Abstract
The energetic efficiency of nutrient uptake and conversion into biomass is a key factor in the ecological behavior of microorganisms. The constraints shaping the metabolic rate-yield trade-off in bacteria are not well understood. To examine whether metabolic rate-yield settings and physiological strategies evolve toward a particular optimum in a constant environment, we studied multiple Escherichia coli isolates evolving in a glucose-limited chemostat population. A major divergence in transport and metabolic strategies was observed, and the isolates included inefficient rate strategists (polluters or cheaters) and yield strategists (conservationists), as well as various hybrid rate-yield strategists and alternative ecotypes (dropouts). Sugar transport assays, strain comparisons based on metabolomics, and Biolog profiling revealed variance to the point of individuality within an evolving population. Only 68 of 177 metabolites assayed were not affected in 10 clonally related strains. The parallel enrichment of rate and yield strategists and the divergence in metabolic phylogenies indicate that bacteria do not converge on a particular rate-yield balance or unique evolutionary solutions. Redundancies in transport and metabolic pathways are proposed to have laid the framework for the multiplicity of bacterial adaptations.
Microorganisms exhibit an immense range of metabolic capabilities, and there are extensive metabolic differences between members of a single bacterial species (29). Metabolic capabilities and the efficiency of metabolism are important in the ecological specialization of all organisms (1). Well-known nutritional strategies involving r- and K-adapted organisms differ in terms of their metabolic rate-yield balances (24). Metabolic efficiency is affected by the energetics of nutrient uptake (31) and is also thought to be governed by the trade-off between the rate and yield of energy metabolism (35). Shifts in this trade-off are poorly documented. Information on what determines metabolic efficiency would help to underpin analyses of the complex architecture of metabolic networks and to determine why, for example, systems biology models of extant organisms exhibit multiple metabolic flux distributions that are equally optimal (26, 38). Advances in modeling the systems biology of bacteria are being made (16), but many important questions still remain. Does metabolic plasticity extend to the rate-yield balance of organisms? How do the environment and evolutionary selection determine the overall efficiency of metabolism? How is the yield-rate trade-off shifted with prolonged nutrient limitation? In this study we examined these questions after perturbing metabolism through continuous culture of Escherichia coli in a glucose-limited environment.
Highly relevant to the questions posed above is that, perhaps paradoxically, one characteristic of bacteria is that microbial growth yields are often 50% less than the optimal yield (48). It is not clear why free-living bacteria like E. coli, which often grow and compete in nutrient-limited environments, evolved to have suboptimal growth yields. In our experiments, a single population of E. coli was subjected to prolonged resource limitation. When a bacterium faces a nutrient-limited environment, the intuitive expectation is that the organism adapts by increasing its efficiency for using the limiting substrate, or “K selection” (24). On the other hand, bacteria in populations may not have the luxury of evolving in this way, and maximization of the growth yield or metabolic efficiency does not appear to have been a dominant factor in the evolution of bacteria like E. coli (48). In situations where there is kinetic growth competition among members of the same culture, perhaps there is more selection for rate (r) than for yield (17). In a recent study MacLean and Gudelj looked at competition between energetically efficient and inefficient yeasts with temporal and spatial variation (25), but researchers have not exhaustively studied how prolonged nutrient limitation in an unstructured environment with a single resource or niche influences metabolic strategies, how the yield-rate trade-off is shifted with long-term selection, and whether redundancy in fitness solutions occurs.
Metabolic individuality in species such as E. coli (29) is, of course, only one facet of the significant intraspecies variation seen in bacteria (11). In most discussions researchers suggest that diversity arises from adaptive radiation into the myriad of structured environmental and biotic niches or from selection for specialization on alternate resources (21, 25, 41). It has also been suggested that the trade-off between the rate and yield of energy metabolism is influenced particularly by how environments are structured, with multiple niches or resources (23, 35). Investigation of metabolic adaptation in simple environments limited by a single resource has been limited to a small number of isolates from long-term cultures, like nutrient-limited chemostat cultures (39, 47). Long-term coexistence of phenotypically distinct clones has been demonstrated (10, 39, 40), and metabolic variance has been described for the creation of cross-feeding consortia partitioning metabolism in a long-term experimental population (36, 43). In general, population heterogeneity determined through phenotypic characterization was found in other studies of experimental evolution with E. coli (15, 18, 32, 34, 42). Recent work has added another dimension to the complexity of unstructured populations and has shown that there is no ecological or periodic selection limitation to phenotypic and genotypic divergence in single bacterial cultures (27). Here we describe the metabolic and energetic consequences of an adaptive radiation in a constant unstructured environment with a single resource.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For the evolution experiment, E. coli K-12 strain BW2952, an MC4100 derivative, was propagated for approximately 600 h (or 90 generations) at a growth rate of 0.1 h−1 in a glucose-limited chemostat exactly as described previously (33). Individuals were isolated from the culture each day, and the nine isolates analyzed were obtained by streaking a 600-h sample on nutrient agar and randomly picking colonies (27). Bacterial isolates chosen for metabolic assays and metabolome analyses were grown in glucose-limited chemostats with minimal medium A (MMA) at a dilution rate of 0.1 h−1 as described previously (33).
Uptake of glucose and oxygen.
The transport of glucose was measured by using the initial rate of uptake of 0.5 μM [U-14C]glucose added to bacteria withdrawn from the chemostat culture. The contributions of the Mgl and PtsG systems were determined from the uptake of 1 μM [U-14C]galactose and 10 μM methyl-α-d-[U-14C]glucoside, respectively. A 50-μl sample was taken directly from the chemostat and mixed with 50 μl MMA containing 1 μM [U-14C]glucose, 2 μM [U-14C]galactose, or 20 μM methyl-α-d-[U-14C]glucoside. Samples (20 μl) were removed at 10-, 20-, 30-, and 40-s intervals and immediately filtered through 0.45-μm cellulose nitrate membrane filters and washed with 10 ml MMA without a carbon source. The filters were counted in liquid scintillant by using a Beckman scintillation counter (LS6500; Beckman, United States), and the initial rate of transport was calculated. The rate of transport was expressed in picomoles of sugar transported per minute per 108 bacteria.
Oxygen uptake was measured with a Clarke-type oxygen electrode (Ranks Brothers Ltd., England) as described in the manufacturer's guide. Briefly, 3 ml of culture from a 24-h-old chemostat was transferred into the oxygen electrode chamber at 37°C. After determination of the oxygen uptake rate due to oxidation of endogenous metabolites, a glucose solution was added (final concentration, 15 μM) using a fine syringe needle. The glucose-dependent rates were based on a solubility of O2 of 6.7 mg/liter at 37°C and atmospheric pressure.
Metabolism of glucose to acetate or CO2.
To analyze the extracellular metabolites of wild-type and chemostat-adapted clones, 5-ml samples from glucose-limited chemostats were immediately filter sterilized by suction through a 0.22-μm MillexGP filter unit (Millipore, United States). Metabolites in the filtrates were analyzed by determination of 1H chemical shifts and their associated correlations using the comprehensive magnetic resonance spectroscopy methods described previously (2). For magnetic resonance spectroscopy, a 550-μl cell-free culture was mixed with 50 μl deuterium oxide and analyzed as described previously (2). Acetate in samples was quantified using an acetic acid UV method assay kit (Boehringer Mannheim/R-Biopharm, Germany).
Rates of complete oxidation of glucose by wild-type and evolved isolates were determined from the conversion of [U-14C]glucose to 14CO2. One milliliter of a culture whose optical density was known was removed from a 24-h-old glucose-limited chemostat culture (growth rate, 0.1 h−1) and transferred to a 5-ml flask with a separate well (diameter, 7 mm; height, 10 mm) containing 50 μl of 0.1 M NaOH. Sixty microliters of glucose (0.65 mM), including 5 μl of [U-14C]glucose (0.65 mM; 306 mCi/mmol), was added to the culture, which was incubated for 5, 10, 15, or 30 min in the sealed flask. The 14CO2 produced was trapped in a strip of Whatman chromatographic paper (thickness, 0.18 mm; width, 5 mm; length, 10 mm) soaked in the 50 μl of 0.1 M NaOH in the well. The strips of paper were transferred into vials containing 4 ml of scintillation fluid (ACS; Amersham, England), and the 14CO2 was measured using a Beckman scintillation counter (LS6500; Beckman, United States).
Physiological parameters.
In the chemostat culture, the dilution rate and thus the specific growth rate were constant (0.1 h−1). Hence, specific glucose consumption and acetate production rates were determined from the difference between the concentration in the feed medium and the concentration in the culture effluent, normalized to the steady-state biomass concentration in each culture.
Biolog assay for substrate utilization screening.
The catabolic activities of the starting strain and the chemostat isolates with 95 substrates were determined using the commercially available Biolog GN2 (Biolog, Hayward, CA) exactly according to the protocol provided by the vendor. Color changes were measured at 600 nm using a microplate reader (Multiskan RC; Labsystem, Finland) after 24 h of incubation at 37°C and were corrected for the no-substrate control. The optical densities at 600 nm were used for cluster analysis after scoring for each substrate as follows: 0, no catabolism; 1, catabolism of the substrate. The cutoff point between negative results and positive results was an optical density at 600 nm of 0.2. Substrates giving values close to the cutoff point were considered positive or negative if there was at least a fourfold difference in replicate readings between the ancestor and isolate. Substrates that gave variable results or substrates whose values hovered around the cutoff point in duplicate assays were considered noninformative (5) and were not used for cluster analysis. There were three such substrates.
Metabolome analysis.
The 14C-labeling and extraction conditions used for bacterial cultures have been described in detail elsewhere (28). For phylogenetic comparisons (29), the extracts were resolved in two dimensions on high-performance thin-layer chromatography (HPTLC) plates (Silica Gel 60; Merck, Germany) using solvent systems A and B (28, 44). Metabolite spots were detected on plates using a Molecular Dynamics PhosphorImager (Typhoon 8600; Amersham Biotech, United States) after 3 to 4 days in contact with a Molecular Dynamics storage phosphor screen. PhosphorImager files were processed using the Molecular Dynamics ImageQuant and Phoretix software (30). The Phoretix parameter settings used for spot detection were as follows: sensitivity, 200; operator size, 31; noise factor, 5; and background, 0. The intensity of each spot was normalized as a percentage of the total pixels on the plate. Data from spot matching with each isolate were used for cluster analysis after they were scored as follows: 1, spot present; 0, spot absent. For quantitative scoring, we used the following scale: 1, least intense spot used in the analysis; 2, two- to threefold difference in intensity from the least intense spot; 3, three- to sixfold difference in intensity from the least intense spot; and 4, more-than-sixfold difference in intensity from the least intense spot. Spots with an intensity that was <0.05% of the total pixel count in the analysis and spots which showed variable results in replicate analysis were considered noninformative and not included in the cluster analysis. At least three independent determinations were used for generation of the metabolome tree. A total of 177 spots were detected in the ancestral strain and the isolates analyzed from the chemostat culture. These spots did not include low-abundance metabolites, so the data were underestimates of the total metabolome (28, 44) but provided information that was more than sufficient for cluster analysis and strain differentiation within a species (29).
Cluster analysis.
We related differences in the metabolome and Biolog profiles of different E. coli isolates, including chemostat isolates, by using the neighbor-joining method for tree construction based on distances estimated by the two-parameter method (37). The reliability of the trees was confirmed by bootstrap analysis of 1,000 replications.
RESULTS
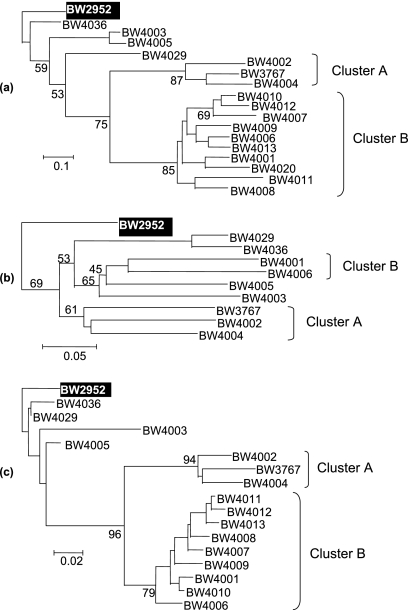
We examined metabolic adaptations to glucose limitation in a previously characterized, rapidly evolving population with high endogenous mutation rates (27, 33), as shown in Table 1. The nine strains included in Table 1 coexisted after 90 generations or 26 days of glucose limitation but belonged to separate phylogenetic clusters within the population when 11 informative phenotypic characteristics were examined (Fig. 1a) (27). Metabolic and energetic characterizations were performed in four distinct ways: first, by analysis of metabolic inputs (changes in glucose and O2 uptake rates); second, by analysis of the end products of energy metabolism (biomass yields, CO2, and acetate); third, by metabolome analysis of intracellular metabolite pools; and fourth, by Biolog screening for changes in the metabolism of 95 extracellular substrates.
TABLE 1.
Properties of the ancestor (BW2952) and isolates coadapted to glucose limitationa
| Strain | Maximal growth rate on glucose (h−1)b | Glucose uptake rate (pmol/min/108 cells)c | Contribution of Mgl to glucose transportd | Contribution of PtsG to glucose transporte | Oxygen uptake rate (nmol O2/min/108 cells)f | Biomass yield (g g−1)g | Acetate production (g g−1)h | Rate of glucose conversion to CO2 (pmol CO2/min/108 cells)i | Fitness relative to ancestorj | Ecostrategyk |
|---|---|---|---|---|---|---|---|---|---|---|
| BW2952 | 0.69 ± 0.01 | 120 ± 19 | 49 ± 13 | 64 ± 1 | 5.9 ± 0.22 | 0.41 ± 0.01 | <0.01 | 180 ± 11 | = | Ancestor |
| BW3767 | 0.77 ± 0.02 | 890 ± 74 | 1600 ± 349 | 100 ± 11 | 8.8 ± 0.33 | 0.52 ± 0.02 | <0.01 | 250 ± 28 | +++ | Mixed K and r |
| BW4001 | 0.53 ± 0.01 | 180 ± 81 | 230 ± 10 | 150 ± 26 | 10.8 ± 0.11 | 0.29 ± 0.01 | <0.01 | 560 ± 44 | +++ | R |
| BW4002 | 0.77 ± 0.03 | 400 ± 190 | 240 ± 34 | 270 ± 100 | 8.3 ± 0.22 | 0.48 ± 0.03 | <0.01 | 210 ± 23 | +++ | Mixed K and r |
| BW4003 | 0.69 ± 0.02 | 130 ± 41 | 37 ± 17 | 86 ± 56 | 7.5 ± 0.11 | 0.40 ± 0.01 | <0.01 | 270 ± 25 | ++ | ? |
| BW4004 | 0.69 ± 0.05 | 960 ± 76 | 1700 ± 189 | 200 | 9.2 ± 0.90 | 0.46 ± 0.01 | <0.01 | 230 ± 21 | +++ | Mixed K and r |
| BW4005 | 0.66 ± 0.01 | 97 ± 6 | 27 ± 32 | 63 ± 23 | 7.2 ± 0.11 | 0.34 ± 0.02 | 0.08 ± 0.01 | 240 ± 22 | − | New ecotype |
| BW4006 | 0.50 ± 0.02 | 370 ± 250 | 149 | 210 ± 145 | 10.1 ± 0.67 | 0.29 ± 0.01 | <0.01 | 530 ± 30 | +++ | r |
| BW4029 | 0.69 ± 0.01 | 140 ± 17 | 67 ± 26 | 85 ± 29 | 4.8 ± 0.22 | 0.52 ± 0.03 | <0.01 | 96 ± 13 | +++ | K |
| BW4036 | 0.69 ± 0.01 | 92 ± 4 | 35 ± 4 | 49 ± 4 | 7.2 ± 0.00 | 0.40 ± 0.02 | <0.01 | 81 ± 8 | = | ? |
The values are means ± standard deviations for replicates (n ≥ 2).
Growth rates were determined from the exponential growth phase for isolates and the wild type in minimal medium with 0.05% (wt/vol) glucose.
Measured using the rate of uptake of [14C]glucose by bacteria growing in a chemostat at a rate of 0.1 h−1. See Materials and Methods for details.
Measured using the Mgl-specific substrate [14C]galactose. The galactose transport rate shown is expressed in pmol/min/108 cells.
Measured using the PtsG-specific analog methyl-α-[14C]glucoside. The methyl-α-glucoside transport rate is expressed in pmol/min/108 cells. The large errors in some of the values reported were due to instability between replicate cultures, probably due to unstable genomic rearrangements affecting transporter genes.
The glucose-dependent respiration rate was measured for cultures transferred to a Clark oxygen electrode. See Materials and Methods for details.
Biomass yields (expressed in grams of cell dry mass produced per gram of glucose) for isolates and the wild type in the chemostat culture were determined from individual dry weight determinations.
Acetate production (expressed in grams of acetate per gram of glucose) by isolates and the wild type was determined by using the culture medium of 48-h-old glucose-limited chemostats grown at a rate of 0.1 h−1.
Rates of [14C]glucose oxidation to 14CO2 were determined as described in Materials and Methods.
Levels of fitness in glucose-limited chemostats were compared by using 50/50 starting population mixtures of the ancestor and an isolate. For ancestor-ancestor competition, a neutral marker was introduced into one competitor as previously described (27). +++, isolate eliminated ancestor within 72 h; =, no change; −, ancestor dominated but did not eliminate isolate; ++, isolate dominated but did not eliminate ancestor within 72 h (27).
The groups are discussed in the text.
FIG. 1.
Phylogenetic relationships among E. coli isolates that evolved in a single population. The sympatric divergence of phenotypic, genotypic, and metabolic characteristics of evolved clones was analyzed by the neighbor-joining method rooted with the ancestral strain, BW2952, as described by Pupo et al. (37). Cluster A contains rpoS mutants, and cluster B strains are low-ppGpp strains (27). The dendrogram (a) was based on the 11 characteristics of 41 isolates described in reference 27. The metabolome tree (b) was based on spot matching and quantitation as described in reference 29. The number of informative spots compared for all isolates was 177. The Biolog tree (c) was based on the 23 informative substrates differentially utilized by the isolates and ancestor shown in Table 2, including all members of cluster B. The bootstrap values at nodes are percentages based on 1,000 replications.
Both the maximal growth rate and the growth yield (or efficiency of glucose conversion into biomass) were distinctly heterogeneous in the isolates under glucose limitation conditions (Table 1), consistent with previously observed differences between individual isolates in other studies (8, 19). Three strains (BW4001, BW4005, and BW4006) were notable in that the yields of all three were strongly reduced and two of them had reduced growth rates at high glucose levels. These strains appeared as “small-colony variants” on agar plates and were previously observed in other experimental evolution studies (19, 45). In addition to this polymorphism, Table 1 shows that the nine isolates did not converge on a single optimum for glucose and O2 uptake rates or in terms of the production of waste products of energy metabolism (CO2 and acetate).
The uptake of a limiting nutrient was assayed using low concentrations of 14C-labeled substrates. Perhaps counterintuitively, the prolonged selection under glucose limitation conditions did not result in uniformly high rates of uptake of glucose in evolved strains. There was variation in both the overall rates and the relative contributions of the two major routes of glucose transport into E. coli (the Mgl and PtsG transport systems [13]) (Table 1). The Mgl and PtsG systems have distinct energetic costs, and differential usage of these systems results in altered ATP balances (13). BW4005 and BW4036 exhibited, if anything, a lower rate of glucose uptake than the ancestral strain. These isolates most likely diverged into distinct ecotypes and evolved to compete for resources other than glucose in the long-term chemostat. The results of experiments examining competition between isolates and between isolates and the ancestor are consistent with this conclusion (Table 1) (27). The other isolates exhibited slightly to very elevated glucose uptake rates; BW4002 and BW4006 were more reliant on PtsG, whereas BW3767 and BW4004 were heavily reliant on MglBAC. The large errors in some of the reported values are due to instability between replicate cultures, probably because of unstable genomic rearrangements affecting transporter genes (unpublished results). Evidently, there were multiple fitness solutions for glucose transport, and the ancestral redundancy in glucose uptake pathways provided the bacteria with alternative evolutionary solutions to the same selection conditions.
Oxygen uptake rates also evolved and were dissimilar in different isolates (Table 1). Except for one strain (BW4029), chemostat-evolved bacteria had elevated respiration rates with glucose as an oxidizable substrate. The low-yield bacteria had particularly elevated respiration rates, raising the possibility of faster energy metabolism and increased glucose throughput to CO2. The elevated respiration rates suggested that there was an emphasis on rate solutions rather than efficiency solutions for competition under nutrient limitation conditions. Consistent with this notion, the rates of CO2 production from glucose were significantly different for different strains, as also shown in Table 1. In the low-yield isolates, such as BW4001 and BW4006, there was significantly elevated conversion of glucose into CO2. Further analysis of culture media using proton nuclear magnetic resonance spectroscopy of fermentation products (2) showed that acetate was a prominent peak in the medium of one strain (BW4005). No major peaks corresponding to soluble carbon products were detected in the media of other isolates. The acetate production by BW4005 was confirmed by an enzymatic assay (Table 1). Interestingly, acetate production was not found in the phenotypically most similar strain, BW4003 (Fig. 1a).
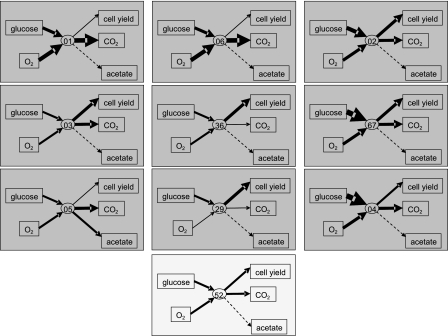
The changes in metabolism adopted by each isolate are shown schematically in Fig. 2; an ecological interpretation is shown in Table 1. The low-yield isolates (BW4001 and BW4006) exhibited significantly elevated levels of respiration and conversion of glucose into CO2; in these strains there was accelerated energy metabolism, but at the cost of a reduced biomass yield (>25% reduction). Despite the stable environment, these strains are evidently inefficient r strategists. BW4005 appears to be similar and produces acetate as well as CO2, but in contrast to BW4001 and BW4006, it was not competitive against the ancestor in reconstituted glucose-limited chemostats (27). This suggested that the evolutionary success of BW4005 was due to its behavior as a new ecotype, dependent on unidentified resources in the evolving population and not necessarily on being effective in glucose utilization. BW4003 was also difficult to assess in terms of ecological strategy, as it had moderately elevated rates of transport, respiration, and fitness but the yield efficiency on glucose was unchanged. More clear-cut was BW4029's solution to fitness; this strain had a >20%-higher growth efficiency than the ancestor and exhibited a decreased loss of resources through respiration. Hence, a K strategy also resulted in competitive fitness under glucose limitation conditions. Mutational changes in the redundant but energetically different respiratory chains of E. coli can increase growth yields (3), so mutations in these components may explain altered respiration. BW4036, like BW4029, exhibited greatly decreased CO2 production, but the transport or growth yields of BW4036 were not particularly altered, so again it was unique. In several other high-biomass strains (BW3767, BW4002, and BW4004) yet another hybrid approach was observed; there were significantly elevated levels of transport, respiration, and CO2 production but distinct 10 to 20% increases in the growth yields. Most strikingly, evolution with at least six different metabolic rate-yield strategies occurred side by side with a single limiting resource. In addition, an additional strategy, the acetate cross-feeding polymorphism noted by Treves et al. (43), could be used if acetate producers such as BW4005 eventually become more common in a population.
FIG. 2.
Heterogeneity of metabolic processes in coevolved bacteria. The thickness of the arrows indicates the magnitude of the measured fluxes in the ancestral strain (light box) and isolates (shaded boxes). The rates of glucose and oxygen uptake and the rates of production of biomass, CO2, and acetate are based on the data in Table 1. The numbers in the ellipses are the last two digits of the strain numbers in Table 1. Dashed lines indicate undetectable rates.
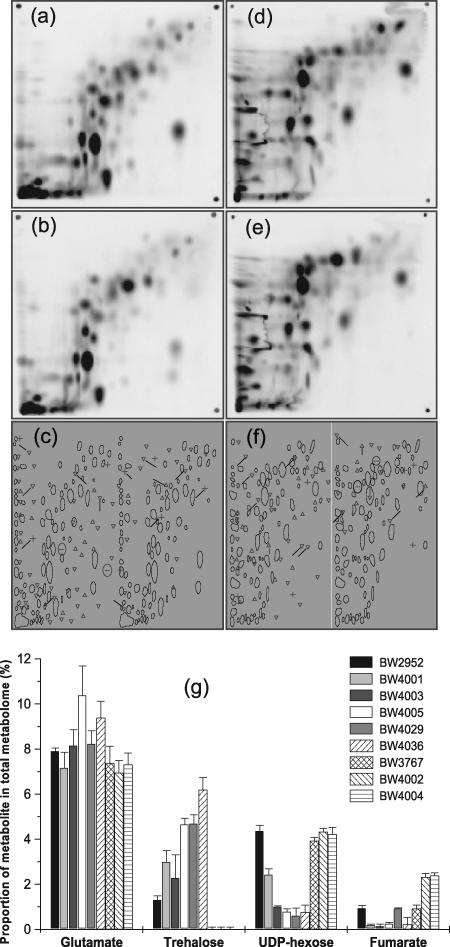
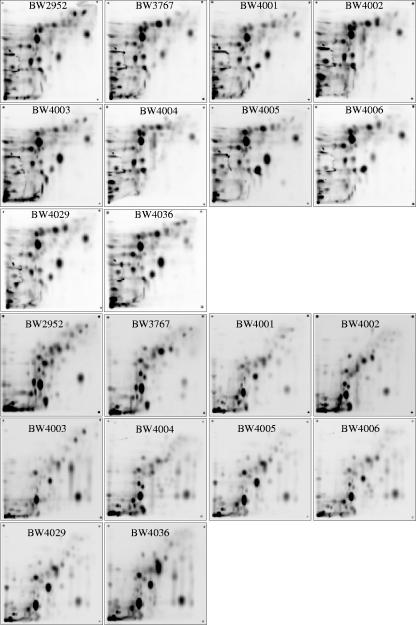
Additional evidence for metabolic divergence was obtained from the analysis of the intracellular metabolite pools in isolates. Metabolome analysis by two-dimensional HPTLC provides a powerful tool for typing bacteria at the subspecies level (29) and, as shown in Fig. 3, also resolves clonally evolved differences. Analysis of intracellular metabolites present in the starting strain and the nine representative isolates (Fig. 4) revealed major differences in metabolism; none of the isolates tested in the various clusters shown in Fig. 1a had a pattern of metabolites (presence or absence of spots or differences in quantity) (Fig. 3a and b and 4) identical to the pattern of another strain or to the pattern of the starting strain when the organisms were grown individually in a glucose-limited chemostat. The changes in the evolved strains were far greater than the changes obtained by repeated analysis of the ancestor strain grown under the selection conditions used (29). Use of the data for 177 metabolite spots reproducibly detected for all the strains to construct a dendrogram (Fig. 1b) showed that the metabolome profiles were very distinctive for the isolates tested. The differences between isolates included both increases and decreases in the amounts of different classes of metabolites, such as UDP-sugars, citric acid cycle intermediates, or trehalose, compared to the amounts in the starting strain (Fig. 3g). After selection, there was a >10-fold difference in the concentrations of common central metabolites, like fumarate, between isolates. The clustering in the metabolome-based dendrogram shown in Fig. 1b was generally consistent with the phenotypic division of the strains shown in Fig. 1a, with both clusters A and B identified by the independent approaches. The other, more dispersed strains in Fig. 1a were related somewhat differently as determined by metabolomics. Metabolomic differences thus largely paralleled phenotypic divergence, but isolates with very similar phenotypic traits (e.g., BW4002 and BW4004 or BW4003 and BW4005) were still differentiated in metabolomic profiles, and there were 34 spot differences (20 differences in the presence of a spot; 14 differences in the quantity of a spot) between BW4002 and BW4004.
FIG. 3.
Metabolome profiles for chemostat-evolved and ancestral clones. The metabolites were extracted (28) from E. coli K-12 strain BW2952 (a and d), the ancestral strain in chemostat experiments (33), and BW3767, a cluster A isolate obtained after 90 generations (b and e). The images are PhosphorImager outputs for 14C-labeled metabolites separated by two-dimensional HPTLC using pairs of solvent systems. In panels a and b the compounds were separated using solvent system A, and panels d and e were obtained using solvent system B (28, 44). Resolved spots in metabolome fingerprints were matched using the Phoretix software (30). (c and f) Results of an analysis of matched spots in the evolved and ancestral metabolomes with solvent systems A and B, respectively. ▿, metabolite spots that were down-regulated; ▵, metabolite spots that were up-regulated; −, metabolite spots that were not present; +, metabolite spots that were unique in the evolved clone. Spots with no symbol were the metabolites whose proportions were the same in the two metabolomes. (g) Variation in the metabolome proportions for four metabolite spots in the isolates. The spots identified as glutamate, UDP-hexose, and fumarate using methods described previously (28, 44) were quantitated in four replicate metabolome estimations. The error bars indicate standard deviations.
FIG. 4.
Metabolome profiles of ancestral and evolved clones. The images are the phosphorimages of 14C-labeled metabolites of ancestral strain BW2952 and isolates BW3767, BW4001 to BW4006, BW4029, and BW4036 that coevolved in the 90-generation chemostat population. Extracts were prepared and separated by two-dimensional HPTLC using solvent systems A (bottom 10 images) and B (top 10 images) as described in the legend to Fig. 3.
The metabolome analysis revealed an even more surprising result: only 68 of the 177 metabolite spots studied were conserved in all 10 strains, indicating that there was major restructuring of metabolism in different isolates. These results demonstrate the huge metabolic plasticity in E. coli, already suggested by the low number of conserved metabolites in diverse members of the species E. coli (29).
Testing the transformation of exogenous substrates provided another perspective on the metabolic variance within the chemostat culture, which was analyzed using the compounds present in 96-well Biolog plates (5). As shown in Table 2 and Fig. 1c, Biolog GN2 profiling strongly differentiated the chemostat isolates from the ancestor. All isolates gained and lost abilities to utilize various substrates during the 90 generations of glucose-limited growth (Table 2), consistent with numerous mutations in the lifetime of the population. Gains in metabolic capabilities due to pleiotropic mutations in rpoS, leading to improved metabolism of poor substrates (22), was evident with rpoS mutants BW3767, BW4002, and BW4004 (27). Altogether, the changes in the 23 informative substrates permitted construction of the dendrogram shown in Fig. 1c. In most respects, including the separate grouping of the rpoS bacteria (cluster A), evolved differences in the metabolism of external substrates gave the same phylogenetic characteristics as phenotypic and metabolomic typing. However, the trees obtained by metabolomic and Biolog analyses were not entirely congruent, suggesting that intracellular metabolite pools and extracellular substrate utilization capabilities do not reflect the same mutational adaptations, particularly in the rpoS+ bacteria that do not belong to cluster A. Biolog profiling was extended to a more detailed analysis of all members of one phenotypic cluster (cluster B), as shown in Fig. 1a. All of the isolates characterized by a reduced growth yield were closely related, but they exhibited individual differences. As shown in Fig. 1c, all nine isolates in cluster B were differentiated. Together with the phenotypic divergence (27), these results are indicative of concurrent and on-going variegation in the culture within a much shorter time scale than previously tested in experimental populations (21, 34, 39). In yeast cultures metabolic properties also change within 250 generations during glucose-limited growth (12), but the extent of intrapopulation divergence and the rapidity of accumulation of the metabolic changes were not suspected previously.
TABLE 2.
Informative substrates from Biolog analysis of evolved isolatesa
| Substrate | Response
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BW2952 | BW3767 | BW4001 | BW4002 | BW4003 | BW4004 | BW4005 | BW4006 | BW4009 | BW4007 | BW4008 | BW4010 | BW4011 | BW4012 | BW4013 | BW4029 | BW4036 | |
| Dextrin | − | + | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Glycogen | − | − | − | − | − | − | − | + | + | + | + | − | − | − | − | − | − |
| Tween 80 | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − |
| Lactulose | − | + | + | + | − | + | − | + | + | + | + | + | + | + | + | − | − |
| Maltose | − | + | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − |
| β-Methyl-d-Glucoside | − | + | − | + | − | + | + | + | − | + | − | − | + | + | + | − | − |
| Turanose | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Pyruvic acid methyl ester | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Succinic acid mono methyl ester | − | + | − | − | − | − | − | − | + | + | + | + | + | − | + | − | − |
| Acetic acid | + | − | + | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| Formic acid | − | − | + | − | − | − | − | + | + | + | + | + | + | + | + | − | − |
| α-Ketoglutaric acid | − | + | + | + | − | + | − | + | + | + | + | + | + | + | + | − | − |
| α-Ketovaleric acid | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Sacchiric acid | − | + | + | + | − | + | − | − | + | + | + | + | + | + | + | − | − |
| Succinic acid | − | + | + | + | − | + | − | + | + | + | + | + | + | + | + | − | − |
| Bromosuccinic acid | − | − | − | − | − | − | − | − | − | + | + | − | − | − | + | − | − |
| d-Alanine | − | + | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − |
| l-Alanyl-glycine | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| l-Asparagine | − | + | + | − | − | + | − | + | + | + | + | + | + | + | + | − | − |
| l-Aspartic acid | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − |
| Glycyl-l-aspartic acid | + | − | + | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| Glycyl-l-glutamic acid | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | − | − |
| l-Threonine | − | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − |
| Thymidine | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| l-Serine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
The cutoff point between negative responses and positive responses in the Biolog assay was an optical density at 600 nm of 0.2. The responses of substrates giving values close to the cutoff point were considered positive or negative if there was at least a fourfold difference in replicate readings between the ancestor and the isolate (see Materials and Methods for details).
DISCUSSION
In this study, we used a domesticated laboratory strain of E. coli and subjected it to prolonged growth in an artificial (chemostat) environment. The population size also may not represent the size of an E. coli population in a natural habitat. Despite these artificial aspects, the extent of biological change observed with a domesticated strain can still tell us a lot about the underlying flexibility of bacterial responses to a new environment. Although one population was analyzed, seven other chemostat populations exhibited the same level of divergence, although they did not always exhibit the same combination of changes (S. Seeto and T. Ferenci, submitted for publication).
Our bioenergetic and metabolomic results are indicative of rapid, multiple metabolic adaptations not previously recognized in experimental bacterial populations (16, 39). The scope of sympatric divergence and the rapidity of accumulation of such changes were also not recognized in yeast cultures during glucose-limited growth (7). Extrapolated to an ecological context, these results obviate the postulated need for multiple niches for bacterial diversification (21) or for adapting energetic efficiency (35). Contrary to predictions (35), derivatives of a single parental strain in a constant environment diverge to take both sides of what was proposed previously to be a fundamental rate-yield trade-off affecting growth energetics. The great plasticity of the yield-rate balance suggests that these characteristics, as well as other metabolic properties (14), are frequently readjusted in bacterial evolution and are not fundamental properties of organisms. The robustness of bacterial evolution is underpinned by the capacity for metabolic adaptive radiation, even in a homogeneous, nearly constant environment. Rapid metabolic divergence in bacteria under suboptimal nutritional conditions, including losses and gains in the utilization of substrates, is undoubtedly a source of diversity and intraspecies heterogeneity (11).
The glucose-limited E. coli population partitioned into populations with several approaches to resource use. As summarized in Table 1, some isolates adopted energetically expensive glucose transport mechanisms (the Mgl system), whereas others adopted the more economical PtsG system (31). Metabolically, some evolved isolates were inefficient, with rapid respiration and production of large amounts of CO2. Another way of looking at this class is that they were “cheaters” depriving other organisms of available resources (25). Concurrently, the conservationist strategy of a minority of isolates involved increased energetic efficiency and reduced waste. Hybrid strategies were particularly common, and these strategies involved various increases in energetic efficiency along with increased rates of substrate transformation. Alternative ecotypes (4) or dropouts no longer competing for the original resource used alternative energy sources not used by the ancestor population. In extending the conclusions from the nine isolates studied to the population as a whole, it is worth noting that of the 41 coevolving isolates studied in previous experiments (27), 9 were wasters, 18 exhibited the opposite adaptation and increased growth yields (mostly with hybrid strategies), and 12 maintained the same yield-rate balance as the parental strain. Only a minority, including the acetate producer, appeared to adapt into an alternative ecotype, and likewise, only one isolate was an outright conservationist. Of course, these proportions are representative of a particular time in a particular culture, and an additional week of selection resulted in an increase in the proportion of isolates derived from polluters (to about 50% of the population) (results not shown).
The diversity in metabolic properties defined in this study must reflect the availability of alternative evolutionary pathways to bacteria for adaptation to a glucose-limited environment. Of course, not all the changes have been proven to contribute to fitness. Some of the changes (e.g., loss of utilization of some substrates or loss of metabolite spots) may have been neutral under the selection conditions used, but other changes were clearly adaptive. Recent studies have suggested that prolonged selection with a single carbon source results in convergent metabolic phenotypes resulting from different transcriptional changes (16). These and other previous studies completely missed the potential for rapid and extensive metabolic diversification in bacterial populations. Metabolomic data and Biolog profiling both point to metabolic divergence to the point of individuality in populations. Bacteria adapt to the same metabolic problem (glucose limitation) by taking both sides of the rate-yield divide and clonally evolve to maintain a very small shared core of metabolite pool characteristics.
The inherent robustness of bacterial processes extends to evolutionary outcomes. This conclusion is consistent with the current view that bacterial metabolic pathways are robust, because many loss-of-function mutations do not eliminate metabolic network capabilities and the ability to grow (9). The same robustness and redundancy of functions probably contribute to the ability of bacteria to adapt their metabolism through alternative beneficial mutations in challenging environments. Our evidence suggests that, as in the case of transport changes, robustness in this system is due to redundancy, although the possibility of distributed robustness cannot be excluded (46). Not only does metabolic and physiological redundancy stabilize already evolved organisms so that they can withstand immediate environmental challenges, but our results also suggest that robustness lays the foundation for providing alternative evolutionary solutions in the presence of prolonged stress.
Finally, what does the seemingly limitless flexibility of metabolic adaptation say about the modeling of metabolism in the systems biology of organisms? There is a growing expectation that computational approaches can explain biological systems (6, 20). It remains to be seen whether the challenge of the explosive radiation represented by our results can be satisfactorily met by in silico models.
Acknowledgments
We thank Bill Bubb for performing the nuclear magnetic resonance analysis.
We thank ARC for past funding support.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Brown, J. H., J. F. Gillooly, A. P. Allen, V. M. Savage, and G. B. West. 2004. Toward a metabolic theory of ecology. Ecology 85:1771-1789. [Google Scholar]
- 2.Bubb, W. A., L. C. Wright, M. Cagney, R. T. Santangelo, T. C. Sorrell, and P. W. Kuchel. 1999. Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: identification of possible virulence factors. Magn. Reson. Med. 42:442-453. [DOI] [PubMed] [Google Scholar]
- 3.Calhoun, M. W., K. L. Oden, R. B. Gennis, M. J. de Mattos, and O. M. Neijssel. 1993. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 175:3020-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, V. S., and R. E. Lenski. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407:736-739. [DOI] [PubMed] [Google Scholar]
- 6.Covert, M. W., E. M. Knight, J. L. Reed, M. J. Herrgard, and B. O. Palsson. 2004. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429:92-96. [DOI] [PubMed] [Google Scholar]
- 7.Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown, F. Rosenzweig, and D. Botstein. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:16144-16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dykhuizen, D., and D. Hartl. 1981. Evolution of competitive ability in Escherichia coli. Evolution 35:581-594. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, J. S., and B. O. Palsson. 2000. Robustness analysis of the Escherichia coli metabolic network. Biotechnol. Prog. 16:927-939. [DOI] [PubMed] [Google Scholar]
- 10.Elena, S. F., and R. E. Lenski. 1997. Long-term experimental evolution in Escherichia coli. 7. Mechanisms maintaining genetic variability within populations. Evolution 51:1058-1067. [DOI] [PubMed] [Google Scholar]
- 11.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483-495. [DOI] [PubMed] [Google Scholar]
- 12.Ferea, T. L., D. Botstein, P. O. Brown, and R. F. Rosenzweig. 1999. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. USA 96:9721-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferenci, T. 1996. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol. Rev. 18:301-317. [DOI] [PubMed] [Google Scholar]
- 14.Ferenci, T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong, S. S., A. R. Joyce, and B. O. Palsson. 2005. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 15:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong, S. S., J. Y. Marciniak, and B. O. Palsson. 2003. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol. 185:6400-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friesen, M. L., G. Saxer, M. Travisano, and M. Doebeli. 2004. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58:245-260. [PubMed] [Google Scholar]
- 19.Helling, R. B., C. N. Vargas, and J. Adams. 1987. Evolution of Escherichia coli during growth in a constant environment. Genetics 116:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420:186-189. [DOI] [PubMed] [Google Scholar]
- 21.Kassen, R., and P. B. Rainey. 2004. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58:207-231. [DOI] [PubMed] [Google Scholar]
- 22.King, T., A. Ishihama, A. Kori, and T. Ferenci. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreft, J. U., and S. Bonhoeffer. 2005. The evolution of groups of cooperating bacteria and the growth rate versus yield trade-off. Microbiology 151:637-641. [DOI] [PubMed] [Google Scholar]
- 24.MacArthur, R. H., and E. O. Wilson. 1967. The theory of island biogeography. Princeton University Press, Princeton, NJ.
- 25.MacLean, R. C., and I. Gudelj. 2006. Resource competition and social conflict in experimental populations of yeast. Nature 441:498-501. [DOI] [PubMed] [Google Scholar]
- 26.Mahadevan, R., and C. H. Schilling. 2003. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5:264-276. [DOI] [PubMed] [Google Scholar]
- 27.Maharjan, R., S. Seeto, L. Notley-McRobb, and T. Ferenci. 2006. Clonal adaptive radiation in a constant environment. Science 313:514-517. [DOI] [PubMed] [Google Scholar]
- 28.Maharjan, R. P., and T. Ferenci. 2003. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 313:145-154. [DOI] [PubMed] [Google Scholar]
- 29.Maharjan, R. P., and T. Ferenci. 2005. Metabolomic diversity in the species Escherichia coli and its relationship to genetic population structure. Metabolomics 1:235-242. [Google Scholar]
- 30.Mahon, P., and P. Dupree. 2001. Quantitative and reproducible two-dimensional gel analysis using Phoretix 2D Full. Electrophoresis 22:2075-2085. [DOI] [PubMed] [Google Scholar]
- 31.Muir, M., L. Williams, and T. Ferenci. 1985. Influence of transport energization on the growth yield of Escherichia coli. J. Bacteriol. 163:1237-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notley-McRobb, L., and T. Ferenci. 1999. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ. Microbiol. 1:33-43. [DOI] [PubMed] [Google Scholar]
- 33.Notley-McRobb, L., S. Seeto, and T. Ferenci. 2003. The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadopoulos, D., D. Schneider, J. Meier-Eiss, W. Arber, R. E. Lenski, and M. Blot. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. USA 96:3807-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeiffer, T., S. Schuster, and S. Bonhoeffer. 2001. Cooperation and competition in the evolution of ATP-producing pathways. Science 292:504-507. [DOI] [PubMed] [Google Scholar]
- 36.Porcher, E., O. Tenaillon, and B. Godelle. 2001. From metabolism to polymorphism in bacterial populations: a theoretical study. Evolution 55:2181-2193. [DOI] [PubMed] [Google Scholar]
- 37.Pupo, G. M., D. K. R. Karaolis, R. T. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed, J. L., and B. O. Palsson. 2004. Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: assessment of correlated reaction subsets that comprise network states. Genome Res. 14:1797-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenzweig, R. F., R. R. Sharp, D. S. Treves, and J. Adams. 1994. Microbial evolution in a simple unstructured environment: Genetic differentiation in Escherichia coli. Genetics 137:903-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rozen, D. E., and R. E. Lenski. 2000. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 155:24-35. [DOI] [PubMed] [Google Scholar]
- 41.Schluter, D. 2000. The ecology of adaptive radiation. Oxford University Press, Oxford, United Kingdom.
- 42.Schneider, D., E. Duperchy, E. Coursange, R. E. Lenski, and M. Blot. 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treves, D. S., S. Manning, and J. Adams. 1998. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol. Biol. Evol. 15:789-797. [DOI] [PubMed] [Google Scholar]
- 44.Tweeddale, H., L. Notley-McRobb, and T. Ferenci. 1998. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 180:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyerman, J., N. Havard, G. Saxer, M. Travisano, and M. Doebeli. 2005. Unparallel diversification in bacterial microcosms. Proc. R. Soc. Lond. B Biol. Sci. 272:1393-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, A. 2005. Distributed robustness versus redundancy as causes of mutational robustness. Bioessays 27:176-188. [DOI] [PubMed] [Google Scholar]
- 47.Weikert, C., U. Sauer, and J. E. Bailey. 1997. Use of a glycerol-limited, long-term chemostat for isolation of Escherichia coli mutants with improved physiological properties. Microbiology 143:1567-1574. [DOI] [PubMed] [Google Scholar]
- 48.Westerhoff, H. V., K. J. Hellingwerf, and K. van Dam. 1983. Thermodynamic efficiency of microbial growth is low but optimal for maximum growth rate. Proc. Natl. Acad. Sci. USA 80:305-309. [DOI] [PMC free article] [PubMed] [Google Scholar]