Abstract
Phosphorus is an essential nutrient, but how phosphates cross the mycobacterial cell wall is unknown. Phosphatase activity in whole cells of Mycobacterium smegmatis was significantly lower than that in lysed cells, indicating that access to the substrate was restricted. The loss of the outer membrane (OM) porin MspA also reduced the phosphatase activity in whole cells compared to that in lysed cells. A similar result was obtained for M. smegmatis that overexpressed endogenous alkaline phosphatase, indicating that PhoA is not a surface protein, contrary to a previous report. The uptake of phosphate by a mutant lacking the porins MspA and MspC was twofold lower than that by wild-type M. smegmatis. Strikingly, the loss of these porins resulted in a severe growth defect of M. smegmatis on low-phosphate plates. We concluded that the OM of M. smegmatis represents a permeability barrier for phosphates and that Msp porins are the only OM channels for the diffusion of phosphate in M. smegmatis. However, phosphate diffusion through Msp pores is rather inefficient as shown by the 10-fold lower permeability of M. smegmatis for phosphate compared to that for glucose. This is likely due to the negative charges in the constriction zone of Msp porins. The phosphatase activity in whole cells of Mycobacterium bovis BCG was significantly less than that in lysed cells, indicating a similar uptake pathway for phosphates in slow-growing mycobacteria. However, porins that could mediate the diffusion of phosphates across the OM of M. bovis BCG and Mycobacterium tuberculosis are unknown.
Phosphorus is indispensable for the biosynthesis of nucleic acids and phospholipids and for the energy supply of any cell. Bacteria employ sophisticated transport mechanisms to acquire phosphorus-containing nutrients from the environment. In gram-negative bacteria, phosphates first need to cross the outer membrane (OM). To this end, Escherichia coli produces the two general porins, OmpF and OmpC, under conditions of phosphate excess. Under phosphate-limiting conditions, these porins are partially replaced by the pore protein PhoE (30), which preferentially allows the diffusion of anions (1), in contrast to the cation preference of OmpF and OmpC (29). Hence, the diffusion of phosphates through PhoE pores is more efficient and is the prevalent pathway for phosphates across the outer membrane under phosphate-limiting conditions (17). While inorganic phosphate is the preferred source of phosphorus, many bacteria can also take up organic phosphates and release phosphate by the action of periplasmic phosphatases such as PhoA. E. coli possesses four transport systems, Pst, Pit, GlpT, and UhpT, that translocate inorganic phosphate across the inner membrane (48). Part of the Pst system is the periplasmic protein PstS, which binds and transfers phosphate to the transmembrane components PstA and PstC. PstB hydrolyzes ATP and delivers energy for the translocation of phosphate across the inner membrane by PstA/PstC. Pst systems bind and transport phosphate with binding constants and apparent transport Km values in the submicromolar range. These systems also exist in gram-positive bacteria (32) and in mycobacteria (5).
Mycobacteria are classified as gram-positive bacteria, but models of their cell walls include asymmetrical outer membranes that are covalently bound to an arabinogalactan-peptidoglycan copolymer (7, 25, 28). These models are consistent with the very efficient permeability barrier established by mycobacterial cell walls towards hydrophobic and hydrophilic compounds (7). The discovery of channel-forming outer membrane proteins in several mycobacteria lends further support to these models (26).
It was conclusively shown that MspA represents the major porin of M. smegmatis (43) and is required for the transport of glucose, serine, and hydrophilic β-lactam antibiotics (44, 45). However, the preference of MspA for cations (27) and the high density of negative charges in the constriction zone of MspA (14) appear to be adverse properties for efficient diffusion of phosphates. Thus, anions may cross the outer membrane of M. smegmatis via PhoE-like porins as in gram-negative bacteria. Importantly, the transport of phosphate across the inner membrane is essential for the growth of Mycobacterium tuberculosis in macrophages (34) and for the survival of mice (31, 38). However, how phosphate is transported across the outer membrane of M. tuberculosis is also unknown.
In this study, we used phoA as a reporter gene and analyzed the uptake of radiolabeled phosphate to examine the uptake of organic phosphate esters and inorganic phosphate by M. smegmatis. We show that the outer membrane represents a permeability barrier for phosphates in both assays, in contrast to previous claims that PhoA and PstS are cell surface proteins (18, 19). Importantly, we demonstrate that Msp porins are required for the efficient uptake of phosphates by M. smegmatis and for the growth of M. smegmatis under low-phosphate conditions.
MATERIALS AND METHODS
Chemicals and enzymes.
Hygromycin B was purchased from Calbiochem. All other chemicals were purchased from Merck, Roth, or Sigma at the highest purities available. Enzymes for DNA restriction and modification were from New England Biolabs, MBI Fermentas, and Boehringer. Oligonucleotides were obtained from MWG Biotech.
Bacterial strains and growth conditions.
E. coli DH5α was used for cloning experiments and routinely grown in Luria-Bertani broth at 37°C. M. smegmatis strains (Table 1) were grown at 37°C in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol and 0.05% Tween 80 or on Middlebrook 7H10 plates supplemented with 0.5% glycerol unless otherwise noted. M. bovis BCG (Strain Institute Pasteur) was grown in Middlebrook 7H9 medium or on Middlebrook 7H10 (Difco) plates supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% OADC (oleic acid-albumin-dextrose-catalase; Remel) unless otherwise noted. When required, the antibiotics hygromycin (200 μg/ml for E. coli; 50 μg/ml for mycobacteria) and kanamycin (30 μg/ml for E. coli; 10 μg/ml for mycobacteria) were added.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | recA1 endA1 gyrA96 thi relA1 hsdR17(rK− mK+) supE44 φ80ΔlacZΔM15 ΔlacZYA-argF UE169 | 36 |
| M. smegmatis SMR5 | mc2155 derivative, not characterized, high transformation efficiency, Smr (rpsL*) | 37 |
| M. smegmatis MN01 | SMR5 derivative mspA::Gmr | 43 |
| M. smegmatis ML10 | SMR5 derivative ΔmspA ΔmspC | 45 |
| M. smegmatis ML40 | mc2155 derivative, attB::pML443, Hygr | This study |
| M. smegmatis ML41 | ML10 derivative, attB::pML443, Hygr | This study |
| M. smegmatis ML42 | ML10 derivative, attB::pML443, Δhyg | This study |
| M. smegmatis ML43 | MN01 derivative, attB::pML443, Hygr | This study |
| M. bovis BCG | Pasteur 35739 | ATCC |
| M. bovis BCG ML44 | M. bovis BCG derivative, attB::pML443 | This study |
| Plasmids | ||
| pMV361 | oriE aph attP int | 47 |
| pMS2 | PAL5000 origin, ColE1 origin, hyg | 16 |
| pMN402 | PAL5000 origin, ColE1 origin, hyg, phsp60-gfp+ | 40 |
| pMN234 | PAL5000 origin, pBR322 origin, aph, rpsL | 46 |
| pML102 | PAL5000 origin, ColE1 origin, aph, sacB, int | This study |
| pML113 | ColE1 origin, bla, FRT-hyg-FRT, attP | This study |
| pML118 | ColE1 origin, bla, FRT-hyg-FRT, attP, phsp60-gfp+ | This study |
| pMN252 | ColE1 origin, bla, FRT-hyg-FRT, rpsL | 46 |
| pML440 | PAL5000 origin, ColE1 origin, hyg, pimyc-phoA | This study |
| pML443 | ColE1 origin, bla, FRT-hyg-FRT, attP, pimyc-phoA | This study |
*, gene encoding a mutated ribosomal protein S12 (K43R) which confers streptomycin resistance (Smr).
Construction of plasmids.
The phoA gene was amplified by PCR from chromosomal DNA of M. smegmatis by using the oligonucleotides phoA_Msfwd01 (5′-GATTACTTAATTAAGCATGCCAGAAAGGAGGTTAATATG CCTGTCAGTACCTATCTCAGAGCGACGGT-3′) and phoA_Msrev (5′-ATATAATTTAAATGCGCTCCGGCACATCGCCAC-3′), introducing the restriction sites SphI and SwaI (underlined). The SphI-digested fragment was cloned into the plasmid pMS2 which was digested with SphI and EcoRV to give the replicative phoA expression vector pML440. To integrate the phoA expression cassette at the genomic attB site of mycobacteria, an integration plasmid, pML443, was constructed as followed. For the construction of the attP site, a 365-bp fragment was amplified from pMV361 (47) by PCR using the oligonucleotides attPbig_fwd (5′-GATTACCTGCAGATCCGCGACGTGCCAACTAG-3′) and attPbig_rev (5′-GATTAGCTCGAGAGCCAGATCAGGGATGCGTTG-3′), introducing the restriction sites PstI and XhoI (underlined). The digested fragment was cloned into pMN252 (46), which was cut with the same restriction endonucleases to give the proficient control vector pML113. The vector pMN402 (40) was digested with ClaI and PmeI, and the fragment was cloned into pML113 digested with the same restriction endonucleases to give pML118. To integrate the phoA expression cassette at the genomic attB site of mycobacteria, pML443 was constructed by cloning the SwaI/PmeI fragment of pML440 into the backbone of pML118 digested with PmeI and EcoRV.
Insertion of a phoA expression cassette into the chromosome of M. smegmatis.
A two-plasmid system derived from mycobacteriophage L5 was used to integrate a phoA expression cassette at the attB site of the mycobacterial chromosome. The replicative vector pML102 carrying the L5 integrase gene (int) was transformed in M. bovis BCG and M. smegmatis. These cells were transformed with the nonreplicative vector pML443 containing the phage attachment site attP and the phoA expression cassette. Since the continued expression of L5 integrase can cause the excision of the integrated vector from the genome, which contributes to plasmid instability (42), cells were plated on 7H10 plates containing hygromycin and 10% sucrose to select for the insertion of pML443 and to counterselect against pML102. Single colonies were plated in parallel on 7H10 plates containing hygromycin and 7H10 plates containing kanamycin to confirm the loss of pML102. The integration of the plasmid resulted in clones resistant to hygromycin and sensitive to kanamycin. Integration into the attB site was confirmed by PCR using the primers attB2 (5′-ACAGGATTTGAACCTGCGGC-3′) and attL01 (5′-TCGCCACGTTCGCCCTAG-3′), while the primers attB1 (5′-ACGTGGCGGTCCCTACCG-3′) and attB2, which only give a PCR product if integration does not occur, were used as controls. The hygromycin resistance marker gene (hyg), flanked by FLP recombination target sites of pML443, was removed from the genome by using the Flp recombinase (46). Competent cells were transformed with the flp expression vector pMN234. After selection on 7H10 plates containing kanamycin, 4 ml of Middlebrook 7H9 medium with kanamycin was inoculated and incubated for 12 h to 20 h at 37°C on a roller drum. In order to get only single cells, the culture was filtrated using a 5.0-μm filter. Several dilutions of the filtrate were plated on 7H10 plates without any antibiotics. Then, single colonies were streaked in parallel on 7H10 plates with and without hygromycin to identify clones which lost the hygromycin resistance gene.
Phosphate-dependent growth and phosphatase activity of M. smegmatis.
In order to examine the dependence of growth and phoA expression on the phosphate concentration, we home-prepared 7H10 medium containing 0.5, 10, and 25 mM inorganic phosphate by adding 1 M Pi buffer (1 M Na2HPO4, adjusted to pH 7 with 1 M KH2PO4). To detect PhoA activity on plates, 60 μg/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and 0.1% Tween 80 were also added. Malachite green was omitted from the self-made 7H10 medium to enhance the visibility of the blue color on the plates. The strains were streaked on Middlebrook 7H10 plates containing 0.1% Tween 80 and incubated at 37°C for 5 to 7 days. Cells were scraped off the plates and resuspended in home-made 7H9 medium containing 0.5 mM Pi buffer. The cell culture was filtered and adjusted to an optical density at 600 nm (OD600) of 0.1. By using an inoculation loop, the cell suspension was streaked on the plates described above. The plates were scanned daily after the 3rd day by using a digital scanner (V700 Photo; Epson). Pictures of single colonies were taken from the same plates by using a stereomicroscope (Stemi 2000-C; Zeiss) and a cooled digital color camera (AxioCam MRc; Zeiss).
Measurement of phosphatase activity of M. smegmatis and M. bovis BCG.
Cells were grown on Middlebrook 7H10 medium. Tween 80 was added to a final concentration of 0.1% to improve the solubility of the cells in liquid medium. The cells were scraped from the plate and suspended in Middlebrook 7H9 medium. This culture was filtrated (pore size, 5 μm; Sartorius) to obtain single cells and diluted with the same medium to an OD600 of 0.5. By using an inoculation loop, the cell suspension was streaked on Middlebrook 7H10 medium containing 60 μg/ml BCIP. Tween 80 (0.1%) was also added to improve the homogeneity of the coloration of the colonies. The plates were put into a sealed plastic bag to keep them moist and incubated in the dark at 37°C. The color of the colonies was examined daily for a period of up to 10 days. Growth and PhoA activity were compared for only strains streaked on the same plate. Pictures of colonies were taken by using a stereomicroscope (Stemi 2000-C; Zeiss) and a cooled digital color camera (AxioCam MRc; Zeiss).
The PhoA activity of M. smegmatis and M. bovis BCG in liquid cultures was measured as described previously (6, 18) with small modifications. Unless otherwise noted, strains were grown in Middlebrook 7H9 medium to an OD600 of about 1.0. Cells were harvested by centrifugation at 3,000 × g for 10 min at 4°C and resuspended in an equal volume of ice-cold 1 M Tris-buffer (pH 8.0). The OD600 of the cell suspension was determined. Cells were kept on ice during all steps. A portion of each cell suspension was lysed by two 20-s pulses at 40 W applied by a sonifier (Branson) using a microtip. PhoA activity was measured in both whole and lysed cells by adding 0.15 ml of samples (Vs) to 1 ml of detection buffer (1 M Tris-HCl, pH 8.0, 4 mM p-nitrophenylphosphate [pNPP]). After 30 to 40 min of incubation at 37°C, 100 μl of 1 M K2HPO4 was added to stop the reaction. The sample was centrifuged in a microcentrifuge (Eppendorf) for 10 min at maximum speed. After centrifugation, 1 ml of the supernatant was used to determine the A405 by using a spectrophotometer (SmartSpec Plus; Bio-Rad). The PhoA activity of whole and lysed cells was calculated using the following formula (18): activity (units) = (1,000 × A405)/(ti × OD600 × Vs), where ti is the incubation time in minutes and Vs is the volume of sample.
Measurement of phosphate uptake by M. smegmatis.
Phosphate uptake measurements were carried out as previously described for glucose (44), with some modifications. Cells were grown on Middlebrook 7H10 plates containing 0.1% Tween 80 and incubated for 5 to 7 days at 37°C. The cells were scraped off the plates, suspended in Middlebrook 7H9 medium, and filtered through a 5.0-μm-pore-size filter (Sartorius). The cell suspension was used to inoculate 100 ml of Middlebrook 7H9 medium to an OD600 of 0.05. The cells were harvested at an OD600 of 1.0 by centrifugation (1,250 × g at 4°C for 10 min), washed twice in uptake buffer [50 mM Tris-HCl (pH 6.9), 15 mM KCl, 10 mM (NH4)2SO4, 0.05% Tween 80], and resuspended in the same buffer. The cell suspension was kept for 15 min at 37°C before [32P]phosphate and nonlabeled phosphate were mixed and added to the cell suspension to obtain final concentrations of 1, 2.5, 5, 10, and 20 μM. The mixtures were incubated at 37°C, and 1-ml samples were removed at the indicated times. The cells were filtered through a 0.45-μm-pore-size filter (Sartorius). The filters were washed with 0.1 M LiCl and water, and their radioactivity levels were measured in a liquid scintillation counter. All experiments were performed in triplicate. The uptake rate was expressed as nanomolars per milligram of cells. The Michaelis-Menten constant Km, the maximal uptake velocity Vmax for the overall transport, and a minimal estimate of the permeability coefficient were determined as described previously (15, 44).
Gel electrophoresis and Western blots.
Protein samples were analyzed in denaturing polyacrylamide gels that were stained with Coomassie blue G250 or silver. All samples were mixed with loading buffer (150 mM Tris, 4% sodium dodecyl sulfate, 30% glycerol, 0.1% bromophenol blue, pH 7.5) and boiled in a water bath for 10 min before being loaded onto a denaturing 10% polyacrylamide gel. The protein gel was blotted (4 h at 150 mA in transfer buffer [10 mM NaHCO3, 3 mM Na2CO3, 20% methanol]) onto a nitrocellulose membrane (Schleicher & Schuell). The presence of PhoA was detected by using a monoclonal mouse antibody against PhoA of E. coli and a secondary rabbit anti-mouse antibody coupled with horseradish peroxidase. Immunoblots were developed by using an ECL Plus kit according to the manufacturer's recommendations (Amersham).
RESULTS
The cell envelope represents a permeability barrier for PhoA substrates in M. smegmatis.
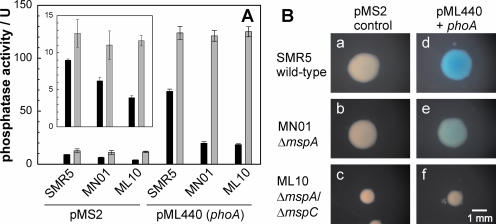
Our first aim was to examine whether the enzymatic activity of the alkaline phosphatase PhoA can be used to analyze the uptake of organic phosphates across the OM of mycobacteria as was shown for E. coli (23). This requires that PhoA be localized in the periplasm of mycobacteria as in that of gram-negative bacteria (10). However, based on the association of PhoA with membranes in subcellular fractions, it was proposed that PhoA is a cell surface protein of M. smegmatis (18). In order to solve the puzzle of the subcellular localization of the M. smegmatis PhoA protein, which is encoded by two identical phoA genes, msmeg1000 and msmeg2294, we examined whether the phosphatase activity was limited by the accessibility to substrates in whole cells. To this end, M. smegmatis SMR5 was grown to end-log phase and the PhoA activity was measured for lysed and whole cells obtained from the same culture. Since the endogenous PhoA activity of M. smegmatis was very low, the difference between whole and lysed cells was small (Fig. 1A). To increase PhoA activity, the vector pML440 was constructed in which one of the phoA genes of M. smegmatis is transcribed from a constitutively active mycobacterial promoter, pimyc (16, 22). The total PhoA activity in lysed M. smegmatis cells carrying pML440 was increased ninefold compared to that in the control vector pMS2 (Fig. 1A). Western blot experiments using an antiserum against PhoA of E. coli revealed a 55-kDa band, which was strongly enhanced in M. smegmatis containing pML440 (not shown), confirming that the increased PhoA activity was due to the enhanced expression of phoA. Importantly, PhoA activity in whole cells was twofold reduced compared to that in lysed cells, indicating that the substrate pNPP is not freely accessible for PhoA. This was also observed for wild-type (wt) M. smegmatis and the porin mutants as evidenced by a small but significantly increased level of PhoA activity in lysed cells compared to that in whole cells (Fig. 1A, inset). This rules out the possibility that the lower PhoA activity level in whole cells compared to that in cell lysates is caused by the overexpression and concomitant mislocation of PhoA. Hence, we concluded that the intact cell envelope of M. smegmatis represents a permeability barrier for the PhoA substrate pNPP.
FIG. 1.
Porin-dependent phosphatase activity of M. smegmatis. (A) The phosphatase activity of M. smegmatis grown in liquid culture was determined using pNPP. Black and gray bars indicate the phosphatase activity in whole and lysed cells, respectively. The inset shows the endogenous phosphatase activity in the parent strains SMR5, MN01, and ML10. Error bars indicate standard deviations. (B) Phosphatase activity of M. smegmatis on Middlebrook 7H10 agar plates containing Tween 80 and BCIP. After 5 days of incubation, pictures of colonies were taken with a Zeiss stereomicroscope Stemi 2000-C using the same magnification for all strains. In both experiments, the strains were SMR5 (wild-type) and the porin mutants MN01 (ΔmspA) and ML10 (ΔmspA ΔmspC), each of which was transformed with the control plasmid pMS2 (control) and the phoA expression plasmid pML440 (pML440+phoA), respectively. The blue color indicates the cleavage of BCIP by PhoA.
Porins mediate the diffusion of PhoA substrates across the OM of M. smegmatis.
The diffusion of the substrate pNPP across the OM is rate limiting for the PhoA activity in E. coli and is mediated mainly by the porins OmpF and OmpC when E. coli is grown in rich medium (24). To analyze whether porins of M. smegmatis play a role in the diffusion of pNPP, PhoA activities were determined for M. smegmatis MN01, which lacks the major porin MspA, and for ML10, which lacks MspA and MspC. The PhoA activities of lysed cells of the wild type and of both porin mutants transformed with pML440 were identical, demonstrating that the deletion of the porin genes did not interfere with phoA expression (Fig. 1A). By contrast, the PhoA activity in whole cells of the ΔmspA mutant MN01 was threefold less than that in wild-type cells. This is consistent with the threefold-reduced number of porin channels in the OM of MN01 (44). A further reduction of the PhoA activity in whole cells for the porin double-mutant ML10 was not observed. These results demonstrate that PhoA activity in M. smegmatis depends on the presence of porins in the OM of M. smegmatis and strongly support the conclusion that PhoA is localized in the periplasm of M. smegmatis.
To test whether any of these strains would produce PhoA in amounts sufficient for detection on plates, M. smegmatis wild type and the porin mutants (MN01 and ML10), carrying the vectors pMS2 and pML440, respectively, were streaked on 7H10 plates containing the chromogenic PhoA substrate BCIP. Colonies of M. smegmatis wild-type-carrying pMS2 appeared white, which was consistent with the very low intrinsic PhoA activity level, while colonies of M. smegmatis with pML440 were blue (Fig. 1B, panels a and d) as a result of increased PhoA expression. Importantly, the intensity of the blue color decreased from the wild type (Fig. 1B, panel d) over MN01 (panel e) to ML10 (panel f). The smaller sizes of the colonies of M. smegmatis ML10 were caused by the reduced growth rates due to the lower number of porins (44). Further incubation of the porin double-mutant ML10 for 2 days increased the sizes of the colonies to those of wt M. smegmatis but did not yield any blue color (not shown). Since the total PhoA activities were equal for all three strains (Fig. 1A), these results supported our previous conclusions that the uptake of BCIP across the OM is mediated by porins and that the PhoA activity is not surface associated in M. smegmatis. Furthermore, the fact that strains with high (M. smegmatis wild type) and low (MN01 and ML10) levels of porin activity are clearly distinguishable on plates containing BCIP showed that PhoA, in principle, is a suitable indicator of the uptake of phosphates across the OM and of the porin activity in M. smegmatis.
Porins are required for growth of M. smegmatis on low-phosphate medium.
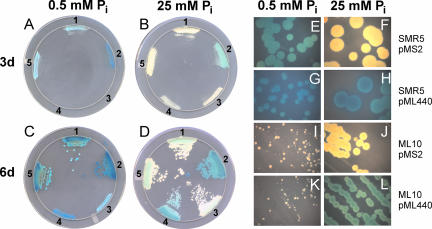
To examine whether porins play a role in the uptake of inorganic phosphate in vivo, the growth levels of M. smegmatis wild type and the porin double-mutant ML10 were examined on self-made 7H10 plates containing 0.5, 10, and 25 mM inorganic phosphate and BCIP. After 3 days of incubation, no growth of ML10 was observed on plates containing the lowest phosphate concentration (Fig. 2A, streaks 3 and 4) in contrast to that of the wild-type strain (Fig. 2A streaks 1 and 2). Even after 6 days of incubation, colonies of the ML10 strain were very small, indicating a severe growth defect of the ML10 strain at 0.5 mM inorganic phosphate (Fig. 2C, streaks 3 and 4). The mspA expression vector pMN016 fully restored the ability of M. smegmatis ML10 to grow on low-phosphate plates (Fig. 2A to D, streaks 5) and demonstrated that the loss of porins caused the growth defect. Surprisingly, a significantly lower level of growth of the porin double-mutant ML10 was also observed on plates containing 10 mM phosphate (not shown), indicating that the level of phosphate uptake is very low in this mutant and limits its growth rate at rather high phosphate concentrations. Taken together, these results show that Msp porins are required for the growth of M. smegmatis under low-phosphate conditions. These results also indicated that the uptake of inorganic phosphate across the OM of M. smegmatis depends on Msp porins.
FIG. 2.
Porin-dependent growth of M. smegmatis on low-phosphate plates. (A to D) M. smegmatis SMR5 (wt) and ML10 (ΔmspA ΔmspC) were streaked on self-made 7H10 agar plates containing different concentrations of phosphate and BCIP. The plates were incubated at 37°C for 6 days and were scanned (V700 Photo, Epson) each day. Scans of plates after 3 (A and C) and 6 days (B and D) of incubation are shown. The strains on the plates are as follows: streak 1, SMR5/pMS2 (control); streak 2, SMR5/pML440 (phoA); streak 3, ML10/pMS2; streak 4, ML10/pML440; and streak 5, ML10/pMN016 (mspA). (E to L) M. smegmatis SMR5 and ML10 were filtered through a 5-μm filter to obtain single cells and plated on self-made 7H10 agar plates containing different concentrations of phosphate and BCIP. Pictures of single colonies after 8 days of incubation at 37°C were taken at a 16-fold magnification using a Zeiss stereomicroscope Stemi 2000-C.
Msp porins are required for uptake of inorganic phosphate by M. smegmatis.
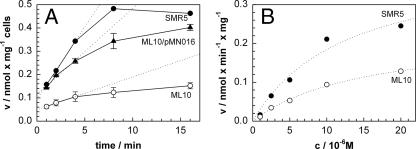
It was shown previously that diffusion through MspA and other Msp porins is the rate-limiting step for the uptake of glucose by M. smegmatis (43, 44). The reduced PhoA activity in whole cells of porin mutants (Fig. 1) and the limited growth of the porin mutant ML10 on plates with low-phosphate concentrations (Fig. 2) strongly indicated that Msp porins are used by M. smegmatis to enable the diffusion of organic and inorganic phosphates across the OM. To provide direct evidence for this conclusion, we examined the uptake of radioactive phosphate by wild-type M. smegmatis and the porin double-mutant ML10. For these experiments, strains were grown in Middlebrook 7H9 medium. The uptake rates were measured at a phosphate concentration of 20 μM because differences in diffusion through porins are more pronounced at solute concentrations in the micromolar range. M. smegmatis SMR5 (wt) took up phosphate with a rate of 0.06 nmol/min per mg cells. The porin double-mutant ML10 showed a 4.5-fold-lower uptake rate of 0.014 nmol/min per mg cells (Fig. 3A). The expression of mspA from the vector pMN016 increased the uptake rate of phosphate by M. smegmatis ML10 to 60% of the wild-type level (Fig. 3A). This demonstrated that the loss of porins in the ML10 strain reduced the ability of M. smegmatis to take up phosphate. We concluded that inorganic phosphate diffuses through the MspA and MspC porins into M. smegmatis.
FIG. 3.
Uptake of phosphate by porin mutants of M. smegmatis. (A) Accumulation of [32P]phosphate by M. smegmatis SMR5 (wild-type) (black circles), the ΔmspA ΔmspC mutant ML10 (white circles), and ML10 complemented with the mspA expression plasmid pMN016 (triangles) was measured. The assay was performed at 37°C at a final phosphate concentration of 20 μM. The uptake rates were determined by a regression analysis of the first 4 min for each strain. The dotted lines represent regression lines. Error bars indicate standard deviations. (B) A series of phosphate uptake measurements was performed with phosphate concentrations ranging from 1 to 20 μM for M. smegmatis SMR5 (wild-type) and ML10 (ΔmspA ΔmspC). For each strain, the uptake rates at different phosphate concentrations were approximated by Michaelis-Menten functions that are shown as regression lines (dotted lines).
Kinetic experiments with phosphate concentrations ranging from 1 μM to 20 μM and time points at 45, 75, 105, 135, 165, and 195 s were performed to determine the apparent permeability coefficients of wild-type M. smegmatis and ΔmspA ΔmspC mutant ML10 for phosphate. The data were fitted well by the Michaelis-Menten equation (Fig. 3B). Data analysis yielded Vmax values of 0.4 and 0.2 nmol min−1 mg−1 and Km values of 12.7 and 15.2 μM for wild-type M. smegmatis and the mutant ML10, respectively. The lower limit of the outer membrane permeability can be estimated by assuming that the phosphate transport across the inner membrane has a very high affinity. This has not been determined for M. smegmatis yet, but Pst systems are highly efficient (Km) and such a system is present in M. smegmatis (2). Thus, the apparent permeability coefficients were calculated to 2.1 × 10−6 and 1 × 10−6 cm s−1 for wild-type M. smegmatis and the porin double-mutant ML10.
The cell envelope of M. bovis BCG represents a permeability barrier for PhoA substrates.
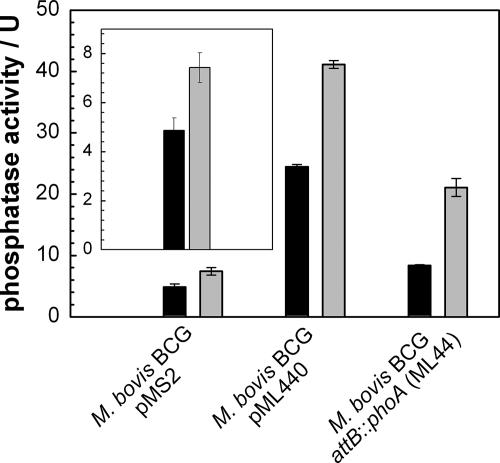
The phosphatase activity in whole cells of M. bovis BCG was significantly reduced compared to that in lysed cells (Fig. 4, inset). This indicated that M. bovis BCG contains at least one endogenous cell-bound phosphatase which hydrolyzes pNPP. The expression of phoA of M. smegmatis from the attB site (strain ML44) or from the replicating plasmid pML440 increased the phosphatase activity level three- or sixfold, respectively. In both recombinant M. bovis BCG strains, the phosphatase activity in whole cells was approximately twofold reduced compared to that in the corresponding cell lysates (Fig. 4). Taken together, these results also demonstrated that the cell envelope of slow-growing mycobacteria represents a permeability barrier for phosphates. This was independent of the type of phosphatase exploited in this assay. Single colonies of M. bovis BCG expressing phoA from the plasmid pML440 at the attB site did not turn blue on plates containing BCIP (not shown). This was due to both a lower level of PhoA and a lower permeability for pNPP as derived from a comparison of cell lysates and whole cells of the recombinant strains of M. bovis BCG and M. smegmatis (Fig. 1A and 4).
FIG. 4.
Expression of phoA in M. bovis BCG. M. bovis BCG strains containing the control plasmid pMS2, the phoA expression plasmid pML440, or a phoA expression cassette integrated at the attachment site of phage L5 were grown in liquid culture. The phosphatase activity in these strains was determined by using pNPP. Black and gray bars indicate the phosphatase activity of whole and lysed cells, respectively. The inset shows the endogenous phosphatase activity of M. bovis BCG. Error bars indicate standard deviations.
Integration of a phoA expression cassette into the chromosome of M. smegmatis.
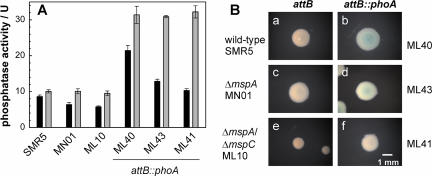
Since porins that mediate the uptake of phosphates in M. tuberculosis and M. bovis BCG are unknown, we attempted to use a phoA-based screening system in M. smegmatis porin mutants to identify them. To this end, the plasmid pML443 carrying the pimyc-phoA cassette was integrated into the attB sites of M. smegmatis and the porin mutants MN01 and ML10. The integration of the phoA expression cassette at the attB site increased the phosphatase activity in lysed cells threefold in all strains compared to that in their parent strains (Fig. 5A). The phosphatase activity in whole cells was significantly reduced compared to that in lysed cells both for the parent strains and for the strains with an integrated recombinant phoA expression cassette. Further, the phosphatase activity in whole cells decreased with a reduced number of Msp porins. These results are consistent with the previous observations that the OM of M. smegmatis represents a permeability barrier for pNPP and that the diffusion of pNPP is porin dependent. However, the colonies of the recombinant strains did not show a clearly detectable blue color on 7H10 agar plates with BCIP (Fig. 5B). We concluded that these M. smegmatis strains are not useful for screening for porins of M. tuberculosis.
FIG. 5.
Integration of phoA into the chromosomes of M. smegmatis wild-type and porin mutants. (A) The phosphatase activity of M. smegmatis grown in liquid culture was determined by using pNPP. Black and gray bars indicate the phosphatase activity of whole and lysed cells, respectively. Error bars indicate standard deviations. (B) Phosphatase activity of M. smegmatis on Middlebrook 7H10 agar plates containing Tween 80 and BCIP. After 5 days of incubation, pictures of colonies were taken with a Zeiss stereomicroscope Stemi 2000-C using the same magnification for all strains. In both experiments, the strains were SMR5 (wild-type) and the porin mutants MN01 (ΔmspA) and ML10 (ΔmspA/mspC) and derivatives with an phoA expression cassette integrated at the attachment site of the phage L5 (attB::phoA). The blue color indicates the cleavage of BCIP by PhoA.
DISCUSSION
Porins are required for the uptake of inorganic and organic phosphates by M. smegmatis.
In this study, we demonstrated that the cell envelope of M. smegmatis represents a permeability barrier for phosphates and that access to both organic phosphates and inorganic phosphate in M. smegmatis is mediated by the porins MspA and MspC. Msp porins are integral pore-forming membrane proteins (14, 27) and are directly accessible on the cell surfaces of M. smegmatis by antibodies (14, 43). It follows that the membrane in which MspA is localized represents the permeability barrier for phosphates. These results are consistent with a model proposed by Minnikin (25) and substantial indirect evidence that the mycolic acids, together with a large variety of extractable lipids, form an unusual outer membrane in addition to the cytoplasmic membrane of mycobacteria (7, 8, 13, 21, 28). These results are similar to those obtained for E. coli: mutations of OmpF and OmpC porins reduced the OM permeability to pNPP by two- to sixfold and concomitantly reduced the phosphatase activity in whole cells (23).
The diffusion of phosphate through Msp porins is quite inefficient compared to that of neutral solutes as shown by the 10-fold lower permeability coefficient of wild-type M. smegmatis for inorganic phosphate compared to that for glucose (44). This observation is not surprising considering the high density of negative charges in the constriction zones of MspA (14) and MspC, the two porins expressed by wild-type M. smegmatis (44). Furthermore, the fact that the deletion of the porins MspA and MspC reduced the OM permeability of ML10 for glucose by 75-fold (44) but only by 2-fold for phosphate compared to that for the isogenic parent strain indicated that the residual porins in ML10 allow a faster diffusion of phosphate than do MspA and MspC to partially compensate for the 15-fold lower number of Msp porins in ML10. We have shown that the expressions of the mspB and mspD genes, which are silent in wild-type M. smegmatis, are induced in ML10 (44). Relative to MspA, in the surface-exposed loop L9, MspB has only two amino acid exchanges (43), which are not likely to alter the solute specificity of this pore. However, the mutation D91G in MspD reduces the number of negative charges in the constriction zone and may increase the diffusion rate of negatively charged solutes relative to MspA.
Growth of M. smegmatis at low-phosphate concentrations depends on Msp porins.
In order to efficiently scavenge phosphate at submillimolar concentrations, E. coli expresses the high-affinity phosphate transport system Pst and the outer-membrane porin PhoE, which preferentially allow the diffusion of anions across the OM (33). Under those conditions, PhoE completely compensates for the loss of the two general porins OmpF and OmpC in E. coli mutants (17). By contrast, the loss of porins MspA and MspC caused a severe growth defect in M. smegmatis on plates containing 0.5 mM phosphate. It is surprising that a twofold reduction of the overall permeability of the porin mutant ML10 for phosphate compared to that of the isogenic parent strain resulted in such a drastic growth defect. A 75-fold-lower level of permeability for glucose was required to produce a significant growth defect of the porin mutant ML10 on regular 7H9 plates containing 22 mM glycerol (44). This demonstrates that no other porin fully compensates for the loss of MspA and MspC in M. smegmatis under those conditions. In this regard, it was striking that the porin mutant already showed a significant growth defect at phosphate concentrations of 10 mM (not shown). Such high concentrations may be necessary to overcome the intrinsically slow diffusion of phosphate through the Msp porins.
Localization of phosphatases in mycobacteria.
The reduced phosphatase activity in whole cells of the recombinant M. smegmatis strains that overexpressed the endogenous alkaline phosphatase (PhoA) to up to 90% of the total phosphatase activity may also be explained by the existence of the OM permeability barrier. However, it may be argued that this effect results from a (partial) mislocalization of PhoA in the cytoplasm due to overexpression and a putative overload of the export apparatus of M. smegmatis. PhoA of M. smegmatis shares almost 60% identical amino acids with PhoA of Escherichia coli, including the conserved residues of the active site and the four cysteines which form essential intrastrand disulfide bridges (41). The efficient formation of these disulfide bonds occurs in only the oxidative milieu of the periplasm of E. coli, explaining why PhoA is unable to fold into an active conformation if retained in the cytoplasm (11). Thus, it is likely that the export of M. smegmatis PhoA is also required for activity. This possibility argues against a contribution of putatively mislocated PhoA to the observed activity. In contrast to its E. coli homolog, PhoA of M. smegmatis has a lipoprotein signal sequence. Cell fractionation experiments of PhoA labeled with [14C]acetate showed indeed that PhoA of M. smegmatis is a membrane-bound lipoprotein (18). Our results strongly suggest that PhoA is not a surface protein as was concluded from the association of PhoA with membranes (18) but rather localized in the periplasm of M. smegmatis as it is for E. coli (23).
Three lipoproteins with phosphatase activity and the secreted acid phosphatase SapM were identified in M. bovis BCG (3) and M. tuberculosis (35), respectively. Our observation that the cell envelope of M. bovis BCG establishes a permeability barrier for phosphates indicates that at least one phosphatase with significant activity is localized in the periplasm. Our experiments do not exclude the possibility that other phosphatases are either located on the surface or secreted. However, the conclusion that proteins are surface proteins based solely on membrane association (4) is premature as demonstrated for PhoA of M. smegmatis.
Phosphate uptake in slow-growing mycobacteria.
Survival inside macrophages requires the adaptation of intracellular pathogens to the phagosomal environment. The transcriptional profiles of M. tuberculosis and Salmonella enterica in infected macrophages revealed that the proteins involved in inorganic phosphate transport are up-regulated (12, 39), indicating that phosphate levels inside phagosomes of macrophages are indeed limited. Consistent with this conclusion, genes encoding efficient phosphate transport systems were found to be essential for the survival of M. tuberculosis in macrophages and mice (34, 38). However, how inorganic or organic phosphates cross the OM of M. tuberculosis is unknown. Since the direct rate of diffusion of phosphates through model lipid membranes is extremely low (permeability coefficient of the monoanion = 5 × 10−12 cm/s [9]), it appears likely that slow-growing mycobacteria also use OM pore proteins for the uptake of phosphate. Indeed, the existence of a porin with anion specificity has been demonstrated (20). This porin still awaits discovery. A screening system based on complementation of the uptake defects for phosphates of a porin mutant of M. smegmatis as described in this study provides a tool to identify such a porin of M. tuberculosis. These experiments are currently being pursued in our laboratory.
Acknowledgments
Preliminary sequence data for Mycobacterium smegmatis were obtained from the Institute for Genomic Research website at http://www.tigr.org.
This work was supported by grant AI063432 from the National Institutes of Health.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Bauer, K., M. Struyve, D. Bosch, R. Benz, and J. Tommassen. 1989. One single lysine residue is responsible for the special interaction between polyphosphate and the outer membrane porin PhoE of Escherichia coli. J. Biol. Chem. 264:16393-16398. [PubMed] [Google Scholar]
- 2.Bhatt, K., S. K. Banerjee, and P. K. Chakraborti. 2000. Evidence that phosphate specific transporter is amplified in a fluoroquinolone resistant Mycobacterium smegmatis. Eur. J. Biochem. 267:4028-4032. [DOI] [PubMed] [Google Scholar]
- 3.Braibant, M., and J. Content. 2001. The cell surface associated phosphatase activity of Mycobacterium bovis BCG is not regulated by environmental inorganic phosphate. FEMS Microbiol. Lett. 195:121-126. [DOI] [PubMed] [Google Scholar]
- 4.Braibant, M., L. De Wit, P. Peirs, M. Kalai, J. Ooms, A. Drowart, K. Huygen, and J. Content. 1994. Structure of the Mycobacterium tuberculosis antigen 88, a protein related to the Escherichia coli PstA periplasmic phosphate permease subunit. Infect. Immun. 62:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braibant, M., P. Lefevre, L. de Wit, P. Peirs, J. Ooms, K. Huygen, A. B. Andersen, and J. Content. 1996. A Mycobacterium tuberculosis gene cluster encoding proteins of a phosphate transporter homologous to the Escherichia coli Pst system. Gene 176:171-176. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., T. I. Griffin, J. I. Kriakov, S. T. Friedman, N. D. Grindley, and W. R. Jacobs, Jr. 2000. Identification of genes encoding exported Mycobacterium tuberculosis proteins using a Tn552′phoA in vitro transposition system. J. Bacteriol. 182:2732-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 8.Camacho, L. R., P. Constant, C. Raynaud, M. A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffé, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti, A. C., and D. W. Deamer. 1992. Permeability of lipid bilayers to amino acids and phosphate. Biochim. Biophys. Acta 1111:171-177. [DOI] [PubMed] [Google Scholar]
- 10.Derman, A. I., and J. Beckwith. 1991. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J. Bacteriol. 173:7719-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derman, A. I., W. A. Prinz, D. Belin, and J. Beckwith. 1993. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262:1744-1747. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 13.Etienne, G., C. Villeneuve, H. Billman-Jacobe, C. Astarie-Dequeker, M. A. Dupont, and M. Daffe. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148:3089-3100. [DOI] [PubMed] [Google Scholar]
- 14.Faller, M., M. Niederweis, and G. E. Schulz. 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189-1192. [DOI] [PubMed] [Google Scholar]
- 15.Jarlier, V., and H. Nikaido. 1990. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J. Bacteriol. 172:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaps, I., S. Ehrt, S. Seeber, D. Schnappinger, C. Martin, L. W. Riley, and M. Niederweis. 2001. Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene 278:115-124. [DOI] [PubMed] [Google Scholar]
- 17.Korteland, J., J. Tommassen, and B. Lugtenberg. 1982. PhoE protein pore of the outer membrane of Escherichia coli K12 is a particularly efficient channel for organic and inorganic phosphate. Biochim. Biophys. Acta 690:282-289. [DOI] [PubMed] [Google Scholar]
- 18.Kriakov, J., S. Lee, and W. R. Jacobs, Jr. 2003. Identification of a regulated alkaline phosphatase, a cell surface-associated lipoprotein, in Mycobacterium smegmatis. J. Bacteriol. 185:4983-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefèvre, P., M. Braibant, L. de Wit, M. Kalai, D. Roeper, J. Grotzinger, J. P. Delville, P. Peirs, J. Ooms, K. Huygen, and J. Content. 1997. Three different putative phosphate transport receptors are encoded by the Mycobacterium tuberculosis genome and are present at the surface of Mycobacterium bovis BCG. J. Bacteriol. 179:2900-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichtinger, T., B. Heym, E. Maier, H. Eichner, S. T. Cole, and R. Benz. 1999. Evidence for a small anion-selective channel in the cell wall of Mycobacterium bovis BCG besides a wide cation-selective pore. FEBS Lett. 454:349-355. [DOI] [PubMed] [Google Scholar]
- 21.Liu, J., E. Y. Rosenberg, and H. Nikaido. 1995. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. USA 92:11254-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mailaender, C., N. Reiling, H. Engelhardt, S. Bossmann, S. Ehlers, and M. Niederweis. 2004. The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis. Microbiology 150:853-864. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, M. B., M. Flickinger, L. Higgins, T. Krick, and G. L. Nelsestuen. 2001. Reduced outer membrane permeability of Escherichia coli O157:H7: suggested role of modified outer membrane porins and theoretical function in resistance to antimicrobial agents. Biochemistry 40:11965-11974. [DOI] [PubMed] [Google Scholar]
- 24.Martinez, M. B., F. J. Schendel, M. C. Flickinger, and G. L. Nelsestuen. 1992. Kinetic properties of enzyme populations in vivo: alkaline phosphatase of the Escherichia coli periplasm. Biochemistry 31:11500-11509. [DOI] [PubMed] [Google Scholar]
- 25.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of the mycobacteria: physiology, identification, and classification, vol. I. Academic Press, London, United Kingdom. [Google Scholar]
- 26.Niederweis, M. 2003. Mycobacterial porins—new channel proteins in unique outer membranes. Mol. Microbiol. 49:1167-1177. [DOI] [PubMed] [Google Scholar]
- 27.Niederweis, M., S. Ehrt, C. Heinz, U. Klöcker, S. Karosi, K. M. Swiderek, L. W. Riley, and R. Benz. 1999. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 33:933-945. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido, H., S. H. Kim, and E. Y. Rosenberg. 1993. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol. Microbiol. 8:1025-1030. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido, H., and E. Y. Rosenberg. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J. Bacteriol. 153:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbeeke, N., and B. Lugtenberg. 1980. Expression of outer membrane protein E of Escherichia coli K12 by phosphate limitation. FEBS Lett. 112:229-232. [DOI] [PubMed] [Google Scholar]
- 31.Peirs, P., P. Lefevre, S. Boarbi, X. M. Wang, O. Denis, M. Braibant, K. Pethe, C. Locht, K. Huygen, and J. Content. 2005. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect. Immun. 73:1898-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi, Y., Y. Kobayashi, and F. M. Hulett. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 179:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao, N. N., and A. Torriani. 1990. Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4:1083-1090. [DOI] [PubMed] [Google Scholar]
- 34.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saleh, M. T., and J. T. Belisle. 2000. Secretion of an acid phosphatase (SapM) by Mycobacterium tuberculosis that is similar to eukaryotic acid phosphatases. J. Bacteriol. 182:6850-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sander, P., A. Meier, and E. C. Bottger. 1996. Ribosomal drug resistance in mycobacteria. Res. Microbiol. 147:59-67. [DOI] [PubMed] [Google Scholar]
- 38.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholz, O., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267:1565-1570. [DOI] [PubMed] [Google Scholar]
- 41.Sone, M., S. Kishigami, T. Yoshihisa, and K. Ito. 1997. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272:6174-6178. [DOI] [PubMed] [Google Scholar]
- 42.Springer, B., P. Sander, L. Sedlacek, K. Ellrott, and E. C. Böttger. 2001. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol. 290:669-675. [DOI] [PubMed] [Google Scholar]
- 43.Stahl, C., S. Kubetzko, I. Kaps, S. Seeber, H. Engelhardt, and M. Niederweis. 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40:451-464. (Author's correction, 57:1509.) [DOI] [PubMed] [Google Scholar]
- 44.Stephan, J., J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailander, H. Engelhardt, and M. Niederweis. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58:714-730. [DOI] [PubMed] [Google Scholar]
- 45.Stephan, J., C. Mailaender, G. Etienne, M. Daffe, and M. Niederweis. 2004. Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:4163-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephan, J., V. Stemmer, and M. Niederweis. 2004. Consecutive gene deletions in Mycobacterium smegmatis using the yeast FLP recombinase. Gene 343:181-190. [DOI] [PubMed] [Google Scholar]
- 47.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 48.van Veen, H. W. 1997. Phosphate transport in prokaryotes: molecules, mediators and mechanisms. Antonie Leeuwenhoek 72:299-315. [DOI] [PubMed] [Google Scholar]