Abstract
The synthesis of structural components and morphogenetic factors required for the assembly of the Bacillus subtilis spore coat is governed by a mother cell-specific transcriptional cascade. The first two temporal classes of gene expression, which involve RNA polymerase sigma σE factor and the ancillary regulators GerR and SpoIIID, are deployed prior to engulfment of the prespore by the mother cell. The two last classes rely on σK, whose activation follows engulfment completion, and GerE. The cotE gene codes for a morphogenetic protein essential for the assembly of the outer coat layer and spore resistance to lysozyme. cotE is expressed first from a σE-dependent promoter and, in a second stage, from a promoter that additionally requires SpoIIID and that remains active under σK control. CotE localizes prior to engulfment completion close to the surface of the developing spore, but formation of the outer coat is a late, σK-controlled event. We have transplanted cotE to progressively later classes of mother cell gene expression. This created an early class of mutants in which cotE is expressed prior to engulfment completion and a late class in which expression of cotE follows the complete engulfment of the prespore. Mutants of the early class assemble a nearly normal outer coat structure, whereas mutants of the late class do not. Hence, the early expression of CotE is essential for outer coat assembly. Surprisingly, however, all mutants were fully resistant to lysozyme. The results suggest that CotE has genetically separable functions in spore resistance to lysozyme and spore outer coat assembly.
The timing and fidelity of several morphogenetic processes in bacterial cells, such as the assembly of the flagellum and other surface appendages, or spore development, often rely on transcriptional cascades that link gene expression to morphogenesis (15, 46). Spores of Bacillus subtilis are encased in a complex proteinaceous structure called the coat, whose formation extends for several hours during the deployment of a mother cell-specific transcriptional cascade (13, 14, 51, 56) (Fig. 1A) and involves the coordinated production and deposition of over 50 protein components (11, 21, 22). The coat, which consists of two main layers, a laminated lightly staining inner layer, and a thick electron-dense outer coat (11, 21, 22), confers protection against lytic enzymes, small toxic molecules, and predation and allows prompt and efficient spore germination (3, 11, 22, 29, 36, 37, 55). Assembly of the coat begins soon after the polar division of the sporulating cell, concurrently with the activation of RNA polymerase sigma factor E (σE) in the larger mother cell compartment (13, 14, 17, 18, 24, 30) (Fig. 1A). Coat assembly proceeds following engulfment of the smaller compartment (or prespore) by the mother cell, with the activation of a second mother cell-specific sigma factor, σK, which governs late stages of development (7, 13, 23, 54, 56) (Fig. 1A). Some coat proteins are produced under the control of σE whereas the synthesis of other relies on σK, but it is only when σK becomes active that the coat layers start gaining their characteristic appearance as viewed by electron microscopy (11, 21, 22).
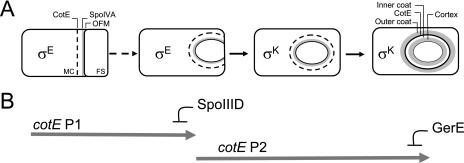
FIG. 1.
Stages in coat formation and CotE expression in B. subtilis in relation to the mother cell line of gene expression. Panel A illustrates the main stages in coat morphogenesis and the periods of activity of the two main mother cell-specific transcriptional regulators σE and σK. σE is activated in the mother cell (MC) soon after polar division of the sporulating cell and drives synthesis of coat morphogenetic proteins SpoIVA and CotE. SpoIVA localizes near the outer forespore membrane (OFM), which faces the mother cell cytoplasm, and CotE localizes at a distance of about 75 nm, in an SpoIVA-dependent manner. The region between CotE and SpoIVA (or matrix) is of unknown composition. σK is activated in the mother cell following engulfment completion and replaces σE. The acquisition of the structural features of the inner and outer coat layers, as viewed by electron microscopy, is a postengulfment, σK-dependent event. The position of CotE is thought to define the site of assembly of the outer coat, whereas the matrix region is converted into the inner layer of the coat. Panel B shows the periods of expression of cotE. CotE is the only coat morphogenetic protein to be produced from tandem promoters, P1 and P2. P1 is under the control of σE and is shut off by SpoIIID prior to the activation of P2, which requires SpoIIID. Expression of cotE is maintained following engulfment completion, under σK control, from P2, and is shut off by GerE during the latest stages of morphogenesis. FS, forespore.
Biogenesis of the spore coat further relies on a class of morphogenetic proteins that act by directing the assembly of the structural components (11, 21, 22, 35). Soon after polar division of the sporangial cell, the morphogenetic protein SpoIVA localizes at or very close to the outer prespore membrane, which faces the mother cell cytoplasm (12, 42, 43) (Fig. 1A). The localization of SpoIVA is required for the assembly of another morphogenetic protein, CotE (10, 56), at a distance of about 75 nm from SpoIVA (12) (Fig. 1A). The gap defined by the positions of SpoIVA and CotE, or matrix, is of unknown composition. When σK becomes active, the matrix is thought to be converted into the inner coat (12). At this postengulfment stage in coat formation, CotE appears to act from the edge of the inner coat region to nucleate assembly of the outer coat, presumably in part by direct interaction with other coat proteins (27, 34). Because spores of a cotE deletion mutant lack the outer coat layer and are highly susceptible to lysozyme, the outer coat was proposed to serve as a barrier against lytic enzymes which would otherwise gain access to the underlying cortex peptidoglycan layer of the spore (55). Additionally, cotE spores are impaired in germination and susceptible to predation (2, 29, 55).
Here, we are concerned with the function of cotE in relation to its time of expression during coat assembly. Consistent with its early assembly, production of CotE relies on tandem promoters, designated P1 and P2, under σE control (55, 56) (Fig. 1B). Transcription from P1 is initiated soon after asymmetric division and is shut off by the repressive action of SpoIIID, a transcription factor that works together with σE prior to activation of transcription from the P2 promoter (13, 33) (Fig. 1B). Transcription from P2 appears to be under the joint control of σE and SpoIIID but remains on after the activation of σK, to be repressed in the final stages of sporulation by the GerE regulatory protein, itself the product of a σK-controlled gene (13, 56) (Fig. 1B). Thus, the cotE gene is expressed both before and after the engulfment process is concluded (Fig. 1B). It has been hypothesized that the early assembly of CotE is a prerequisite for its role in guiding the assembly of the outer coat components once σK becomes active (12), but it is not known whether expression of cotE following engulfment completion is sufficient for outer coat assembly.
To test the possibility that the assembly of the outer coat requires the early expression of cotE and to examine the contribution of the cotE promoters for the assembly and functionality of the outer coat structure, we have fused cotE to either its P1 or P2 promoter, to the σK-dependent gerE promoter, and to the GerE-dependent cotG promoter. These fusions created two main classes of mutants, an early class in which cotE is expressed prior to engulfment completion and a late class in which expression takes place following engulfment completion. The early, but not the late, mutants are able to assemble a nearly normal outer coat structure. Hence, the early expression of cotE is essential for the late stages in outer coat formation. However, spores of all mutants were resistant to lysozyme. The results indicate that CotE has a function in lysozyme resistance that is genetically separable from its role in promoting assembly of the outer coat structure.
MATERIALS AND METHODS
Bacterial strains, media, and general techniques.
The B. subtilis strains used in this study derive from the Spo+ strain MB24 (Table 1). Plasmids were constructed using Escherichia coli strain DH5α (Bethesda Research Laboratories). Luria-Bertani (LB) medium was used for routine growth of E. coli and B. subtilis strains, with appropriate antibiotic selection when needed. Sporulation was induced by growth and nutrient exhaustion in Difco sporulation medium (DSM), and the titers of heat- or lysozyme-resistant spores were measured 24 h after sporulation onset, as described previously (39). Genetic manipulations of B. subtilis were as previously described (9).
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| AH1835 | BL21(DE3) (pLysS) (pMS17) | This work |
| MB24 | trpC2 metC3; Spo+ | Laboratory stock |
| AOB68 | trpC2 metC3 ΔsafA::sp; Spr | Laboratory stock |
| IS105 | ΔcotE::cat; Cmr | 56 and BGSCa |
| AH2835 | trpC2 metC3 ΔcotE::cat; Cmr | This work |
| AH2914 | trpC2 metC3 ΔcotE::cat ΔamyE::PgerE-cotE; Cmr Nmr | This work |
| AH2915 | trpC2 metC3 ΔcotE::cat ΔamyE::PcotG-cotE; Cmr Nmr | This work |
| AH2920 | trpC2 metC3 ΔcotE::cat ΔamyE::PcotEP2-cotE; Cmr Nmr | This work |
| AH2921 | trpC2 metC3 ΔcotE::cat ΔamyE::PcotEP1-cotE; Cmr Nmr | This work |
| AH2942 | trpC2 metC3 ΔcotE::cat ΔamyE::PcotEP1P2-cotE; Cmr Nmr | This work |
| AH2654 | trpC2 metC3 SPβ::cotA-lacZ; Cmr Ermr | This work |
| AH4113 | trpC2 metC3 ΔcotE::cat ΔamyE::PgerE-cotE SPβ::cotA-lacZ; Cmr Ermr Nmr | This work |
| AH4114 | trpC2 metC3 ΔcotE::cat ΔamyE::PcotG-cotE SPβ::cotA-lacZ; Cmr Ermr Nmr | This work |
| AH4116 | trpC2 metC3 ΔcotE::cat SPβ::cotA-lacZ; Cmr Ermr | This work |
Bacillus Genetic Stock Center.
Insertion of cotE at amyE.
A PCR product obtained with primers ymcA664D and cotE761R (the sequences of all primers are available on request), including the cotE gene and promoter region, was cut with HindIII-EcoRI and inserted between the same sites of pMLK83 (26), producing pTC84. In this and all constructions described below, the absence of mutations in the cloned inserts was verified by sequencing. Transformation of a cotE null mutant, AH2835 (ΔcotE::cat), generated by transforming MB24 with DNA from strain 1S105 (55) (Table 1), with PstI-digested pTC84, produced the Nmr AmyE− strain AH2942 (ΔcotE::cat ΔamyE::PcotEP1P2-cotE) (Table 1).
Expression of cotE from its P1 or P2 promoters.
First, primers ymcA664D and cotE499R were used for PCR amplification of a fragment encompassing the cotE promoter region and part of its coding region, which was doubly digested with HindIII and AatII, and the product was cloned between the same sites of pAH256 (19) to yield pTC18. Second, we used pTC18 and primers cotE178D and cotE222R to create a 6-bp deletion of the −10 region of the cotEP2 promoter using the QuickChange system (Stratagene). Third, a 611-bp DNA fragment encompassing the entire ymcA coding region was PCR amplified using primers ymcA74D and ymcA681R, digested with BglII and NsiI, and cloned between the NsiI and BglII sites of pAH256 (19) to yield pTC20. Lastly, a 633-pb XhoI-BglII ymcA fragment obtained from pTC20 was cloned between XhoI and BglII sites of pTC19 to yield pTC21 which, after ScaI-linearization, was used to transform MB24 to spectinomycin resistance (Spr). A double crossover at the cotE locus (verified by PCR) generated AH1936, in which deletion of the −10 region of cotEP2 (herein called the PcotEP1-cotE allele) was confirmed by sequencing. To insert PcotEP1-cotE at amyE in a cotE null mutant, we used DNA from AH1936 to PCR amplify the mutant promoter with primers ymcA664D and cotE761R. The resulting product was cloned between the HindIII and EcoRI sites of pMLK83 (26) to create pTC63. AH2835 was transformed with PstI-linearized pTC63, resulting in the neomycin-resistant (Nmr) Cmr AmyE− strain AH2921 (ΔcotE::cat ΔamyE::PcotEP1-cotE) (Table 1).
A cotE promoter fragment excluding its P1 promoter was amplified by PCR with primers cotE123D and cotE761R and inserted between the HindIII and EcoRI sites of pMLK83 (26) to create pTC56. AH2835 (see above) was transformed to Nmr with PstI-linearized pTC56 to yield the Cmr AmyE− strain AH2920 (ΔcotE::cat ΔamyE::PcotEP2-cotE) (Table 1).
Expression of cotE from the gerE or cotG promoters.
A 314-bp PCR product obtained with primers gerE1D and gerE314R, encompassing the gerE promoter and part of its coding sequence (7), was cut with HindIII and SpeI and inserted between the same sites of pMLK83 (26) to form pTC35. A 587-bp fragment encompassing the cotE ribosome binding site, and the coding region was obtained by PCR with primers cotE174D and cotE761R, doubly digested with SpeI and EcoRI and inserted between the same sites of pTC35 to yield pTC36. PstI-linearized pTC36 was used to transform a B. subtilis strain to Nmr, from which chromosomal DNA was prepared and then used to transform AH2835 to Nmr, yielding the Cmr AmyE− strain AH2914 (ΔcotE::cat ΔamyE::PgerE-cotE) (Table 1).
For the expression of cotE from the cotG promoter (47), a fragment carrying the cotG promoter region, extending 173 bp and 88 bp upstream and downstream, respectively, from its transcriptional start site was PCR amplified with primers cotG28D and cotG288R and digested with HindIII and SpeI. Next, the cotG fragment and the 587-bp SpeI- and EcoRI-digested cotE fragment generated with primers cotE174D and cotE761R (see above) were ligated with HindIII- and EcoRI-cleaved pMLK83 (26), yielding pTC69. PstI-linearized pTC69 transformed AH2835 to Nmr, yielding the Cmr Amy− strain AH2915 (ΔcotE::cat ΔamyE::PcotG-cotE) (Table 1).
Overproduction and purification of a six-His-S-tagged CotE for antiserum production.
The cotE coding sequence was amplified by PCR with primers cotE224D and cotE808R, and the resulting product was cut with EcoRI and XhoI and inserted between the same sites of pET-30b(+) (Novagen). The resulting pMS17 was transformed into BL21(DE3) (pLysS) (Novagen) to create AH1835, in which a six-His-S-tagged CotE fusion protein could be produced from the T7lac promoter (Table 1). AH1835 was grown in LB to an optical density at 600 nm (OD600) of 0.3 and induced for 3 h with 1 mM isopropyl-β-d-thiogalactopyranoside. The fusion protein, mostly present in inclusion bodies, was solubilized in 20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, 10 mM imidazole, and 8 M urea and purified over a HisTrap column as described by the manufacturer (Amersham Biosciences). Gel-purified CotE was sent to Eurogentec (Belgium) for antibody production in rabbits.
Whole-cell lysates and immunoblot analysis.
Whole-cell lysates were prepared from samples of DSM cultures using a French press, and proteins were separated and immunoblotted as described previously (6, 49, 58). The anti-CotE antibody was used overnight at 4°C at a dilution of 1:1,000 in phosphate-buffered saline-Tween 20 containing 0.5% low-fat milk.
Preparation of prespore fractions.
Prespore fractions of sporulating cells were isolated as described before (25) except that whole-cell lysates were prepared as described above. The whole-cell lysates were then fractionated by centrifugation (20 min at 13,000 rpm in a microcentrifuge), and samples (5 μg) of the cell sediment (prespore fraction) were subjected to immunoblot analysis with the anti-CotE antibody (see above).
Purification of spores and extraction of coat proteins.
Spores were collected from DSM cultures 24 h after the onset of sporulation and purified on Gastrografin gradients, and the coat proteins were extracted and separated as described previously (6, 19, 20, 49, 58). Twenty OD580 units of the purified spore suspension was subjected to an alkaline spore decoating procedure prior to lysozyme susceptibility testing, as described before (41, 44). One OD580 unit of decoated spores was tested for heat or lysozyme resistance (see above).
Enzyme assays.
Chromosomal DNA of AH2654 (cotA-lacZ) (Table 1) was used to transform strains AH2914 (PgerE), AH2915 (PcotG), and AH2835 (cotE), yielding strains AH4113, AH4114, and AH4116, respectively (Table 1). The activity of β-galactosidase was determined with the substrate ο-nitrophenyl-β-d-galactopyranoside (6, 40).
Microscopy.
Samples (0.6 ml) from DSM cultures of MB24 and of its congenic derivatives AH2835, AH2921, AH2920, AH2914, AH2915, and AH2942 (Table 1) were collected 2, 3, 4, 6, and 8 h after the initiation of sporulation, resuspended in 0.2 ml of phosphate-buffered saline supplemented with 200 nM MitoTracker Green (MTG; Molecular Probes, Eugene, OR) for visualization of membranes. Aliquots were applied to agarose-coated microscope slides and visualized with a standard filter for fluorescence, as described before (6, 45). For electron microscopy, spores were collected from 24 h DSM cultures of the same strains, purified on Gastrografin gradients (see above), and processed as described before (22). Electron microscopy analysis and photography were conducted on a Phillips EM301, operated at 80 keV.
Protein identification by peptide mass fingerprinting.
Proteins were excised from Coomassie brilliant blue R250-stained gels and subjected to matrix-assisted laser desorption-ionization time-of-flight analysis as previously described (6).
RESULTS
Morphogenesis in a series of cotE heterochronic mutants.
To begin addressing the function of CotE in relation to its time of expression during sporulation, we engineered strains in which cotE was placed under the control of either its P1 (strain AH2921; PcotEP1) or P2 (strain AH2920; PcotEP2) promoters (Table 1). Note that AH2921 bears a 6-bp deletion of the −10 region of the downstream cotE P2 promoter and that cloning of a cotE fragment extending just 82 bp upstream of the P2 transcriptional start site allowed elimination of the upstream cotE P1 promoter in strain AH2920 (see Material and Methods) (56). In addition, we have produced strains expressing cotE from the σK-dependent gerE promoter (strain AH2914; PgerE) (7) or from the σK- and GerE-dependent cotG promoter (AH2915; PcotG) (47). All the promoter fusions were inserted at the nonessential amyE locus of a strain bearing a deletion of cotE (AH2835) at its normal locus. To control for any effects of expressing cotE at amyE, we constructed AH2942, a derivative of AH2835 that bears a wild-type copy of cotE (under the control of both P1 and P2 [PcotEP1P2]) at amyE (Table 1). Results described below indicate that expression of cotE from both P1 and P2 at amyE (strain AH2942) largely mimics the expression pattern and functionality of cotE at its normal locus. We first wanted to test whether under our conditions any of the cotE alleles interfered with the normal course of morphogenesis. To do this, we used the hydrophilic membrane dye MTG and fluorescence microscopy to quantify the kinetics of appearance of asymmetric septa, curved septa, and fully engulfed prespores, and we used phase contrast microscopy to quantify the accumulation of phase-gray and phase-bright spores and released mature spores in the various strains. The results in Fig. 2A indicate that the various cotE promoter mutants did not cause any discernible effect on the timing of the main events that take place during spore morphogenesis. In particular, in all the strains examined, the engulfment process is concluded in the majority of the population 4 h after the onset of the sporulation (Fig. 2A). Engulfment completion represents the signal for σK activation and triggers expression of a large subset of coat genes, among which is cotA, coding for an abundant outer coat component (48). The results in Fig. 2B show that expression of cotA-lacZ is induced in the PgerE and PcotG strains, as in the wild type or in the cotE null mutant, at hour 4 of sporulation. Expression of cotA-lacZ was also unaltered in the PcotEP1P2, PcotEP1, and PcotEP2 strains (not shown in Fig. 2B for clarity).
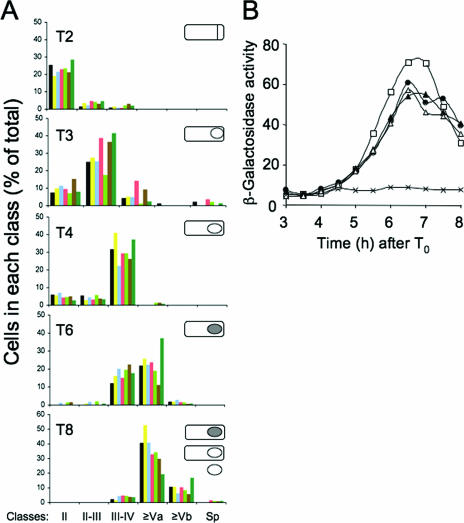
FIG. 2.
The course of morphogenesis in the various cotE mutants. Panel A represents the evolution of various morphological classes during sporulation of the different mutants. Samples from sporulating cultures were collected at various times (T) in hours (2, 3, 4, 6, and 8) after the onset of sporulation, as indicated. Strains are represented as follows: MB24 (wild type), black; AH2942 (PcotEP1P2), yellow; AH2921 (P cotEP1), blue; AH2920 (P cotEP2), magenta; AH2914 (PgerE), light green; AH2915 (PcotG), brown; and AH2835 (cotE), dark green. Classes were defined as follows: II, straight septa; II-III, curved septa; III-IV, cells in which engulfment is completed; Va, phase-gray spores; Vb, phase-bright spores; Sp, free mature spores. Scoring was based on fluorescence microscopy following staining with the vital membrane stain MTG to visualize septa and on phase-contrast microscopy for the visualization of phase-gray, phase-bright, and released spores (Material and Methods). The inserts in each panel represent the main morphological class found at each time: straight septa (hour 2), curved septa (hour 3), engulfed prespores (hour 4), and phase-gray spores (hour 6 and 8). The hour 8 panel also includes phase-bright spores, as well as free spores. Phase-bright spores were the predominant class at hour 10 of sporulation (not represented). Panel B shows the levels of β-galactosidase activity (expressed in Miller units) resulting from expression of a cotA-lacZ transcriptional fusion at the indicated times after the onset of sporulation (T0) in the following strains: MB24 (wild type), crosses; AH2654 (cotA-lacZ), open squares; AH4113 (PgerE-cotE cotA-lacZ) closed circles; AH4114 (PcotG-cotE cotA-lacZ), closed triangles; and AH4116 (ΔcotE cotA-lacZ), open triangles.
Synthesis and assembly of CotE.
Next, we wanted to investigate the time of synthesis and assembly of CotE in the series of promoter mutants. Cells of the wild-type strain MB24 and of the various cotE mutants were harvested at various times during sporulation and lysed, and the presence of CotE in whole-cell extracts or in spore fractions was examined by immunoblot analysis using a CotE-specific antibody (25) (see also Material and Methods). The presence of CotE in whole-cell extracts reflects its time of synthesis, whereas its detection in the spore fractions indicates the time of its recruitment to the surface of the forming spore (25). CotE was first detected in whole-cell extracts from hour 2 of sporulation onward in MB24, AH2942 (PcotEP1P2), and AH2921 (PcotEP1) and in the prespore fraction of the same strains from hour 3 onward (Fig. 3). Note, however, that the absence of CotE in the prespore fraction prior to hour 3 may result in part from the inefficiency with which this fraction is prepared by centrifugation at this time in development (25, 39, 58). On the whole, our observations are consistent with previous studies in which CotE was found to be targeted to the surface of the developing spore at an early stage in development (12, 42, 53). In strain AH2920 (PcotEP2), CotE was only detected from hour 3 onward in both whole-cell and prespore fractions (Fig. 3). However, this strain differed from the wild type or the two strains in which expression of cotE at amyE involved the P1 promoter, as the level of CotE assembled appeared lower (Fig. 3). Indeed, three times more protein had to be analyzed for this strain in order to detect CotE in the prespore fractions (Fig. 3, legend).
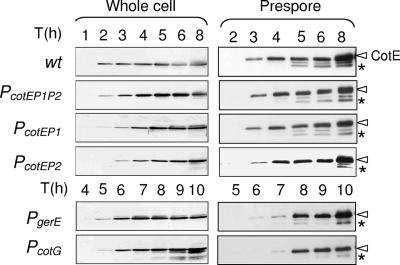
FIG. 3.
Assembly of CotE. Panels A and B represent the accumulation of CotE in the whole-cell and prespore extracts of sporulating cells in strains MB24 (wild type), AH2942 (PcotEP1P2), AH2921 (PcotEP1), AH2920 (PcotEP2), AH2914 (PgerE), and AH2915 (PcotG), as indicated. Samples were collected at the indicated times (in hours) after the onset of sporulation. Proteins (30 μg for the whole-cell lysates or 5 μg for the prespore fraction) were resolved on 12.5% SDS-PAGE gels and transferred to nitrocellulose membranes, which were then probed with an anti-CotE polyclonal antibody. Please note that in case of the PcotEP2 prespore fraction, 15 μg of protein was used, as the mutant accumulates reduced levels of CotE. The arrowheads indicate the positions of the main CotE antigen, and the asterisks indicate the positions of a presumptive processed product of about 21 kDa.
In strain AH2914 (PgerE), CotE was detected in whole-cell lysates at hour 5 and in the prespore fraction at hour 6 (Fig. 3). In AH2915 (PcotG), CotE was found in the cell extracts at hour 5, albeit weakly, and more noticeably at hour 6, but its association with the spore fraction was only clearly seen at hour 7 of sporulation (Fig. 3). Together, these results show that when produced from P1 or P2, CotE is assembled essentially at the same time of sporulation as in the wild type and with little if any delay relative to its time of synthesis. In contrast, the postengulfment synthesis of CotE (from either the gerE or cotG promoters) appears to result in a delay of 1 to 2 h in the assembly of CotE, relative to its time of synthesis.
Levels of CotE in purified spores.
The results described in the two preceding sections indicated that transplanting cotE to the σK regulon interfered with the assembly of CotE (Fig. 3). However, these experiments do not allow a direct comparison of the levels of CotE among the various mutants. This is because the samples were analyzed by loading an equal amount of total protein and because prespores of the various mutants (which predominate in the hour 8 sample) may have different protein contents (19, 20). We therefore compared the levels of CotE in equal numbers of purified spores of the various mutants. The results shown in Fig. 4A indicate that CotE is extracted from wild-type spores not only as a main band of about 24 kDa (black arrowhead) which is the size of monomeric CotE but also as several multimeric forms, in agreement with previous immunoblot analyses (2) and with previous results in which mass spectrometry techniques have been used to analyze the coat composition (31, 32). The multimeric forms of CotE comprise five main species, labeled a through e in Fig. 4A (white arrowheads and lane 1). Essentially the same levels of monomeric CotE and of species a through e are seen in the PcotEP1P2 control strain (Fig. 4A, lanes 1 and 2). In the P1 and P2 strains, however, bands a and b are considerably reduced (Fig. 4A, lanes 3 and 4), whereas in PgerE and PcotG (lanes 5 and 6) bands a to e are almost undetectable. In all strains, the levels of monomeric CotE remains very similar (Fig. 4A). Together, the results suggest that the late expression of cotE drastically reduces the multimerization of CotE at the spore surface but has no proportional effect on the level of monomeric CotE.
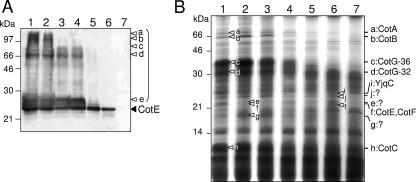
FIG. 4.
Coat polypeptide composition in spores of the various cotE mutants. Panel A shows the accumulation of CotE in purified mature spores of strains MB24 (wild type; lane 1), AH2942 (PcotEP1P2; lane 2), AH2921 (PcotEP1; lane 3), AH2920 (PcotEP2; lane 4), AH2914 (PgerE; lane 5), AH2915 (PcotG; lane 6), and AH2835 (cotE; lane 7). Coat-extractable proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and probed with an anti-CotE antibody. The position of the CotE monomer is indicated by the black arrowhead, and the parenthesis indicates multimeric forms of CotE (bands a to e, white arrowheads). The positions of molecular mass markers (in kDa) are also shown. Panel B shows a Coomassie-stained gel of electrophoretically resolved coat proteins extracted from purified spores of the same strains indicated in panel A. White arrowheads in lane 1 (wild type) point to bands reduced or missing in the late or the null cotE mutants (lanes 5 to 7); they are identified on the right side of the panel. Bands designated as e, f, g, i, and j were excised and subjected to mass spectrometry analysis (see Material and Methods); the identification of bands i and f is indicated on the right side of the panel. No identification was possible for bands e, j, and g. The positions of molecular mass markers (in kDa) are shown on the left side of the panel.
The overall composition of the coat.
To examine the overall composition of the coat in the various mutants, coat protein extracts were prepared from equivalent numbers of purified spores of the various strains and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were stained with Coomassie (see Material and Methods). Overall, the coat polypeptide composition of AH2942 (PcotEP1P2) and AH2921 (PcotEP1) spores was very similar to that of wild-type spores (Fig. 4B, lanes 1 to 3), except for two species above the 21-kDa marker band that appeared reduced (Fig. 4B, lane 2, bands e and f) and another below the 21-kDa marker (band g) that was more extractable. Band f was identified by mass spectrometry as containing CotE and CotF species (see below). Attempts to identity species present on bands e and g were inconclusive (not shown). In contrast to the PcotEP1 strain, the representation of several coat components was altered in spores produced by AH2920 (PcotEP2) relative to AH2942 (PcotEP1P2) spores (Fig. 4B, lanes 2 and 4). The use of mutants in conjunction with immunoblot analysis indicated that CotA (band a), CotB (band b), two forms of CotG of 32 (band c) and 36 kDa (band d), and CotC (band h) were reduced in PcotEP2 spores (not shown) (10, 33, 58). All these proteins have been assumed to reside mainly in the spore outer coat because they are greatly reduced or missing from the coats of a cotE null mutant (55) (Fig. 4B, lane 7). It seems that proper coat assembly relies to a greater extent upon the expression of cotE from P1 than from P2 and that expression solely from P2 results in spores with reduced levels of some outer coat proteins. Nevertheless, the coat protein profile of AH2920 (PcotEP2) spores resembled more that of wild-type spores (Fig. 4B, lane 1) than that of cotE spores (Fig. 4B, lane 7). Strikingly, the coat protein profiles of AH2914 (PgerE) and AH2915 (PcotG) spores appeared very similar to the profile of cotE spores (Fig. 4B, lanes 5 to 7). In particular, outer coat proteins CotA, CotB, CotC, and CotG (10, 47, 56, 58) were greatly reduced or undetected. The results suggest that spores of the PgerE and PcotG mutants have a compromised outer coat structure. The results further advocate that the early expression of cotE is important for proper outer coat assembly.
Early expression of cotE is essential for assembly of the outer coat structure.
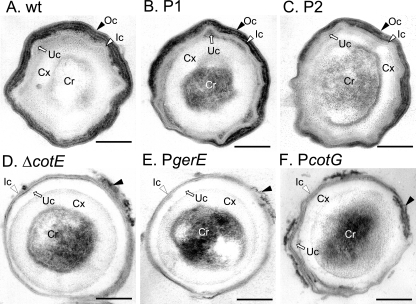
To assess the status of the coat layers, purified spores were examined by thin-section electron microscopy. The coats of MB24 (wild type) spores showed the typical organization of the coat layers, with a lamellar inner coat and a thick, electron-dense striated outer coat closely apposed to the inner coat (Fig. 5A). Spores of AH2942 (PcotEP1P2) (not shown) and those of AH2921 (PcotEP1) (Fig. 5B) were indistinguishable from wild-type spores. Spores of AH2920 (PcotEP2) also showed no major alterations in the structure of the coat layers (Fig. 5, compare panels A and C). Thus, expression of cotE from either P1 or P2 still permitted the formation of spores essentially with a normal ultrastructural organization.
FIG. 5.
Electron microscopy of wild-type and various cotE mutant spores. Spores were collected 24 h after the initiation of sporulation from DSM cultures of MB24 wild-type (wt) strain (A), AH2921 (PcotEP1) (B, P1), AH2920 (PcotEP2) (C, P2), AH2835 (cotE) (D, ΔcotE), AH2914 (PgerE) (E), and AH2915 (PcotG) (F). The spores were purified and processed for electron microscopy analysis as described in Material and Methods. The white and black arrowheads point to the inner and outer coat structures, respectively, and the arrow points to the undercoat (the slightly more electro-dense region between the inner coat and the cortex) or undercoat region. Ic, inner coat; Oc, outer coat; Uc, under coat; Cx, cortex peptidoglycan; Cr, spore core. Bar, 0.2 μm.
In contrast, electron micrographs of AH2914 (PgerE) (Fig. 5E) and AH2915 (PcotG) (Fig. 5F) spores revealed a seemingly normal inner coat but the almost complete absence of the outer coat layer (Fig. 5E and F). An expanded undercoat region is also particularly evident for AH2914 (PgerE) spores (Fig. 5E). These morphological features are characteristic of spores of a cotE null mutant (Fig. 5D) (2, 49, 55). Occasionally, remnants of electro-dense material were observed in association with the defective spores of AH2914 or AH2915 (Fig. 5E and F). However, this was also observed for cotE spores (Fig. 5D). Thus, spores of AH2914 or AH2915 did not differ structurally in any discernible way from spores produced by a cotE null mutant. Hence, expression of cotE under the control of the gerE or cotG promoters, while causing no discernible structural alterations at the level of inner coat, does not permit assembly of a visible outer coat structure.
Spore resistance to lysozyme can be uncoupled from outer coat formation.
Spores of a cotE null mutant are unable to assemble the outer coat layer and are highly susceptible to lysozyme treatment (55). Because shifting cotE to late classes of mother cell-specific gene expression blocked outer coat assembly, we wanted to determine whether spores of the various cotE promoter mutants retained resistance to lysozyme. Spores produced by the various promoter mutants were purified 24 h after the onset of sporulation and assayed for their resistance to lysozyme and heat (see Material and Methods). The cotE null mutant AH2835 formed only 106 lysozyme-resistant spores per ml of culture (Table 2). To our surprise, all the cotE promoter mutants presented close to parental levels of lysozyme resistance, about 108 CFU/ml (Table 2). We noted that the resistance levels observed for the AH2915 (PcotG) spores were reproducibly lower than for the other mutants but still close to 108 CFU/ml (Table 2). To exclude the possibility that lysozyme was inactivated by a coat-associated protease exposed due to abnormal coat assembly in the mutants, we treated spores of the cotE null mutant with the lysozyme solutions recovered after treatment of AH2914 and AH2915 spores and found the same spore titer as with a freshly prepared solution (106 CFU/ml) (data not shown). In all the promoter mutants, formation of lysozyme-resistant spores still required expression of the morphogenetic loci spoIVA, spoVID, and safA (11, 21, 22) (not shown). None of the cotE promoter mutations affected the resistance of spores to heat treatment (Table 2).
TABLE 2.
Spore resistance properties of various cotE mutants
| Strain | Relevant genotype | Cell count (CFU/ml)a
|
Cell count after decoating (CFU/ml)a
|
||||
|---|---|---|---|---|---|---|---|
| Viable | HeatR | LysR | Viable | HeatR | LysR | ||
| MB24 | Wild type | 7.9 × 108 | 6.4 × 108 | 5.8 × 108 | 3.4 × 107 | 1.9 × 107 | 5.0 × 103 |
| AH2835 | ΔcotE | 3.3 × 108 | 3.7 × 108 | 1.5 × 106 | 2.6 × 108 | 1.3 × 108 | ≤103 |
| AH2914 | ΔcotEΔamyE::PgerE-cotE | 6.1 × 108 | 6.4 × 108 | 3.5 × 108 | 9.8 × 107 | 5.1 × 107 | 1.0 × 104 |
| AH2915 | ΔcotEΔamyE::PcotG-cotE | 4.2 × 108 | 6.2 × 108 | 9.9 × 107 | 7.9 × 107 | 7.7 × 107 | ≤103 |
| AH2920 | ΔcotE ΔamyE::PcotEP2-cotE | 6.9 × 108 | 6.0 × 108 | 1.7 × 108 | 6.0 × 107 | 6.3 × 107 | 5.0 × 103 |
| AH2921 | ΔcotE ΔamyE::PcotEP1-cotE | 3.0 × 108 | 2.2 × 108 | 1.0 × 108 | 9.5 × 107 | 6.5 × 107 | 1.1 × 104 |
| AH2942 | ΔcotE ΔamyE::PcotEP1P2-cotE | 1.9 × 108 | 2.0 × 108 | 1.2 × 108 | 6.6 × 107 | 2.0 × 107 | 2.9 × 104 |
The titer of viable cells and heat- and lysozyme-resistant (HeatR and LysR) spores was measured 24 h after the onset of sporulation in DSM (see Material and Methods).
Remarkably, while spore resistance to lysozyme demands expression of the cotE gene, it is largely independent of the time of expression of cotE. Because the “late” mutants fail to assemble the outer coat but form lysozyme-resistant spores, it follows that outer coat assembly is not a strict requirement for spore resistance to lysozyme.
CotE and spore resistance to lysozyme.
We presume that the lysozyme resistance of spores produced by the various cotE promoter mutants, including those unable to assemble the outer coat, is due to the assembly of coat components around the developing spore and not to any other alteration of the spore not apparent by transmission electron microscopy. To investigate this, we subjected purified wild-type and cotE spores, as well as spores of the various cotE promoter mutants, to a standard alkaline decoating regime (4, 41) and assayed for lysozyme resistance. For all strains the decoating regime reduced the lysozyme-resistant cell count to about 0.01% of the level obtained prior to decoating (Table 2). In contrast, spores of all strains remained heat resistant following decoating (Table 2), indicating that the spore cortex was not affected by the treatment. Moreover, the SDS-PAGE profile of the proteins extracted by the alkaline treatment (not shown) resembled that obtained by the SDS-dithiothreitol extraction of the coat proteins from spores of the same strains (Fig. 4B). This suggests that the alkali treatment removed most of the coat proteins present or remaining in spores of the various cotE promoter mutants.
Since species of about 26 kDa (Fig. 4B, band i in lane 6), 24 kDa (band j), and 22 kDa (band f) that were still extracted from AH2914 or AH2915 spores (lysozyme resistant) were missing from coat extracts of the cotE null mutant (lysozyme susceptible), we attempted to identify them by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Note that band e is also seen in AH2914 or AH2915 spores but absent from the null mutant, but this species is also reduced in the lysozyme-resistant spores of AH2921 (PcotEP1) (see above). In any case, no clear identification by peptide mass fingerprinting was possible for bands e (above) or j. Coat proteins CotE and CotF were identified in band f (see above). The identification of CotE (21 kDa) is in agreement with the immunodetection of CotE in spores of AH2915, but the 19-kDa product of the cotF gene is normally proteolytically converted into forms of about 5 and 8 kDa by the YabG protease (8). The 19-kDa CotF band may reflect abnormal assembly of YabG or, alternatively, a cross-linked product of mature CotF (31). In any event, CotF is not required for spore resistance to lysozyme (8). Lastly, and in good agreement with its predicted size, we found band i to contain the YjqC protein (31.2 kDa). YjqC was recently identified as a spore-associated protein in the proteomics study of Kuwana et al. (31), and yjqC was identified as a σK-dependent gene (13, 51). YjqC appears to be an Mn-dependent catalase related to inner coat protein CotJC (20, 50). In contrast to CotJC, inactivation of which has no major impact on spore properties (20, 50), we found that a yjqC insertional mutant produced heat- and lysozyme-sensitive spores (about 103 CFU/ml) (data not shown). Heat sensitivity implies the absence or deficiency of the cortex peptidoglycan layer (16). Undoubtedly, YjqC plays an important role in sporulation but does not appear to have a specific role in spore protection against lysozyme.
DISCUSSION
The early localization of CotE close to the forespore outer membrane in an spoIVA-dependent manner has been hypothesized to help establish a topological plan important for guiding the assembly of the outer coat when, following engulfment completion, σK becomes active in the mother cell (12) (Fig. 1A). The position of CotE, which forms a ring around the forespore after the completion of engulfment, has been viewed as the site of outer coat assembly, whereas the region interior to the CotE ring is presumably converted into the inner coat (11, 12, 21, 22).
However, cotE is expressed both before and after engulfment completion, and since the inner coat forms essentially in a CotE-independent manner (12, 55), it would seem plausible that CotE produced in the postengulfment sporangium could still localize to the edge of the matrix region and still nucleate deposition of the outer coat. An important finding of our study is that a class of “early” mutants, in which cotE is expressed from either P1 or P2, is able to form the outer coat, whereas a class of “late” mutants, expressing cotE under the control of σK-dependent promoters, fails to assemble a visible outer coat structure (Fig. 4 and 5). It thus appears that the early expression of cotE (as part of the σE regulon) is essential for outer coat assembly. The incapacity of the late mutants to support assembly of the outer coat structure is not likely to be due to changes in expression levels inherent to the various promoters used because the greatly reduced levels of CotE that accumulate from the weak P2 promoter, turned on just before engulfment completion (13, 14, 51, 56), supported outer coat assembly while the much higher levels that accumulate from the gerE promoter, activated just after engulfment completion (7, 13, 14, 51), did not (Fig. 3). P2 has an atypical −35 promoter element, which could explain the need for SpoIIID for its utilization in vivo (56), and was found to be active during late stages of development, under σK control (13). The capacity of P2 to support outer coat assembly (in contrast to the stronger gerE promoter) underscores the importance of cotE expression prior to engulfment completion for outer coat assembly.
Why the late cotE mutants fail to nucleate normal coat assembly is unclear. When made under σK control, CotE assembly appears reduced (Fig. 4), and it seems plausible that CotE is only productively assembled at the edge of the matrix region prior to the stage (following engulfment completion) when this region begins to be converted into a visible inner coat. In this view, assembly of CotE produced under σK control would be abnormal. We note that in the early class of mutants as in the wild type, CotE appears to undergo extensive multimerization at the spore surface (2, 31, 32) (Fig. 4). In contrast, in the late mutants, multimerization of CotE, which may represent formation of SDS-dithiothreitol-resistant cross-linked species (2, 31, 32), is greatly reduced, in spite of the fact that the level of the monomeric form of CotE does not vary much among the early or late mutants (Fig. 4). Reduced multimerization could be due to decreased assembly of proteins with which CotE interacts, but it is also tempting to speculate that multimerization of CotE is a factor in its ability to nucleate assembly of the outer coat.
Because spores of a cotE deletion mutant lack the outer coat and are sensitive to lysozyme treatment, the outer coat has been viewed as a barrier to the access of lytic enzymes to the spore cortex peptidoglycan (55). However, the observation that the late cotE mutants, which lack the outer coat, are still resistant to lysozyme suggests that CotE has genetically separable functions in lysozyme resistance and outer coat assembly. The apparent lack of correlation between outer coat assembly and spore resistance to lysozyme has been documented before. For example, a cotE mutant with a deletion of residues 58 to 75 failed to assemble the outer coat but showed a small decrease in lysozyme resistance (22% of the wild type, compared to 6% for a cotE null mutant) (5). A modest decrease in lysozyme resistance along with the absence of an outer coat was also seen for other deletions in the N or C termini of CotE (5). Also, in Bacillus anthracis in which a highly similar CotE ortholog is present, disruption of the gene for the Cotα protein strongly interferes with assembly of the darkly staining outer coat but not with lysozyme resistance (28).
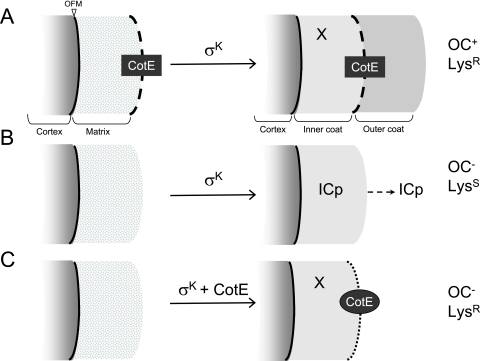
The basis for the lysozyme resistance of the PgerE and PcotG mutants is not presently known. One possibility is that CotE per se is sufficient to confer lysozyme resistance. It could be that sufficient CotE is assembled so as to form a shell around the spore surface capable of blocking access of lysozyme to the spore cortex (Fig. 6). However, we cannot exclude the possibility that in the PgerE and PcotG mutants residual assembly of one or several CotE-controlled proteins mainly associated with the outer coat layer. However, CotE may also control the assembly of inner coat proteins important for lysozyme resistance (Fig. 6, X). Deletion of cotE also affects the inner coat (although not sufficiently to cause any alteration visible by electron microscopy), from which some proteins are missing or tend to be lost when the spore is released into the environment (1, 6, 44, 52) (Fig. 6). The involvement of the inner coat layers in lysozyme resistance has also been suggested before (38, 57).
FIG. 6.
Dual function of CotE in spore resistance to lysozyme and assembly of the outer coat. Morphogenetic protein SpoIVA localizes at or close to the outer forespore membrane (OFM). The localization of SpoIVA is required for the assembly of CotE (black box and dashed line) at the edge of a matrix of unknown composition, which is adjacent to the cortex. Following engulfment completion and the activation of σK, the matrix is transformed into the inner coat. In wild-type cells (A), CotE remains at the edge of the changing matrix from where it nucleates outer coat assembly (Oc+), and the resulting spores are resistant to lysozyme (LysR). In the absence of CotE (B), the matrix still develops into an inner coat, but the resulting spores lack the outer coat (OC−) and are susceptible to lysozyme (LysS). Some inner coat proteins (ICp) are lost from these spores. The expression of cotE under σK control (C) causes abnormal assembly of CotE (black oval and dotted line; CotE is represented at the edge of the inner coat, but it may also be deposited within the inner coat). The resulting spores lack an outer coat (OC−) yet are fully resistant to lysozyme (LysR). Inner coat proteins that may contribute to spore lysozyme resistance are represented by X.
Our analysis of the species present in the coats of spores of the PcotG strain (lysozyme resistant) but absent from cotE spores (lysozyme sensitive) did not lead to the identification of proteins (other than CotE itself) with roles in lysozyme resistance. Also, we note that no CotE-controlled protein is presently known to have a key role in lysozyme resistance (11, 21, 22). However, recent studies suggest the existence of a number of as-yet-uncharacterized proteins that may associate with the coat layers, and some of these could be important for spore resistance to lysozyme (13, 14, 31, 32). Together, our results indicate that assembly of the outer coat structure is not a strict requirement for spore resistance to lysozyme and raise the possibility that CotE itself or as-yet-uncharacterized CotE-controlled proteins have a key role in resistance to this agent.
Acknowledgments
We thank Hong Yi (Emory Microscopy Core Facility) for sample processing and Sérgio Gulbenkian (Instituto Gulbenkian de Ciência, Imaging Facility) for use of the electron microscope.
This work was supported by European Union grant QLK5-CT-2001-01729 to A.O.H., by NIH grant GM54395 to C.P. Moran, Jr., and by grants from the Max-Planck-Institute for Terrestrial Microbiology (Marburg) and the Bundesministerium für Bildung und Forschung to U.V. T.C. holds a Ph.D. fellowship (PRAXIS XXI/BD/1167/00) from “Fundação para a Ciência e a Tecnologia.”
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Abe, A., H. Koide, T. Kohno, and K. Watabe. 1995. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology 141:1433-1442. [DOI] [PubMed] [Google Scholar]
- 2.Aronson, A. I., L. Ekanayake, and P. C. Fitz-James. 1992. Protein filaments may initiate the assembly of the Bacillus subtilis spore coat. Biochimie 74:661-667. [DOI] [PubMed] [Google Scholar]
- 3.Aronson, A. I., and P. Fitz-James. 1976. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 40:360-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer, T., S. Little, A. G. Stöver, and A. Driks. 1999. Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 181:7043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa, T., L. Steil, L. O. Martins, U. Völker, and A. O. Henriques. 2004. Assembly of an oxalate decarboxylase produced under σK control into the Bacillus subtilis spore coat. J. Bacteriol. 186:1462-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 8.Cutting, S., L. B. Zheng, and R. Losick. 1991. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J. Bacteriol. 173:2915-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. V. Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, England.
- 10.Donovan, W., L. B. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 13.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLOS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The sigma E regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 15.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, S. J., and D. L. Popham. 2001. Structure and synthesis of the cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, DC.
- 17.Halberg, R., and L. Kroos. 1994. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J. Mol. Biol. 243:425-436. [DOI] [PubMed] [Google Scholar]
- 18.Halberg, R., V. Oke, and L. Kroos. 1995. Effects of Bacillus subtilis sporulation regulatory protein SpoIIID on transcription by sigma K RNA polymerase in vivo and in vitro. J. Bacteriol. 177:1888-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriques, A. O., T. Costa, L. O. Martins, and R. Zilhão. 2004. Functional architecture and assembly of the spore coat, p. 34-52. In E. Ricca, A. O. Henriques, and S. M. Cutting (ed.), Bacterial spores: probiotics and emerging applications. Horizon Scientific Press, London, United Kingdom.
- 22.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 23.Ichikawa, H., R. Halberg, and L. Kroos. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322-8327. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 25.Isticato, R., G. Esposito, R. Zilhão, S. Nolasco, G. Cangiano, M. De Felice, A. O. Henriques, and E. Ricca. 2004. Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J. Bacteriol. 186:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69-74. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487-502. [DOI] [PubMed] [Google Scholar]
- 28.Kim, H. S., D. Sherman, F. Johnson, and A. I. Aronson. 2004. Characterization of a major Bacillus anthracis spore coat protein and its role in spore inactivation. J. Bacteriol. 186:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klobutcher, L. A., K. Ragkousi, and P. Setlow. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 103:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroos, L., B. Kunkel, and R. Losick. 1989. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science 243:526-529. [DOI] [PubMed] [Google Scholar]
- 31.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971-3982. [DOI] [PubMed] [Google Scholar]
- 32.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Z., and P. J. Piggot. 2001. Development of a two-part transcription probe to determine the completeness of temporal and spatial compartmentalization of gene expression during bacterial development. Proc. Natl. Acad. Sci. USA 98:12538-12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 42:1107-1120. [DOI] [PubMed] [Google Scholar]
- 35.McPherson, D. C., H. Kim, M. Hahn, R. Wang, P. Grabowski, P. Eichenberger, and A. Driks. 2005. Characterization of the Bacillus subtilis spore morphogenetic coat protein CotO. J. Bacteriol. 187:8278-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moir, A. 1981. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 146:1106-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moir, A., E. H. Kemp, C. Robinson, and B. M. Corfe. 1994. The genetic analysis of bacterial spore germination. J. Appl. Bacteriol. 77:9S-16S. [PubMed] [Google Scholar]
- 38.Naclerio, G., L. Baccigalupi, R. Zilhão, M. De Felice, and E. Ricca. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 178:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. John Wiley & Sons, Ltd., Chichester, England.
- 40.Ozin, A. J., T. Costa, A. O. Henriques, and C. P. Moran, Jr. 2001. Alternative translation initiation produces a short form of a spore coat protein in Bacillus subtilis. J. Bacteriol. 183:2032-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogliano, K., E. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 43.Price, K. D., and R. Losick. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181:781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragkousi, K., P. Eichenberger, C. van Ooij, and P. Setlow. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 185:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Real, G., S. Autret, E. J. Harry, J. Errington, and A. O. Henriques. 2005. Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol. Microbiol. 55:349-367. [DOI] [PubMed] [Google Scholar]
- 46.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72:367-394. [DOI] [PubMed] [Google Scholar]
- 47.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandman, K., L. Kroos, S. Cutting, P. Youngman, and R. Losick. 1988. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J. Mol. Biol. 200:461-473. [DOI] [PubMed] [Google Scholar]
- 49.Serrano, M., R. Zilhão, E. Ricca, A. J. Ozin, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25:955-966. [DOI] [PubMed] [Google Scholar]
- 51.Steil, L., M. Serrano, A. O. Henriques, and U. Völker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399-420. [DOI] [PubMed] [Google Scholar]
- 52.Takamatsu, H., Y. Chikahiro, T. Kodama, H. Koide, S. Kozuka, K. Tochikubo, and K. Watabe. 1998. A spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of σK and GerE during development and is located in the inner coat layer of spores. J. Bacteriol. 180:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webb, C. D., A. Decatur, A. Teleman, and R. Losick. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J. Bacteriol. 177:5906-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226:1037-1050. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]
- 57.Zilhão, R., G. Naclerio, A. O. Henriques, L. Baccigalupi, C. P. Moran, Jr., and E. Ricca. 1999. Assembly requirements and role of CotH during spore coat formation in Bacillus subtilis. J. Bacteriol. 181:2631-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]