Abstract
Plasmid pIP501 has a very broad host range for conjugative transfer among a wide variety of gram-positive bacteria and gram-negative Escherichia coli. Functionality of the pIP501 transfer (tra) genes in E. coli was proven by pIP501 retrotransfer to Enterococcus faecalis (B. Kurenbach, C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann, Plasmid 50:86-93, 2003). The 15 pIP501 tra genes are organized in a single operon (B. Kurenbach, J. Kopeć, M. Mägdefrau, K. Andreas, W. Keller, C. Bohn, M. Y. Abajy, and E. Grohmann, Microbiology 152:637-645, 2006). The pIP501 tra operon is negatively autoregulated at the transcriptional level by the conjugative DNA relaxase TraA. Three of the 15 pIP501-encoded Tra proteins show significant sequence similarity to the Agrobacterium type IV secretion system proteins VirB1, VirB4, and VirD4. Here we report a comprehensive protein-protein interaction map of all of the pIP501-encoded Tra proteins determined by the yeast two-hybrid assay. Most of the interactions were verified in vitro by isolation of the protein complexes with pull-down assays. In conjunction with known or postulated functions of the pIP501-encoded Tra proteins and computer-assisted prediction of their cellular location, we propose a model for the first type IV-secretion-like system encoded by a conjugative plasmid from gram-positive bacteria.
Conjugative plasmid transfer is one of the most important mechanisms for the spread of antibiotic resistance genes and thereby the emergence of multiple resistant pathogenic bacteria. pIP501 is a 30,599-bp plasmid with the broadest known host range for a conjugative plasmid originating from gram-positive (G+) bacteria. pIP501 can self-transfer to a variety of G+ bacteria, including pathogens and nosocomial pathogens such as streptococci, staphylococci, enterococci, listeria, multicellular Streptomyces lividans, and also to gram-negative (G−) Escherichia coli (32). The pIP501 tra region is organized in an operon comprising almost half of the plasmid genome. It bears 15 genes, all of which are putatively involved in conjugative plasmid transfer. Cotranscription of all 15 tra genes was shown by reverse transcription-PCR in Enterococcus faecalis JH2-2 (33). The tra genes are transcribed throughout the growth cycle of E. faecalis, and their expression level remains constant independent of the growth phase. The pIP501 tra operon is negatively autoregulated at the transcriptional level by the first gene product of the operon, the TraA relaxase (33). The TraA relaxase was biochemically characterized as the enzyme attacking a specific dinucleotide within oriT, thereby initiating the directed transfer of the plasmid single strand to the recipient. The TraA relaxase and its amino-terminal relaxase domain TraAN246 (the first 246 amino-terminal amino acids) were shown to bind to oriT and to the tra operon promoter, Ptra, which partially overlaps with oriT (30, 33).
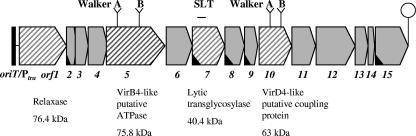
Three of the 15 tra gene products show significant similarity with type IV secretion system (T4SS) components required for conjugative DNA transport, DNA transformation, and transfer of effector proteins from G− bacteria to eukaryotic hosts (for recent reviews, see references 14, 15, 34, and 46). Orf5pIP501 is a putative VirB4-like ATPase, Orf7pIP501 is a VirB1-type lytic transglycosylase, and Orf10pIP501 is a putative VirD4-like coupling protein (Fig. 1). These proteins are also conserved in the pIP501-related plasmids pRE25 from E. faecalis, pSK41/pGO1 from Staphylococcus aureus, and pMRC01 from Lactococcus lactis (in pMRC01, the Orf7 ortholog is missing). Details on the modular structure of the tra regions of these plasmids and their similarities have been summarized by Grohmann et al. (26). The Orf7pIP501 protein has been shown to efficiently cleave peptidoglycan isolated from E. faecalis as well as from E. coli (C. Söllü and E. Grohmann, unpublished data). Functional characterization of the two putative ATPases/ATP binding proteins is in progress.
FIG. 1.
Organization of the pIP501 tra region. The overlapping oriT and Ptra regions are indicated (not to scale). The operon (orf1 to orf15) is terminated by a putative strong rho-independent termination signal (hairpin). Hatched segments indicate genes encoding proteins similar to those identified in G− bacterial T4SSs. The Orf5 protein shows conserved features, a nucleotide binding site motif A (Walker A box 250GLSGGGKT257) and a motif B (Walker B box 509DEFHFLL515), of proteins belonging to the VirB4 family of nucleoside triphosphate-binding proteins (COG3451). Orf10 is a member of the pfam02534 family of TraG/TrwB/TraD/VirD4 coupling proteins. It shows the P-loop motif (Walker A box) and a Walker B motif for nucleotide binding. Orf7 (a VirB1 homolog of the Agrobacterium T-DNA transfer system) contains the SLT domain present in bacterial lytic transglycosylases and was shown to cleave peptidoglycan isolated from E. faecalis and E. coli in an in vitro muramidase assay (Söllü and Grohmann, unpublished data). Open reading frames whose corresponding gene products contain potential signal peptide sequences are marked with a black wedge within the segment.
The T4SSs from G− bacteria have been studied in some detail on genetic, biochemical, and structural-biological levels, resulting in several models for the assembly and functioning of the secretion machinery (1, 14, 34). Many important data for the construction of the T4SS models and verification of genetic and biochemical data have been derived from protein-protein interaction studies performed by use of the yeast two-hybrid assay, the bacterial two-hybrid assay, and in vitro protein-protein interaction studies (17, 18, 23, 27, 35, 47, 49, 53). To analyze the putative type IV-secretion-like system (T4SLS) encoded by a G+ plasmid, we tested all of the 15 pIP501-encoded Tra proteins in a LexA yeast two-hybrid system for interaction with themselves and each other. Qualitative and quantitative interaction data were obtained and led to a comprehensive protein-protein interaction map of the pIP501-encoded Tra proteins. The major Tra protein complexes were verified in vitro by pull-down assays. On the basis of the interaction data, computer predictions of the localization of the Tra proteins, and functions of some of the Tra proteins, we constructed an architectural model for the structure of the pIP501-encoded T4SLS.
MATERIALS AND METHODS
Strains and growth conditions.
Genotypes and relevant features of all used strains are listed in Table 1. E. faecalis JH 2-2 (29) harboring pIP501 was cultivated in brain heart infusion medium (Becton Dickinson, Sparks, MD) supplemented with 20 μg chloramphenicol/ml at 37°C. E. coli JM109 (Promega, Mannheim, Germany), E. coli XL10 (Stratagene, Amsterdam, The Netherlands), and BL21-CodonPlus(DE3)-RIL (Stratagene, Amsterdam, The Netherlands) harboring the expression plasmids or the yeast two-hybrid plasmids (Table 2) were grown in LB medium supplemented with 100 μg ampicillin/ml at 37°C. Saccharomyces cerevisiae L40ccU was grown in yeast extract-peptone-dextrose medium or in SD medium (Synthetic Drop out; Becton Dickinson, Sparks, MD), lacking the respective amino acids, at 30°C, as described below (see “Yeast two-hybrid assay”).
TABLE 1.
Bacterial and yeast strains used in this work
| Strain | Genotype/relevant features | Reference/source |
|---|---|---|
| Escherichia coli | ||
| XL 10 Gold | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10(Tetr) Amy Cmr] | Stratagene |
| BL21-CodonPlus(DE3)-RIL | F−ompT hsdS(rB− mB−) dcm+ Tetrgal λ (DE3) endA Hte [argU ileY leuW Cmr] | Stratagene |
| JM109 | recAl endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB.lacIqlacZΔM15] | Promega |
| Enterococcus faecalis | ||
| JH 2-2 | Rifr Fusr | 29 |
| Saccharomyces cerevisiae | ||
| L40ccU | Derived from L40c (54) uracil prototroph | E. Wanker, MDC, Berlin, Germany |
TABLE 2.
Plasmids used in this work
| Plasmid | Replicon | Size (kb) | Relevant features and antibiotic resistancea | Reference/source |
|---|---|---|---|---|
| pIP501 | pIP501 | 30.6 | tra+ Cmr MLSr | 20 |
| pBTM117c | ColE1/2μm | 9.3 | PADH1lexA TRP1 AprCAN1 | E. Wanker, MDC, Berlin, Germany |
| pGAD426 | ColE1/2μm | 7.8 | PADH1GAL4 LEU2 CYH2r Apr | E. Wanker, MDC, Berlin, Germany |
| pQTEV | pMB1/ColE1 | 4.8 | Pt4lacIq His7 Apr | 44 |
| pGEX-6P-2 | pBR322 | 4.9 | PtaclacIq GST Apr | Amersham BioSciences |
| pMAL-c2x | pMB1/M13 | 6.7 | PtaclacIqmalE Apr | New England BioLabs |
MLS, macrolide, lincosamide, streptogramin B; Cm, chloramphenicol; Ap, ampicillin; CAN1, S. cerevisiae CAN1 gene; CYH2r, resistance to cycloheximide.
DNA preparation and transformation.
Extraction and purification of plasmid DNA from E. coli were performed using a QIAGEN kit (QIAGEN, Hilden, Germany) or a Gen Elute plasmid miniprep kit (Sigma-Aldrich, Taufkirchen, Germany). Restriction endonucleases were purchased from Promega (Mannheim, Germany) and New England Biolabs (Frankfurt am Main, Germany), T4 DNA ligase and shrimp alkaline phosphatase were from Roche Diagnostics (Mannheim, Germany), Gen Therm DNA polymerase was from Rapidozym (Berlin, Germany), and Taq DNA polymerase was from GenScript (Scotch Plains, NJ). Synthetic oligonucleotides were purchased from Sigma-Aldrich (Taufkirchen, Germany) or MWG Biotech (Ebersberg, Germany). The enzymes were used as specified by the suppliers. PCR fragments for cloning experiments were purified by use of a Wizard SV gel and a PCR Clean-Up system (Promega, Mannheim, Germany). Preparation of competent cells and E. coli transformations with plasmid DNA were performed by standard methods (43). Yeast transformations were carried out by the lithium acetate method (45).
Cloning of tra genes for protein-protein interactions. (i) Yeast two-hybrid assay.
The pIP501 tra genes were amplified by PCR using the primers listed in Table 3. They were inserted as SalI/NotI DNA fragments into SalI/NotI-cut pBTM117c and pGAD426 plasmids. The nucleotide sequences of the insertions were verified by dideoxy chain termination sequencing in an automated sequencer (ABI prism 310; Perkin Elmer, Rodgau, Jügesheim, Germany) performed by Services in Molecular Biology (Berlin, Germany).
TABLE 3.
Oligonucleotides used in this work
| Function and name | Sequence (5′-3′)c | Position |
|---|---|---|
| Insertion into pBTM117c, pGAD426, pQTEV, and pGEX6P-2 | ||
| orf1-SalI-fw | CTCGGGTCGACAAGAGAGGTGATACAATTGG | 1378-1397a |
| orf1-NotI-rev | CGCTCGCGGCCGCTTTATACACCTCTTGTTT | 3385-3402a |
| orf2-SalI-fw | GGCGTCGACAGAGGTGTATAAAATGA | 3390-3406a |
| orf2-NotI-rev | CCTGCGGCCGCCTTCTCTATTAAGCAA | 3728-3743a |
| orf3-SalI-fw | GCGGTCGACGAGAAGGGAGTTAGTTATG | 3738-3756a |
| orf3-NotI-rev | TGGCGCGGCCGCACAATTAAATCACCAC | 4142-4157a |
| orf4-SalI-fw | GCGGTCGACAAGCGATACGATGAAAGA | 4201-4218a |
| orf4-NotI-rev | CTTGCGGCCGCTCCTAACTATTCAAAAC | 4769-4785a |
| orf5-SalI-fw | GCCTCGTCGACAATAGTTAGGAGCGTTAAA | 4775-4793a |
| orf5-NotI-rev | TGCTCGCGGCCGCTCTCCCTTCTATTGAATTT | 6745-6763a |
| orf6-SalI-fw | GGCGTCGACATTCAATAGAAGGGAGAAA | 6747-6765a |
| orf6-NotI-rev | TTCGCGGCCGCAATCACCAACCTTCCTA | 8119-8135a |
| orf7-SalI-fw | CCGTCGACATTTCATATCA | 33-43b |
| orf7-NotI-rev | CTCGCGGCCGCAACTCCATTTCTT | 1160-1172b |
| orf8-SalI-fw | GCGGTCGACGGAAGAAATGGAGTTTGA | 1158-1175b |
| orf8-NotI-rev | GGCGCGGCCGCTTCTACTCCTCTCCTA | 1703-1718b |
| orf9-SalI-fw | GGCGGTCGACAAGCATGGCGAAGAAGA | 1717-1733b |
| orf9-NotI-rev | GCTTGCGGCCGCCTAATAAACTAGTCA | 2146-2160b |
| orf10-SalI-fw | GGCCTGTCGACGGGAAAAATGACTAGTTTATTAGC | 2138-2161b |
| orf10-NotI-rev | TGCTTGCGGCCGCCATTTGATTTCCTCCGATCT | 3801-3820b |
| orf11-SalI-fw | GGCGTCGACCGGAGGAAATCAAATGAA | 3805-3822b |
| orf11-NotI-rev | GCTTGCGGCCGCTAAATCCATTAGTAAA | 4734-4749b |
| orf12-SalI-fw | GCTTGTCGACGAGGTGTTTACTAATGG | 4728-4744b |
| orf12-NotI-rev | GCTTGCGGCCGCCTCTCTTATTTTCTGA | 5663-5678b |
| orf13-SalI-fw | GGCCGTCGACATTGTCTTATTATTTTG | 5689-5705b |
| orf13-NotI-rev | CCGGCGGCCGCTTTAGCGTATTTCAGTT | 6653-6669b |
| orf14-SalI-fw | GCGGTCGACGAGTGCTGAAACAATGGGA | 6674-6692b |
| orf14-NotI-rev | GGCGCGGCCGCAATATGCTTTATCTGA | 7048-7063b |
| orf15-SalI-fw | GGCGTCGACAGGAGAGAAGAAAATGAAA | 7102-7120b |
| orf15-NotI-rev | GCTGCGGCCGCCTTAATTAGATTCTCTT | 7951-7968b |
| Insertion into pMAL-c2X | ||
| orf4-EcoRI-fw | GCCGAATTCAGCGATACGAGGAAAGA | 4202-4218a |
| orf4-SalI-rev | GCCGTCGACCTCCTAACTATTCAAAAC | 4769-4786a |
| orf5-EcoRI-fw | GCGGAATTCGTTAAA ACG GAG AAG ATA | 4788-4805a |
| orf5-SalI-rev | GGCGTCGACTCCCTTCTATTGAAT TT | 6745-6760a |
| orf7-EcoRI-fw | TCCGAATTCTTTCATATCATGGGA | 34-48b |
| orf7-SalI-rev | CTCGTCGACAACTCCATTTCTTCCT | 1157-1172b |
| orf7Δtmh-EcoRI-fw | TCCGAATTCCTAGCAACAGAA | 178-189b |
| orf10-EcoRI-fw | GCGGAATTCATGACTAGTTTA | 2145-2156b |
| orf10-SalI-rev | GCGGTCGACTTAAAATGGTAA | 3789-3800b |
| Sequencing primers | ||
| pBTM-seq-fw | TCGTAGATCTTCGTCAGCAG | 956-976 |
| pBTM-seq-rev | AGCAACCTGACCTACAGG | 1201-1218 |
| pGAD-seq-fw | TACCACTACAATGGATGATGT | 758-778 |
| pGAD-seq-rev | GCACAGTTGAACTGAACTTGC | 899-919 |
| pQTEV-seq-fw | CCCGAAAAGTGCCACCTG | 4712-4729 |
| pQTEV-seq-rev | GTTCTGAGGTCATTACTGG | 277-295 |
| pGEX-seq-fw | GGTCTGGCAAGCCACGTTTG | 869-888 |
| pGEX-seq-rev | CCGGGAGCTGCATGTGTCAGAGG | 1035-1057 |
| pMAL-seq-fw | ACGCGCAGACTAATTCGAGC | 2618-2637 |
| pMAL-seq-rev | AAGGCGATTAAGTTGGGTAACG | 2778-2799 |
| orf1-550-seq-fw | TGTTTCTGAAATTCGTAAAG | 1960-1979a |
| orf1-1125-seq-fw | AAGTTAGAGCAATGGTTAAT | 2541-2560a |
| orf5-center-seq-fw | GCTGTTGAGGTATGCTAAAT | 5236-5282a |
| orf5-center-seq-rev | CACGTTGTATCGCAAGTGGA | 6241-6260a |
| orf10-center-seq-fw | ATCCACGCTATAACGAAGAAG | 2650-2670b |
| orf10-center-seq-rev | GCTTTCTGACTTACTTCCGCT | 3318-3338b |
(ii) Pull-down assays.
The pIP501-borne tra genes were amplified by PCR using the primers listed in Table 3. They were inserted as SalI/NotI DNA fragments into the SalI/NotI-cut seven-histidine-tag plasmid pQTEV (a gift from K. Büssow, Max Planck Institute for Molecular Genetics, Berlin) and the glutathione S-transferase tag plasmid pGEX-6P-2. orf1 was inserted as a BamHI/KpnI fragment into the BamHI/KpnI-cut six-histidine tag plasmid pQE30 (QIAGEN, Hilden, Germany). orf1N246, encoding the first 246 amino acids of Orf1, was cloned into the unique BamHI and HindIII sites of pQE30 (30). In the maltose binding protein (MBP) fusion vector pMAL-c2X, the tra genes were inserted as EcoRI/SalI fragments. The nucleotide sequences of the insertions were verified by dideoxy chain termination sequencing in an automated sequencer (ABI prism 310; Perkin Elmer, Rodgau, Jügesheim, Germany).
Yeast two-hybrid assay.
A general description of the two-hybrid system has been detailed elsewhere (21). Before being used in the two-hybrid assay, all 15 pBTM117c-tra plasmids were transformed into the yeast L40ccU strain (a kind gift of E. Wanker, MDC Berlin, Germany), and the resulting transformants were tested for the absence of autoactivation of the lacZ and HIS3 reporter genes. In order to test for protein-protein interaction, the pBTM117c and pGAD426 plasmids carrying the respective tra genes were cotransformed into the yeast L40ccU strain and plated on SD minimal medium lacking leucine and tryptophan. After incubation at 30°C for 2 to 3 days, the transformants were replica plated on SD medium lacking leucine, tryptophan, and histidine and with galactose and raffinose as sugar sources (SD-leu-tryp-his/gal/raff medium) and incubated at 30°C for 3 to 10 days for selection of interacting proteins. From the SD-leu-tryp-his/gal/raff plates, replica filters were made and cells were permeabilized by freezing in liquid nitrogen (10 s) and thawing at room temperature. Filters were transferred onto Whatman 3MM paper saturated with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid) solution (10) and incubated at 37°C. β-Galactosidase-positive clones were tested further by a quantitative β-galactosidase assay with CPRG (chlorophenol red-β-d-galactopyranoside) as the substrate. A single colony from the SD-leu-tryp-his/gal/raff plate was inoculated into 5 ml SD-leu-tryp-his/gal/raff medium and grown overnight at 30°C. A 1-ml aliquot of this preculture was used to inoculate 5 ml of fresh SD-leu-tryp-his/gal/raff medium. The cells were grown to an A600 of 0.5 to 0.8; the exact A600 was recorded. A 1.5-ml aliquot of this culture was centrifuged at 14,000 × g for 30 s to harvest the cells. The cells were resuspended in 0.3 ml of buffer 1 (HEPES, 2.38%; NaCl, 0.9%; l-aspartate, 0.065%; bovine serum albumin, 1%; Tween 20, 0.05% [pH 7.3]; with a concentration factor of 5). A 0.1-ml aliquot of the cells was permeabilized by two freeze/thaw cycles in liquid nitrogen and a 37°C water bath. A solution of 0.7 ml of buffer 2 (2.23 mM CPRG in buffer 1) was added and vigorously mixed. Cells were incubated at 37°C until the samples developed a red color. To stop color development, 3 mM ZnCl2 (0.5 ml) was added. Samples were centrifuged at 14,000 × g for 1 min to pellet cell debris. The A578 was measured, and the number of β-galactosidase units was calculated with the following formula: β-galactosidase units = 1,000 × [A578/(t × V × A600)] (36, 37), where t is the elapsed time (in min) of incubation, V is 0.1× concentration factor (5), and A600 is the value for 1 ml of sample.
Expression of fusion proteins.
E. coli XL10 or BL21-CodonPlus(DE3)-RIL harboring the recombinant expression plasmids was inoculated into 10 ml of LB broth containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol and grown overnight at 37°C. The preculture was used to inoculate 200 ml of fresh LB broth containing the same antibiotics. The cells were grown at 37°C to A600s of 0.3 (7×His-Orf8, 22.4 kDa), 0.6 (MBP-Orf7ΔTMH, 78.5 kDa; 6×His-Orf1, 78.6 kDa; 6×His-Orf1N246, 31.6 kDa; and 7×His-Orf15, 33.6 kDa), 0.8 (MBP-Orf4, 65.5 kDa; 7×His-Orf10, 66.2 kDa; 7×His-Orf12, 36.1 kDa; and GST-Orf14, 42.2 kDa), or 1.0 (MBP-Orf5, 119.2 kDa; MBP-Orf10, 106.4 kDa; 7×His-Orf5, 77.8 kDa; and 7×His-Orf7, 44.1 kDa) and induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). Gene expression was induced overnight, with the exception of Orf7 fusions, which were induced only for 3 h due to the cell toxicity of the protein. MBP-Orf7ΔTMH expression was induced by the addition of IPTG to a concentration of 0.5 mM. Protein synthesis of all other proteins was induced with 1 mM IPTG. The solubility of each protein or protein fragment was assessed by harvesting the cells, resuspending them in 10 ml of lysis buffer [100 mM K2HPO4/KH2PO4, 50 mM (NH4)2SO4, 1% Triton X-100 (pH 7)], lysing them by the addition of lysozyme (1 mg/ml), and administering ultrasonic treatment. The cell debris was pelleted by centrifugation, and the supernatant was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining. Partial solubility of 7×His-Orf10 was achieved by the following procedure: the cell pellet was resuspended in 20 mM Tris-HCl, 300 mM NaCl, 1 mM EDTA (pH 7.5). Lysozyme was added to a final concentration of 1 mg/ml, and the samples were incubated on ice for 20 min and centrifuged at 16,000 × g at 4°C for 20 min. The pellet was washed twice with 50 mM EDTA (pH 8.0) and centrifuged at 16,000 × g at 4°C for 10 min. The pellet was partially dissolved in buffer A (8 M urea, 20 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole [pH 7.5]) and centrifuged at 30,000 × g at 4°C for 30 min. The supernatant was loaded onto a Ni2+ charged HiTrap chelating column (Amersham Biosciences, Freiburg, Germany) equilibrated with buffer A. The 7×His-Orf10 protein was eluted in a 10-column-volume gradient with buffer B (8 M urea, 20 mM Tris-HCl, 500 mM NaCl, 250 mM imidazole [pH 7.5]) and concentrated in a Centricon centrifugal filter unit (Millipore, Schwalbach, Germany) with a 30-kDa cutoff. The concentrated protein solution was loaded onto a HiTrap desalting column (Amersham Biosciences, Freiburg, Germany) equilibrated with 30 mM Tris-HCl, 300 mM NaCl, 20 mM MgCl2 (pH 7.5). To assess the integrity and purity of the 7×His-Orf10 protein, an aliquot of the purified protein was loaded onto a 10% SDS polyacrylamide gel, followed by Coomassie blue staining. 7×His-Orf10 was purified to approximately 80% homogeneity (J. Kopeć and E. Grohmann, unpublished data).
In vitro binding experiments.
E. coli BL21-CodonPlus(DE3)-RIL lysate or E. coli XL10 lysate with the expression plasmid (pMAL-c2X, pQTEV/pQE30, or pGEX-6P-2) containing the inserted traX gene was mixed with the putative interaction partner (E. coli lysate with pQTEV-/pQE30-, pGEX-6P-2-, or pMAL-c2X-traY or partially purified TraY protein) and incubated for complex formation for 30 to 60 min at room temperature. The complex was loaded onto amylose magnetic beads (New England Biolabs, Frankfurt am Main, Germany) and purified as specified by the manufacturer. Alternatively, the mixture of two E. coli lysates was loaded onto Ni-nitrilotriacetic acid (NTA) spin columns (QIAGEN, Hilden, Germany), and the protein complex was eluted as specified by the manufacturer, mixed with SDS sample buffer, and after heat denaturation, loaded onto SDS-polyacrylamide gels with the appropriate percentage of acrylamide (6 to 12%). Two gels were made per complex: one was stained with Coomassie brilliant blue (Merck, Darmstadt, Germany), and the other was used for Western blotting. The separated proteins were blotted onto nitrocellulose membranes (Bio-Rad, München, Germany) using liquid transfer for 1.5 h at 90 mA (Mini ProteanIII system; Bio-Rad, München, Germany). The membranes containing the transferred proteins were initially incubated in the blocking solution (QIAGEN, Hilden, Germany). The seven-histidine-tag fusion protein (six-histidine-tag fusion protein for Orf1) was detected by incubating the membrane with 5 ml of five-histidine horseradish peroxidase (HRP) conjugate (QIAGEN; dilution, 1:5,000), the MBP fusion protein was detected by incubation with 5 ml of anti-MBP HRP conjugate (New England Biolabs; dilution, 1:5,000), and the GST fusion protein was detected by incubation with 5 ml of anti-GST HRP conjugate (Roche Diagnostics; 1:5,000 dilution) for 1 h at room temperature. The signal was visualized by using the ECL Western blot detection kit (Pierce, Perbio Science, Bonn, Germany) followed by autoradiography.
Cross-linking experiment with 7×His-Orf10.
Cross-linking of 7×His-Orf10 was performed as described by Kopeć et al. (30) with minor modifications. The reaction volume of 50 μl consisted of 0.5 mg/ml protein, 100 mM Bicine (pH 7.5), 300 mM NaCl, 1 mM dithiothreitol, and various concentrations of glutaraldehyde (0.002%, 0.003%, 0.004%, 0.005%, 0.006%, 0.007%, 0.008%, and 0.01% [vol/vol]). The reaction was stopped after 15 min by the addition of 1 M glycine (pH 8.0), to a final concentration of 140 mM. The samples were incubated for another 5 min. The proteins were precipitated with 400 μl of cold acetone for 2 h at −20°C and centrifuged at 15,000 × g for 15 min at room temperature. Prior to being loaded onto a 10% (wt/vol) polyacrylamide gel in the presence of SDS, the pellets were dissolved in loading buffer consisting of 50 mM Tris-HCl (pH 6.8), 2 mM EDTA, 2% (wt/vol) SDS, 0.1% (wt/vol) bromophenol blue, 10% (vol/vol) glycerol, and 150 mM β-mercaptoethanol and heated to 95°C for 5 min. The samples were electrophoresed at a constant voltage of 180 V and stained with Coomassie brilliant blue.
RESULTS
To determine protein-protein interactions of the pIP501-encoded Tra proteins, we selected the yeast two-hybrid method that detects proteins capable of interaction with a known protein. The method has the advantage of immediate availability of the cloned genes for the interaction proteins. The interactions are identified in vivo in Saccharomyces cerevisiae through reconstitution of the activity of a transcriptional activator (13). The yeast two-hybrid screen identified 18 protein-protein interactions of pIP501-encoded Tra proteins in total; seven of these interactions were strong, resulting in more than 20 β-galactosidase units (Table 4).
TABLE 4.
pIP501 Tra protein-protein interactions detected by the yeast two-hybrid system
| Bait | Prey | Growtha | ß-Galactosidase units | Fold increase in ß-galactosidase activityb | Pull-down assay |
|---|---|---|---|---|---|
| Orf1 | Orf1 | + | 9.4 | 39.1 | |
| Orf1 | Orf10 | + | 1.1 | 4.5 | √ |
| Orf4 | Orf5 | ++ | 91.2 | 380 | √ |
| Orf5 | Orf5 | +++ | 111.8 | 465.8 | √ |
| Orf5 | Orf7 | ++ | 5.9 | 24.5 | √ |
| Orf6 | Orf3 | + | 1.1 | 4.5 | |
| Orf6 | Orf10 | +++ | 95.3 | 397 | √ |
| Orf7 | Orf2 | + | 19.2 | 80 | |
| Orf7 | Orf5 | + | 0.75 | 3.1 | √ |
| Orf7 | Orf7 | ++ | 31.8 | 132.5 | √ |
| Orf7 | Orf10 | ++ | 34 | 141.6 | √ |
| Orf7 | Orf14 | + | 13.8 | 57.5 | √ |
| Orf9 | Orf3 | +++ | 34.4 | 143.3 | |
| Orf14 | Orf5 | ++ | 7.8 | 32.5 | √ |
| Orf14 | Orf8 | ++ | 35.7 | 148.7 | √ |
| Orf14 | Orf12 | ++ | 7.2 | 30 | √ |
| Orf14 | Orf14 | ++ | 0.8 | 3.3 | |
| Orf15 | Orf12 | + | 0.9 | 3.8 |
Growth on SD-leu-tryp-his/gal/raff medium was observed after 3 days (+++), after 4 to 7 days (++), or after more than 7 days (+).
Increase of ß-galactosidase units compared with bait/prey plasmids without inserts.
Protein-protein interactions of T4SLS proteins.
The TraA DNA relaxase (Orf1) was shown to dimerize by in vitro cross-linking experiments (30). TraA self-interaction was confirmed by the yeast two-hybrid screen (Table 4).
The putative ATPase Orf5 interacts with itself and with Orf4, Orf7, and Orf14.
The VirB4-like putative ATPase Orf5 (653 amino acids) was shown to strongly interact with itself (111.8 β-galactosidase units) (Table 4). It also interacts with two non-T4SLS proteins, Orf4 and Orf14, which are predicted to be located at least partially in the cytoplasm. Orf5 also bound to the lytic transglycosylase Orf7. This interaction could possibly help recruit the putative energizing protein Orf5 to its location in the transfer complex.
The lytic transglycosylase Orf7 interacts with itself and with Orf2, Orf5, Orf10, and Orf14.
Orf7 (369 amino acids) demonstrated five different interactions with pIP501-encoded Tra proteins. The protein was shown to interact with all T4SLS proteins encoded by the pIP501 tra operon. It bound to the putative ATPase Orf5, formed homodimers, and bound to the putative coupling protein Orf10. Orf7 self-interaction is in agreement with dimerization of the lytic transglycosylase VirB1 of the Agrobacterium transfer DNA (T-DNA) transfer system shown by Ward et al. (53). Putative intermediate complex formation between Orf7 and Orf10 could aid in transporting the coupling protein to its location in the T4SLS complex. The Orf7-Orf10 interaction is an interaction specific for G+ bacteria which has not shown before for any of the T4SSs from G− bacteria. Orf7 also bound to the predicted cytoplasmic membrane protein Orf2 and to the Orf14 protein.
The putative coupling Orf10 binds to Orf1, Orf6, and Orf7.
Orf10 was excluded as a bait from the screen, as it showed autoactivation when orf10 was cloned into the bait plasmid pBTM117-c. But when Orf1, Orf6, or Orf7 was used as a bait, Orf10 associated with these proteins, showing a very strong interaction with the Orf6 protein (95.3 β-galactosidase units) (Table 4). The proposed interaction with the TraA relaxase Orf1 was not convincingly shown by the genetic screen (Table 4) and therefore had to be verified by the in vitro pull-down assay (see below) (Table 4).
Protein-protein interactions of non-T4SLS proteins.
Most interactions were shown for the Orf14 protein, five interactions in total, including those with the T4SLS proteins Orf5 and Orf7. Orf14 proved to self-associate and to bind to the putative membrane-associated Orf8 protein and the cytoplasmic membrane protein Orf12 (six or seven predicted transmembrane helices [TMH]). Orf6 bound weakly to the Orf3 protein (one or two predicted TMH). The predicted cytoplasmic membrane protein Orf9 (two TMH) interacted strongly with the Orf3 protein. Orf15 is postulated to be located in the cell wall associated with the cytoplasmic membrane protein Orf12. This interaction could enable formation of the outermost part of the DNA/protein secretion machinery.
Most of the in vivo-detected protein-protein interactions were verified by in vitro pull-down assays with differently (six-histidine-/seven-histidine-, GST-, or MBP-) tagged Tra proteins. All in vitro protein-protein interaction data are summarized in Fig. 2 and Table 4.
FIG. 2.
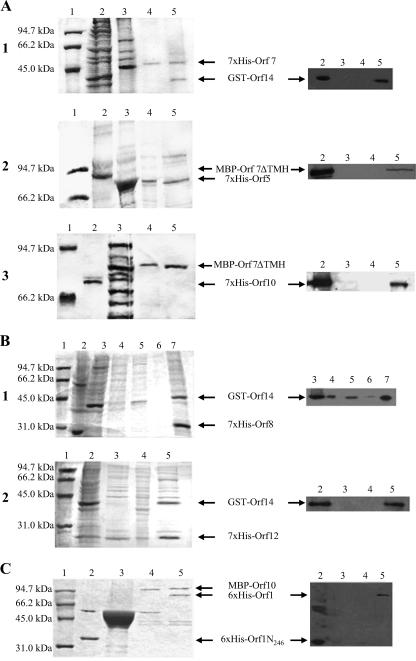
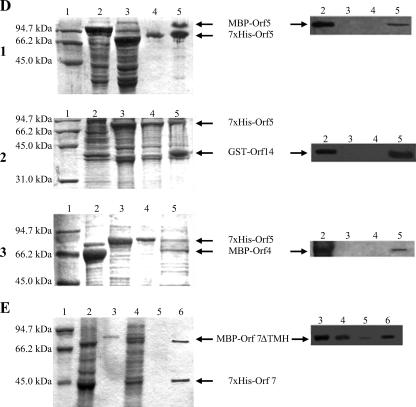
In vitro binding assays. (A to E) SDS-PAGE gels (left panels) and corresponding Western blots (right panels). (A) Interactions probably involved in recruitment of Tra proteins to the T4SLS complex. (A1) 12% SDS-PAGE. Lane 1, low-range protein standard (Bio-Rad); lane 2, GST-Orf14 lysate; lane 3, 7×His-Orf7 lysate; lane 4, negative control 7×His-Orf7/GST; lane 5, eluate of the GST-Orf14-7×His-Orf7 complex. Corresponding Western blot with anti-GST antibodies. (A2) 6% SDS-PAGE. Lane 1, low-range protein standard; lane 2, MBP-Orf7ΔTMH lysate; lane 3, 7×His-Orf5 lysate; lane 4, negative control 7×His-Orf5/MBP; lane 5, eluate of the 7×His-Orf5-MBP-Orf7ΔTMH complex. Corresponding Western blot with anti-MBP antibodies. (A3) 8% SDS-PAGE. Lane 1, low-range protein standard; lane 2, 7×His-Orf10; lane 3, MBP-Orf7ΔTMH lysate; lane 4, negative control MBP-Orf7ΔTMH/pQTEV lysate; lane 5, eluate of the MBP-Orf7ΔTMH-7×His-Orf10 complex. Corresponding Western blot with anti-Penta-His antibodies.(B) Interactions of postulated core complex components. (B1) 12% SDS-PAGE. Lane 1, low-range protein standard; lane 2, 7×His-Orf8 lysate; lane 3, GST-Orf14 lysate; lane 4, proteins not bound to Ni-NTA column; lanes 5 and 6, wash steps after binding of the protein complex; lane 7, eluate of the 7×His-Orf8-GST-Orf14 complex. Corresponding Western blot with anti-GST antibodies. (B2) 12% SDS-PAGE. Lane 1, low-range protein standard; lane 2, 7×His-Orf12 lysate; lane 3, GST-Orf14 lysate; lane 4, negative control 7×His-Orf12/GST; lane 5, eluate of the 7×His-Orf12-GST-ORF14 complex. Corresponding Western blot with anti-GST antibodies. (C) Interactions of the putative coupling protein Orf10 in a 12% SDS-PAGE gel. Lane 1, low-range protein standard; lane 2, 6×His-Orf1N246; lane 3, negative control MBP/6×His-Orf1N246; lane 4, eluate of the MBP-Orf10-6×His-Orf1N246 complex; lane 5, eluate of the MBP-Orf10-6×His-Orf1 complex. Corresponding Western blot with anti-Penta-His antibodies. (D) Interactions of the putative ATPase ORF5. (D1) 12% SDS-PAGE. Lane 1, low-range protein standard; lane 2, MBP-Orf5 lysate; lane 3, 7×His-Orf5 lysate; lane 4, negative control 7×His-Orf5/MBP; lane 5, eluate of the 7×His-ORF5-MBP-ORF5 complex. Corresponding Western blot with anti-MBP antibodies. (D2) 12% SDS-PAGE. Lane 1, low-range protein standard; lane 2, GST-Orf14 lysate; lane 3, 7×His-Orf5 lysate; lane 4, negative control 7×His-Orf5/GST; lane 5, eluate of the 7×His-Orf5-GST-Orf14 complex. Corresponding Western blot with anti-GST antibodies. (D3) 10% SDS-PAGE. Lane 1, low-range protein standard; lane 2, MBP-Orf4 lysate; lane 3, 7×His-Orf5 lysate; lane 4, negative control 7×His-Orf5/MBP; lane 5, eluate of the 7×His-Orf5-MBP-Orf4 complex. Corresponding Western blot with anti-MBP antibodies. (E) Homotypic interaction of the lytic transglycosylase Orf7 in a 12% SDS-PAGE. Lane 1, low-range protein standard; lane 2, 7×His-Orf7 lysate; lane 3, purified MBP-Orf7ΔTMH; lane 4, proteins not bound to Ni-NTA column; lane 5, wash step after binding of the protein complex; lane 6, eluate of the 7×His-Orf7-MBP-Orf7ΔTMH complex. Corresponding Western blot with anti-MBP antibodies.
Interactions probably involved in recruitment of Tra proteins to the T4SLS complex (Fig. 2A).
The interaction of the lytic transglycosylase Orf7 with the putative core complex protein Orf14 was verified by the isolation of a 7×His-Orf7-GST-Orf14 complex, association of Orf7 with the putative ATPase Orf5 by isolation of a 7×His-Orf5-MBP-Orf7ΔTMH complex. Binding of MBP-Orf7ΔTMH to the putative coupling protein Orf10 was confirmed by pulling down a complex consisting of MBP-Orf7ΔTMH and 7×His-Orf10 from the amylose magnetic beads.
Interactions of postulated core complex components.
Association of Orf8 with Orf14, thought to constitute an important part of the translocation complex in the cytoplasmic membrane, was confirmed by complex formation between 7×His-Orf8 and GST-Orf14 (Fig. 2B). Interaction of the cytoplasmic membrane protein Orf12 (six or seven predicted TMH) with Orf14 has been shown by isolation of a 7×His-Orf12-GST-Orf14 complex.
Interactions of the putative coupling protein Orf10.
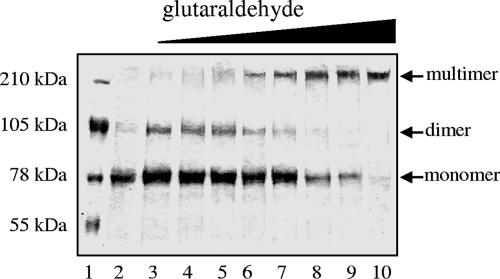
The weak interaction in the yeast system between the Orf1 (TraA relaxase) and the Orf10 coupling protein was proven in vitro by complex formation between 6×His-Orf1 and MBP-Orf10 (Fig. 2C). This interaction is a further hint for the putative coupling role of Orf10. Orf10 is thought to link the relaxosome consisting of Orf1 bound to single-stranded pIP501 DNA with the T4SLS complex. 6×His-Orf1N246, containing the 246 amino-terminal amino acids of the relaxase (the relaxase domain), did not support complex formation with MBP-Orf10. The postulated Orf10 self-association was convincingly confirmed by glutaraldehyde cross-linking. At glutaraldehyde concentrations between 0.002% and 0.005%, predominantly dimeric 7×His-Orf10 forms appeared in the 10% SDS polyacrylamide gel, whereas at a glutaraldehyde concentration of 0.01%, only multimeric forms were detectable (Fig. 3). Preliminary gel filtration experiments with purified 7×His-Orf10 confirmed the oligomeric structure of Orf10 (M. Saleh, W. Keller, and E. Grohmann, unpublished data). With the yeast two-hybrid system, Orf10 dimerization could not be tested due to autoactivation of Orf10 in fusion with the BD domain of pBTM117c. Orf10 also interacted with the lytic transglycosylase Orf7 (see above).
FIG. 3.
Glutaraldehyde cross-linking of 7×His-Orf10. Samples of 0.5 mg/ml of 7×His-Orf10 were incubated with increasing glutaraldehyde concentrations. The products were loaded onto a 10% SDS polyacrylamide gel, electrophoresed at a constant voltage of 180 V, and stained with Coomassie brilliant blue. Lane 1, SeeBlue plus2-prestained protein standards (Invitrogen, Karlsruhe, Germany); lane 2, no glutaraldehyde; lanes 3 to 10, glutaraldehyde at 0.002% (lane 3). 0.003% (lane 4), 0.004% (lane 5), 0.005% (lane 6), 0.006% (lane 7), 0.007% (lane 8), 0.008% (lane 9); and 0.01% (lane 10).
Interactions of the putative ATPase Orf5.
The most important Orf5 protein interactions could be proven by biochemical assays. Orf5 was shown to dimerize (Orf5-Orf5 was one of the strongest in vivo interactions); to interact with the soluble protein Orf4 and the lytic transglycosylase Orf7; and to associate with one of the postulated core component proteins, the Orf14 protein (Fig. 2D).
Homotypic interaction of the lytic transglycosylase Orf7 (Fig. 2E).
ORF7 could be isolated as a 7×His-Orf7-MBP-Orf7ΔTMH complex from a Ni affinity column. Orf7 could fulfill a bifunctional role in the T4SLS process: its amino-terminal specific lytic transglycosylase (SLT) domain was shown to efficiently cleave peptidoglycan (Söllü and Grohmann, unpublished data), and its carboxy-terminal portion is postulated to be released as an immunoreactive Orf7* protein. The Orf7 amino acid sequence shows significant similarity with VirB1 in the region where VirB1 is processed to VirB1* (4), which leads us to the speculation that, similarly, Orf7 could be processed to VirB1. One possible role of the processed protein could be participation in direct interactions with the recipient cell.
DISCUSSION
We have generated a comprehensive protein-protein interaction map of the pIP501-encoded Tra proteins based upon the data obtained by two different approaches, the yeast two-hybrid assay and in vitro pull-down assays. The interactions crucial for building up a speculative model of the T4SLS encoded by pIP501 were proven by both methods. On the basis of these data, together with the predicted localization of the proteins and functional characterization of some of them, we propose a possible scenario for assembly of the pIP501-encoded T4SLS (Fig. 4): the Orf7 lytic transglycosylase for which peptidoglycan cleavage activity has been demonstrated on murein substrates from different origins (Söllü and Grohmann, unpublished observations) should play a crucial role in local opening of the peptidoglycan, thereby facilitating assembly of the transenvelope transport complex. The role of Orf7 should be more pronounced than that of lytic transglycosylases encoded by T4SSs of G− bacteria because of the greater thickness and the multilayered structure of the peptidoglycan in G+ bacteria. In T4SSs of G− bacteria, such as the prototype Agrobacterium transfer DNA (T-DNA) transfer system, tumor formation was attenuated (38), and RSF1010 transfer capacity was reduced approximately 10-fold (9) in a virB1 mutant. In the F-like R1-19 transfer system, conjugative transfer frequency was reduced 5- to 10-fold (5, 6) in mutants affecting the lytic transglycosylase P19. In none of the mutants was transfer proficiency completely abolished, indicating that lytic transglycosylases encoded on the bacterial chromosome might have replaced the plasmid-encoded enzyme. The chromosomes of G+ bacteria encode only a limited number of muramidases, which potentially could replace the plasmid-encoded lytic transglycosylase activity. This hypothesis is currently being tested on a pIP501 orf7 knockout mutant. Orf7 associates with itself and interacts with other T4SLS components, such as the putative ATPase Orf5 and the putative Orf10 coupling protein. The latter interaction seems to be specific for G+ bacteria, as it has not been shown for any T4SS coupling protein from G− bacteria. Furthermore, Orf7 interacts with the postulated cytoplasmic membrane protein Orf2 and the Orf14 protein. For the Orf2 protein, this association is the only interaction demonstrated until now. By interaction with Orf5, Orf10, and Orf14, Orf7 might recruit these proteins and enable their incorporation in the transport apparatus. At the N terminus, Orf7 possesses a potential signal peptide sequence (SignalP 3.0) (8), and at the C terminus, it shows high similarity with the COG3942 family of surface antigens (score, 108; E value, 1e−24). Several secreted or possibly secreted proteins from Streptococcus and Staphylococcus spp. belong to this family. This observation fits well with putative secretion of the C terminus of Orf7 (Orf7*) after cleavage at a processing site similar to that in VirB1, located between the SLT domain and the carboxyl terminus with similarity to COG3942 family proteins.
FIG. 4.
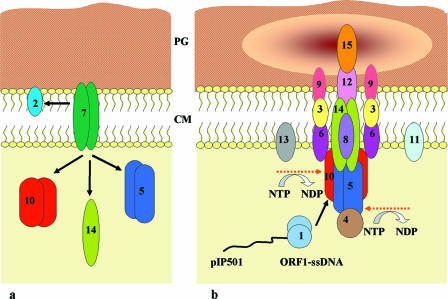
Working model for pIP501 conjugative transfer. The postulated DNA secretion complex is assembled in a manner reminiscent of a simplified T4SS. Arrows indicate protein-protein interactions determined with the yeast two-hybrid system. Protein localization is consistent with computer predictions made by using Psort (22), PHDhtm (42), HMMTOP (50, 51), TMPred (28), TMAP (39, 40), and TopPred (16, 52) (available at www.expasy.org). Decreased shading of peptidoglycan (PG) symbolizes Orf7-mediated local opening of PG. (a) Protein-protein interactions detected for Orf7. (b) Assembly of the putative pIP501 transport apparatus. Numbers refer to proteins specified by the pIP501 tra region. Dashed arrows mark putative ATPases. CM, cytoplasmic membrane; ssDNA, single-stranded DNA; NTP, nucleoside triphosphate; NDP, nucleoside diphosphate.
The putative coupling protein Orf10, which interacts with the relaxase TraA (Orf1), could link the relaxosome, consisting of TraA bound at oriT of the pIP501 T strand, with the transport apparatus. The TraA relaxase was shown to be a dimer in solution and to preferentially bind in a dimeric form to its cognate oriT (30). Orf10 oligomerization could be shown by glutaraldehyde cross-linking and gel filtration (data not shown). A three-dimensional structure prediction of Orf10 based upon the known TrwB structure (coupling protein of R388) (24, 25) showed significant similarity of the two structures (J. Kopeć, W. Keller, and E. Grohmann, unpublished data).
One possible candidate for delivering energy by ATP hydrolysis for establishment of the DNA transfer machinery and for the transport process would be the VirB4-like Orf5 protein. Alternatively, energy could be supplied by the putative coupling protein Orf10. ATP-binding and hydrolysis tests are in progress for both proteins to investigate this assumption. Preliminary experiments showed ATP-binding and ATPase activities for both Orf5 and Orf10 (R. Salih, M. Saleh, M. Abajy, J. Kopeć, and E. Grohmann, unpublished results). A possible transenvelope structure could be built up by the Orf8, Orf14, Orf12, and Orf15 proteins. Orf8 and Orf15 contain potential signal peptides at their N termini; for Orf8, a TMH in the N-terminal portion was postulated. Depending on the algorithm applied, for ORF15, one or two TMH were found (one at the amino terminus and another one at the carboxy terminus). The PSORTb v.2.0 program (22) suggested cell wall localization for the Orf15 protein. Therefore, Orf15 could possibly make up the outermost portion of the transport complex. For Orf12, five to seven TMH have been postulated. Orf12 and Orf14 were analyzed with the Secretome 2.0 prediction tool (7) (www.cbs.dtu.dk/services/SecretomeP-2.0/), which predicts nonclassically secreted proteins (proteins without a signal peptide). For both proteins, a high SecP score for nonclassically secreted proteins was obtained (threshold, 0.5), namely, 0.94 for Orf12 and 0.84 for Orf14.
The proteins Orf3, Orf6, and Orf9, which all possibly contain TMH, could build part of the scaffold structure of the secretion apparatus. Orf9 also contains a possible signal peptide sequence with a high probability of cleavage (0.92). Orf6 showed strong interaction (95.3 β-galactosidase units) with the putative coupling protein Orf10. For two proteins with postulated TMH, Orf11 and Orf13, no interactions could be detected so far. Both proteins show no significant similarity to any characterized protein in the data bank. The soluble protein Orf4, with no significant match in the data bank, showed high affinity (91.2 β-galactosidase units, Table 4) for the Orf5 protein in the yeast two-hybrid screen. A possible physiological significance of this association has to be further investigated.
Comparisons of the plasmid-encoded conjugative transfer systems in G+ bacteria revealed two general mechanisms governing conjugative plasmid transfer, namely (i) a T4SLS mechanism such as that proposed for the broad-host-range plasmids pIP501, pRE25, pMRC01, and pSK41/pGO1 (26, 33, 34); the Clostridium perfringens plasmid pCW3 (3); and the E. faecalis sex pheromone plasmids as exemplified by pCF10 (31); and (ii) a completely distinct mechanism exerted by the multicellular G+ bacteria (26, 41).
In G+ bacteria, the close contact between donor and recipient cells preceding conjugative transfer is thought to be established without the help of pili. With the exception of the Enterococcus sex pheromone plasmids, the mechanism of establishing physical contact between G+ donor and recipient cells is not known (reviewed in references 11 and 19). Interestingly, last year, pili were characterized in all three of the principal streptococcal pathogens that cause invasive disease in humans, Streptococcus pyogenes, S. agalactiae, and S. pneumonia (reviewed in reference 48). In G− bacteria, pili are typically formed by noncovalent interactions between pilin subunits. By contrast, the recently discovered pili in G+ pathogens are formed by covalent polymerization of adhesive pilin subunits. Pili of G+ pathogens are likely to have a function similar to that in G− bacteria, where they play a key role in the adhesion and invasion process and in pathogenesis. In G− bacteria, only type IV pili were shown to allow the transfer of genetic material (12). Type IV pilin-like proteins have also been demonstrated to be involved in genetic transformation of G+ bacteria (reviewed in reference 2). Although no pilin-like genes have been found on conjugative plasmids with a G+ bacterial origin, participation of pilus-like structures in the conjugative transfer process of G+ bacteria cannot be excluded to date.
In summary, we have presented a first model for a T4SL transfer system encoded by the broad-host-range plasmid pIP501 with self-transmission capability to a wide variety of G+ bacteria and to G− E. coli. The further elucidation of the pIP501 conjugative transfer mechanism should also aid in deciphering the transfer process of the related staphylococcal and enterococcal antibiotic resistance plasmids, enabling the development of specific agents inhibiting T4SLS processes in G+ pathogens.
Acknowledgments
We thank E. Lanka for critical reading of the paper; K. Büssow, Max Planck Institute for Molecular Genetics, Berlin, Germany, for the gift of plasmid pQTEV; and Ralf Sopora for skillful technical assistance. We acknowledge M. Saleh and W. Keller for the gel filtration experiments.
M.Y.A. is a recipient of a doctoral fellowship from the University of Aleppo, Syria. J.K. received a scholarship from Berliner Programm zur Förderung der Chancengleichheit für Frauen in Forschung und Lehre. This work was partially supported by the Austrian Science Foundation (FWF project P15040).
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averhoff, B. 2004. DNA transport and natural transformation in mesophilic and thermophilic bacteria. J. Bioenerg. Biomembr. 36:25-33. [DOI] [PubMed] [Google Scholar]
- 3.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 188:4942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, C., M. Llosa, S. Zhou, and P. C. Zambryski. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J. Bacteriol. 179:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, M., R. Eferl, G. Zellnig, K. Teferle, A. Dijkstra, G. Koraimann, and G. Högenauer. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentheiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of P19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183:3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., L. Kiemer, A. Fausboll, and S. Brunak. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. J. Mol. Biol. 340:783-795.15223320 [Google Scholar]
- 9.Bohne, J., A. Yim, and A. N. Binns. 1998. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc. Natl. Acad. Sci. USA 95:7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50:643-650. [DOI] [PubMed] [Google Scholar]
- 11.Chandler, J. R., and G. M. Dunny. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 25:1377-1388. [DOI] [PubMed] [Google Scholar]
- 12.Chen, I., and D. Dubnau. 2003. DNA transport during transformation. Front. Biosci. 8:5544-5556. [DOI] [PubMed] [Google Scholar]
- 13.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie, P. J., and E. Cascales. 2005. Structural and dynamic properties of bacterial type IV secretion systems (review). Mol. Membr. Biol. 22:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 17.de Paz, H. D., F. J. Sangari, S. Bolland, J. M. Garcia-Lobo, C. Dehio, F. de la Cruz, and M. Llosa. 2005. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiology 151:3505-3516. [DOI] [PubMed] [Google Scholar]
- 18.Ding, Z., Z. Zhao, S. J. Jakubowski, A. Krishnamohan, W. Margolin, and P. J. Christie. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 184:5572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunny, G. M., M. H. Antiporta, and H. Hirt. 2001. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides 22:1529-1539. [DOI] [PubMed] [Google Scholar]
- 20.Evans, R. P., Jr., and F. L. Macrina. 1983. Streptococcal R plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives. J. Bacteriol. 154:1347-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields, S., and R. Sternglanz. 1994. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 10:286-292. [DOI] [PubMed] [Google Scholar]
- 22.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 23.Gilmour, M. W., J. E. Gunton, T. D. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 24.Gomis-Ruth, F. X., and M. Coll. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33:839-843. [DOI] [PubMed] [Google Scholar]
- 25.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 26.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris, R. L., and P. M. Silverman. 2004. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J. Bacteriol. 186:5480-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann, K., and W. Stoffel. 1993. TMBASE—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 29.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopeć, J., A. Bergmann, G. Fritz, E. Grohmann, and W. Keller. 2005. TraA and its N-terminal relaxase domain of the Gram-positive plasmid pIP501 show specific oriT binding and behave as dimers in solution. Biochem. J. 387:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozlowicz, B. K., M. Dworkin, and G. M. Dunny. 2006. Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity? Int. J. Med. Microbiol. 296:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurenbach, B., C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann. 2003. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 50:86-93. [DOI] [PubMed] [Google Scholar]
- 33.Kurenbach, B., J. Kopeć, M. Mägdefrau, K. Andreas, W. Keller, C. Bohn, M. Y. Abajy, and E. Grohmann. 2006. The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology 152:637-645. [DOI] [PubMed] [Google Scholar]
- 34.Llosa, M., and F. de la Cruz. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 156:1-6. [DOI] [PubMed] [Google Scholar]
- 35.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Mushegian, A. R., K. J. Fullner, E. V. Koonin, and E. W. Nester. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. USA 93:7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persson, B., and P. Argos. 1994. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237:182-192. [DOI] [PubMed] [Google Scholar]
- 40.Persson, B., and P. Argos. 1996. Topology prediction of membrane proteins. Protein Sci. 5:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuther, J., C. Gekeler, Y. Tiffert, W. Wohlleben, and G. Muth. 2006. Unique conjugation mechanism in mycelial streptomycetes: a DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol. Microbiol. 61:436-446. [DOI] [PubMed] [Google Scholar]
- 42.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Scheich, C., F. H. Niesen, R. Seckler, and K. Büssow. 2004. An automated in vitro protein folding screen applied to a human dynactin subunit. Protein Sci. 13:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 46.Schröder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 47.Shamaei-Tousi, A., R. Cahill, and G. Frankel. 2004. Interaction between protein subunits of the type IV secretion system of Bartonella henselae. J. Bacteriol. 186:4796-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 49.Terradot, L., N. Durnell, M. Li, D. Li, J. Ory, A. Labigne, P. Legrain, F. Colland, and G. Waksman. 2004. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol. Cell. Proteomics 3:809-819. [DOI] [PubMed] [Google Scholar]
- 50.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 51.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 52.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 53.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]