Abstract
Streptomyces coelicolor A3(2) does not have a canonical cell division cycle during most of its complex life cycle, yet it contains a gene (ftsKSC) encoding a protein similar to FtsK, which couples the completion of cell division and chromosome segregation in unicellular bacteria such as Escherichia coli. Here, we show that various constructed ftsKSC mutants all grew apparently normally and sporulated but upon restreaking gave rise to many aberrant colonies and to high frequencies of chloramphenicol-sensitive mutants, a phenotype previously associated with large terminal deletions from the linear chromosome. Indeed, most of the aberrant colonies had lost large fragments near one or both chromosomal termini, as if chromosome ends had failed to reach their prespore destination before the closure of sporulation septa. A constructed FtsKSC-enhanced green fluorescent protein fusion protein was particularly abundant in aerial hyphae, forming distinctive complexes before localizing to each sporulation septum, suggesting a role for FtsKSC in chromosome segregation during sporulation. Use of a fluorescent reporter showed that when ftsKSC was deleted, several spore compartments in most spore chains failed to express the late-sporulation-specific sigma factor gene sigF, even though they contained chromosomal DNA. This suggested that sigF expression is autonomously activated in each spore compartment in response to completion of chromosome transfer, which would be a previously unknown checkpoint for late-sporulation-specific gene expression. These results provide new insight into the genetic instability prevalent among streptomycetes, including those used in the industrial production of antibiotics.
Bacteria of the gram-positive genus Streptomyces are of great interest for their mycelial growth and complex development, as well as being major producers of antibiotics. On solid media, Streptomyces spores germinate and grow into a vegetative mycelium from which aerial hyphae emerge, eventually developing into chains of unigenomic spores. Vegetative hyphae have only sparse septation without actual cell separation; each hyphal compartment contains several unsegregated copies of the chromosome (16). In contrast, during sporulation of aerial hyphae, regularly spaced septa form synchronously in long multigenomic compartments, each receiving one chromosome, and cell separation eventually takes place. Unlike the situation in dividing bacteria such as Escherichia coli, initiation of sporulation septation in Streptomyces often occurs over unsegregated chromosomes, which do not separate until very late in septation (31, 39). This raises the question of how Streptomyces clears chromosomal DNA from sites of septal closure to ensure faithful segregation of the replicated linear chromosomes to progeny spores, avoiding guillotining by septal closure. Completion of chromosome transfer to daughter cells in E. coli is carried out by a septum-located multifunctional protein, FtsK, which is deposited at the FtsZ ring and couples closure of the septum (cytokinesis) with resolution of chromosome dimers (1, 5, 52). In addition, FtsK interacts with the ParC subunit of topoisomerase IV to activate chromosome decatenation (15) and functions as a DNA translocase that pumps DNA through the closing septum, thereby ensuring that the sister chromosomes are fully partitioned into daughter cells (34). The closest homologue of FtsK in Bacillus subtilis is SpoIIIE; the two proteins have similar C termini but different N termini (50). SpoIIIE is essential for sporulation (41), localizing to the asymmetric sporulation septum, where it translocates trapped chromosomal DNA into the prespore compartment (4, 7, 40). SpoIIIE-mediated chromosome transfer is needed to activate the sigma-factor-mediated cascade of gene regulation underpinning forespore development (14).
FtsK homologues are present in most, but not all, bacteria (24). The Streptomyces coelicolor A3(2) ftsK-like gene SCO5750 (6), termed ftsKSC here, encodes a 929-amino-acid (aa) protein with 45% identity in a 552-aa overlap with FtsK of E. coli. Streptomyces avermitilis also contains such a gene (84% amino acid identity to FtsKSC) in a conserved genetic context (23). If FtsKSC were to be involved in the completion of chromosome transfer during septation, one might predict that some spores of an ftsKSC mutant would receive incomplete chromosomes. This could be relevant to the long history of reports of genetic, and associated phenotypic, instability as a major, universal, and industrially inconvenient feature of Streptomyces, which is commonly associated with large deletions in the end regions of the linear chromosomes (11, 44). Such deletions can result in linear chromosomes becoming circular, and displaying further instability in the offspring, without major effects on viability.
In this study, we used mutations and gene fusions to obtain evidence that FtsKSC plays an important role in the maintenance of an intact genome through sporulation but is not required for cell division. We also show that the late-sporulation gene expression program is at least partially spore compartment specific and is impaired in some spore compartments of ftsKSC null mutants.
MATERIALS AND METHODS
Bacterial strains and media.
The strains used in this study are listed in Table 1. Streptomyces strains were grown on mannitol soy flour agar (MS) minimal medium (MM), and R2YE agar and in yeast extract-malt extract liquid medium at 30°C (27). E. coli DH5α was used to construct recombinant plasmids (21), and ET12567/pUZ8002 was used to drive conjugative transfer of nonmethylated DNA from E. coli to S. coelicolor and to prepare plasmids for transformation of S. coelicolor protoplasts (18). E. coli BW25113/pIJ790 was used for PCR-targeted disruptions, and BT340 was used for the construction of in-frame-deleted mutants (20). Apramycin (50 μg ml−1), thiostrepton (10 μg ml−1), chloramphenicol (25 μg ml−1), ampicillin (100 μg ml−1), and kanamycin (50 μg ml−1) were used for selection if required.
TABLE 1.
Bacterial strains and oligonucleotides
| Strain or oligonucleotide | Characteristics | Source or reference | ||
|---|---|---|---|---|
| Strains
|
||||
| E. coli
|
||||
| DH5α | supE44 lacU169(80lacZ M15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 21 | ||
| BW25113/pIJ790 | K-12 derivative: ΔaraBAD, ΔrhaBAD/λ-Red(gam bet exo) cat araC rep101(Ts) | 20 | ||
| BT340 | DH5α/pCP20 | 20 | ||
| ET12567/pUZ8002 | dam, dcm, hsdS, cat, tet, tra, neo, RP4 | 18 | ||
| Streptomyces coelicolor
|
||||
| M145 | SCP1- SCP2- | 6 | ||
| WL1 | M145 ΔftsK:: acc(3)IV-oriT | This work | ||
| WL11 | M145 ΔftsKSC in-frame deletion mutant | This work | ||
| WL12 | M145 ΔSCO5751 in-frame deletion mutant | This work | ||
| Oligonucleotide primers for PCR amplification
|
||||
| ftsk1 | 5′GAGGGGCGACACGAACGGGTGAAGCGGTAGGCACACGTGATTCCGGGGATCCGTCGACC | |||
| ftsk2 | 5′GGCGGATCGTCGGCGGAACCCTTCTCCTACCCGCCCCTATGTAGGCTGGAGCTGCTTC | |||
| oriT-1 | 5′GCGTCGCTTGGTCGGTCATTTCGAACCCCAGAGTCCCGCATTCCGGGGATCCGTCGACC | |||
| oriT-2 | 5′CGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGATGTAGGCTGGAGCTGCTTC | |||
| WL4-3 | 5′CGTCAACTGACGTCGGGAGT | |||
| WL4-5 | 5′TCAGGGGATTTGCCGTTGGA | |||
| SC7C706-1 | 5′CCAGCACAGGCGGCTAAACGCTCGAAAGGCGCCCCCGTGATTCCGGGGATCCGTCGACC | |||
| SC7C706-2 | 5′CCGGCCGACCCCGCGTGAACGCGGTCCCCGCACCGCCTATGTAGGCTGGAGCTGCTTC | |||
| SC7C706-3 | 5′GCACCAAGCAGAAGAAGGAG | |||
| SC7C706-4 | 5′TTCAGGCATGGGCTCAAGAC | |||
| ftsKn-1 | 5′GGGCAGGGCGCCAAGAACCTCGACCCGGCCCACCGCAAGATTCCGGGGATCCGTCACC | |||
| ftsKn-2 | 5′GCCGAGCCGCACCCCGAGCTGCCGCAGCCGCTGCGGGATTGTAGGCTGGAGCTGCTTC | |||
| ftsKL-1 | 5′TGGCGCGAGGCGCTGCCCGCCCGCCCCCGCAAGCGCGCCATTCCGGGGATCCGTCACC | |||
| ftsKL-2 | 5′GGCGTCGTTGGCCGCGCTGCGCGCCTTGCCGGGTCCGCCTGTAGGCTGGAGCTGCTTC | |||
| ftsKc-1 | 5′GCCTTCGGCAAGGACGTCGAGGGCGGCTACGTCATGCACATTCCGGGGATCCGTCACC | |||
| ftsKc-2 | 5′CCCTTCAGACTCCCCACGGATCACCGCGAGCACGCCGTCTGTAGGCTGGAGCTGCTTC | |||
| egfp1 | 5′GACGGCGTGCTCGCGGTGATCCGTGGGGAGTCTGAAGGGGCCATGGTGAGCAAGGGCGA | |||
| egfp6 | 5′GGTCGACGGATCCCCGGAATTTACTTGTACAGCTCGTCCAT | |||
| egfp2 | 5′ATGGACGAGCTGTACAAGTAAATTCCGGGGATCCGTCGACC | |||
| egfp3 | 5′CGTCGGCGGATCGTCGGCGGAACCCTTCTCCTACCCGCCCTGTAGGCTGGAGCTGCTTC | |||
| J11-40-1 | 5′GAGCAACGCATTTCACAACA | |||
| J11-40-2 | 5′AACCACCATCGACCTGGTAA | |||
| 8E7-egfp-7 | 5′AGCCGTTCGCAACCTCAGTG | |||
| 8E7-egfp-8 | 5′TCAGGGTGGTCCGTGGTGTC | |||
| CmlR2-1 | 5′TTCCGCTGTACCTGCTCGCC | |||
| CmlR2-2 | 5′AGAGGACCCGCGTGATCAGC | |||
| 9H11-4-3 | 5′GAACACGACGGAACGGCAC | |||
| 9H11-4-4 | 5′GACGGGTGACTCCCACTCG | |||
| ArgG-1 | 5′CCACTCACGAGGCCAAGAC | |||
| ArgG-2 | 5′GTGCCGATGAGACCGAGAC | |||
PCR amplification.
Taq DNA polymerase (Biostar) and the high-fidelity DNA polymerase Pyrobest (Takara) were used in PCR amplification. The oligonucleotides used for PCR amplification are listed in Table 1.
Construction of ftsKSC null mutants.
An ftsKSC gene replacement mutant (WL1) and an in-frame deletion mutant (WL11) were generated by homologous recombination between modified cosmids carrying mutant alleles and the chromosome carrying the wild-type ftsKSC allele. First, the supercos1-derived cosmid SC7C7 carrying ftsKSC and long flanking regions (36) was modified by λRED PCR targeting (20) using the oligonucleotide primers ftsk1 and ftsk2, resulting in the modified cosmid pHL51, in which the ftsKSC coding sequence was replaced by the acc(3)IV-oriT cassette, leaving only the ftsKSC translational start and stop codons. Then, pHL51 was conjugated into M145, followed by screening for apramycin-resistant (Aprr) and kanamycin-sensitive (Kans) exconjugants, resulting in the gene replacement strain WL1. The gene replacement was confirmed by PCR using primers WL4-3 and WL4-5 and by Southern blotting (Roche Blot Kit).
To generate the ftsKSC in-frame deletion mutant, pHL51 was further modified by FLP-mediated recombination by passing it through BT340 (20) to remove the acc(3)IV-oriT cassette, leaving an 81-bp scar sequence, to generate cosmid pHL59. Because pHL59 did not have an RK2-derived oriT fragment to mediate E. coli-Streptomyces conjugation, it was further modified using λRED PCR targeting of an aac(3)IV-oriT cassette amplified by the primers oriT-1 and oriT-2, resulting in pHL66. pHL66 was introduced into M145 (Aprr selection), and the exconjugants were further cultured nonselectively. Spores were harvested and plated out to give separate colonies. Aprs Kans colonies were identified by replication and screened for double-crossover events by PCR using WL4-3 and WL4-5, resulting in the in-frame deletion mutant WL11. The deletion in WL11 was confirmed by Southern blotting.
A strain, WL12, carrying an in-frame deletion of SCO5751, a gene downstream of ftsKSC, was generated by the same method using the primers SC7C706-1 and SC7C706-2. The deletion in WL12 was confirmed by PCR with the primers SC7C706-3 and SC7C706-4.
Construction of plasmids to complement the ftsKsc mutants.
To construct plasmids for complemention tests (Fig. 1), restriction fragments originating from cosmid SC7C7 were subcloned into the appropriate sites of either of two vectors: pHJL401, a shuttle vector with an intermediate copy number in Streptomyces (27), and pHL58, a derivative of pSET151 (27) into which the 2.7-kb PstI fragment containing the ΦC31 integration system from pSET152 (27) had been inserted at the PstI site. pHL52, pHL53, pHL60, and the vector pHJL401 were introduced into WL11 by protoplast transformation, and pHL56, pHL57, and pHL58 were introduced by conjugation from E. coli, as described previously (27).
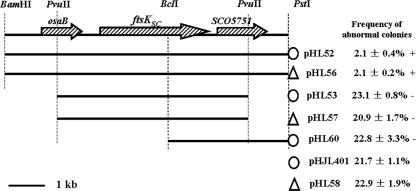
FIG. 1.
Complementation of the ΔftsKSC in-frame deletion mutant WL11 by constructs with various inserts. The restriction sites at the top of the diagram were used to clone these constructs. pHL52, pHL53, and pHL60 were from the SCP2*-derived moderate-copy-number vector pHJL401 (marked with an open circle). The open triangles represent the ΦC31 integration function in pHL56 and pHL57, derived from pSET152. The 6.1-kb BamHI/PstI fragment in pHL52/pHL56 and the 4.2-kb PvuII fragment in pHL53/pHL57 were excised directly from cosmid SC7C7. To construct pHL60, the 2.3-kb BclI/PstI fragment from pHL52 was inserted into pHJL401. +, colony heterogeneity restored to wild-type level; −, colony heterogeneity not restored to wild-type level.
Construction of truncated ftsKSC genes.
The plasmids pHLW20, pHLW21, pHLW22, and pHLW29 carrying truncated ftsKSC genes were derived from pHL52. The desired regions of ftsKSC in pHL52 were first replaced with an acc(3)IV-oriT cassette by λRED PCR targeting (20) to generate pHLW14, pHLW15, pHLW16, and pHLW28 as follows. Three pairs of primers, ftsKn-1 and ftsKn-2, ftsKL-1 and ftsKL-2, and ftsKc-1 and ftsKc-2, were designed with their 5′ ends homologous to desired regions of ftsKSC and their 3′ ends homologous to the acc(3)IV-oriT cassette from pIJ773, which was used as a PCR template. The PCR products were electroporated into BW25113/pIJ790/pHL52, with apramycin selection. Then, FLP-mediated recombination was used to remove the cassette from the resulting plasmids, giving pHLW20, pHLW21, and pHLW22. Primers ftskL-1 and ftsKc-2 were used to construct pHLW29 by the same method. All the four truncated constructions were confirmed by sequencing.
Construction of an ftsKSC-egfp fusion gene.
Primers egfp1 and egfp6 were used to amplify the enhanced green fluorescent protein gene (egfp) in pIJ8655 (42). Primers egfp2 and egfp3 were used to amplify the aac(3)IV-oriT cassette of pIJ773. The two PCR products shared 40-bp homology at one end and were spliced by overlap extension PCR using egfp1 and egfp3 to generate a 2.1-kb egfp-aac(3)IV-oriT fragment. This fragment was used in PCR-targeted recombination with pHL52 in BW25113/pIJ790, giving rise to pHL64, in which egfp was fused in frame to the ftsKSC coding sequence immediately before the stop codon of ftsKSC. Similarly, egfp was fused to the truncated ftsKSC in pHLW20, in which the coding sequence for the transmembrane domain had been removed, by PCR targeting using the same egfp-aac(3)IV-oriT fragment, generating pHLW23.
Investigation of chromosomal deletions by PCR.
Six PCR experiments were performed. In PCR1, WL4-3 and WL4-5 were used to amplify a 710-bp fragment covering the scar region resulting from the ΔftsKSC in-frame deletion of WL11. J11-40-1 and J11-40-2 were used to amplify a 737-bp fragment in SCO0111 in PCR2. 8E7-egfp-7 and 8E7-egfp-8 were used to amplify a 759-bp intergenic fragment between SCO7813 and SCO7814 in PCR3. CmlR2-1 and CmlR2-2 were used to amplify a 939-bp fragment, including most of cmlR (SCO7662) in PCR4. 9H11-4-3 and 9H11-4-4 were used to amplify a 658-bp fragment of SCO7350 in PCR5. ArgG-1 and ArgG-2 were used to amplify a 693-bp fragment in argG (SCO7036) in PCR6.
Fluorescence microscopy.
A previously described method (27) was used with minor modification. Samples for fluorescence microscopy were incubated on MS agar against inserted coverslips and grown for 1 to 3 days. The coverslips with attached samples were covered with methanol, air dried, washed in distilled water, air dried, and mounted in 50% glycerol on microscope slides. For DNA staining, the 50% glycerol contained 1.25 μg ml−1 DAPI. An Olympus BX51 microscope was used.
Computer analysis.
The TMHMM program (http://www.cbs.dtu.dk/services/TMHMM/) was used for the prediction of transmembrane domains in proteins, Jpred (http://www.compbio.dundee.ac.uk/∼www-jpred/submit.html) for consensus secondary-structure prediction, and SMART (http://smart.embl-heidelberg.de/) for signaling-domain searches.
RESULTS
Morphological heterogeneity of colonies arising from restreaking of ftsKSC mutant.
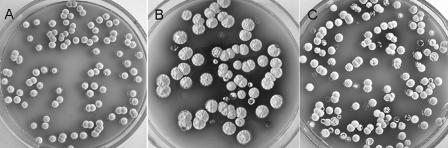
To investigate the role of ftsKSC, two ftsKSC null mutants were constructed [WL1, containing an aac(3)IV gene replacement, and WL11, an in-frame deletion mutant]. Lawns of three WL1 and two WL11 isolates showed apparently normal vegetative growth, but all produced more blue pigment and fewer spores than the parent strain, M145, on solid MS or minimal medium. When the mutants were streaked out to give separate colonies, high heterogeneity was observed: 21.5 ± 2.0% (mean ± standard error) of colonies were small and strongly blue pigmented, and many of them developed poorly, some sporulating 2 to 4 days late and others producing no aerial mycelium (Fig. 2A to C). On restreaking, spores from morphologically normal colonies again gave highly heterogeneous colonies. Under the same conditions, M145 showed about 1.4% abnormal colonies. To verify that the unstable phenotype resulted from the deletion of ftsKSC, we introduced plasmids containing ftsKSC with various flanking regions (Fig. 1) into WL11. Complementation was observed with some of the ftsKSC-containing plasmids but required the presence of more than 451 bp of upstream DNA, including osaB, a gene involved in aerial-hyphal formation under conditions of high osmolarity, and 167 bp of noncoding DNA between osaB and ftsKSC (9). These results confirmed that deletion of ftsKSC was indeed implicated in colony heterogeneity and implied that sequences a considerable distance upstream of ftsKSC are needed for its efficient expression (ftsKSC and osaB may well be cotranscribed). The involvement of the downstream gene SCO5751 in the high-heterogeneity phenotype was excluded because a SCO5751-disrupted mutant, WL12, did not show the phenotype.
FIG. 2.
Inactivation of ftsKSC leading to progeny colonies with heterogeneous morphology. MS agar cultures show colony heterogeneity of WL1 (B) and WL11 (C) compared with M145 (A).
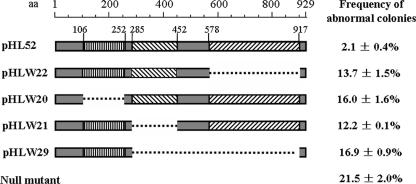
Bioinformatic analysis indicated that, like other FtsK/SpoIIIE family members, FtsKSC consists of three main parts. An N-terminal domain, predicted to contain four transmembrane segments (from aa 109 to 250) and another less strongly predicted one (aa 77 to 94), was separated from a C-terminal conserved ATP-dependent DNA translocase domain (aa 485 to 783) by a linker region (aa 250 to 485), including 22 prolines and 29 alanines in the aa 300 to 450 region. To investigate the functions of these three regions, four truncated FtsKSC versions were constructed and conjugated into WL11 to study their abilities to complement the mutant. The segments deleted were aa 106 to 252, including the four main transmembrane segments; aa 285 to 452, removing the proline/alanine-rich linker segment; aa 578 to 916, removing the DNA translocase domain; and aa 285 to 916, leaving only the first 284-aa fragment. High frequencies of abnormal colonies (12.2 to 16.9%) were observed in all four strains (Fig. 3), suggesting that the three parts are all important for genetic stability.
FIG. 3.
Functional dissection of the FtsKSC protein. More information is provided in the text.
Another ftsK-related gene is present in S. coelicolor (SCO4508; 33.1% identity in a 245-aa overlap with E. coli FtsK) (6). We also disrupted this gene but observed no genetic instability or any other distinct phenotype compared with the parent wild-type strain. Informatics analysis of functional domains suggests that the SCO4508 product has a coiled-coil domain and three ATPase domains, so it may be a YukA-like transporter protein, a crucial component of the gram-positive type IV secretion system (33).
Most abnormal colonies of ftsKSC mutants were associated with large deletions from the chromosome.
The high frequency of abnormal colonies observed with the ftsKSC mutants was reminiscent of previous observations of genetic instability in various streptomycetes, in which instability involving colony morphology and other attributes, including antibiotic production and resistance, was often associated with spontaneous chromosomal deletions or amplifications (11, 44). In S. coelicolor and its close relative Streptomyces lividans, poorly growing, highly pigmented colonies are often chloramphenicol sensitive (Cmls) because of loss of a chloramphenicol resistance gene (cmlR) as part of large chromosomal deletions (17). In the ftsKSC null mutant WL11, we found that Cmls colonies made up 10.6 ± 0.5% of all colonies (among which 13% of the colonies were also arginine auxotrophs [see below]) compared with 0.4 ± 0.1% in the parent strain, M145. Most of the Cmls colonies were small, highly pigmented, and poorly sporulating. Using PCR primers designed to amplify the cmlR coding sequence (SCO7662), 18 of 20 Cmls colonies (90%) were found to have lost this locus.
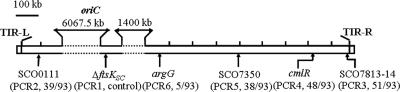
To investigate chromosomal deletions in the ftsKSC null mutant, another two pairs of PCR primers (in PCR2 and -3) were designed to amplify the fragments near the chromosome ends, avoiding terminal inverted repeats and repeated sequences in the subterminal region (Fig. 4). DNA preparations from 93 abnormal colonies, randomly selected from WL11, were used as templates for more PCR experiments. Using the PCR2 primers to amplify a segment 92 kb from the left end, 41.9% (39/93) strains gave no amplified products, suggesting loss of the left terminal fragment, and using the PCR3 primers to amplify a segment 32 kb from the right end, 54.8% (51/93) of strains gave no amplification, suggesting loss of the right chromosomal terminal fragment. Twenty-four of these strains had apparently lost both ends. Overall, 71.0% (66/93) of abnormal colonies appeared to have lost one or both of the terminal fragments (some abnormal colonies [27/93] showed no evidence of deletion in the PCR test). In these experiments, PCR controls using the PCR1 primers to amplify the 710-bp segment covering the ΔftsKSC deletion position were always positive.
FIG. 4.
The positions of the amplified segments in PCR1 to -6 on the S. coelicolor chromosome. The locations of the six amplified segments on the linear chromosome are indicated by arrows below. The numbers of strains that gave no amplification product in the selected 93 abnormal colonies are presented in parentheses. oriC, replication origin; TIR, terminal inverted repeats.
To find out the range of lengths of the deletions at the right end of the chromosome, three more primer pairs (PCR4, -5, and -6) were used with the 93 template preparations, to amplify segments in cmlR and argG that had previously been extensively used in characterizing chromosomal deletions (17, 47) and another segment located almost at their midpoint (Fig. 4). The previously mentioned segment in cmlR (SCO7662, 178 kb from the right end; PCR4) failed to be amplified from 48/93 templates; 38/93 appeared to lack the segment in SCO7350 (503 kb from the right end; PCR5), and 5/93 even lacked a segment 841 kb from the right end, in argG (SCO7036; PCR6). With very few exceptions, the absence of any one segment was associated with the absence of all the segments closer to the end. Thus, most abnormal colonies of the ftsKSC mutant were associated with large terminal-fragment deletions, often from both ends.
An FtsKSC-enhanced green fluorescent protein (EGFP) fusion protein was up-regulated during the development of aerial hyphae and showed two distinct consecutive patterns of distribution.
In order to obtain clues about the reasons for the frequent deletion of chromosome ends in ftsKSC mutants, we examined the location and abundance of FtsKSC at different stages of mycelial growth and development. An ftsKSC-egfp fusion plasmid (pHL64) was introduced into the ΔftsKSC mutant WL11. The resulting strain gave rise to 2.4% ± 0.4% abnormal colonies, indicating near-complete complementation and suggesting that the fusion protein was functional.
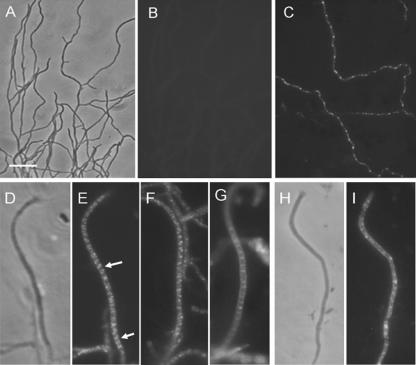
Cultures of WL11/pHL64 were analyzed by fluorescence microscopy (Fig. 5). Early in vegetative growth (28 h), green fluorescent foci were absent (Fig. 5B). By 40 h, many hyphae showed green fluorescence, either diffuse or as dispersed or clumped foci (Fig. 5C). Fluorescence was often seen at tips and near branch points. In aerial hyphae, distinguishable from vegetative hyphae by their larger diameter and absence or near absence of branching, green fluorescence was brighter than in substrate hyphae and appeared to be distinctly organized. Initially, angled bands of foci extended along the full length of sporogenic aerial-hyphal tip compartments in an arrangement that was possibly helical, while subapical compartments showed a less organized fluorescence (Fig. 5D to F). In young spore chains, where prespore compartments were already established and constriction between compartments had begun, FtsKSC-EGFP was clearly located at sporulation septa (Fig. 5G). As prespores matured, the localized fluorescence disappeared. Overall, the cytological analysis indicated a special role for FtsKSC during sporulation, perhaps analogous to the role played by SpoIIIE in completing chromosome translocation into the Bacillus subtilis prespore (4, 7, 41). This suggests that FtsKSC might interact with both chromosome arms during sporulation to ensure that they are transferred efficiently into prespore compartments and that, in the absence of FtsKSC, the chromosome ends are sometimes excluded from prespore compartments, giving rise to the observed deletions. Other mechanisms exist to ensure that the central region of the chromosome is correctly partitioned (25). Presumably, guillotining of chromosomes during septation could also occur during vegetative septation.
FIG. 5.
Spatial and temporal expression of FtsKSC and its truncated forms visualized by FtsKSC-EGFP fusion proteins. In vegetative mycelium up to 28 h old, there was no sign of green fluorescent foci (B), but obvious foci appeared in 40-h mycelium (C). Long sporogenic aerial hyphae often contained many fluorescent foci in angled bands (arrows) that appeared to form a long spiral (D to F). As septal constriction initiated, the spiral-like bands disappeared, and fluorescence was clearly located at septa (G). For an EGFP fusion with a truncated FtsKSC lacking the four main transmembrane segments, diffuse and occasionally bright fluorescent foci were observed in prespore hyphae instead of regular spiral-like bands (I). Panels A, D, and H are corresponding phase-contrast images. Scale bar, 5 μm.
Organized patterns of fluorescence were not seen when the fusion protein lacked the N-terminal transmembrane domain (from aa 106 to 252 in WL11/pHLW23) (Fig. 5I). WL11/pHLW23, like WL11/pHLW20 (Fig. 3), showed severe genetic instability (14.5% ± 2.1% abnormal colonies), consistent with the correct location of FtsKSC being important for genetic stability.
The assembly and stability of FtsZ rings in both E. coli and Caulobacter crescentus is affected by loss of FtsK (48, 52). This did not appear to be the case for S. coelicolor, since sporulation septa formed apparently normally in the ftsKSC mutants. To further confirm this, we introduced pKF41 carrying an FtsZ-EGFP translational fusion gene (19) into M145 and WL11. Both strains showed diffuse green fluorescence in early aerial hyphae; then, regularly spaced ringlike foci appeared and were maintained until septal constriction (data not shown).
ftsKSC deletion abolishes expression of sigF in some prespore compartments.
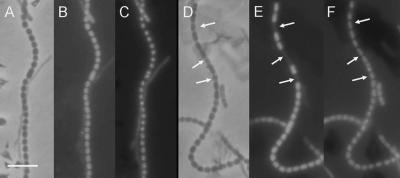
The SpoIIIE-mediated transfer of a complete chromosome into the B. subtilis prespore is necessary for the proper activation of the prespore program of gene expression, which involves the sigma factor SigF (14). We wondered whether a similar checkpoint operated in S. coelicolor sporulation, even though the two kinds of sporulation are quite different. It has been shown that a member of the same sigma factor subfamily, coincidentally also called SigF, plays a role in late spore development in streptomycetes, since sigF disruption in S. coelicolor results in defective spore maturation (26, 32, 35, 37). Fusion of egfp to the sigF promoter sigFp in the construct pIJ8635 was previously used to show that the expression of sigF is synchronous and confined to immature spore chains (42). We therefore used pIJ8635 to visualize sigF expression in the ΔftsKSC mutant WL11. The mycelial samples were also stained with 4′,6′-diamidino-2-phenylindole (DAPI) to visualize DNA. In WL11/pIJ8635, spore compartments with obvious DAPI foci but no EGFP foci were readily observed (EGFP− DAPI+; 6.5% ± 0.3%; 3,545 spores examined) (Fig. 6E). Such compartments were much rarer in the M145/pIJ8635 control strain (0.2%; 2/910 spores) (Fig. 6B).When the ftsKSC-complementing plasmid pHL52 was introduced into WL11/pIJ8635, the frequency of EGFP− DAPI+ spores fell to 1.8% ± 0.5% (5,624 spores examined), showing that the compartment-specific effect on sigF was associated with the loss of ftsKSC. The fact that most separate EGFP− DAPI+ compartments were flanked on both sides by normal ones provides rather direct evidence that sigF expression is autonomously regulated within individual prespore compartments. The finding that ftsKSC deletion increases the frequency of sigF-nonexpressing compartments, as well as of chromosomal deletions, suggests that the mechanisms for these two phenotypic effects may be connected. We did not try to evaluate the extent to which further maturation of these SigF-deficient prespores was prevented.
FIG. 6.
Inactivation of FtsKSC results in occasional compartment-specific elimination of sigF expression. Expression of sigFp-egfp was examined in M145 (B) and the ΔftsKSC in-frame deletion mutant WL11 (E). The arrows show prespore compartments without green fluorescence but staining strongly with DAPI. Panels A and C and panels D and F are corresponding phase-contrast and DAPI fluorescent images. About 50-hour-old cultures on MS agar were sampled for these images. Scale bar, 5 μm.
DISCUSSION
Genomic instability of various Streptomyces species, involving large deletions, has been documented by an extensive catalogue of papers over the last 3 decades (11, 44) and is a significant problem for the maintenance of high-yielding strains used for the commercial production of antibiotics. It is often increased by classic mutagenic treatments and is even elicited by circular and end-to-end linear fusion forms of the chromosome (47, 49). Previous work identified several genes responsible for genetic stability in Streptomyces. For example, in Streptomyces ambofaciens, antibiotics that interact with DNA gyrase stimulated high levels of genomic rearrangment, suggesting that DNA gyrase might play an important part in maintaining genetic instability (45, 46). Mutation of recA in S. lividans led to large deletions in the chromosome termini, often including the cmlR (chloramphenicol resistance) and argG loci (43). The terminal binding proteins Tap and Tpg are also both required to maintain chromosome linearity (2, 3). Nevertheless, despite decades of research, the molecular mechanism underlying instability has remained unknown. Here, we have discovered that a range of deletions closely resembling those described in studies of instability are generated at high frequency in S. coelicolor as a consequence of eliminating part or all of a gene resembling ftsK of gram-negative bacteria and spoIIIE of B. subtilis.
FtsK/SpoIIIE proteins from different bacteria function in chromosome segregation and translocation (5, 10, 48, 50). For example, in E. coli, FtsK is able to glide along DNA in opposite directions for 30 to 50 kb on either side of the dif site involved in decatenation of interwound circular daughter chromosomes near the replication terminus (29). Short FtsK-orienting skewed sequences in the chromosome direct such movement (8, 28). Candidate equivalent sequences (architecture-imparting sequences) have also been found by bioinformatic analysis toward the ends of the S. coelicolor linear chromosome (22), and it was speculated that these architecture-imparting sequences might direct FtsKSC movement. Our observations suggest that FtsKSC, like other members of the FtsK/SpoIIIE family, pumps any replication-terminal parts of the chromosome trapped by ingrowing septa through to the compartment containing the origin-proximal part, with the consequence that about 15 to 20% of the spores of ftsKSC mutants have deletions of one or both chromosome ends. Some of these deletions remove more than 10% of the chromosome. We found no evidence that elimination of ftsKSC affected vegetative-mycelial growth, during which the relatively infrequent septation results in hyphal compartments having several copies of the chromosome. Taking this together with the cytological analysis of FtsKSC, we speculate that many of the chromosomal deletion events occur during sporulation septation, a view consistent with the spore compartment-specific effects of ftsKSC deletion on sigF expression (see below).
In summary, we propose the following model. The origin-proximal regions of chromosomes in fully grown aerial-hyphal tip compartments form complexes with the ParB protein, which are located roughly midway between incipient sporulation septa, ensuring correct partitioning of the origin region (25). However, the fact that DNA is usually observably continuous between adjacent incipient prespore compartments at a late stage in cytokinesis (31) implies that the chromosome ends often move into their final prespore destinations very late in septation. We suggest that FtsKSC, which is particularly abundant in aerial hyphae and localizes at young sporulation septa, provides part of the machinery for this and that the occasional failure of FtsKSC to complete sporulation-associated DNA transfer results in the occurrence of deletions in spores of wild-type strains. Since many spores of the ftsKSC deletion mutant appear to retain intact chromosomes, we suspect that some other less efficient mechanism contributes to the completion of chromosome partitioning. Cytological studies also suggest that chromosomal condensation accompanies sporulation septation (31), so this could pull the bulk of each chromosome to its final destination, with FtsKSC serving to complete the process. This model does not straightforwardly account for the increased frequency of chromosome deletions induced by DNA-damaging agents.
The role suggested for FtsKSC in ensuring that spores receive a complete copy of the chromosome raises further questions. Although the origin region appears to have a well-defined location in its destination spore compartment (25), there is no information on the positions of the corresponding chromosome ends. They could be attached to each other (11, 51; note that the latter paper did not incorporate any analysis of aerial hyphae) or separate and could be far away from, or on either side of, their destination compartment: the extended length of each chromosome arm is about 1,000 times longer than a spore, and aerial hyphae often form chains of 50 or so spores. In the most complicated scenario, multiple copies of the chromosome could be moving through FtsKSC channels in some near-complete septa, some copies in one direction, some in the other. The likely difficulty of knowing which chromosome ends are attached to any particular origin may prove to complicate the interpretation of data obtained by fluorescence in situ hybridization during differentiation.
A striking finding in this work was the failure of some, but not all, spore compartments to show any sigF promoter activity in the ftsKSC deletion mutant. This is the first evidence of a prespore compartment-autonomous pattern of gene expression in Streptomyces and implies that genes that depend directly or indirectly on SigF will remain unexpressed in such compartments. One such gene set is an operon of whiE genes involved in making the gray spore pigment (26, 32). What causes this compartment-specific failure of sigFp expression? Clearly there is not a direct dependence on FtsKSC, since the majority of ftsKSC mutant spores do express sigF. As a working hypothesis, we propose that a genetic element close to a chromosome end must be transferred into its destination compartment in order to activate sigFp. Compartments failing to express sigFp would be those that have not received this element because of chromosome guillotining. Since non-sigFp-expressing compartments were somewhat less frequent than aberrant progeny colonies in the ftsKSC deletion mutant, the proposed element may be at an intermediate point along the deletable part of one or the other chromosome arm.
In addition to chromosome translocation, the C terminus of FtsK in E. coli participates in resolution and decatenation of replicated circular chromosomes through a XerCD-mediated site-specific resolution system (1) and topoisomerase IV (15). Streptomycetes, however, have no xerCD-like genes (6, 23), though they do have a topoisomerase IV biochemically able to catalyze ATP-dependent decatenation (6, 23, 38). Whether this enzyme plays a role in chromosome decatenation during sporulation and/or interacts with FtsKSC is yet to be revealed. FtsK is essential for cell division in E. coli and C. crescentus (13, 48), and in E. coli, the recruitment of the essential division protein FtsQ to FtsZ-rings relies on FtsK (12). In contrast, in the S. coelicolor ftsKSC deletion mutant no defects in septation or growth were observed, and FtsZ rings formed apparently normally during sporulation, so neither the formation nor the closure of septa depends on FtsKSC. Consistent with this, although ftsQSC is essential for cell division of S. coelicolor (30), the mutant with ftsKSC deleted was unimpaired in sporulation septation.
It was unexpected to find that FtsKSC-EGFP foci were organized into angled bands, possibly a spiral, along the full length of each sporulating aerial hypha before becoming closely associated with sporulation septa. This initial organization appeared to require the N-terminal domain, which is likely to attach the protein to the membrane, since an FtsKSC-EGFP N-terminal deletion protein accumulated in a delocalized manner (although the deletion might also have interfered with the conformation of the remainder of the protein). We are not aware of previous descriptions of this kind of behavior of FtsK-like proteins.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (NSFC no. 30200005) and from the Youth Chengguang Project of Science and Technology of Wuhan City of China to M. Tao and by a Joint Project grant from the Royal Society and NSFC to K. F. Chater and Z. X. Deng.
We thank Helen M. Kieser for cosmid SC7C7, Bertolt Gust for the λRED-mediated PCR-targeting kit, Maureen J. Bibb for pIJ8655 and pIJ8635, and Klas Flärdh for pKF41.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Aussel, L., F. X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108:195-205. [DOI] [PubMed] [Google Scholar]
- 2.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao, K., and S. N. Cohen. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 17:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. [DOI] [PubMed] [Google Scholar]
- 5.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. Assembly of the SpoIIIE DNA translocase depends on chromosome trapping in Bacillus subtilis. Curr. Biol. 13:2196-2200. [PubMed] [Google Scholar]
- 8.Bigot, S., O. A. Saleh, C. Lesterlin, C. Pages, M. El Karoui, C. Dennis, M. Grigoriev, J. F. Allemand, F. X. Barre, and F. Cornet. 2005. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 24:3770-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop, A., S. Fielding, P. Dyson, and P. Herron. 2004. Systematic insertional mutagenesis of a Streptomycetes genome: a link between osmoadaptation and antibiotic production. Gen. Res. 14:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chary, V. K., D. W. Hilbert, M. L. Higgins, and P. J. Piggot. 2000. The putative DNA translocase SpoIIIE is required for sporulation of the symmetrically dividing coccal species Sporosarcina ureae. Mol. Microbiol. 35:612-622. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. W., C. H. Huang, H. H. Lee, H. H. Tsai, and R. Kirby. 2002. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18:522-529. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for colocalization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 13.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errington, J. 1996. Determination of cell fate in Bacillus subtilis. Trends Genet. 12:31-34. [DOI] [PubMed] [Google Scholar]
- 15.Espeli, O., C. Lee, and K. J. Marians. 2003. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 278:44639-44644. [DOI] [PubMed] [Google Scholar]
- 16.Flärdh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564-571. [DOI] [PubMed] [Google Scholar]
- 17.Flett, F., and J. Cullum. 1987. DNA deletions in spontaneous chloramphenicol-sensitive mutants of Streptomyces coelicolor A3(2) and Streptomyces lividans 66. Mol. Gen. Genet. 207:499-502. [DOI] [PubMed] [Google Scholar]
- 18.Gewain, K. M., J. L. Occi, F. Foor, and D. J. MacNeil. 1992. Vectors for generating nested deletions and facilitating subcloning G+C-rich DNA between Escherichia coli and Streptomyces sp. Gene 119:149-150. [DOI] [PubMed] [Google Scholar]
- 19.Grantcharova, N., U. Lustig, and K. Flärdh. 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 187:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hendrickson, H., and J. G. Lawrence. 2006. Selection for chromosome architecture in bacteria. J. Mol. Evol. 62:615-629. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 24.Iyer, L. M., K. S. Makarova, E. V. Koonin, and L. Aravind. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic. Acids Res. 32:5260-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakimowicz, D., B. Gust, J. Zakrzewska-Czerwinska, and K. F. Chater. 2005. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 187:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation. Norwich, United Kingdom.
- 28.Levy, O., J. L. Ptacin, P. J. Pease, J. Gore, M. B. Eisen, C. Bustamante, and N. R. Cozzarelli. 2005. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc. Natl. Acad. Sci. USA 102:17618-17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., B. Youngren, K. Sergueev, and S. Austin. 2003. Segregation of the Escherichia coli chromosome terminus. Mol. Microbiol. 50:825-834. [DOI] [PubMed] [Google Scholar]
- 30.McCcormick, J. R., and R. Losick. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 178:5295-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miguélez, E. M., B. Rueda, C. Hardisson, and M. B. Manzanal. 1998. Nucleoid partitioning and the later stages of sporulation septum synthesis are closely associated events in the sporulating hyphae of Streptomyces carpinensis. FEMS Microbiol. Lett. 159:59-62. [Google Scholar]
- 32.Novakova, R., J. Bistakova, and J. Kormanec. 2004. Characterization of the polyketide spore pigment cluster whiESa in Streptomyces aureofaciens CCM3239. Arch. Microbiol. 182:388-395. [DOI] [PubMed] [Google Scholar]
- 33.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 34.Pease, P. J., O. Levy, G. J. Cost, J. Gore, J. L. Ptacin, D. Sherratt, C. Bustamante, and N. R. Cozzarelli. 2005. Sequence-directed DNA translocation by purified FtsK. Science 307:586-590. [DOI] [PubMed] [Google Scholar]
- 35.Potuckova, L., G. H. Kelemen, K. C. Findlay, M. A. Lonetto, M. J. Buttner, and J. Kormanec. 1995. A new RNA polymerase sigma factor, sigma F, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 17:37-48. [DOI] [PubMed] [Google Scholar]
- 36.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 37.Rezuchova, B., I. Barak, and J. Kormanec. 1997. Disruption of a sigma factor gene, sigF, affects an intermediate stage of spore pigment production in Streptomyces aureofaciens. FEMS Microbiol. Lett. 153:371-377. [DOI] [PubMed] [Google Scholar]
- 38.Schmutz, E., S. Hennig, S. M. Li, and L. Heide. 2004. Identification of a topoisomerase IV in actinobacteria: purification and characterization of ParYR and GyrBR from the coumermycin A1 producer Streptomyces rishiriensis DSM 40489. Microbiology 150:641-647. [DOI] [PubMed] [Google Scholar]
- 39.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25:847-858. [DOI] [PubMed] [Google Scholar]
- 40.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharp, M. D., and K. Pogliano. 2003. The membrane domain of SpoIIIE is required for membrane fusion during Bacillus subtilis sporulation. J. Bacteriol. 185:2005-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, J., G. H. Kelemen, J. M. Fernández-Abalos, and M. J. Bibb1. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 43.Volff, J. N., and J. Altenbuchner. 1997. Influence of disruption of the recA gene on genetic instability and genome rearrangement in Streptomyces lividans. J. Bacteriol. 179:2440-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volff, J. N., and J. Altenbuchner. 1998. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27:239-246. [DOI] [PubMed] [Google Scholar]
- 45.Volff, J. N., D. Vandewiele, and B. Decaris. 1994. Stimulation of genetic instability and associated large genomic rearrangements in Streptomyces ambofaciens by three fluoroquinolones. Antimicrob. Agents Chemother. 38:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volff, J. N., D. Vandewiele, J. M. Simonet, and B. Decaris. 1993. Stimulation of genetic instability in Streptomyces ambofaciens ATCC 23877 by antibiotics that interact with DNA gyrase. J. Gen. Microbiol. 139:2551-2558. [DOI] [PubMed] [Google Scholar]
- 47.Volff, J. N., P. Viell, and J. Altenbuchner. 1997. Artificial circularization of the chromosome with concomitant deletion of its terminal inverted repeats enhances genetic instability and genome rearrangement in Streptomyces lividans. Mol. Gen. Genet. 253:753-760. [DOI] [PubMed] [Google Scholar]
- 48.Wang, S. C., L. West, and L. Shapiro. 2006. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J. Bacteriol. 188:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wenner, T., V. Roth, G. Fischer, C. Fourrier, B. Aigle, B. Decaris, and P. Leblond. 2003. End-to-end fusion of linear deleted chromosomes initiates a cycle of genome instability in Streptomyces ambofaciens. Mol. Microbiol. 50:411-425. [DOI] [PubMed] [Google Scholar]
- 50.Wu, L. J., and J. Errington. 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 16:2161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 183:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, X. C., A. H. Tran, Q. Sun, and W. Margolin. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol. 180:1296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]