Abstract
Mycoplasma pulmonis possesses a cassette of genes that are predicted to code for type III restriction and modification (R-M) enzymes. Transposon disruption of a gene predicted to code for the endonuclease subunit of the enzyme resulted in loss of R-M activity. Genomic data indicate that the cassette was acquired by horizontal gene transfer and possibly located on a mobile element.
Despite having small genomes, mycoplasmas devote considerable coding capacity to genes coding for restriction and modification (R-M) systems. Both type I and type II R-M systems are common in mycoplasmas (15). The genome of the murine pathogen Mycoplasma pulmonis strain CT is 960 kb and codes for fewer than 800 proteins (4). About 2% of the genome (20 kb) consists of hsd genes coding for type I R-M enzymes. Type I enzymes are multisubunit complexes. The S subunit dictates the specificity of the DNA sequence that is recognized. The M subunit catalyzes the DNA methyltransferase (MTase) reaction that modifies the DNA recognition sequence to protect the DNA from the enzyme's restriction endonuclease (REase) activity. The R subunit is required for the DNA helicase and REase activities of the holoenzyme that result in cleavage of DNA at essentially random sites that can be as far as several kilobases from the DNA recognition sequence. The type I enzymes of M. pulmonis were among the first of the phase-variable R-M systems to be described (16). On/off switching occurs as a result of DNA inversion. Each locus has multiple recombination sites at which DNA inversion can occur within the hsdS coding regions to generate a variety of R-M enzymes of differing specificities (14). The DNA inversions occur at a high frequency, and cultures of M. pulmonis are remarkably heterogeneous with respect to the phenotype of the type I R-M enzymes that are produced.
Type III R-M enzymes are similar to type I but have just two subunits, Res and Mod. Both the Res and Mod subunits are required for REase activity, with the DNA recognition specificity dictated by Mod. The Mod subunit catalyzes the MTase reaction and can function independently of Res. Type III restriction enzymes have not been described previously in mycoplasmas, but several species including M. pulmonis contain genes predicted to code for Mod subunits. Described here is a 15-kb region of the M. pulmonis genome that codes for a type III R-M system with features suggestive of a mobile element. The R-M system is complex, with genes coding for one Res and three Mod proteins. Each mod gene has dinucelotide repeat regions that could expand or contract by slipped-strand mispairing to alter the sequence of the Mod proteins. Thus, as is the case for the type I R-M enzymes of this species, M. pulmonis may generate cell populations that are heterogeneous with respect to the activity of its type III R-M enzymes. Such variability in R-M activity may contribute to the overall fitness of a mycoplasmal population.
MATERIALS AND METHODS
Mycoplasmas and phage.
M. pulmonis strain CT was originally isolated from a mouse lung (7). The location of transposon Tn4001T in library members of M. pulmonis strain CT has been cataloged using an inverse PCR strategy described previously (26). CT228 is a previously described mutant of CT that has the transposon disrupting the hvsR gene (23). The HvsR recombinase catalyzes DNA inversions within the hsd and vsa loci that are responsible for the phase-variable production of the Hsd and Vsa proteins. Hence, CT228 is phase locked, lacking type I R-M activity and producing the VsaA surface protein. CT269 is a transposon mutant with Tn4001T inserted at nucleotide position 466151 of the complete genome sequence (4), disrupting the MYPU_3950 gene. Strain KD735-15 is a derivative of strain UAB 6510, originally isolated from a rat lung and unrelated to CT (2). Mycoplasmas were cultured in mycoplasma broth and assayed for colony-forming units on mycoplasma agar as previously described (10). Subclones of mycoplasma strains were obtained by filter cloning (17). The orientation of the DNA-invertible elements encoding the hsd genes of M. pulmonis strains was determined by using previously described PCR primers and amplification conditions (14). Stocks of the M. pulmonis P1 phage were prepared and assayed for plaque-forming units as previously described (13, 16).
Mini-transposon construction and mapping of transposon insertion sites in transformed mycoplasma.
Plasmid pMT85 contains a previously described Tn4001 mini-transposon (28), but its gentamicin resistance marker does not function in M. pulmonis. A chloramphenicol resistance marker (cat) driven by the promoter of the M. pulmonis vsa gene was inserted into the mini-transposon by a method similar to that described for the construction of Tn4001C (11). A 400-bp fragment containing the vsa promoter was amplified from M. pulmonis genomic DNA using the forward primer TGTTTGGATCCAGTTTTTTTGAACC and the reverse primer TGCATGGTACCTCCTATTTTAAAATTATG (underlined sequences refer to KpnI and BamHI restriction sites introduced into the primers for convenient cloning). The cat coding region (660 bp) was amplified from Tn4001C template using the forward primer GGAAGGTACCATGGAGAAAAAAATCAC and the reverse primer GAGGATCCACTTCTCGAGGCGTAGCACCAGG. The two PCR products were purified from an agarose gel using QiaQuick (QIAGEN), digested with KpnI, and ligated at room temperature for 30 min with T4 DNA ligase (New England Biolabs). One microliter of the ligation served as the template in a second PCR using the vsa forward primer and cat reverse primer. The resulting PCR product was gel purified, digested with BamHI, and ligated into the BamHI site of the pMT85 mini-transposon. The resulting plasmid was designated pTF86-VC. All PCRs were performed with i-Proof DNA polymerase (Bio-Rad) per the manufacturer's recommendations. Plasmids were transformed into TOP10 Escherichia coli (Invitrogen) and maintained with 15 μg of chloramphenicol/ml.
M. pulmonis was transformed with pTF86-VC by using the polyethylene glycol-mediated procedure developed for this species, selecting for transformants using 15 μg of chloramphenicol/ml (8, 11, 12). Plasmid pTF86-VC does not replicate in mycoplasmas, and transformants are obtained only when the mini-transposon integrates into the chromosome. It was determined by inverse PCR amplification of the junction between the transposon and the mycoplasmal genome that the mini-transposon inserted at diverse sites in the M. pulmonis genome. The protocol for mapping the transposon insertion sites was similar to that used for the mapping of Tn4001T sites in M. pulmonis (26), except that Sau3A was the restriction enzyme used to digest genomic DNA and the primers for PCR amplification were CTGATTCTGTGGATAACCGTATTACCGC and CAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTA.
PCR amplification and Southern analysis.
Genomic DNA isolation and conditions for PCR amplification of genes were as described previously (26). For Southern analysis, a 1.8-kb PCR product (Fig. 1) was radiolabeled with 32P and used as a probe of HindIII-digested genomic DNA. The PCR primers used to obtain the probe were CCACATGTACCGGTATTGGTATTGACCG and TGTGGATCCTTTTTCACTTGGACCTCCACG. Conditions for radiolabeling, agarose gel electrophoresis, blotting, and hybridization were as described previously (14).
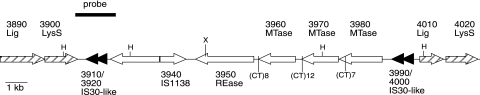
FIG. 1.
Schematic of a 22-kb region of M. pulmonis strain CT containing the type III R-M locus. Arrows point in the direction of transcription. Black arrows indicate the two identical IS30-like elements, and hatched arrows indicate flanking gene duplication of 100% nucleotide sequence identity. X refers to the position of Tn4001T in CT269.FC2. The letter H refers to HindIII sites flanking the IS30 elements that give rise to the DNA fragments shown in Fig. 3 with the IS30-like probe indicated by the solid bar. (CT)8, (CT)12, and (CT)7 refer to the positions and number of CT dinucleotide repeats (on the noncoding strands of MYPU_3960, MYPU_3970, and MYPU_3980, respectively) with the number of repeats indicated in parenthesis.
Sequence analysis.
BLAST analysis was performed at www.ncbi.nlm.nih.gov. ClustalW and phylogenetic reconstructions (best tree by the neighbor-joining method) were obtained using the default settings of the MacVector software package (Accelrys).
RESULTS AND DISCUSSION
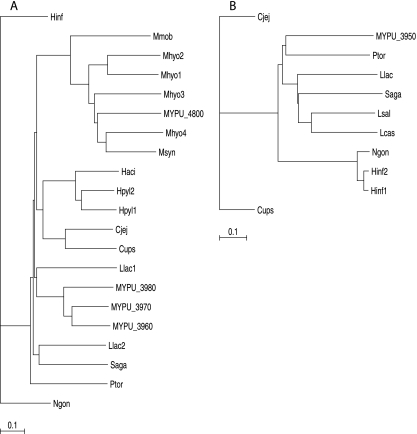
Most of the M. pulmonis genes annotated as coding for type III MTases are located in the 22-kb region illustrated in Fig. 1 (annotations of the M. pulmonis genome are provided at http://genolist.pasteur.fr/MypuList/). The open reading frames (ORFs) MYPU_3960, MYPU_3970, and MYPU_3980 are predicted to code for type III MTases based on BLAST analysis and Clusters of Orthologous Groups database assignments. MYPU_3950 was originally annotated as coding for an MTase, but this annotation must have been inadvertent. The predicted gene product lacks motifs indicative of an MTase, and BLAST analysis reveals a high level of amino acid sequence similarity to type III Res subunits rather than Mod subunits. An additional type III MTase predicted from the genome sequence is the product of MYPU_4800. The top hits obtained by BLAST analysis of MYPU_3960, MYPU_3970, MYPU_3980, and MYPU_4800 were analyzed by ClustalW. The MYPU_4800 MTase clusters with several other type III MTases of mycoplasmal origin, while MYPU_3960, MYPU_3970, and MYPU_3980 are more similar to type III MTases from other bacteria (Fig. 2A). A similar analysis of the top hits obtained by BLAST analysis of MYPU_3950 is provided in Fig. 2B. All of the sequences analyzed in Fig. 2B are REases from bacteria other than mycoplasmas because MYPU_3950 is the only known (putative) mycoplasmal type III REase.
FIG. 2.
Phylogenetic reconstruction of type III MTases (A) and REases (B) most closely related to those of M. pulmonis. The scale indicates the expected number of substitutions per site. The name of the bacterial species (and accession number) from which each enzyme sequence was obtained is as follows in panel A: Cjej, Campylobacter jejuni (AAW35760); Cups, Campylobacter upsaliensis (ZP_00371642); Haci, Helicobacter acinonychis (YP_665417); Hinf, Haemophilus influenzae (AAC22721); Hpyl1, Helicobacter pylori (AAD06869); Hpyl2, H. pylori (ABF85395); Llac1, Lactococcus lactis subsp. cremoris (ZP_00382503); Llac2, L. lactis (AAD15792); Mhyo1, Mycoplasma hyopneumoniae (AAZ253758); Mhyo2, M. hyopneumoniae (AAZ44509); Mhyo3, M. hyopneumoniae (AAZ44470); Mhyo4, M. hyopneumoniae (YP_115842); Mmob, Mycoplasma mobile (AAT27947); MYPU_3960, M. pulmonis (NP_326227); MYPU_3970, M. pulmonis (NP_326228); MYPU_3980, M. pulmonis (NP_326229); MYPU_4800, M. pulmonis (NP_326311); Msyn, Mycoplasma synoviae (AAZ43581); Ngon, Neisseria gonorrhoeae (AAW89368); Ptor, Psychroflexus torques (ZP_0125993); and Saga, Streptococcus agalactiae (ZP_00787820). In panel B, the species (and accession numbers) are as follows: Cjej, C. jejuni (YP_178743); Cups, C. upsaliensis (ZP_00371643); Hinf1, H. influenzae (AB185526); Hinf2, H. influenzae (ZP_00155673); Lcas, Lactobacillus casei (ZP_00386539); Llac, L. lactis (AAD15793); Lsal, Lactobacillus salivarius subsp. salivarius (YP_536647); MYPU_3950, M. pulmonis (NP_326226); Ngon, N. gonorrhoeae (YP_207779); Ptor, P. torques (ZP_0125992); and Saga, S. agalactiae (ZP_00787826).
Assessment of R-M activity.
To assess type III R-M activity, the phenotypes of mutants with a transposon disrupting MYPU_3950 was studied. Transposon library members that had disruptions in MYPU_3950 were screened to identify those that were susceptible to infection by mycoplasma virus P1. P1 adsorbs only to cells that produce the phase-variable lipoprotein VsaA (9), and cells producing an alternative Vsa protein (the full repertoire of Vsa proteins in strain CT includes VsaA, -C, -E, -F, -G, -H, and -I) are resistant to the phage (17). Strain CT269 was identified as a phage-susceptible library member that has the transposon inserted in MYPU_3950 at a site (nucleotide position 466151 in the CT genome) that would truncate 152 (17%) of 897 amino acids from the gene product (Fig. 1).
To assess type III R-M activity in CT269, cell stocks were prepared that lacked type I R-M activity. The hsdR and hsdM genes are transcribed as part of an operon, and no type I R-M activity is detected when hsdR and hsdM are not transcribed. Transcription of hsdR and M occurs in only one orientation of the hsd invertible element. Subclones of CT269 were screened by PCR to identify those that had the hsd loci oriented such that the hsdR and hsdM genes would not be transcribed. Filter clone 2 (CT269.FC2) was identified as having the hsd loci in the off orientation, lacking type I R-M activity.
The R-M activity of CT269.FC2 was compared to that of controls. Strain CT228 is a transposon mutant from the same CT parent strain as CT269.FC2, but the transposon in CT228 is inserted into the gene coding for the HvsR recombinase and is phase-locked with the hsd loci oriented such that type I R-M activity is absent (23, 24). R-M activity was also assessed for KD735-15, another strain of M. pulmonis that lacks detectable R-M activity because its hsd loci are in the off orientation (16). When P1 was propagated on CT269.FC2 (P1 · CT269.FC2), the virus readily infected CT269.FC2 and KD735-15 but had a low titer on CT228, indicative of restriction (Table 1). CT228 can also modify the virus because when P1 was propagated on CT228 (P1 · CT228), the virus had essentially equal titers on all strains, and no restriction was evident. When P1 was propagated on KD735-15 (P1 · KD735-15), the virus was restricted by CT228 but not by CT269.FC2. Collectively, the data indicate that CT228 possesses an R-M system that is absent in both CT269.FC2 and KD735-15. The data also indicate that neither KD735-15 nor CT269.FC2 has R-M activity that is absent in the other strain. Because CT228 is phase-locked in respect to type I R-M activity, the R-M activity detected in CT228 must be the type III system that is inactivated in CT269.FC2 by the transposon inserted into MYPU_3950.
TABLE 1.
R-M properties of M. pulmonis
| Virus stock used for infection | No. of PFU on indicated M. pulmonis strain (relative titer)a
|
||
|---|---|---|---|
| KD735-15 | CT228 | CT269.FC2 | |
| P1 · KD735-15 | 2 × 107 (1.0)b | 1 × 106 (0.05) | 2 × 107 (1.0) |
| P1 · CT228 | 3 × 107 (1.5) | 2 × 107 (1.0) | 2 × 107 (1.0) |
| P1 · CT269.FC2 | 3 × 106 (1.5) | 8 × 104 (0.04) | 2 × 106 (1.0) |
Similar results were obtained in three independent experiments. The relative titer of each phage stock when assayed on its parental host strain was assigned a value of 1.0.
Another approach for assessing R-M activity is to compare transformation frequencies. From five independent experiments, the average frequency for transformation of M. pulmonis with plasmid pTF86-VC was 2 × 10−6 per CFU for CT269.FC2 and 4 × 10−7 per CFU for CT228. The transformation frequency for CT269.FC2 was comparable to previously reported transformation frequencies for strain KD735-15 (11), consistent with the data on plaque-forming units indicating that KD735-15 and CT269.FC2 lack R-M activity.
Evidence for motility of the M. pulmonis type III R-M genes.
The lack of R-M activity detectable in KD735-15 suggested that this strain lacks the MYPU_3950 REase. PCR primers were designed to amplify MYPU_3950 (Table 2). PCR products of the expected size were obtained when CT genomic DNA was used as template, but no product was obtained from KD735-15 DNA template.
TABLE 2.
PCR amplification of type III R-M genes and flanking sequences
| Primer pair | Product
|
|
|---|---|---|
| KD735-15 template | CT template | |
| ATGGTGGAAAAGGTGATTCGC | None | 250-bp region of MYPU_3950 |
| TCAGGCTCAAATCTTTCTCCTGC | ||
| ATTGATATTCCTGAAATAGTTCAAAGAGG | None | 2,401-bp region flanking MYPU_3950 |
| CTTGTTTTAATAGATGTGATCCCTTGG | ||
| TGCTTCTTTTCAAAGGGGCTC | None | 178-bp region of MYPU_3910 |
| TGGTCAAAAAACTTCAGCAGCG | ||
| CGGAATTTATGAACGAGTTACTGCTC | None | 177-bp region of MYPU_3920 |
| CTCTCTTTAATTTTCGAGTGCGTTG | ||
The type III R-M genes shown in Fig. 1 are flanked by potential insertion sequence (IS) elements. Not only do MYPU_3910 and MYPU_3920 have the same nucleotide sequence as MYPU_3990 and MYPU_4000, respectively, but the intervening sequences between these pairs of ORFs are also identical. Each of these genes is annotated as a fragment of the M. pulmonis insertion sequence element IS1138 (3). However, the top hits obtained by BLAST analysis of the product of these genes was not the transposase of IS1138 but, instead, transposases from a variety of other mycoplasmas. If these ORFs code for functional transposases, the structure is unusual. The two genes of each pair of ORFs are in the same reading frame. Thus, there is no possibility of a single protein being coded by each pair as a result of translational frameshifting, as is found for transposases of the IS3 family (5). The products of MYPU_3910 and MYPU_3990 and of MYPU_3920 and MYPU_4000 contain 146 and 186 amino acids, respectively, which is about half the size of a typical transposase. The top hit by BLAST obtained for MYPU_3920 and MYPU_4000 corresponded to the amino terminal half of the IS30-like element of Mycoplasma bovis (accession number AAK94953). Likewise, the top BLAST hit for MYPU_3910 and MYPU_3990 was the carboxyl terminal half of the same M. bovis IS-like element. Perhaps each pair of the M. pulmonis ORFs codes for a heteromeric transposase, as found for Tn7 (22).
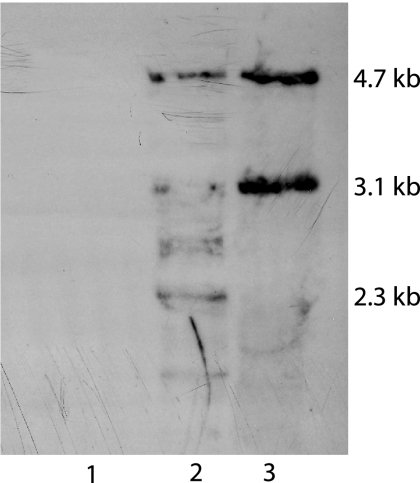
To investigate whether MYPU_3910 and MYPU_3920 (and MYPU_3990 and MYPU_4000) are common to all strains of M. pulmonis, PCR primers were designed to amplify these sequences (Table 2). Products of the expected size were obtained when the template was CT genomic DNA, but no product was obtained from KD735-15 DNA template. This result was confirmed by Southern analysis. A 1.8-kb PCR product (Fig. 1) encompassing MYPU_3910 and MYPU_3920 hybridized to DNA fragments of the expected size from CT but not from KD735-15, indicating that KD735-15 lacks not just MYPU_3950 but also MYPU_3910 and MYPU_3920 (Fig. 3). DNA from two different stocks of CT was examined. One stock has been maintained in the laboratory with a minimum amount of passaging. The hybridization profile obtained for this stock was, for the most, part limited to the two HindIII fragments corresponding to MYPU_3910 and MYPU_3920 and to MYPU_3990 and MYPU_4000. The other stock of CT has been passaged frequently in the laboratory and yielded a more complicated hybridization profile that included several minor bands in addition to the two expected bands. The minor bands may have resulted from transposition of the IS30-like element within some of the CT cells.
FIG. 3.
Southern analysis of HindIII-digested M. pulmonis DNA probed with the IS30-like element. Lane 1, KD735-15 DNA; lane 2, DNA from a highly passaged stock of strain CT; lane 3, DNA from a low-passage stock of strain CT. The 4.7-kb and 3.1-kb DNA fragments are of the predicted size for the HindIII fragments containing MYPU_3990 and MYPU_4000 and for MYPU_3910 and MYPU_3920, respectively.
We propose that a cassette of type III R-M genes bounded by IS elements corresponding to MYPU_3910 and MYPU_3920 and to MYPU_3990 and MYPU_4000 was acquired by horizontal gene transfer from another bacterium. This gene transfer event may have involved the duplication of the DNA ligase (MYPU_3890 and MYPU_4010) and lysyl tRNA synthetase (MYPU_3900 and MYPU_4020) genes (Fig. 1). The duplication event must have been relatively recent because there are no nucleotide substitutions in the duplicated region. Once acquired, the transposable element was parasitized by IS1138 (MYPU_3940). Most bacteria use UGA as a stop codon, but UGA codes for tryptophan in mycoplasmas. The IS30-like elements each have 5 UGA codons, and MYPU_3950 to MYPU_3980 have a total of 21 UGA codons. Thus, the cassette would not be functional in a nonmycoplasmal bacterium, making another mycoplasma the likely donor of the cassette to M. pulmonis. Although not thought to be capable of natural transformation or supporting replication of plasmids other than those that contain oriC, M. pulmonis can likely obtain transposable elements by mating events involving cell fusion (17). The cassette has evidently been in mycoplasmas for a long time, but surprising nucleotide similarity to type III R-M genes of other bacteria still exists. The top hit identified by nucleotide BLAST analysis of MYPU_3950 was the res gene of Lactobacillus plasmid pMP118 (accession number CP000234). The top hits obtained by nucleotide BLAST analysis of MYPU_3960, MYPU_3970, and MYPU_3980 included virtually all of the Haemophilus influenzae mod genes identified by Bayliss and coworkers (1) and also mod genes from several other gram-negative bacteria and plasmid pMP118.
It is not clear whether the M. pulmonis type III R-M cassette is a composite transposon. The stability of oriC plasmids indicates that M. pulmonis has only a low level of homologous recombination (6), and a model for integration of the cassette that requires homologous recombination between a chromosomal copy of IS30 and a plasmid-borne copy of IS30 adjacent to the cassette is unattractive. Attempts to identify inverted repeats, to which a transposase may bind, at the ends of the IS30-like elements or at the ends of the cassette were not successful. Such repeats may be difficult to identify until an empty allele lacking the cassette is available for sequence comparison. The duplication of the lig and lysS genes flanking the cassette is large, 2.6 kb. Transposable elements that can generate long duplications in target DNA upon integration have been described previously in some mycoplasmas and other bacteria (18, 20, 27). The size of the long duplications associated with transposition of these elements is usually on the order of 100 to 500 bp, which is significantly smaller than the direct repeats flanking the type III R-M cassette of M. pulmonis. In some cases, long direct repeats are associated with R-M genes that are not flanked by apparent transposase genes, giving rise to a recombinase-independent model by which mobility of the R-M genes involves an attack of the chromosome by the REase (19). With the flanking IS30-like elements, it is improbable that such a model would apply to integration of the R-M cassette of M. pulmonis.
The type III Res subunit encoded by MYPU_3950 could likely partner with any of the three Mod subunits coded for by MYPU_3960, MYPU_3970, and MYPU_3980 and possibly with MYPU_4800. Alignment of the M. pulmonis Mod subunits with one another reveals extensive amino acid sequence diversity, especially in the central portion of the proteins, reminiscent of the hypervariable region identified in Mod subunits of H. influenzae (1). Thus, three or four type III R-M enzymes, each with a different DNA sequence specificity, may be operative. As previously noted (21), the coding regions of MYPU_3960, MYPU_3970, and MYPU_3980 contain a dinucleotide repeat region (Fig. 1) that could vary in length by slipped-strand mispairing, leading to alterations in the encoded amino acid sequence that might affect the DNA sequence specificity of the Mod subunit. Thus, cultures of M. pulmonis may be heterogeneous in regard to not only the repertoire of type I R-M enzymes that are produced but also the repertoire of type III enzymes. The phase-variable type I R-M enzymes may contribute to the fitness of the mycoplasmal population in vivo, as evidenced by the relatively high percentage of cells that produce active type I R-M enzymes in the respiratory tract of infected animals (17). The type III R-M systems of M. pulmonis may also contribute to the fitness of the population, as studies in Haemophilus indicate that type III R-M systems can affect the expression of multiple genes and that selection pressure exists to generate diversity in Mod subunits (1, 25).
Acknowledgments
This work was supported by Public Health Services grants AI63909 and AR44252.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Bayliss, C. D., M. J. Callaghan, and E. R. Moxon. 2006. High allelic diversity in the methyltransferase gene of a phase-variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res. 34:4046-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhugra, B., and K. Dybvig. 1992. High-frequency rearrangements in the chromosome of Mycoplasma pulmonis correlate with phenotypic switching. Mol. Microbiol. 6:1149-1154. [DOI] [PubMed] [Google Scholar]
- 3.Bhugra, B., and K. Dybvig. 1993. Identification and characterization of IS1138, a transposable element from Mycoplasma pulmonis that belongs to the IS3 family. Mol. Microbiol. 7:577-584. [DOI] [PubMed] [Google Scholar]
- 4.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. C. Rocha, and A. Blanchard. 2001. The complete genome of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 6.Cordova, C. M. M., C. Lartigue, P. Sirand-Pugnet, J. Renaudin, R. A. F. Cunha, and A. Blanchard. 2002. Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J. Bacteriol. 184:5426-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denison, A. M., B. Clapper, and K. Dybvig. 2005. Avoidance of the host immune system through phase variation in Mycoplasma pulmonis. Infect. Immun. 73:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dybvig, K., and J. Alderete. 1988. Transformation of Mycoplasma pulmonis and Mycoplasma hyorhinis: transposition of Tn916 and formation of cointegrate structures. Plasmid 20:33-41. [DOI] [PubMed] [Google Scholar]
- 9.Dybvig, K., J. Alderete, and G. H. Cassell. 1988. Adsorption of Mycoplasma pulmonis virus P1 to host cells. J. Bacteriol. 170:4373-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dybvig, K., and G. H. Cassell. 1987. Transposition of gram-positive transposon Tn916 in Acholeplasma laidlawii and Mycoplasma pulmonis. Science 235:1392-1394. [DOI] [PubMed] [Google Scholar]
- 11.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 182:4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dybvig, K., G. E. Gasparich, and K. W. King. 1995. Artificial transformation of mollicutes via polyethylene glycol- and electroporation-mediated methods, p. 179-184. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. I. Academic Press, Orlando, FL. [Google Scholar]
- 13.Dybvig, K., A. Liss, J. Alderete, R. M. Cole, and G. H. Cassell. 1987. Isolation of a virus from Mycoplasma pulmonis. Isr. J. Med. Sci. 23:418-422. [PubMed] [Google Scholar]
- 14.Dybvig, K., R. Sitaraman, and C. T. French. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl. Acad. Sci. USA 95:13923-13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dybvig, K., and L. L. Voelker. 1996. Molecular biology of mycoplasmas. Annu. Rev. Microbiol. 50:25-57. [DOI] [PubMed] [Google Scholar]
- 16.Dybvig, K., and H. Yu. 1994. Regulation of a restriction and modification system via DNA inversion in Mycoplasma pulmonis. Mol. Microbiol. 12:547-560. [DOI] [PubMed] [Google Scholar]
- 17.Gumulak-Smith, J., A. Teachman, A.-H. Tu, J. W. Simecka, J. R. Lindsey, and K. Dybvig. 2001. Variations in the surface proteins and restriction enzyme systems of Mycoplasma pulmonis in the respiratory tract of infected rats. Mol. Microbiol. 40:1037-1044. [DOI] [PubMed] [Google Scholar]
- 18.Lubys, A., J. Lubiene, S. Kulakauskas, K. Stankevicius, A. Timinskas, and A. Janulaitis. 1996. Cloning and analysis of the genes encoding the type IIS restriction-modification system HphI from Haemophilus parahaemolyticus. Nucleic Acids Res. 24:2760-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobusato, A., I. Uchiyama, S. Ohashi, and I. Kobayashi. 2000. Insertion with long target duplication: a mechanism for gene motility suggested from comparison of two related bacterial genomes. Gene 259:99-108. [DOI] [PubMed] [Google Scholar]
- 20.Plikaytis, B. B., J. T. Crawford, and T. M. Shinnick. 1998. IS1549 from Mycobacterium smegmatis forms long direct repeats upon insertion. J. Bacteriol. 180:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocha, E. P. C., and A. Blanchard. 2002. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarnovsky, R. J., E. W. May, and N. L. Craig. 1996. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 15:6348-6361. [PMC free article] [PubMed] [Google Scholar]
- 23.Sitaraman, R., A. M. Denison, and K. Dybvig. 2002. A unique, bifunctional site-specific DNA recombinase from Mycoplasma pulmonis. Mol. Microbiol. 46:1033-1040. [DOI] [PubMed] [Google Scholar]
- 24.Sitaraman, R., and K. Dybvig. 1997. The hsd loci of Mycoplasma pulmonis: organization, rearrangements and expression of genes. Mol. Microbiol. 26:109-120. [DOI] [PubMed] [Google Scholar]
- 25.Srikhanta, Y. N., T. L. Maguire, K. J. Stacey, S. M. Grimmond, and M. P. Jennings. 2005. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. USA 102:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teachman, A. M., C. T. French, H. Yu, W. L. Simmons, and K. Dybvig. 2002. Gene transfer in Mycoplasma pulmonis. J. Bacteriol. 184:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilei, E. M., J. Nicolet, and J. Frey. 1999. IS1634, a novel insertion element creating long, variable-length direct repeats which is specific for Mycoplasma mycoides subsp. mycoides small-colony type. J. Bacteriol. 181:1319-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmerman, C.-U., and R. Herrmann. 2005. Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol. Lett. 253:315-321. [DOI] [PubMed] [Google Scholar]