Abstract
We have identified the following events during the late stage in the mother cell in Bacillus subtilis: spore detachment from the polar site of the mother cell, membrane rupture, cell wall collapse, and release of the free spore. The membrane rupture was followed by mother cell lysis. Moreover, we found that NucB, an extracellular nuclease, is involved in DNA degradation after mother cell lysis.
Programmed cell death is a phenomenon of widespread biological importance and is recognized to play a role in morphogenesis by eliminating unnecessary cells. Most studies on cell death have been carried out with vertebrates (5), and little attention has been given to the process of programmed cell death in unicellular organisms, although cell suicide responses have been reported to occur in prokaryotic and unicellular eukaryotic cells (6).
The gram-positive bacterium Bacillus subtilis possess two characteristics which make it an attractive microorganism for the study of cell death at the prokaryote level: first, like Escherichia coli, it continuously divides by binary fission into two functionally and structurally identical daughter cells; second, it has a simple developmental cycle called sporulation (1, 2). In response to nutrient deprivation, B. subtilis forms dormant and environmentally resistant spores. The spore attains a state of dormancy, which is maintained until the return to a favorable environment induces spore germination and further growth. Formation of a spore takes about 8 h at 37°C and is controlled by a genetically regulated process. During sporulation, the timing of spore maturation is coordinated by continuous close collaboration between two cells, the mother cell and forespore. The mother cell engulfs the future daughter cell and eventually actively lyses prior to release of the spore. However, previous studies, which have focused on cell wall lysis in the mother cell death process, have provided only a limited understanding of the process of mother cell death during sporulation. Therefore, the initial goal of this study was to characterize the mother cell death process by cytological studies using time-lapse fluorescence microscopy.
Mother cell death observed in liquid culture.
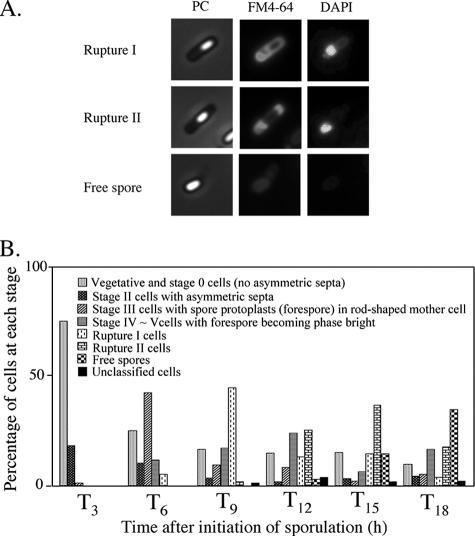
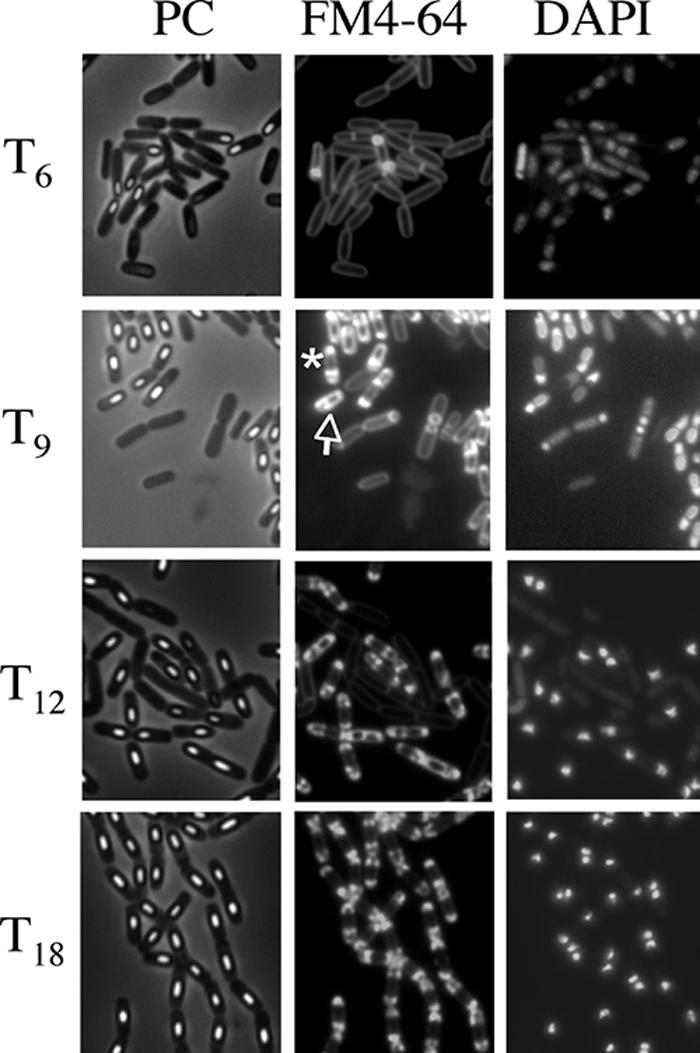
The morphological characteristics of mother cell death in B. subtilis 168 were observed when the cells were allowed to develop in shaking liquid culture at the late stage of sporulation. At 3 to 18 h after initiation of sporulation (T3 to T18), the cultures were collected, and then the membranes and DNA were stained with dyes FM4-64 (a membrane-impermeative fluorescent lipophilic styryl dye) (2.5 μg/ml for 10 min) and DAPI (4′,6′-diamidino-2-phenylindole) (a membrane-permeative fluorescent dye) (1 μg/ml for 10 min), respectively (11). Cells with asymmetric septa and cells with engulfing sporangia within the rod-shaped mother cells at T3 and T6 were visualized using the fluorescence of FM4-64 (Fig. 1B and E). The forespores in sporangia that had completed engulfment were not stained by FM4-64. However, fluorescence in the outline of the forespore and (intense) dot-like fluorescence appeared in the mother cell compartment of sporangia containing a phase-bright forespore at T9 to T12 (Fig. 1H, K, and N). We interpret this to mean that the mother cell membrane ruptured and that this event allowed FM4-64 into the mother cell cytosol, where it most probably interacted with the aggregated membrane, which caused it to fluoresce. These cells could be divided into two major types: (i) a cell with a forespore localized to one pole of the cell and with (intense) dot-like fluorescence at the other side (rupture I) and (ii) a cell with a forespore at the center of the mother cell and with intense fluorescence on both sides in the cell (rupture II). Typical cells and the total fractions of these types of cells are shown in Fig. 2. The fraction of rupture II cells tended to increase with a decreasing amount of rupture I cells (Fig. 2B). The features of theses two types of cells might be explained by supposing that membrane rupture first occurs at the opposite side of the mother cell from where the forespore is localized to one pole of the cell. The forespore may prevent the observation of fluorescence at that end of the cell until it detaches from the pole of the cell.
FIG. 1.
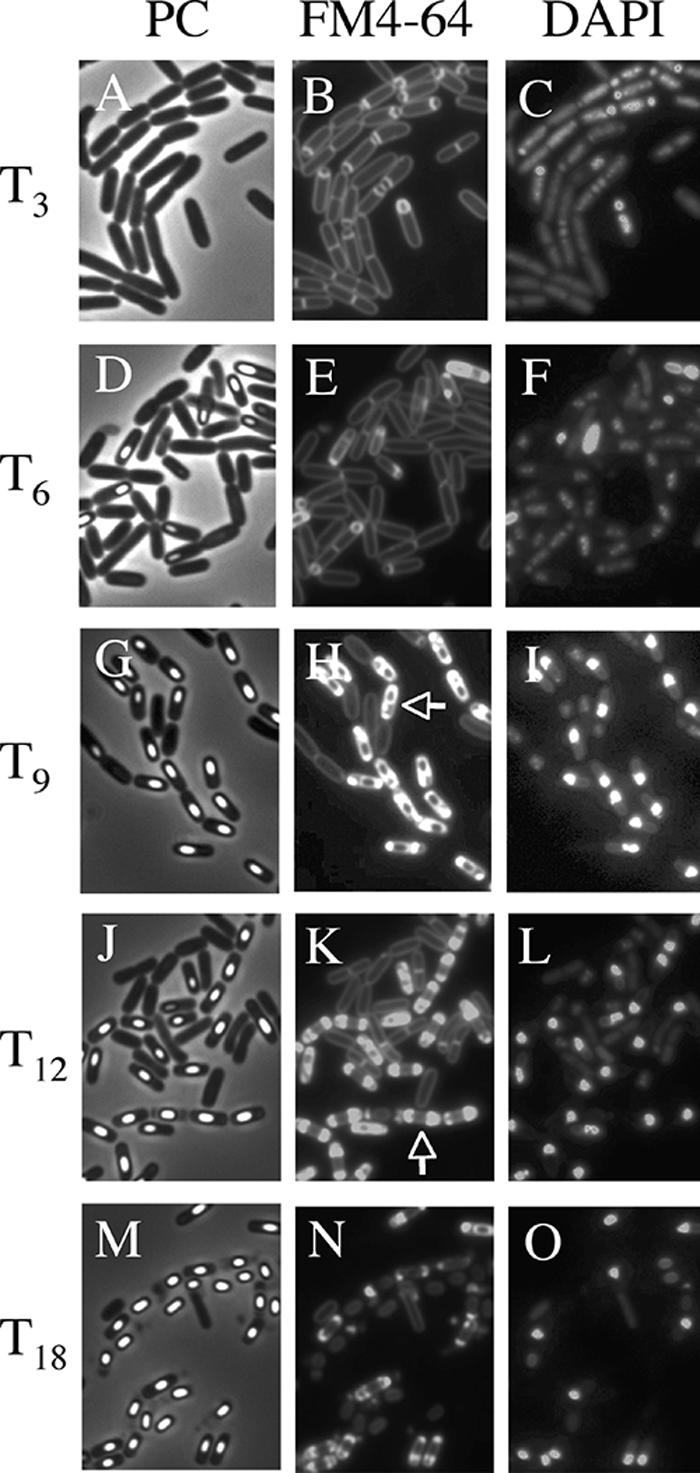
Images of mother cell in cells from liquid culture at the late stages of sporulation. Cells from strain 168 were grown to T3 (A, B, and C), T6 (D, E, and F), T9 (G, H, and I), T12 (J, K, and L), and T18 (M, N, O) in resuspension medium (8, 14). Panels A, D, G, J, and M show phase-contrast (PC) images; panels B, E, H, K, and N and panels C, F, I, L, and O show the corresponding FM4-64 staining and DAPI staining images, respectively. DAPI fails to stain the developed forespore (bright spore) DNA, but it can stain the mother cell DNA. Representative cells with intense fluorescence on one side (rupture I) (H) and both sides (rupture II) (K) are indicated with arrows.
FIG. 2.
Accumulation of membrane-disrupted cells at the late stages of sporulation. Cells from strain 168 were grown to T3, T6, T9, T12, and T18 in resuspension medium. (A) Images of typical mother cell-lysed cells. In rupture I, the cell contained the developed forespore fixed on the polar site in the mother cell, and disrupted membrane was detected on the other side in the mother cell. In rupture II, the cell contained the developed forespore detached on the polar site in the mother cell, and disrupted membrane was detected on both sides in the mother cell. PC, phase-contrast. (B) Quantification of morphological categories in the cell population at the late stages of sporulation in resuspension medium. Altogether, over 800 cells were counted.
Mother cell death observed in solid culture by time-lapse microscopy.
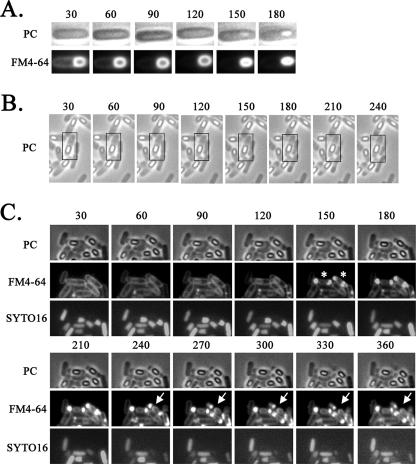
To monitor the mother cell death process at the single-cell level without shaking, the process was observed in microculture by time-lapse microscopy. Late sporulating cells were incubated with vital staining dyes FM4-64 and SYTO16 (a DNA staining dye, i.e., a fluorescent DNA binding probe that permeates the cell membrane) in shaking liquid culture and then were transferred to thin agar containing the same staining dyes overlaid on a microscope slide beneath a coverslip. The cells were then monitored over time in the microculture by fluorescence microscopy. The cells were grown at 30°C, resulting in slower sporulation than that which occurred under the conditions shown in Fig. 1 (37°C). As a control, under the conditions of growth in the microculture, spore development was successfully monitored by phase-contrast microscopy, revealing the conversion of the phase-dark forespore to a phase-bright (refractive) forespore (Fig. 3A). We next monitored stage IV and V cells containing a phase-bright forespore in the microculture transferred from liquid culture. Interestingly, as shown in Fig. 3B, we observed cells containing the phase-bright forespore moving to the center of the mother cell from one pole of the cell, a phenomenon we call detachment. In order to monitor the integrity of the cell membrane and DNA in this process, we also observed the cells stained with FM4-64 and SYTO16 for a longer period (360 min). As shown in Fig. 3C, at 150 min, cells with intense fluorescence on both sides in the cell (rupture II) were observed. Coincidentally, disappearance of the SYTO16 fluorescence was observed in such cells. Eighty-eight percent (36/41) of stage IV and V cells were shown to undergo such a process (Fig. 3C). After 270 min, the intense fluorescence appeared to decay, as if the ruptured membrane materials “leaked” out from the cell poles (Fig. 3C and Fig. 4). These results suggest that the membrane rupture and DNA disappearance occur before cell wall disruption (we call this cell wall collapse), which causes a definitive distortion of the cell shape. The manner of the membrane rupture of the cells in the microculture on the slide glass (for time-lapse studies) was slightly different from that of the cells incubated in the shaking liquid culture. In the case of cells incubated in the microculture, bright spores appeared to be detached from the membrane of the mother cell before the membrane rupture event, whereas in the shaking liquid culture, bright spores appeared to be detached from the mother cell membrane after the membrane rupture event (Fig. 2A, rupture II). This membrane rupture prior to the detachment may have been caused by the shaking stress of the culture during incubation. We therefore suppose that the mother cell death process observed in the microculture more closely resembles the natural process than that observed in the shaking liquid culture. On the other hand, in the microculture, the DNA fluorescence after staining with SYTO16 disappeared following the membrane rupture, whereas the fluorescence caused by staining with DAPI in the ruptured mother cell was still observed in the shaking liquid culture even after the rupture event. However, we observed a significant decrease in the intensity of DNA fluorescence upon staining with SYTO16 even in the membrane-ruptured cells incubated in the shaking liquid culture (data not shown). Therefore, the difference in the behavior of the DNA in shaken and unshaken cultures could be due to the use of two different DNA stains (DAPI and SYTO16) in the experiments using the two different culture conditions. The disappearance of DNA fluorescence with SYTO16 staining may have been due to some conformational change of DNA followed by membrane rupture affecting the affinity to SYTO16 in these experimental conditions.
FIG. 3.
Time-lapse microscopy of the mother cell death process. Strain 168 was monitored beginning at 3 h (A), 9 h (B), and 13 h (C) after the start of sporulation. Time-lapse intervals are indicated in minutes from the first observation time. For each time-lapse sequence, phase-contrast (PC) images are shown alone (B) or in the upper row (A and C), corresponding fluorescence images obtained with the membrane-staining dye FM4-64 are shown in the lower row (A) or middle row (C), and images obtained with the DNA-staining dye SYTO16 are shown in the lower row in panel C. SYTO16 fails to stain the developed forespore (bright spore) DNA, but it can stain the mother cell DNA. Time-lapse microscopy was performed essentially as described by Price and Losick (12). Cells were grown and induced to sporulate at 37°C by the resuspension method, in which the resuspension medium contained FM4-64 (final concentration, 0.5 μg/ml) and SYTO16 (final concentration, 0.1 μM). Portions (20 μl) of each culture were mounted on an agarose bed containing FM4-64 and SYTO16. Phase-contrast and fluorescence images were acquired every 30 min over the course of the time-lapse experiments. The time-lapse experiments were carried out at 30°C; representative cell containing the phase-bright forespore moving to the center of the mother cell from one pole of the cell are shown in boxes (B). Rupture II cells at 150 min are indicated with asterisks, and the positions at which to look for leakage of FM4-64 staining are indicated with arrows (C).
FIG. 4.
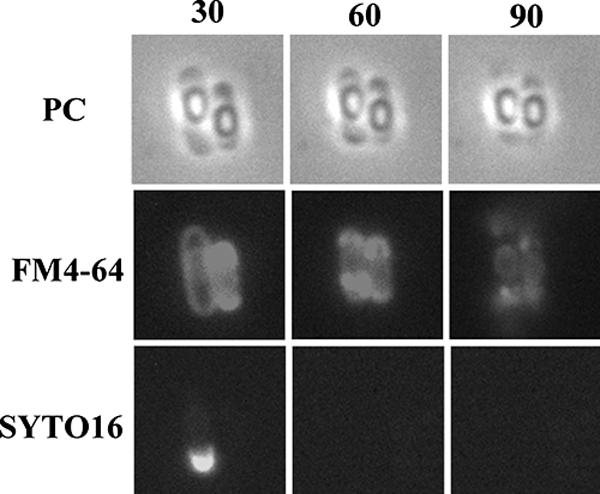
Final process of mother cell death. Typical cells in which the mother cell membrane appeared to have burst are shown. Strain 168 was monitored beginning at T13. Time-lapse intervals are indicated in minutes from the first observation time. Phase-contrast (PC) images are shown in the upper row, and corresponding fluorescence images with membrane-staining dye FM4-64 and DNA-staining dye SYTO16 are shown in the middle and lower rows, respectively.
We next investigated the relationship between the membrane rupture and the cell wall lysis in the mother cell death process. Previously, Nugroho et al. (9) showed that a cwlB (lytC) cwlC cwlH triple mutation led to a significant decrease of mother cell lysis. The products of these genes are N-acetylmuramoyl-l-alanine amidases involved in cleaving peptidoglycans. In particular, cwlC and cwlH are transcribed by EσK, which directs late mother cell-specific gene expression. We observed morphological changes in the triple mutant at late stages of sporulation. Although the triple mutant (strain CWLBCH) (Table 1) cells that produced a phase-bright spore had a bright spore in the mother cell, rupture I and rupture II cells were observed among T9 to T15 cells (Fig. 5). Moreover, trapped DAPI DNA fluorescence in the thick cell wall was also observed in the mutant, and the appearance of membrane rupture observed in cwlBCH mutant cells coincided with those in the wild-type strain (Fig. 5), suggesting that the mother cell membrane is ruptured before the cell wall lysis.
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Source or constructiona |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| CWLBn | trpC2 cwlB::pUCNcwlB | pUCNcwlBb→168 |
| CWLCc | trpC2 cwlC::pCAcwlC | pCAcwlCc→168 |
| CWLHd | trpC2 cwlH::pMUTincwlH | pMUTincwlHd→168 |
| CWLBC | trpC2 cwlB::pUCNcwlB cwlC::pCAcwlC | CWLBn→CWLCc |
| CWLBCH | trpC2 cwlB::pUCNcwlB cwlC::pCAcwlC cwlH::pMUTincwlH | CWLHd→CWLBC |
| NUCBs | trpC2 nucB::pUCSnucB | pUCSnucBe→168 |
Arrows indicate transformation from donor DNA to the recipient strain.
First, to obtain the vector pUCN192 harboring the bla and neo genes, a PstI/EcoRI DNA fragment carrying the neo gene of pBEST501 (4) was blunted by treatment of the DNA with T4 DNA polymerase in the presence of deoxynucleoside triphosphates and then ligated with DNA containing the T4 DNA polymerase-treated NdeI site of pUC19 (17). pUCNcwlB is a derivative of an integrational plasmid, pUCN192, carrying a 188-bp internal segment of cwlB that was generated with primers cwlBF (5′-CCCAAGCTTAATGTGTTTTCTGGGGC-3′ [underlining indicates a restriction site]) and cwlBR (5′-CGCGGATCCTAGTGTAAAGCAATGGC-3′).
pCAcwlC is a derivative of an integrational plasmid, pCA191 (7), carrying a 1,176-bp internal segment of cwlC that was generated with primers cwlCF (5′-CGCGGATCCTTTTATTGATCCTGGCC-3′) and cwlCR (5′-TGCTCTAGAAATCTGCTCCCCAGTTA-3′).
pMUTincwlH is a derivative of an integrational plasmid, pMUTinT3 (10), carrying a 162-bp internal segment of cwlH that was generated with primers cwlHF (5′-AAGAAGCTTTAATCGTCCTGGCTATG-3′) and cwlHR (5′-GGAGGATCCATTGTTTTCCTTCATCA-3′).
pUCSnucB is a derivative of an integrational plasmid, pUCS192 (3), carrying a 93-bp internal segment of nucB that was generated with primers nucBF (5′-CCCAAGCTTCCTGTTTCTTGCTGCAG-3′) and nucBR (5′-CGCGGATCCCTAATATGACTGCCGGT-3′).
FIG. 5.
Membrane disruption in the cwlBCH mutant. Cells from strain CWLBCH were grown to T6, T9, and T12 in resuspension medium. Representative cells with intense fluorescence on one side (rupture I) or both sides (rupture II) at T9 are indicated with an arrow and an asterisk, respectively. PC, phase-contrast.
Fate of mother cell DNA after mother cell death.
We next investigated the fate of chromosomal DNA after mother cell death in the wild-type strain. NucB is an extracellular DNase whose transcription is regulated by the mother cell-specific sigma factor σE (16). We first measured the DNase activity in the culture media of T9 wild-type and nucB mutant cells. As shown in Fig. 6A, the DNase activity was detected in the culture medium from the wild-type cells but not in that from the nucB mutant. This result shows that NucB acts as a major extracellular DNase during the late stage of sporulation. Next, to determine if the nucB nuclease has a role as a DNase that degrades DNA from the mother cell, we tried to detect the mother cell-specific release of DNA from the membrane-ruptured cells. At a late stage in the mother cell, DNA rearrangement is catalyzed by a site-specific recombinase, SpoIVCA, and creates a new composite gene, sigK, by the fusion of the spoIIIC and spoIVCB genes (13, 15). We examined the presence of the rearranged sigK gene in released mother cell DNA by PCR using two primers, sigKF (5′-GCAGAGGACTTAATGTCC-3′, +226 [the 3′-end position corresponding to the nucleotide numbers from the initiation codon of sigK]) and sigKR (5′-GTATCTATCACGTCTTC-3′, +476), whose sequences are located in spoIVCB and spoIIIC, respectively. As shown in Fig. 6B, a small amount of amplified DNA was detected in the culture medium of the wild-type strain. We interpret this to mean that partial lysis occurred in some T9 cells, allowing mother cell-derived DNA into the cell supernatant, since no free spores were observed at this time (Fig. 2B). However, a significantly larger amount of amplified DNA was detected in the culture medium of the nucB mutant (Fig. 6B). We also determined by using quantitative PCR that the ratio of the amount of DNA in the nucB mutant culture to that in the wild-type culture was 1,000 (data not shown). These results showed that the released mother cell DNA was digested mainly by NucB nuclease, indicating that this is a major extracellular DNase during sporulation. As mentioned above, a cwlB cwlC cwlH triple mutation significantly blocks the cleavage of the peptidoglycan of the mother cell during sporulation. To examine the presence of DNA in the culture medium from the triple mutant, we performed PCR assays, but no amplified sigK gene derived from the mother cell was detected (Fig. 6B). It seems likely that the reason no mother cell-derived DNA was detected in the supernatant of the triple cwlBCH mutant was the failure of these cells to lyse. It is interesting to note that the expression of nucB is dependent on the mother cell-specific sigma factor σE (16), showing that the NucB protein is produced before the time of mother cell death. We further investigated whether the disappearance of DNA fluorescence in the membrane-ruptured cells was caused by the NucB nuclease. The disappearance of SYTO16-stained signals occurred even in the membrane-ruptured cells of the nucB mutant (data not shown). This finding indicated that the intensity of SYTO16 fluorescence was decreased in accordance with the membrane rupture and was independent of NucB DNase activity. However, it is obvious that NucB has an important role in the degradation of mother cell DNA after cell lysis (Fig. 6A and B). It is intriguing that the mother cell DNA after cell lysis is digested by the nucB product, which is secreted prior to mother cell lysis. The resulting digested nucleotides may be utilized for growth of the nonsporulated cells in the culture.
FIG. 6.
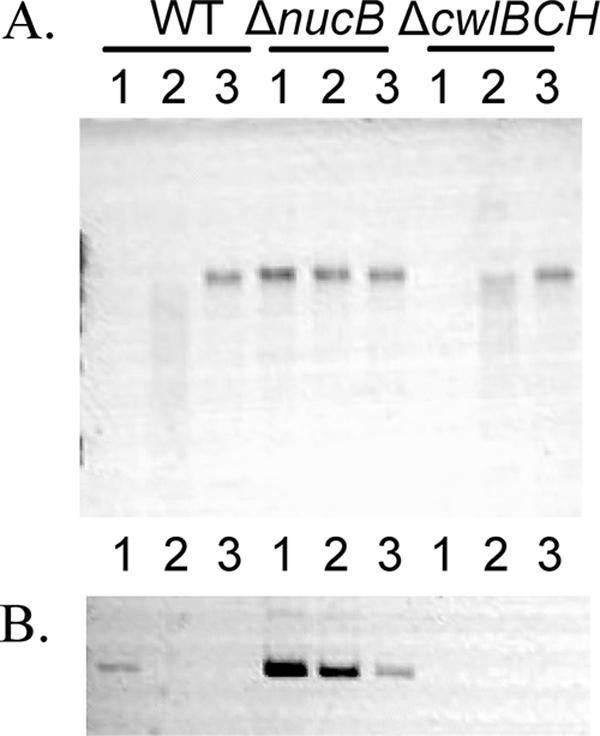
Mother cell DNA after cell lysis. (A) DNase activity of T9 cell culture medium from strains 168 (wild type [WT]), NUCBs (ΔnucB), and CWLBCH (ΔcwlBCH). Culture of each strain was centrifuged, and cells in the supernatant were removed by filtration (0.22 μm). B. subtilis chromosomal DNA (0.6 μg) was added to 8 μl of each filtered supernatant (lane 1) or diluted supernatant (1:10, lane 2; 1:100, lane 3) isolated from T9 cultures, incubated at 37°C for 1 h, and then analyzed by ethidium bromide-stained 0.8% agarose electrophoresis. (B) Detection of mother cell DNA after lysis. The mother cell-specific DNA (the sigK gene) was amplified by PCR with primers sigKF and sigKR and analyzed by 0.8% agarose electrophoresis. The template DNA sample consisted of 1, 1/10, or 1/100 μl of the filtered supernatant (WT, ΔnucB, and ΔcwlBH). PCR amplification was performed with 30 cycles of denaturation at 93°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min.
The ability to sporulate depends on the availability of sufficient energy reserves to complete the developmental process, and this energy source may run out in the mother cell at the time of spore maturation. Therefore, the timing of mother cell death might be determined by the level of energy reserves at the initiation of sporulation. Although the large and complex regulatory network system of sporulation has been well documented by many molecular biologists, very little quantitative data on mother cell death are available. Further analysis of the relationship between spore formation and mother cell death will provide another milestone on the road to the comprehensive description of the program of this cellular differentiation phenomenon.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan and by a grant from Takano Life Science Research Foundation (Japan).
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 2.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosoya, S., K. Asai, N. Ogasawara, M. Takeuchi, and T. Sato. 2002. Mutation in yaaT leads to significant inhibition of phosphorelay during sporulation in Bacillus subtilis. J. Bacteriol. 184:5545-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson, M. D., M. Weil, and M. C. Raff. 1997. Programmed cell death in animal development. Cell 88:347-354. [DOI] [PubMed] [Google Scholar]
- 6.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami, T., K. Haga, M. Takeuchi, and T. Sato. 2002. Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J. Bacteriol. 184:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom.
- 9.Nugroho, F. A., H. Yamamoto, Y. Kobayashi, and J. Sekiguchi. 1999. Characterization of a new sigma-K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J. Bacteriol. 181:6230-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogasawara, N. 2000. Systematic function analysis of Bacillus subtilis genes. Res. Microbiol. 151:129-134. [DOI] [PubMed] [Google Scholar]
- 11.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. R. Perez, Y.-L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price, K. D., and R. Losick. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181:781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato, T., Y. Samori, and Y. Kobayashi. 1990. The cisA cistron of Bacillus subtilis sporulation gene spoIVCA encodes a protein homologous to a site-specific recombinase. J. Bacteriol. 172:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stragier, P., B. Kunkel, L. Kroos, and R. Losick. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507-512. [DOI] [PubMed] [Google Scholar]
- 16.van Sinderen, D., R. Kiewiet, and G. Venema. 1995. Differential expression of two closely related deoxyribonuclease genes, nucA and nucB, in Bacillus subtilis. Mol. Microbiol. 15:213-223. [DOI] [PubMed] [Google Scholar]
- 17.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]