mutant inside macrophages may relate to its inability to mount the multiple defenses normally required for intracellular survival (16, 18, 20, 83). For example, lack of FNR may result in reduced expression of genes whose products are required for protection against the antimicrobial defenses utilized by macrophages (61, 84). Therefore, we compared the virulence of the fnr mutant and the WT in iNOS−/− and gp91phox−/− mice, which are deficient in producing RNS and ROS, respectively. The data showed that the fnr mutant is attenuated in the iNOS− mice (data not shown); however, it was able to kill the gp91phox−/− mice and survive in macrophages isolated from gp91phox−/− mice (Fig. 8). Indeed, restoration of virulence to the fnr mutant in the gp91phox−/− mice but not in the iNOS−/− mice suggested that the mutant is more sensitive to ROS than to RNS generated in the phagosomes. Our microarray data have shown that the lack of FNR resulted in a threefold increase in the expression of hmpA (coding for flavohemoglobin, Hmp), required for protection against NO. Interestingly, overexpression of Hmp in Salmonella increases its sensitivity to hydrogen peroxide (4). On the other hand, the expression of other genes required for protection against ROS (i.e., sodA, sodB, sodC1, and sodC2 [coding for superoxide dismutases] and katG and katE [coding for hydroperoxidases]) were not significantly affected by the lack of FNR (see Table S1 in the supplemental material). However, upon further examination of our array data, two candidate genes (i.e., ftnB [coding for a ferritin-like protein] and ynaF [coding for a universal stress protein]) whose expression levels were dramatically reduced (i.e., 21- and 116-fold, respectively) in the fnr mutant were identified (see Table S1 in the supplemental material). The gene products of ftnB and ynaF could provide protection against ROS generated inside the phagosomes and thus an explanation for the inability of the mutant to survive inside the macrophages. Studies are in progress to determine the roles of FtnB and YnaF in Salmonella pathogenesis.
Our findings also suggest that FNR is functional inside the phagosome. This conclusion seems to be at odds with the current dogma that macrophages are highly aerobic and therefore that FNR is inactive. However, in E. coli, full expression of FNR-regulated genes (e.g., frd, dms) has been shown to take place in a medium with a redox potential (Eh) of approximately +400 mV (81) or at [O2] in the gas phase of ≤2% (56, 80). It is also known that a steep pO2 gradient exists between the media in which the macrophages are suspended and the inside of the phagosome (i.e., the pO2 drops from ∼210 μM in the medium to 79.8 ± 1.6 to 32.6 ± 1.7 μM in the phagosome, depending on the density of the macrophages) (28, 36, 37). In addition, the production of ROS and oxygen consumption by the macrophages as well as oxygen consumption by the phagocytosed Salmonella cells are expected to further decrease the pO2 in the phagosome. Furthermore, the pO2 inside macrophages cultured in the laboratory at atmospheric oxygen is most likely different from the pO2 inside macrophages residing in the host (2). Indeed, this is a complex topic, and further work is needed to directly measure pO2 available to bacterial cells residing inside activated macrophages in culture as well as in the host. In spite of this lack of knowledge, we must conclude from the present findings that the intraphagosomal pO2 allows for the expression of FNR-dependent genes. Indeed, other reports support this conclusion: (i) the expression of Brucella suis genes, normally associated with low pO2, while residing inside the phagosome (38, 49, 58) supports the notion of low intraphagosomal pO2, and (ii) the up-regulation of the FNR-dependent nipAB promoters of serovar Typhimurium in the activated macrophage-like cells RAW 264.7 (45) supports the notion that FNR is functional inside the phagosome.
In conclusion, this is the first report demonstrating the role of FNR in coordinating anaerobic metabolism, flagellar biosynthesis, motility, chemotaxis, and virulence in serovar Typhimurium. Also, recent reports that appeared during the preparation and submission of the manuscript for publication clearly corroborate our findings and demonstrate the important roles of FNR in the virulence of other pathogens (i.e., Neisseria meningitides [5] and serovar Typhimurium [69]) and of ANR (an FNR orthologue) in Pseudomonas aeruginosa (21).
Supplementary Material
Acknowledgments
This work was supported in part by the North Carolina Agricultural Research Service (to H.M.H.); by NIH grants R01AI034829, R01AI022933, R21AI057733, and R01AI52237 and generous gifts from Sidney Kimmel and Ira Lechner (to M.M. and S.P.); by NIH grants AI054959 and RR16082 (to A.V.-T. and J.J.); and by NIH grant AI48622 (to S.J.L.).
We are grateful to Valerie Knowlton, Carlos Santiviago, Nabil Arrach, and Pui Cheng for assistance. Special thanks go to Gabriele Gusmini and Russell Wolfinger for assistance with the statistical analyses/SAS software.
Footnotes
Published ahead of print on 12 January 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adkins, J. N., H. M. Mottaz, A. D. Norbeck, J. K. Gustin, J. Rue, T. R. W. Clauss, S. O. Purvine, K. D. Rodland, F. Heffron, and R. D. Smith. 2006. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol. Cell. Proteomics 5:1450-1461. [DOI] [PubMed] [Google Scholar]
- 2.Atkuri, K. R., L. A. Herzenberg, and L. A. Herzenberg. 2005. Culturing at atmospheric oxygen levels impacts lymphocyte function. Proc. Natl. Acad. Sci. USA 102:3756-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Bang, I.-S., L. Liu, A. Vazquez-Torres, M.-L. Crouch, J. S. Stamler, and F. C. Fang. 2006. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin Hmp. J. Biol. Chem. 281:28039-28047. [DOI] [PubMed] [Google Scholar]
- 5.Bartolini, E., E. Frigimelica, S. Giovinazzi, G. Galli, Y. Shaik, C. Genco, J. A. Welsch, D. M. Granoff, G. Grandi, and R. Grifantini. 2006. Role of FNR, and FNR-regulated, sugar fermentation genes in Neisseria meningitidis infection. Mol. Microbiol. 60:963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beinert, H., and P. J. Kiley. 1999. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 3:152-157. [DOI] [PubMed] [Google Scholar]
- 7.Bilous, P. T., and J. H. Weiner. 1988. Molecular cloning and expression of the Escherichia coli dimethyl sulfoxide reductase operon. J. Bacteriol. 170:1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279:20327-20338. [DOI] [PubMed] [Google Scholar]
- 10.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattopadhyay, S., Y. F. Wu, and P. Datta. 1997. Involvement of FNR and ArcA in anaerobic expression of the tdc operon of Escherichia coli. J. Bacteriol. 179:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 14.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K-12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4815. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, P. A., S. Darie, and R. P. Gunsalus. 1992. The effect of iron limitation on expression of the aerobic and anaerobic electron-transport pathway genes in Escherichia coli. FEMS Microbiol. Lett. 100:227-232. [DOI] [PubMed] [Google Scholar]
- 16.Crawford, M. J., and D. E. Goldberg. 1998. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273:12543-12547. [DOI] [PubMed] [Google Scholar]
- 17.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. Weblogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeGroote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. S. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type-1 fimbriae, and growth phase are factors that affect invasion of Hep-2 cells by Salmonella typhimurium. Infect. Immun. 58:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. S. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filiatrault, M. J., K. F. Picardo, H. Ngai, L. Passador, and B. H. Iglewski. 2006. Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finlay, B. B., and S. Falkow. 1989. Salmonella as an intracellular parasite. Mol. Microbiol. 3:1833-1841. [DOI] [PubMed] [Google Scholar]
- 23.Frye, J., J. E. Karlinsey, H. R. Felise, B. Marzolf, N. Dowidar, M. McClelland, and K. T. Hughes. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galan, J. E., and R. Curtiss. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. Brooks Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 26.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Phys. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 28.Grinberg, O. Y., P. E. James, and H. M. Swartz. 1998. Are there significant gradients of pO2 in cells? Adv. Exp. Med. Biol. 454:415-423. [DOI] [PubMed] [Google Scholar]
- 29.Gunsalus, R. P. 1992. Control of electron flow in Escherichia coli—coordinated transcription of respiratory pathway genes. J. Bacteriol. 174:7069-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunsalus, R. P., and S. J. Park. 1994. Aerobic-anaerobic gene regulation in Escherichia coli—control by the arcAB and fnr regulons. Res. Microbiol. 145:437-450. [DOI] [PubMed] [Google Scholar]
- 31.Hassan, H. M., and L. W. Schrum. 1994. Roles of manganese and iron in the regulation of the biosynthesis of manganese superoxide dismutase in Escherichia coli. FEMS Microbiol. Rev. 14:315-323. [DOI] [PubMed] [Google Scholar]
- 32.Hassan, H. M., and H. C. H. Sun. 1992. Regulatory roles of FNR, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:3217-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, G. L., R. A. Shankar, M. Chzhan, A. Samouilov, P. Kuppusamy, and J. L. Zweier. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. USA 96:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (New York) 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 35.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 36.James, P. E., O. Y. Grinberg, G. Michaels, and H. M. Swartz. 1995. Intraphagosomal oxygen in stimulated macrophages. J. Cell Physiol. 163:241-247. [DOI] [PubMed] [Google Scholar]
- 37.James, P. E., O. Y. Grinberg, and H. M. Swartz. 1998. Superoxide production by phagocytosing macrophages in relation to the intracellular distribution of oxygen. J. Leukoc. Biol. 64:78-84. [DOI] [PubMed] [Google Scholar]
- 38.Jimenez de Bagues, M. P., S. Loisel-Meyer, J.-P. Liautard, and V. Jubier-Maurin. 2007. Different roles of the two high-oxygen-affinity terminal oxidases of Brucella suis: cytochrome c oxidase, but not ubiquinol oxidase, is required for persistence in mice. Infect. Immun. 75:531-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, B. D., and S. Falkow. 1994. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect. Immun. 62:3745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser, M., and G. Sawers. 1997. Overlapping promoters modulate FNR- and ArcA-dependent anaerobic transcriptional activation of the focA pfl operon in Escherichia coli. Microbiology 143:775-783. [DOI] [PubMed] [Google Scholar]
- 43.Kang, Y., K. D. Weber, Y. Qiu, P. J. Kiley, and F. R. Blattner. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 45.Kim, C. C., D. Monack, and S. Falkow. 2003. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect. Immun. 71:3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein, J. R., T. F. Fahlen, and B. D. Jones. 2000. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko, M., and C. Park. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303:371-382. [DOI] [PubMed] [Google Scholar]
- 48.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohler, S., F. Porte, V. Jubier-Maurin, S. Ouahrani-Bettache, J. Teyssier, and J. P. Liautard. 2002. The intramacrophagic environment of Brucella suis and bacterial response. Vet. Microbiol. 90:299-309. [DOI] [PubMed] [Google Scholar]
- 50.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633-1645. [DOI] [PubMed] [Google Scholar]
- 51.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 52.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 89:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee, H. S., Y. S. Lee, H. S. Kim, J. Y. Choi, H. M. Hassan, and M. H. Chung. 1998. Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: roles of fnr, arcA, and fur. Free Radic. Biol. Med. 24:1193-1201. [DOI] [PubMed] [Google Scholar]
- 56.Levanon, S. S., K. Y. San, and G. N. Bennett. 2005. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol. Bioeng. 89:556-564. [DOI] [PubMed] [Google Scholar]
- 57.Liu, X. Q., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588-12597. [DOI] [PubMed] [Google Scholar]
- 58.Loisel-Meyer, S., M. P. Jimenez de Bagues, E. Basseres, J. Dornand, S. Kohler, J. P. Liautard, and V. Jubier-Maurin. 2006. Requirement of norD for Brucella suis virulence in a murine model of in vitro and in vivo infection. Infect. Immun. 74:1973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mastroeni, P., A. Vazquez-Torres, F. C. Fang, Y. S. Xu, S. Khan, C. E. Hormaeche, and G. Dougan. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McClelland, M., L. Florea, K. Sanderson, S. W. Clifton, J. Parkhill, C. Churcher, G. Dougan, R. K. Wilson, and W. Miller. 2000. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 28:4974-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mills, D. M., V. Bajaj, and C. A. Lee. 1995. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15:749-759. [DOI] [PubMed] [Google Scholar]
- 64.Miron, M., O. Z. Woody, A. Marcil, C. Murie, R. Sladek, and R. Nadon. 2006. A methodology for global validation of microarray experiments. BMC Bioinformatics 7:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park, S. J., G. Chao, and R. P. Gunsalus. 1997. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode α-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J. Bacteriol. 179:4138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Porwollik, S., R. M. Wong, S. H. Sims, R. M. Schaaper, D. M. DeMarini, and M. McClelland. 2001. The delta uvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat. Res. 483:1-11. [DOI] [PubMed] [Google Scholar]
- 67.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B-12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robison, K., A. M. McGuire, and G. M. Church. 1998. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 284:241-254. [DOI] [PubMed] [Google Scholar]
- 69.Rollenhagen, C., and D. Bumann. 2006. Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 74:1649-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12—the effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 71.Satterthwaite, F. E. 1946. An approximate distribution of estimates of variance components. Biometrics Bull. 2:110-114. [PubMed] [Google Scholar]
- 72.Sawers, R. G. 2005. Expression of fnr is constrained by an upstream IS5 insertion in certain Escherichia coli K-12 strains. J. Bacteriol. 187:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schiemann, D. A., and S. R. Shope. 1991. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect. Immun. 59:437-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spiro, S., and J. R. Guest. 1991. Adaptive responses to oxygen limitation in Escherichia coli. Trends Biochem. Sci. 16:310-314. [DOI] [PubMed] [Google Scholar]
- 75.Stojiljkovic, I., A. J. Baumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strauch, K. L., J. B. Lenk, B. L. Gamble, and C. G. Miller. 1985. Oxygen regulation in Salmonella typhimurium. J. Bacteriol. 161:673-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swords, W. E., B. M. Cannon, and W. H. Benjamin, Jr. 1997. Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect. Immun. 65:2451-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 80.Tseng, C. P., J. Albrecht, and R. P. Gunsalus. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Unden, G., M. Trageser, and A. Duchene. 1990. Effect of positive redox potentials (greater than +400 mv) on the expression of anaerobic respiratory enzymes in Escherichia coli. Mol. Microbiol. 4:315-319. [DOI] [PubMed] [Google Scholar]
- 82.van Helden, J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 31:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vazquez-Torres, A., and F. C. Fang. 2001. Salmonella evasion of the NADPH phagocyte oxidase. Microb. Infect. 3:1313-1320. [DOI] [PubMed] [Google Scholar]
- 84.Vazquez-Torres, A., J. Jones-Carson, P. Mastroeni, H. Ischiropoulos, and F. C. Fang. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 86.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 87.Wei, Y., and C. G. Miller. 1999. Characterization of a group of anaerobically induced, fnr-dependent genes of Salmonella typhimurium. J. Bacteriol. 181:6092-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, W. Helmberg, D. L. Kenton, O. Khovayko, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, J. U. Pontius, K. D. Pruitt, G. D. Schuler, L. M. Schriml, E. Sequeira, S. T. Sherry, K. Sirotkin, G. Starchenko, T. O. Suzek, R. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2005. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 33:D39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilmes-Riesenberg, M. R., J. W. Foster, and R. Curtiss III. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Worley, M. J., K. H. L. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.