Abstract
The periplasmic α-carbonic anhydrase of Helicobacter pylori is essential for buffering the periplasm at acidic pH. This enzyme is an integral component of the acid acclimation response that allows this neutralophile to colonize the stomach. Transcription of the HP1186 α-carbonic anhydrase gene is upregulated in response to low environmental pH. A binding site for the HP0166 response regulator (ArsR) has been identified in the promoter region of the HP1186 gene. To investigate the mechanism that regulates the expression of HP1186 in response to low pH and the role of the HP0165-HP0166 two-component system (ArsRS) in this acid-inducible regulation, Northern blot analysis was performed with RNAs isolated from two different wild-type H. pylori strains (26695 and 43504) and mutants with HP0165 histidine kinase (ArsS) deletions, after exposure to either neutral pH or low pH (pH 4.5). ArsS-dependent upregulation of HP1186 α-carbonic anhydrase in response to low pH was found in both strains. Western blot analysis of H. pylori membrane proteins confirmed the regulatory role of ArsS in HP1186 expression in response to low pH. Analysis of the HP1186 promoter region revealed two possible transcription start points (TSP1 and TSP2) located 43 and 11 bp 5′ of the ATG start codon, respectively, suggesting that there are two promoters transcribing the HP1186 gene. Quantitative primer extension analysis showed that the promoter from TSP1 (43 bp 5′ of the ATG start codon) is a pH-dependent promoter and is regulated by ArsRS in combating environmental acidity, whereas the promoter from TSP2 may be responsible for control of the basal transcription of HP1186 α-carbonic anhydrase.
Helicobacter pylori is a gram-negative neutralophile that shares some cytoplasmic pH regulatory systems with other neutralophiles, such as Salmonella enterica serovar Typhimurium, Vibrio cholera, and pathogenic strains of Escherichia coli (20, 21, 23). These bacteria can survive in acidic environments by maintaining the cytoplasmic pH at ∼4.0 to 5.0 but do not grow and therefore cannot colonize the stomach. However, H. pylori has developed additional unique acid acclimation mechanisms that enable this organism not only to survive in but also to colonize the acidic environment of the human stomach (21, 23). This is due to its ability to buffer its periplasmic pH in acidic environments to levels at which both the cytoplasmic pH and membrane potential are compatible with growth (22). Several systems may be involved in maintenance of periplasmic pH which form the basis for acid acclimation and growth in the gastric environment.
The urease system is one of the best-studied processes in H. pylori which maintain periplasmic and cytoplasmic pH near neutrality when the organism is exposed to acidity (36, 37). Urease is a nickel-containing hexameric heterodimer, composed of subunits UreA and UreB, which hydrolyzes urea, leading to the formation of ammonia and carbon dioxide, which buffer the cytoplasm and periplasm of the bacterium (34, 39, 40). The enzymatic activity of intrabacterial urease is controlled by the inner membrane pH-gated channel UreI, which regulates the access of the substrate, urea, to urease in the bacterial cell in response to acidic pH (33, 38, 53). Both urease and the pH-gated channel protein UreI are essential for colonization in several animal infection models (11, 12, 41, 47).
Although these studies have clarified the important role of UreI and urease in the acid acclimation processes of H. pylori, it would be extremely unlikely that they are the only means of combating gastric acidity for colonization of the mammalian stomach by the organism (22, 37, 40). Even though urease produces ammonia (NH3), which rapidly diffuses into the periplasm, its pKa of 9.2 shows that it would be a very weak buffer at the desired relatively neutral periplasmic pH. The other product of urease activity is H2CO3, which is rapidly converted to carbon dioxide (CO2) by the bacterial cytoplasmic β-carbonic anhydrase, allowing rapid diffusion of this gas into the periplasm. The periplasmic α-carbonic anhydrase (HP1186) converts this periplasmic CO2 to HCO32−, providing a buffer with a pKa of 6.1. This enzyme has been shown to be important in buffering the bacterial periplasm (22, 42). Wild-type H. pylori maintains a periplasmic pH of ∼6.1 in the presence of strongly buffered acidic medium between pH 3.5 and 6.2 (38), partially due to the activity of UreI that admits urea into the bacterial cytoplasm (39, 53). However, in an H. pylori mutant strain with α-carbonic anhydrase deletion (22), the addition of urea no longer restored membrane potential in acidic medium, as had been found in ureI deletion mutants, although UreI and urease remained fully functional in the carbonic anhydrase deletion strains in that there was a normal increase of bacterial urease activity with decreasing pH. At low pH, the elevation of cytoplasmic and periplasmic pH with urea was abolished in the absence of α-carbonic anhydrase activity (22). Furthermore, acid survival was significantly impaired, either with deletion of HP1186 or by addition of the carbonic anhydrase inhibitor acetazolamide (22). These results suggest that the role of periplasmic α-carbonic anhydrase in acid acclimation is to generate HCO32− in the periplasm both to neutralize entering acid and to buffer the periplasm to a level, pH 6.1 (the pKa of HCO32−), at which a viable inner membrane potential is maintained. Therefore, the periplasmic α-carbonic anhydrase activity of H. pylori is essential for acid acclimation.
In recent transcriptome studies, the gene expression of urease (including ureAB and ureIEFGH), as well as some other ammonia-producing enzymes (including aliphatic amidase amiE, asparaginase ansB, and arginase rocF), which might be considered pH homeostatic genes, were found to be induced upon exposure of H. pylori to low pH (5, 26, 49, 55). One of these studies also showed a nearly fivefold increase in expression of HP1186 α-carbonic anhydrase upon exposure to acidic pH, and the low-pH-induced expression was confirmed by real-time PCR analysis (55).
Better understanding of the mechanisms that regulate gene expression in response to the acidic conditions in H. pylori, especially for the acid acclimation genes, would eventually facilitate the development of novel eradication therapies for this carcinogenic gastric pathogen. Transcription of the ureAB genes was found to be positively regulated by the NikR protein in response to increasing concentrations of Ni2+ ions in the surrounding medium (50, 51), through which the environmental acidity might be indirectly sensed (52). More recent studies have shown that transcriptional induction of urease genes (ureAB and ureI) in response to low pH is mediated mainly by the HP0165-HP0166 two-component system (ArsRS), since the effect was largely abolished in an ArsS (HP0165)-deficient mutant (29, 31, 32). The phosphorylated HP0166 response regulator ArsR (ArsR∼P) was found to bind to extended regions overlapping the PureA and PureI promoters (31). In addition, it appeared that ArsRS may also be involved in the pH-responsive transcriptional regulation of some other ammonia-producing genes (amiE, amiF, and rocF) (30). It has been suggested that ArsRS plays a prominent role in the control of acid-responsive gene expression during infection (29, 31, 32). However, very little is known about the regulation of gene expression for HP1186 α-carbonic anhydrase in response to low pH.
The two-component system ArsRS is an acid-responsive signaling system that perceives low-pH stimuli by the periplasmic domain of its sensor histidine kinase (ArsS) and regulates acid-responsive gene expression by the cognate response regulator (ArsR), which directly interacts with the promoter region in H. pylori (29-32, 54). A recent electrophoretic mobility shift assay (EMSA) study (54) with a recombinant HP0166-His6 showed that ArsR binds directly to the promoter region of HP1186 α-carbonic anhydrase, and EMSA studies with a mutated nonphosphorylatable protein (HP0166-D52N) suggested a phosphorylation-dependent binding. Given that the expression of the HP1186 gene is acid inducible and that the phosphorylated form of ArsR can directly bind to the promoter region of HP1186 α-carbonic anhydrase, it is possible that acid-induced expression of HP1186 α-carbonic anhydrase requires the two-component system ArsRS.
In the current study, we demonstrate that the two-component system ArsRS directly regulates low-pH-induced expression of HP1186 α-carbonic anhydrase. Using 5′-rapid amplification of cDNA ends (5′-RACE) and primer extension analysis, we also identify two potential transcriptional start points (TSP1 and TSP2) in the 5′-upstream region of the HP1186 α-carbonic anhydrase gene, indicating the presence of two promoters that control the expression of HP1186 α-carbonic anhydrase. We provide evidence that one of the two promoters is pH dependent and controlled by ArsRS in response to low environmental pH, and the other one may be involved in maintaining basal transcription of HP1186 α-carbonic anhydrase.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strains 26695 and 43504 were obtained from the American Type Culture Collection. An arsS-deficient mutant, 26695/HP0165::cat, with an insertion of the chloramphenicol acetyltransferase (cat) gene in the HP0165 open reading frame (ORF) (13), was a gift from M. H. Forsyth's laboratory. An arsS-deficient mutant, 43504/HP0165::km, and an H. pylori strain with deletion of the ArsR-binding site within the promoter region of the HP1186 α-carbonic anhydrase gene were constructed by allelic exchange using a kanamycin resistance gene as described below. Primary plate cultures of H. pylori were grown from glycerol stocks on blood agar medium for 2 to 3 days in a microaerobic environment (5% O2, 10% CO2, and 85% N2) at 37°C. In preparation for an experiment, bacteria were scraped from the plates, suspended in 1 mM phosphate HP buffer (138 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, 1 mM glutamine), pH 7.0, and transferred to fresh plates for 24 h. For exposure to experimental low-pH conditions, the overnight cultures of H. pylori strains on trypticase soy agar plates supplemented with 5% sheep blood were suspended in brain heart infusion medium (Difco) to an optical density at 600 nm of 0.20 to 0.25. The pH of brain heart infusion medium was adjusted to pH 7.4 or pH 4.5 using concentrated HCl followed by filtration to remove any precipitate. H. pylori was then incubated in the presence of 5 mM urea with shaking (120 rpm) under microaerobic conditions at 37°C for 30 min before further extraction of RNA or protein. Escherichia coli strains were grown in Luria-Bertani broth. When necessary, antibiotics were added to the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 10 to 30 μg/ml.
Construction of mutant H. pylori strains (43054/HP0165:km and 43504/ΔPHP1186) by allelic exchange mutagenesis.
An isogenic arsS-deficient mutant, 43054/HP0165::km, was obtained by transforming H. pylori strain 43504 with a pBluescript II vector (Stratagene) carrying the Tn903 kanamycin resistance gene (45) flanked by a 650-bp fragment comprising the 3′ region of the HP0166 gene and a 463-bp fragment comprising the 5′ region of the HP0163 gene. The DNA fragments were obtained by PCR performed on H. pylori chromosomal DNA with primer pairs HP0166-5′p/HP0166-3′p and HP0163-5′p/HP0163-3′p (Table 1), respectively. The resulting mutant had the entire HP0165-0164 coding sequence replaced by the kanamycin resistance gene.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′)a | Siteb | Strand | Positionc |
|---|---|---|---|---|
| HP0166-5′p | CATGTAACCAATctaGATGAGCCATATACCGGC | XbaI | − | 174345-174377 |
| HP0166-3′p | CTTTAAAAAAGATAGAGgtcgacAAAACCCCTTAACTC | SalI | + | 173728-173765 |
| HP0163-5′p | CCCGAAAATTTGAGAtctGTGAGCGGAATGAAGGG | BglII | − | 172448-172482 |
| HP0163-3′p | GCCCATGGTCGGTaccTTCACAAAAACACAAATCCGC | Acc65I | + | 172020-172056 |
| HP1185-5′p | attgggtaccCGCGACACGAATGACTAAAGAAGC | KpnI | + | 1255108-1255131 |
| HP1185-3′p | gcaggaattcGGTAACAACTCCCTTAAATTAATGC | EcoRI | − | 1255613-1255638 |
| HP1186-5′p | tagtggatccAATCTATTTTTTGTAACTGCGGTCA | BamHI | + | 1255678-1255702 |
| HP1186-3′p | tgttgagctcTCACAAACCATGCCACCCCCTCTGT | SacI | − | 1256356-1256380 |
| P1186-5′ (479-502) | TTCAAATACGCCGCTTCTAAACCT | + | 1255979-1256002 | |
| P1186-3′ (875-856) | AACCATGCCACCCCCTCTGT | − | 1256356-1246375 | |
| P1186-3′ (481-504) | TTAGGTTTAGAAGCGGCGTATTTG | − | 1255981-1256004 | |
| P1186-3′ (308-327) | TCAGCGCCTACAAGAGAAGC | − | 1255808-1255827 | |
| P1186-5′ (1-20) | TTAAATCTTAAGGGGTTGAA | + | 1255501-1255520 | |
| P0547-5′ (3728-3750) | AGCTAGCCCTGAACCCATTTACG | + | 582815-582837 | |
| P0547-3′ (4150-4127) | TGCGCAACTATCTTATCATTCACG | − | 583214-583237 | |
| P0071-5′ (249-272) | GGCAATGCTAGGACTTGTATTGTT | − | 75315-75338 | |
| P0071-3′ (821-840) | TCACACCCAGTGTTGGATAA | + | 74747-74756 |
Sequences in uppercase letters are derived from the genome sequences of H. pylori 26695 (46). Sequences introduced for cloning purposes are given in lowercase letters, and restriction recognition sites are underlined.
Restriction recognition site.
Nucleotide positions refer to the genome sequence of H. pylori 26695 (46).
To construct the H. pylori strain with deletion of the ArsR-binding site within the promoter region of the HP1186 α-carbonic anhydrase gene, a pBluescript II vector (Stratagene) carrying a kanamycin resistance cassette flanked by a 531-bp fragment comprising 393 bp of the 3′ region of the HP0185 ORF and 138 bp of the intergenic region between HP1185 and HP1186 and a 703-bp fragment comprising 94 bp of the upstream region and 609 bp of the complete ORF of HP1186 was introduced into H. pylori strain 43504 by natural transformation. The DNA fragments were obtained by PCR amplified from chromosomal DNA of H. pylori 26695 with primer pairs HP1185-5′p/HP1185-3′p and HP1186-5′p/HP1186-3′p (Table 1), respectively. The resulting strain (43504/ΔPHP1186) had the ArsR-binding site ranging from position −98 to −136 (with respect to the translational start codon) within the promoter region of the HP1186 α-carbonic anhydrase gene replaced with a kanamycin resistance cassette in a way that the transcriptional direction of the Km gene was divergent from HP1186.
RNA preparation.
Total RNA was isolated from H. pylori strains using TRIzol reagent (Invitrogen) combined with RNeasy columns (QIAGEN). The bacterial pellet was resuspended in 500 μl of TRIzol reagent (Invitrogen) and lysed at room temperature for 5 min before 100 μl of chloroform was added. After spinning at 12,000 × g for 10 min at 4°C, the supernatant was mixed with 250 μl ethanol and applied to an RNeasy spin column (QIAGEN). RNA purification was processed following the manufacturer's instructions (beginning with the application to the column). RNA concentration was quantified by absorbance at 260 nm, and the quality was evaluated by capillary electrophoresis using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano Assay kit (Agilent Technologies).
Northern blot analysis.
Fifteen μg of total RNA from different H. pylori samples was fractionated on 1% agarose-formaldehyde gels and transferred to Nytran membranes (Schleicher & Schuell, Keene, NH). The RNA on the gel was visualized by ethidium bromide staining and photographed before the RNA was transferred to Nytran membranes.
Internal fragments of the α-carbonic anhydrase (HP1186), ureI (HP0071), and cagA (HP0547) genes were PCR amplified from genomic DNA of H. pylori 26695 with the primer pairs P1186-5′(479-502)/P1186-3′ (875-856), P0071-5′(249-272)/P0071-3′ (821-840), and P0547-5′ (3728-3750)/P0547-3′ (4150-4127) (listed in Table 1) and used as probes. The probes were radiolabeled with [α-32P]dCTP using a random primer labeling kit (Stratagene, La Jolla, CA) to specific activities of 1 × 108 to 10 × 108 cpm/μg. The blot was hybridized (9) with radiolabeled probe overnight at 65°C in a buffer containing 0.45 M sodium phosphate (pH 7.2), 7% sodium dodecyl sulfate (SDS), 1% bovine serum albumin, and 20 mM EDTA. The hybridized blot was washed with 0.1× SSC-0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C and autoradiographed using a PhosphorImager 445 SI (Molecular Dynamics, Sunnyvale, CA). The RNA hybridization bands were normalized based on 16S and 23S rRNA band intensities.
Western blot analysis.
To isolate membrane proteins from H. pylori strains (wild types and arsS deletion mutants) treated with different pHs, the bacteria pellet was resuspended in ice-cold 25 mM sodium phosphate buffer, pH 7.4, and passed three times through a French press at 20,000 lb/in2. The lysate was centrifuged at 3,000 × g for 10 min to pellet any remaining whole cells. The supernatant was removed and centrifuged at 10,000 × g for 10 min. This supernatant was layered onto 5 ml of 20% sucrose and centrifuged at 100,000 × g for 1 h. The sucrose layer prevented soluble cytoplasmic proteins from contaminating the membrane pellet. The supernatant was removed from the top of the sucrose layer and concentrated by using a centrifugal filter device with a 10-kDa cutoff (Centricon, Inc.). The membrane pellet was resuspended in 25 mM sodium phosphate buffer, pH 7.4. Protein concentration was determined by using bicinchoninic acid (Pierce).
The proteins from the membrane fraction were fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using 4 to 12% NuPage Bis-Tris gradient gels (Invitrogen) and transferred onto nitrocellulose (Bio-Rad). Western blot analysis was carried out using an antibody against α-carbonic anhydrase (22). Immunolabeling was detected with the SuperSignal West Pico chemiluminescence detection kit (Pierce). A Western blot analysis by a rabbit polyclonal antibody (NCL-HPp) reactive with heat-stable, somatic antigens of the whole H. pylori organism (Novocastra Laboratories Ltd., United Kingdom) with the same preparation of the membrane proteins was used as a loading control.
Characterization of the HP1186 promoter region.
The BD Smart RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA) was used for characterization of the HP1186 promoter region from H. pylori by RACE-PCR. According to the manufacturer's manual, 1 μg of total RNA isolated from the H. pylori strain 26695 was reverse transcribed with a gene-specific reverse primer, P1186-3′ (481-504) (Table 1), which is 232 bp downstream of the translation start codon ATG in the ORF of the HP1186 gene, and the BD PowerScript reverse transcriptase (RT; BD Biosciences Clontech, Palo Alto, CA), while the BD SMART II A oligonucleotide was also included in the RT reaction mixture. When the RT reaches the 5′ end of the RNA template, the SMART II A oligonucleotide attaches to the first-strand cDNA tail and serves as an extended template for the RT, resulting in a complete cDNA copy of the original RNA with the additional SMART sequence at the 5′ end (24, 56). A nest gene-specific reverse primer, P1186-3′ (308-327) (Table 1), which is 55 bp downstream of the translation start codon ATG in the ORF of HP1186, was used for 5′-RACE-PCR with the SMART universal primer. The RACE-PCR products were cloned into the T/A cloning vector pCR4-TOPO (Invitrogen, Carlsbad, CA). The authenticity was confirmed by DNA sequencing in both directions with T7 and M13 reverse primers.
Primer extension analysis.
The gene-specific reverse primer P1186-3′ (308-327), was 5′-end labeled in the presence of [γ-33P]ATP (5,000 Ci/mmol; GE Healthcare Biosciences, Piscataway, NJ) and T4 polynucleotide kinase (Promega, Madison, WI). Labeled oligonucleotide (1 pmol) was then coprecipitated with 35 μg of H. pylori total RNA and resuspended in 6 μl of H2O and 5 μl of 2× avian myeloblastosis virus (AMV) primer extension buffer (Primer Extension system; Promega). The reaction mixture was incubated for 20 min at 58°C followed by 10 min at room temperature, then 5 μl of 2× AMV primer extension buffer, 1.4 μl of 40 mM sodium pyrophosphate, and 1 μl of AMV reverse transcriptase (1 U/μl) were added, and reverse transcription in a total volume of 20 μl was carried out by incubating at 42°C for 30 min. The sample was then ethanol precipitated and resuspended in 5 μl of sequencing loading buffer. After denaturing at 90°C for 2 min, samples were subjected to 6% urea-polyacrylamide gel electrophoresis and autoradiographed using a PhosphorImager 445 SI (Molecular Dynamics, Sunnyvale, CA). To ensure correct mapping of the extension products, the HP1186 promoter region was cloned with the primer pair P1186-5′ (1-20)/P1186-3′ (308-327) (Table 1) from the chromosomal DNA of H. pylori 26695 and sequenced in parallel with the same primer used for primer extension.
RESULTS
Low-pH-induced transcription of HP1186 α-carbonic anhydrase is dependent on the two-component system ArsRS.
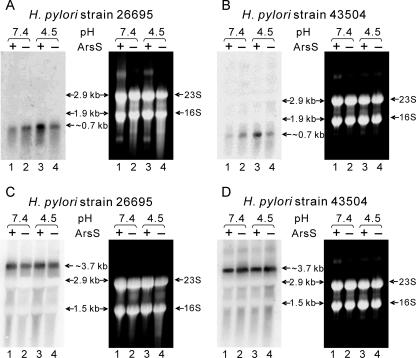
With microarray analysis, the HP1186 α-carbonic anhydrase gene was found to be induced under low-pH conditions (55). In our previous EMSA studies, the promoter region for the HP1186 α-carbonic anhydrase gene was found to interact with HP0166-His6, likely due to a phosphorylation-dependent binding (54). To investigate the two-component ArsRS system dependency of the low-pH-induced up-regulation on HP1186 α-carbonic anhydrase transcription, Northern blot analysis was performed with equal amounts of total RNAs extracted from H. pylori strain 26695 and the arsS-deficient mutant 26695/HP0165::cat (13) which had been exposed to either neutral pH (pH 7.4) or low pH (pH 4.5) for 30 min. From the genome organization of H. pylori it can be deduced that the HP1186 α-carbonic anhydrase gene is monocistronically transcribed with a 609-bp ORF. Northern blot analysis showed a single band at ∼700 bp (Fig. 1A). In the 26695 wild-type strain, transcription of HP1186 α-carbonic anhydrase was strongly induced at pH 4.5 (a two- to fourfold increase compared to the level at pH 7.4). At neutral pH, the transcription of HP1186 α-carbonic anhydrase in the arsS-deficient mutant was slightly higher than in the 26695 wild-type strain. However, no change in transcription level of HP1186 α-carbonic anhydrase in the arsS-deficient mutant was observed when the bacteria were exposed to pH 4.5 (Fig. 1A).
FIG. 1.
Low-pH-induced transcription of HP1186 α-carbonic anhydrase is ArsS dependent. Total RNAs were extracted from wild-type strains 26695 (A and C) and 43504 (B and D) and the ArsS mutant strains 26695/HP0165::cat (A and C) and 43504/HP0165::km (B and D), which were exposed to either neutral pH (pH 7.4, lanes 1 and 2) or low pH (pH 4.5, lanes 3 and 4) for 30 min. RNAs were analyzed by Northern blotting with HP1186 α-carbonic anhydrase (A and B) and the HP0547 cagA (C and D) gene-specific probes. The same blot with RNAs from H. pylori strain 43504 was hybridized twice separately with different probes (B and D). Single transcripts of ∼0.7 kb (A and B) and ∼3.7 kb (C and D) were detected, which correspond to the sizes of HP1186 α-carbonic anhydrase and the HP0547 cagA transcripts. Ethidium bromide staining of the gel showed that equal amounts of RNA were analyzed. The size standards are the 16S and 23S rRNA species.
In order to understand the low-pH-induced transcription of the HP1186 α-carbonic anhydrase gene in a different H. pylori strain, an arsS-deficient mutant was constructed in H. pylori strain 43504 (43504/HP0165::km). By homologous recombination with a suicide vector, the entire HP0165/HP0164 coding sequence of H. pylori strain 43504 was replaced with a kanamycin resistance gene. Correct replacement of the wild-type sequence with the antibiotic resistance cassette was verified by means of PCR using primers complementary to regions flanking the insertion site (data not shown). A similar Northern blot analysis was performed comparing transcription levels of HP1186 α-carbonic anhydrase from the mutant and wild-type H. pylori strain 43504 after exposing the cells to pH 7.4 and pH 4.5. Very similar results were obtained (Fig. 1B) as found in strain 26695 (Fig. 1A). These results suggest that the acid-responsive transcriptional regulation of the HP1186 α-carbonic anhydrase gene is mediated by ArsRS in both strains 26695 and 43504 under our experimental conditions, since the arsS-deficient mutants were unable to induce the transcription at pH 4.5 to the levels found in the wild-type strains.
Northern blot analysis on transcription of the cagA (HP0547) gene from both H. pylori strains was included as a control (Fig. 1C and D) that is pH and ArsS independently transcribed to eliminate the possibility that the observed ArsS-dependent acid-responsive change in HP1186 α-carbonic anhydrase transcription is caused by global changes in transcription activity. Our previous microarray studies have shown that the expression level of cagA (HP0547) did not significantly change under the same experimental conditions (55).
Low-pH-induced protein expression of HP1186 α-carbonic anhydrase is dependent on the two-component system ArsRS.
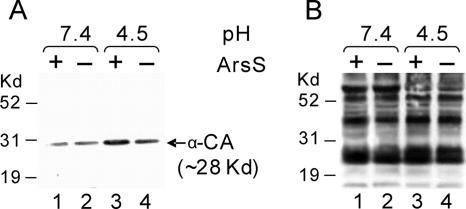
To confirm the regulatory role of ArsS in HP1186 α-carbonic anhydrase expression in response to low-pH conditions, we carried out Western blot analysis on membrane protein fractions to compare the levels of HP1186 α-carbonic anhydrase protein between the H. pylori wild-type strain 26695 and the isogenic mutant strain 26695/HP0165::cat. As shown in Fig. 2A, the protein expression of HP1186 α-carbonic anhydrase in H. pylori wild-type strain 26695 was increased by about fourfold at low pH (pH 4.5) compared to the level at pH 7.4. However, no significant increase of protein expression of HP1186 α-carbonic anhydrase at pH 4.5 was detected in H. pylori mutant strain 26695/HP0165::cat, suggesting that the protein level of HP1186 α-carbonic anhydrase is up-regulated in response to low-pH conditions, and the low-pH-induced protein expression of HP1186 α-carbonic anhydrase depends on ArsRS. The same preparation of membrane proteins was also used for Western blot analysis with a polyclonal antibody (NCL-HPp) against whole H. pylori to ensure that similar amounts of protein samples were loaded (Fig. 2B).
FIG. 2.
Western blot analysis of membrane protein from H. pylori. Membrane fractions were prepared from wild-type H. pylori strain 26695 (lanes 1 and 3) and ArsS mutant strains 26695/HP0165::cat (lanes 2 and 4), which were exposed to either neutral pH (pH 7.4, lanes 1 and 2) or low pH (pH 4.5, lanes 3 and 4) for 30 min. The proteins from the membrane fractions were fractionated by SDS-PAGE and probed with polyclonal antibody against HP1186 α-carbonic anhydrase (A). Western blot analysis of the same preparation of the membrane proteins with a polyclonal antibody (NCL-HPp) against whole H. pylori showed that similar amounts of membrane protein from H. pylori strains were analyzed (B). The MultiMark MultiColored standard (Invitrogen) was used as a size marker.
Deletion of the ArsR-binding site in the promoter region of the HP1186 α-carbonic anhydrase gene abolishes low-pH induction of HP1186 transcription.
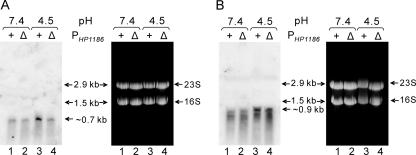
In our previous studies, we identified a binding site for the response regulator ArsR in a region between 95 bp and 145 bp upstream of the translation start codon (ATG) for the HP1186 gene (54). To further verify the role of the ArsR-binding site in low-pH regulation of the HP1186 promoter, we generated a mutant H. pylori strain (26695/ΔPHP1186) with a deletion of the ArsR-binding site in the promoter region of the HP1186 α-carbonic anhydrase gene and analyzed the transcription of HP1186 in response to low pH by Northern blotting with an HP1186 probe in this strain. As shown in Fig. 3A, no increase in transcription of HP1186 was observed when the mutant 26695/ΔPHP1186 was exposed to pH 4.5 for 30 min, which is similar to what was found for the arsS-deficient mutants. As a control, ureI (HP0071) transcription, which has been shown to be up-regulated by low pH in an ArsRS-dependent manner (31), retained the low-pH induction in the mutant strain 26695/ΔPHP1186 (Fig. 3B). This demonstrates that removal of the ArsR-binding site specifically abolishes the low-pH induction of the HP1186 promoter, confirming the role of the in vitro-identified ArsR-binding site.
FIG. 3.
Deletion of the ArsR-binding site in the promoter region of the HP1186 α-carbonic anhydrase gene abolishes low-pH induction of HP1186 transcription. Total RNAs from wild-type strain 43504 and mutant strain 43504/ΔPHP1186, which were exposed to either neutral pH (pH 7.4, lanes 1 and 2) or low pH (pH 4.5, lanes 3 and 4) for 30 min, were analyzed by Northern blotting with HP1186 α-carbonic anhydrase (A) and the HP0071 ureI (B) gene-specific probes. The ∼0.9-kb band corresponds to the size of the ureIE transcript. Ethidium bromide staining of the gel showed that equal amounts of RNA were analyzed. The size standards are the 16S and 23S rRNA species.
Analysis of the promoter region of HP1186 (α-carbonic anhydrase) revealed two possible TSPs.
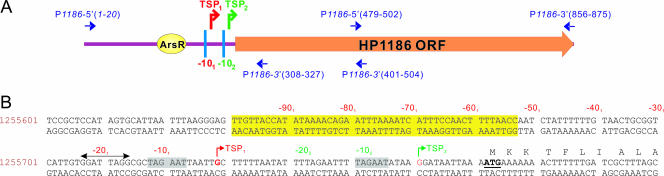
While the binding site for the response regulator ArsR has been identified for the HP1186 gene (54), the promoter region of the HP1186 gene has not been systematically studied. To experimentally identify the transcription start point of the HP1186 α-carbonic anhydrase gene, a reverse primer, P1186-3′ (308-327), which is 55 bp downstream of the translation start codon ATG in the ORF of HP1186, was used for 5′-RACE with RNA isolated from the H. pylori 26695 strain. Twenty clones of the 5′-RACE product that had been cloned into the pCR4-TOPO vector were randomly picked, sequenced, and compared with the genomic sequence of the HP1186 gene in H. pylori strain 26695 (46). Among the 5′-RACE clones examined, about 55% contained a 5′-RACE product of 99 bp in length, indicating a transcription start point (TSP1) at a G nucleotide located 43 bp 5′ of the translation initiation site (unpublished data). However, about 45% of examined clones contain a 66-bp RACE product, suggesting a possible second transcription start point (TSP2) at a G nucleotide located 11 bp 5′ of the ATG start codon (unpublished data).
A −10 consensus sequence (TAGAAT) was found to be present upstream from each of the two separate TSPs (Fig. 4), indicating that the sites are compatible for binding of the H. pylori σ80. Compared with E. coli σ70-recognized promoters (18), this sequence shows one nucleotide mismatch (underlined) in the −10 consensus element sequence (TATAAT). No −35 consensus sequence (TTGACA) (48) with a typical 17 ± 1-bp spacer region resembling that of E. coli was identified for either TSP. A mirror repeat (GGATTAGG), possibly representing a site for binding of regulatory elements, was present in the sequence upstream of the −10 region of both TSPs (−15 to −23 of TSP1 or −47 to −54 of TSP2).
FIG. 4.
(A) Schematic representation of the genomic structure of the HP1186 α-carbonic anhydrase gene. The promoter region is shown with two transcription start points (TSP1 and TSP2) and their −10 consensus sequences (vertical bars). An HP0166 response regulator binding site is also shown as ArsR. The primers used for preparation of the Northern probe and for 5′-RACE of the HP1186 α-carbonic anhydrase gene are indicated at their corresponding regions of the HP1186 ORF. (B) Genomic sequence of HP1186 α-carbonic anhydrase promoter region. A 200-bp (position 1255601-1255800) 5′-flanking region sequence of the HP1186 α-carbonic anhydrase gene from the H. pylori 26695 genome (GenBank accession number NC_000915) (46) is shown. Two transcription start points (TSP1 and TSP2) are indicated by bent arrows, and their −10 consensus sequences are highlighted by gray shading. The translational start codon is underlined. The binding site of response regulator ArsR is highlighted in yellow. A mirror repeat (GGATTAGG) is indicated by a double-arrowed line above the double-stranded sequence. Two sets of numbers (in red and green) above the sequence indicate the nucleotide position with respect to the corresponding transcriptional start points (TSP1 and TSP2).
Our previous EMSA and DNase I footprinting studies (54) had shown that ArsR binds directly to the HP1186 promoter region that is located between −52 and −98 of TSP1 (Fig. 4). This ArsR-binding region is located immediately upstream of (and may partially overlap with) an AT-rich sequence, possibly a UP element for TSP1 (between −40 and −60 of TSP1).
These results suggest that there are two promoters transcribing the HP1186 gene, and both promoters may be regulated by RNA polymerase containing σ80, the H. pylori homologue of the vegetative sigma factor σ70 from E. coli.
Effect of low pH on TSP utilization in the HP1186 promoter.
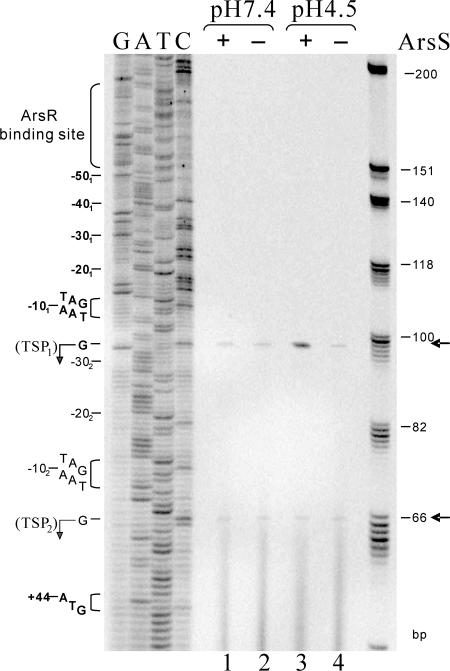
A study by McGowan et al. (25) identified multiple TSPs for several acid-inducible genes in H. pylori strain 60190 and suggested the existence of alternative promoters that transduce signals under either neutral or acidic pH conditions in H. pylori. In order to investigate the possible relative transcriptional responses from the two identified TSPs of the HP1186 promoter to the low-pH condition that is signaled by the two-component system ArsRS, we analyzed the transcription of HP1186 α-carbonic anhydrase by quantitative primer extension using a reverse primer, P1186-3′ (308-327) and equal amounts (35 μg) of RNA samples from H. pylori strains 26695 and 26695/HP0165::cat (13) which had been exposed to either neutral pH (pH 7.4) or low pH (pH 4.5) for 30 min. To ensure correct mapping of the extension products, we cloned and sequenced in parallel the HP1186 promoter region from the chromosomal DNA of H. pylori 26695. In agreement with the 5′-RACE results, two extension products were detected from either the wild-type or the 26695/HP0165::cat RNAs (the transcriptional start sites are unchanged in the HP0165− background), which place the transcription start sites 43 bp and 11 bp, respectively, upstream from the HP1186 translational start codon. As shown in Fig. 5, the TSP1 (99-bp elongated primer product) was used preferentially at low pH in the wild-type 26695 strain, whereas the low-pH-induced transcription from TSP1 could not be detected in the histidine kinase ArsS mutant 26695/HP0165::cat. This suggests that the HP1186 promoter with TSP1 is responsible for pH-dependent regulation signaled by the two-component system ArsSR. The second transcription start point (TSP2) of the HP1186 promoter (shown as a 66-bp elongated primer product in Fig. 5) appeared unaffected by pH and ArsS, since it was used equally under all pH conditions in both wild-type and 26695/HP0165::cat strains, indicating a possible role in maintaining the basal transcription level of HP1186 α-carbonic anhydrase independently of the acid-responsive signaling system in H. pylori.
FIG. 5.
Primer extension analysis of transcription from the HP1186 α-carbonic anhydrase promoter in H. pylori strains exposed to neutral and low pH. Primer extension experiments using the 33P-labeled reverse primer P1186-3′ (308-327) were performed on equal amounts of RNAs isolated from wild-type strain 26695 (lanes 1 and 3) and the ArsS mutant strain 26695/HP0165::cat (lanes 2 and 4), which were exposed to either neutral pH (pH 7.4, lanes 1 and 2) or low pH (pH 4.5, lanes 3 and 4) for 30 min. The same primer used for the primer extension analysis was used for sequencing reactions (lanes G, A, T, and C). The arrows indicate the primer extension products corresponding to TSP1 and TSP2. The −10 consensus sequences for each of the TSPs and the nucleotide positions with respect to the corresponding TSPs are given on the left. The binding site for the response regulator ArsR is also marked on the left. The size standards are 33P-labeled ΦX174 HinfI markers provided in the Primer Extension system (Promega).
DISCUSSION
The carbonic anhydrase of the human gastric pathogen H. pylori has been studied in some detail because of its possible involvement in acid acclimation that allows colonization and thence virulence (7, 8, 22, 37, 44). These studies have resulted in a large amount of biochemical and physiological data concerning the specific roles of H. pylori carbonic anhydrase but have left analysis of the transcriptional regulation of the carbonic anhydrase gene almost untouched. We provide evidence that the HP1186 gene, encoding the periplasmic α-carbonic anhydrase of H. pylori, is transcribed by the vegetative sigma factor σ80 and regulated by the two-component system ArsSR in response to low-pH conditions.
Evidence demonstrating that ArsRS acts as a transcriptional regulator of α-carbonic anhydrase in response to acidic pH comes from the observation that the mRNA levels of the HP1186 gene were increased about fourfold in wild-type H. pylori strains when treated at pH 4.5, whereas no such upregulation of α-carbonic anhydrase mRNA was detected in the ArsS− background (Fig. 1). Western blot analysis of membrane fractions confirmed the regulatory role of ArsS in HP1186 α-carbonic anhydrase expression in response to low-pH conditions (Fig. 2). EMSA and DNase I footprinting experiments with purified HP0166-His6 (54) showed that the response regulator ArsR protein binds directly to the promoter of the HP1186 gene. When a mutated response regulator, HP0166-D52N, in which the aspartic acid residue at position 52 was replaced with asparagine, was used for EMSA, no binding was observed for the promoter of HP1186 (54), demonstrating that the phosphorylated form of ArsR is necessary for efficient binding of the promoter of HP1186. Consistent with these observations, we also show that deletion of the ArsR-binding site in the HP1186 promoter region abrogates the acid induction of HP1186 transcription. These data also strongly support the conclusion that the acid-induced activation of the HP1186 promoter is regulated by ArsRS.
The acid-responsive expression of HP1186 α-carbonic anhydrase has been controversial in different studies. Microarray studies by Merrell et al. (26) found a down-regulation of the HP1186 gene at pH 5 after 60 and 120 min of exposure. Other studies (1, 2, 5) did not find a significant change in the expression level of the HP1186 gene in response to low pH. The discrepancies between different studies may be explained by the different bacterial strains used in different studies and the experimental conditions, including the nature of acid exposure, the growth phase, the growth medium used, and the duration of acid exposure, which varied from study to study. Two different H. pylori strains (26695 and 43504) have been used in this study, and very similar results were obtained, suggesting that the acid-responsive transcriptional regulation of the HP1186 α-carbonic anhydrase gene is mediated by ArsRS in both 26695 and 43504 strains under our experimental conditions.
Very little is known about the basic processes of gene transcription and transcriptional regulation in H. pylori, particularly the regulation of gene expression in response to acid stress. The association of appropriate alternative sigma factors with core RNA polymerase provides a mechanism for cellular responses through redirection of transcription initiation, which leads to the regulated expression of virulence genes and virulence-associated genes in many bacterial pathogens (19). However, an analysis of the H. pylori genome performed at The Institute for Genomic Research (46) has shown that this organism probably has only three types of σ factors (σ80, σ54, and σ28), suggesting that H. pylori may use different mechanisms to regulate gene expression in response to environmental changes, such as low pH. Recent studies have shown that the two-component system ArsRS is one of the mechanisms responsible for regulation of many genes in H. pylori in response to an acidic environment, which regulates most of the acid acclimation genes identified so far (29-32, 54). In H. pylori, the environmental pH is probably sensed directly by the histidines (pKa, ∼5.9) in the periplasmic domain of the histidine kinase sensor protein ArsS, which autophosphorylates in response to pH changes and transfers the phosphoryl group to its cognate response regulator, ArsR. The phosphorylated ArsR may function both as an activator and repressor of pH-responsive target genes by interacting with the promoter regions (30, 32). It is clear from the current study that the phosphorylated ArsR induced by low pH acts as an activator in interacting with one of the promoters of the HP1186 α-carbonic anhydrase gene.
Using 5′-RACE and primer extension assays (Fig. 4 and 5), we found two TSPs for the H. pylori HP1186 α-carbonic anhydrase gene, suggesting that two promoters are present for the HP1186 α-carbonic anhydrase gene. The presence of two promoters may allow differential regulation of the gene in response to environmental conditions. The use of multiple promoters therefore provides an efficient means of controlling the expression of a number of genes, particularly when the promoters are differentially utilized. In Caulobacter crescentus, two functional promoters (P1 and P2) were found to precede the dnaK/dnaJ operon (16). P1 was a heat shock-inducible promoter, with characteristics of a σ32 promoter, and P2 was an adjacent σ70 promoter. Transcription initiating at the P2 promoter was under temporal control and linked to the onset of DNA replication. McGowan et al. (25) recently identified multiple promoters from several acid-inducible genes in H. pylori and found that certain promoters were expressed preferentially under acidic conditions, whereas others either showed maximal expression at pH 7 or showed no differential expression at any pH. These studies provide evidence for pH-dependent promoter regulation in H. pylori and demonstrate the existence of alternative promoters that transduce signals under either neutral or acidic pH conditions.
As seen in Fig. 4, the promoters from TSP1 and TSP2 are very similar to each other, with an identical −10 element, indicating that both of them may be regulated by RNA polymerase containing σ80, the H. pylori homologue of the vegetative sigma factor σ70 from E. coli. The observation that the promoter from TSP1 responds strongly in response to low pH even though there is no good −35 sequence for both promoters suggests the involvement of positive transcriptional control by a trans-acting factor. The lack of the typical E. coli −35 consensus is consistent with previous analysis of transcription in H. pylori (4, 14, 25, 28, 43), and this usually provides evidence for a physiological basis for activator function (17). Functioning as an activator, ArsR interacts with a binding site that is located between −52 and −98 of TSP1 (54), which partially overlaps with a potential UP element (15, 35) found only for TSP1 (an AT-rich sequence between −40 and −60 of TSP1). This may lead to different transcription levels of HP1186 α-carbonic anhydrase in response to low-pH conditions.
Similar to class I activators that recruit RNA polymerase via a direct protein-protein interaction (3), it may be that ArsR binds to an upstream location (between −52 and −98) (Fig. 4B) of the HP1186 α-carbonic anhydrase promoter from TSP1, usually near the carboxy-terminal domain of the RNA polymerase α-subunit (αCTD). This interaction recruits αCTD, and hence the rest of the RNA polymerase, to the promoter (6), and therefore initiates the transcription from TSP1. Although the location of activator binding at promoters subject to class I activation is variable, because the linker between αCTD and the α-amino-terminal domain is sufficiently flexible to permit the binding of αCTD at different positions, the promoter from TSP2 appears to be at a location that renders αCTD too distant for ArsR to reach from its binding site (between −84 and −122 of TSP2) (Fig. 4B).
The discovery of multiple promoters responsible for the expression of HP1186 α-carbonic anhydrase provides new insight regarding the transcriptional organization of the HP1186 α-carbonic anhydrase gene. The presence of the multiple promoters for HP1186 may facilitate differential expression of the HP1186 α-carbonic anhydrase gene in response to environmental pH. Using quantitative primer extension analysis (Fig. 5), we demonstrated that the promoter from TSP1 is a pH-dependent promoter and is regulated by ArsRS, whereas the promoter from TSP2 may be involved in control of the basal (or housekeeping) transcription of HP1186 α-carbonic anhydrase or regulated by a different mechanism yet to be identified.
It seems that a basal level of carbonic anhydrase is maintained, irrespective of external acidic pH, and this housekeeping function may maintain periplasmic pH within viable limits at around neutral pH between pH 7.0 and 8.0 in the face of basal urease activity. Since H. pylori is killed at pH 8.2 (10, 27), bicarbonate generation at pH levels between 7.4 and 8.0 may prevent lethal alkalinization by UreI-independent basal urease activity. However, when faced with an acidic environment, the organism requires higher levels of this periplasmic carbonic anhydrase to maintain its periplasmic pH at levels compatible with not only survival but also growth in the gastric niche it inhabits. With the acid-responsive regulation by the ArsRS two-component system, the combination of urease, UreI, and α-carbonic anhydrase comprise an important component of its acid acclimation mechanisms.
Acknowledgments
This work was supported by the U.S. Veterans Administration, NIH grants DK46917, 53462, and 58333, and the Pilot and Feasibility grant from The CURE Digestive Diseases Research Center at UCLA.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 4.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 7.Chirica, L. C., B. Elleby, and S. Lindskog. 2001. Cloning, expression and some properties of alpha-carbonic anhydrase from Helicobacter pylori. Biochim. Biophys. Acta 1544:55-63. [DOI] [PubMed] [Google Scholar]
- 8.Chirica, L. C., C. Petersson, M. Hurtig, B. H. Jonsson, T. Boren, and S. Lindskog. 2002. Expression and localization of alpha- and beta-carbonic anhydrase in Helicobacter pylori. Biochim. Biophys. Acta 1601:192-199. [DOI] [PubMed] [Google Scholar]
- 9.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clyne, M., A. Labigne, and B. Drumm. 1995. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 63:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero, R. L., V. Cussac, P. Courcoux, and A. Labigne. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 174:4212-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 184:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsyth, M. H., and T. L. Cover. 1999. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J. Bacteriol. 181:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredrick, K., and J. D. Helmann. 1997. RNA polymerase sigma factor determines start-site selection but is not required for upstream promoter element activation on heteroduplex (bubble) templates. Proc. Natl. Acad. Sci. USA 94:4982-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes, S. L., J. W. Gober, and L. Shapiro. 1990. Expression of the Caulobacter heat shock gene dnaK is developmentally controlled during growth at normal temperatures. J. Bacteriol. 172:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gralla, J. D. 1996. Activation and repression of E. coli promoters. Curr. Opin. Genet. Dev. 6:526-530. [DOI] [PubMed] [Google Scholar]
- 18.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 21.Malaty, H. M., A. El-Kasabany, D. Y. Graham, C. C. Miller, S. G. Reddy, S. R. Srinivasan, Y. Yamaoka, and G. S. Berenson. 2002. Age at acquisition of Helicobacter pylori infection: a follow-up study from infancy to adulthood. Lancet 359:931-935. [DOI] [PubMed] [Google Scholar]
- 22.Marcus, E. A., A. P. Moshfegh, G. Sachs, and D. R. Scott. 2005. The periplasmic α-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J. Bacteriol. 187:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 24.Matz, M., D. Shagin, D. Shagin, O. Britanova, S. Lukyanov, L. Diatchenko, and A. Chenchik. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27:1558-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGowan, C. C., A. S. Necheva, M. H. Forsyth, T. L. Cover, and M. J. Blaser. 2003. Promoter analysis of Helicobacter pylori genes with enhanced expression at low pH. Mol. Microbiol. 48:1225-1239. [DOI] [PubMed] [Google Scholar]
- 26.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer-Rosberg, K., D. R. Scott, D. Rex, K. Melchers, and G. Sachs. 1996. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology 111:886-900. [DOI] [PubMed] [Google Scholar]
- 28.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537-1548. [DOI] [PubMed] [Google Scholar]
- 29.Pflock, M., P. Dietz, J. Schar, and D. Beier. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol. Lett. 234:51-61. [DOI] [PubMed] [Google Scholar]
- 30.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pflock, M., S. Kennard, I. Delany, V. Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pflock, M., S. Kennard, N. Finsterer, and D. Beier. 2006. Acid-responsive gene regulation in the human pathogen Helicobacter pylori. J Biotechnol. 126:52-60. [DOI] [PubMed] [Google Scholar]
- 33.Rektorschek, M., A. Buhmann, D. Weeks, D. Schwan, K. W. Bensch, S. Eskandari, D. Scott, G. Sachs, and K. Melchers. 2000. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol. Microbiol. 36:141-152. [DOI] [PubMed] [Google Scholar]
- 34.Rektorschek, M., D. Weeks, G. Sachs, and K. Melchers. 1998. Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology 115:628-641. [DOI] [PubMed] [Google Scholar]
- 35.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 36.Sachs, G., D. L. Weeks, K. Melchers, and D. R. Scott. 2003. The gastric biology of Helicobacter pylori. Annu. Rev. Physiol. 65:349-369. [DOI] [PubMed] [Google Scholar]
- 37.Sachs, G., D. L. Weeks, Y. Wen, E. A. Marcus, D. R. Scott, and K. Melchers. 2005. Acid acclimation by Helicobacter pylori. Physiology (Bethesda) 20:429-438. [DOI] [PubMed] [Google Scholar]
- 38.Scott, D. R., E. A. Marcus, D. L. Weeks, A. Lee, K. Melchers, and G. Sachs. 2000. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect. Immun. 68:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 40.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 41.Skouloubris, S., J. M. Thiberge, A. Labigne, and H. De Reuse. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 66:4517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335-366. [DOI] [PubMed] [Google Scholar]
- 43.Spohn, G., D. Beier, R. Rappuoli, and V. Scarlato. 1997. Transcriptional analysis of the divergent cagAB genes encoded by the pathogenicity island of Helicobacter pylori. Mol. Microbiol. 26:361-372. [DOI] [PubMed] [Google Scholar]
- 44.Stahler, F. N., L. Ganter, K. Lederer, M. Kist, and S. Bereswill. 2005. Mutational analysis of the Helicobacter pylori carbonic anhydrases. FEMS Immunol. Med. Microbiol. 44:183-189. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 47.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanet, A., L. Marsan, A. Labigne, and M. F. Sagot. 2000. Inferring regulatory elements from a whole genome. An analysis of Helicobacter pylori σ80 family of promoter signals. J. Mol. Biol. 297:335-353. [DOI] [PubMed] [Google Scholar]
- 49.van Vliet, A. H., E. J. Kuipers, J. Stoof, S. W. Poppelaars, and J. G. Kusters. 2004. Acid-responsive gene induction of ammonia-producing enzymes in Helicobacter pylori is mediated via a metal-responsive repressor cascade. Infect. Immun. 72:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Vliet, A. H., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Vliet, A. H., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Vliet, A. H., F. D. Ernst, and J. G. Kusters. 2004. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 12:489-494. [DOI] [PubMed] [Google Scholar]
- 53.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 54.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2006. Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori. J. Bacteriol. 188:1750-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, Y. Y., E. M. Machleder, A. Chenchik, R. Li, and P. D. Siebert. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30:892-897. [DOI] [PubMed] [Google Scholar]