Abstract
Resistance to chemotherapy is a major complication during treatment of cancer patients. Hypermethylation of the MGMT gene alters DNA repair and is associated with longer survival of glioblastoma patients treated with alkylating agents. Therefore, MGMT promoter methylation plays an important role as a predictive biomarker for chemotherapy resistance. To adopt this established correlation into a molecular diagnosis procedure, we compared and optimized three experimental techniques [combined bisulfite restriction analysis, a primer extension- and denaturing high-performance liquid chromatography-based method named SIRPH (SNuPE ion pair-reverse phase high-performance liquid chromatography), and pyrosequencing] with regard to their accuracy of detecting MGMT promoter methylation. Initially, bisulfite sequencing was used to obtain a comprehensive methylation profile of the MGMT promoter region in 22 glioblastoma samples and in three normal brain controls. Next, we statistically identified CpG sites that best discriminate between methylated and unmethylated MGMT promoters. These results were then used to design optimal combined bisulfite restriction analysis, SIRPH, and pyrosequencing assays for accurate and cost-efficient assessment of MGMT promoter methylation. We compared all three techniques with regard to their reliability and reproducibility on well-characterized tumor samples. The optimized pyrosequencing assay performed best and provides a sensitive, robust, and easy-to-use method for quantitative assessment of MGMT methylation, for both snap-frozen and paraffin-embedded specimens.
The human MGMT (O6-methylguanine DNA methyltransferase, EC 2.1.1.63) gene encodes a protein with DNA repair activity. This protein removes alkyl groups from the O6-position of guanine1,2 by an irreversible transfer of the alkyl group to a cysteine residue at its active site.3,4 Guanine in the DNA is thereby restored and MGMT sentenced to proteasome-mediated degradation. Because of the stoichiometry of this reaction and the unavoidable fate of the MGMT protein, the repair capacity of a cell depends on the amount of MGMT molecules in the nucleus and the cell’s capability of resynthesis. Failure to repair the O6-alkylguanine (O6-AG) DNA adducts increases mutagenic potential during replication. This is because O6-AG can be mistaken for adenine and mismatched with thymine, giving rise to a G:C to A:T transition mutation. In addition, the adducts show a cytotoxic potential by causing DNA double-strand breaks. Both effects frequently induce apoptosis.5
The MGMT expression level and its activity vary widely between different tissues, cell types, and, in particular, between different tumors.6 Brain tumors show low expression, whereas the activity of MGMT is increased relative to the surrounding normal tissue.7,8 Expression of MGMT is (partially) regulated by methylation of the MGMT promoter region. This important epigenetic mechanism contributes to loss of MGMT expression in human tumors in vivo as first described by Esteller and colleagues.9
Resistance to chemotherapy is a major complication during treatment of cancer patients with alkylating agents. The epigenetically mediated silencing of the MGMT gene in tumors has been associated with an increased mean survival time in glioma patients that were treated with alkylating agents.10,11 The high repair activity in tumors with a transcriptionally active MGMT gene is believed to protect tumor cells against the cytotoxic effect of these anticancer drugs.12 Recently, a phase I clinical trial showed that presence of DNA methylation in the 5′-region of the MGMT gene is a predictive biomarker of favorable outcome in patients with glioblastoma treated with the alkylating agent temozolomide.13 This drug mediates its cytotoxic effect by forming O6-methylguanine (O6-MeG) DNA adducts, and it induces strong apoptotic response to O6-MeG DNA adducts in MGMT-deficient glioma cells.14 Therefore, MGMT promoter methylation may represent an important epigenetic biomarker for chemotherapy sensitivity.
Most of the publications dealing with the detection of MGMT methylation use a variant of methylation-specific polymerase chain reaction (MSP),15,16 which was first adapted for MGMT by Esteller and colleagues.9 This method enables cost-efficient analysis of MGMT promoter methylation. However, it is nonquantitative and bears a significant risk of false-positive or false-negative results, especially when DNA quality and/or quantity is low, which is often the case in a clinical setting in which samples are typically obtained from formalin-fixed, paraffin-embedded (FFPE) specimens. Alternative techniques for methylation analysis, such as bisulfite sequencing of multiple clones, are more tolerant toward low sample quality than MSP, are semiquantitative, and are widely used in basic research. However, they are neither cost-effective nor fast enough to be implemented for routine clinical diagnosis.
In this study, we adapted and optimized the analysis of MGMT promoter methylation for clinical settings to make this epigenetic biomarker available for routine diagnosis. To that end, we first identified positions in the MGMT promoter that are reliably correlated with the overall methylation state of the promoter and are accessible to at least one of three experimental techniques (all of which fulfill the basic requirements of clinical settings, such as robustness, cost efficiency, and ease of use): COBRA (combined bisulfite restriction analysis),17 SIRPH [SNuPE ion pair-reverse phase high-performance liquid chromatography (HPLC)],18 and pyrosequencing.19,20,21 Second, we systematically optimized each method for robust determination of MGMT promoter methylation and tested its performance on well-characterized tumor samples. Finally, we discuss our results with respect to reliability, expenditure, and applicability for molecular diagnostics.
Materials and Methods
DNA Samples
Tissue samples were collected from 22 patients with primary glioblastoma multiforme (World Health Organization IV) treated at the Departments of Neurosurgery at the Medical Centers in Bonn and Düsseldorf, Germany. The histological typing of the tissues was performed according to the World Health Organization grading system of brain tumors using standard histological and immunohistological methods.22 Tissues were selected for extraction of DNA after careful examination on hematoxylin and eosin staining of corresponding sections to exclude contaminating necrotic debris or normal brain tissue. Molecular genetic analyses were performed on samples showing an estimated tumor cell content of at least 80%. Genomic DNA was extracted from snap-frozen tumor tissues using standard proteinase K digestion and phenol/chloroform extraction,23 whereas for FFPE samples, the QIAamp DNA mini kit (Qiagen, Valencia, CA) was used in accordance to the manufacturer’s instructions. Three white matter biopsies served as normal brain controls. All patients gave written informed consent for these studies.
Bisulfite Treatment
Three hundred ng of genomic DNA (FFPE, 400 to 500 ng) was subjected to bisulfite conversion with the EpiTect bisulfite kit (Qiagen) according to the manufacturer’s instructions. Cytosine and its counterpart 5-methylcytosine show a different behavior to the treatment with sodium bisulfite on single-stranded DNA. Although cytosine residues react with this reagent and are converted to uracil, 5-methylcytosine stays inert under the same conditions. In a subsequent polymerase chain reaction (PCR), the uracil residues are transcribed to thymine and 5-methylcytosine to cytosine. After cloning and sequencing of the amplicons a comparison with the genomic sequence reveals that a formerly unmethylated CpG-dinucleotide appears as a TpG, whereas a methylated one remains as a CpG.
PCR
Pipetting steps for PCR reactions were performed in a DNA-workstation L020-GC (Kisker, Steinfurt, Germany) and designated working environments for steps before and after PCR were used to prevent cross-contamination.
Bisulfite Sequencing
The primers used for amplification of bisulfite-treated DNA were MGMT-Bis forward, 5′-GGATATGTTGGGATAGTT-3′24; and MGMT-Bis reverse, 5′-AAACTAAACAACACCTAAA-3′ and do not amplify untreated genomic DNA (data not shown). The amplified region corresponds to GenBank accession number AL355531, nucleotides 46891 to 47156. PCR was performed in a 200-μl PCR tube and with a final volume of 30 μl, containing 6 pmol of each primer, 200 μmol/L of each dNTP, 1.5 U of HOT FIREPol DNA polymerase (Solis BioDyne, Tartu, Estonia) in buffer B containing 2.5 mmol/L MgCl2 and 2 μl of bisulfite-treated DNA as template. The initial denaturation (97°C, 15 minutes) was followed by 37 cycles of 1 minute at 95°C, 1 minute at 47.5°C, 1 minute at 72°C, and a final extension step at 72°C for 10 minutes.
PCR products were resolved on a 4% agarose gel, the specific band excised and purified with the QIAquick gel extraction system (Qiagen). The purified PCR products were cloned by using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and at least eight clones were subjected to sequencing using the BigDye V.1.1 cycle sequencing chemistry (Applied Biosystems, Foster City, CA) and separated on a 3130 Genetic Analyzer (Applied Biosystems). Single clone sequences were analyzed with the BiQ Analyzer software (Max-Planck-Institut für Informatik, Saarbrücken, Germany).25
COBRA
For amplification of bisulfite-treated DNA, we used a two-step PCR approach. The primers of the first step contained a nonmatching M13 tail. The second step used M13 primers labeled with FAM (forward) and JOE (reverse). The primers used for the first step were MGMT-CO-1 forward, 5′-CACGACGTTGTAAAACGACGATATGTTGGGATAGTT-3′ and MGMT-CO-1 reverse, 5′-GGATAACAATTTCACACAGGCCCAAACACTCACCAAA-3′. M13-primers used for the second step were MGMT-CO-2 forward, 5′-(6-FAM)-CACGACGTTGTAAAACGAC-3′ and MGMT-CO-2 reverse, 5′-(JOE)-GGATAACAATTTCACACAGG-3′. Amplification on untreated genomic DNA resulted in no product corresponding to the expected size (data not shown). The amplified region corresponds to GenBank accession number AL355531, nucleotides 46892 to 46988. The first PCR was performed in a 200-μl PCR tube and with a final volume of 30 μl, containing 6 pmol of each primer, 200 μmol/L of each dNTP, 1.5 U of HOT FIREPol DNA polymerase (Solis BioDyne) in buffer B containing 2.5 mmol/L MgCl2, and 2 μl of bisulfite-treated DNA as template. The initial denaturation (97°C, 15 minutes) was followed by 25 cycles of 1 minute at 95°C, 1 minute at 48°C, 1 minute at 72°C, and a final extension step at 72°C for 10 minutes. The second PCR uses the same reaction setup except that 1 μl of the first PCR reaction was used as template. The initial denaturation (97°C, 15 minutes) was followed by 25 cycles of 1 minute at 95°C, 1 minute at 54°C, 1 minute at 72°C, and a final extension step at 72°C for 10 minutes.
PCR products were resolved on a 4% agarose gel; the specific bands were excised and purified using the QIAquick gel extraction system (Qiagen). The elution was done with 50 μl of H2O. Subsequently the eluate was carefully evaporated with a Savant SC110 Speed Vac concentrator (Thermo Electron Corporation, Waltham, MA) and finally redissolved in 16 μl of H2O. Digestion of 3.5 μl of the purified PCR product was done by using the restriction endonucleases TaqαI and BstUI (New England Biolabs, Ipswich, MA) in a final volume of 10 μl. The optimized reaction conditions for TaqαI were 5 U in NEBuffer 3, bovine serum albumin, and an incubation time of 4 hours at 65°C, whereas for BstUI, 10 U were used in NEBuffer 2 and an incubation time of 16 hours at 60°C. The solutions were mixed with 2× loading buffer (formamide and ethylenediaminetetraacetic acid (25 mmol/L) containing dextran blue as a marker) in a ratio of 1:1, and 1.5 μl were loaded onto an ABI 377 DNA sequencer (Applied Biosystems). The electropherograms were analyzed with the Gene Scan 3.1 software (Applied Biosystems). Methylation levels were calculated according to the following formula: methylation [%] = (AFi/sumAFi + AND) × 100. AFi represents the integral of fragment Fi, sumAFi represents the sum of the integrals of all fragments, and AND is the integral of the undigested product. Each sample was analyzed in two separate PCR reactions using the same bisulfite preparation as template. Reproducibility of bisulfite modification has been evaluated by MethyLight.26 Both amplification products were treated as described above and analyzed in duplicates.
SIRPH
Conditions used for generating the appropriate PCR product are identical to those described for bisulfite sequencing. Four μl of PCR product was treated with 1.6 μl of ExoSAP-IT (GE Healthcare, Little Chalfont, Buckinghamshire, UK) at 37°C for 15 minutes, heating at 80°C for 15 minutes, and then added to the primer extension mix. The primer used for primer extension reaction was 5′-GTGAGTGTTTGGGT-3′. The reaction was performed in a final volume of 20 μl, containing 60 pmol of primer, 100 μmol/L each of ddCTP and ddTTP, 1 U of TermiPol (Solis BioDyne) in buffer C containing 5 mmol/L MgCl2. The initial denaturation (95°C, 5 minutes) was followed by 50 cycles of 30 seconds at 94°C, 30 seconds at 43°C, and 1.5 minutes at 60°C.
For the separation of the extended primers, an aliquot of 19 μl of the SNuPE reaction was loaded onto a denaturing HPLC machine (WAVE DNA fragment analysis system by Transgenomics, San Jose, CA). The oven temperature was set to 50°C, and elution was done with a gradient of acetonitrile (20 to 40% for 15 minutes) made by mixing buffers A and B, consisting of 0.1 mol/L triethylammonium acetate (TEAA) buffer and 0.1 mol/L TEAA buffer with 25% acetonitrile, respectively. The column was re-equilibrated by 90% buffer B for 1 minute. The DNA was detected with a UV detector at 260-nm wavelength. Qualitative information for methylation is calculated as the ratio: Q = (hC/hC + hT), where hC and hT represent the peak high of the signal contributed to the ddCTP-extended primer and the ddTTP-extended primer, respectively. Each sample was analyzed in two separate PCR reactions using the same bisulfite preparation as template. Both amplification products were treated as described above and analyzed in duplicates.
Pyrosequencing
The primers used for amplification of bisulfite-treated DNA were MGMT-Py forward, 5′-biotin-GGATATGTTGGGATAGTT-3′ (GenBank accession number AL355531, nucleotides 46891 to 46908) and MGMT-Bis reverse, 5′-AAACTAAACAACACCTAAA-3′ (GenBank accession number AL355531, nucleotides 47138 to 47156), both of which do not amplify untreated genomic DNA (data not shown). PCR was performed in a 200-μl PCR tube and with a final volume of 50 μl, containing 10 pmol of each primer, 200 μmol/L of each dNTP, 2.5 U of HOT FIREPol DNA polymerase (Solis BioDyne) in buffer B containing 2.5 mmol/L MgCl2, and 3 μl of bisulfite-treated DNA as template. The initial denaturation (97°C, 15 minutes) was followed by 38 cycles (FFPE, 40 cycles) of 1 minute at 95°C, 1 minute at 47.5°C, 1 minute at 72°C, and a final extension step at 72°C for 10 minutes.
Forty μl of the PCR product was subjected to pyrosequencing. The primer used for primer extension reaction was 5′-CCCAAACACTCACCAAA-3′, which belongs to the sequence context: 5′-TCRCAAACRATACRCACCRC-3′. The sequencing reaction was performed on an automated PSQ 96MA System (Biotage, Uppsala, Sweden) using the Pyro Gold reagents kit (Biotage). Purification and subsequent processing of the biotinylated single-strand DNA was done according to the manufacturer’s instructions. Resulting data were analyzed and quantified with the PSQ 96MA 2.1 software (Biotage). Each tumor sample was analyzed in triplicates and each control sample in duplicates by individual PCR reactions using the same bisulfite preparation as template.
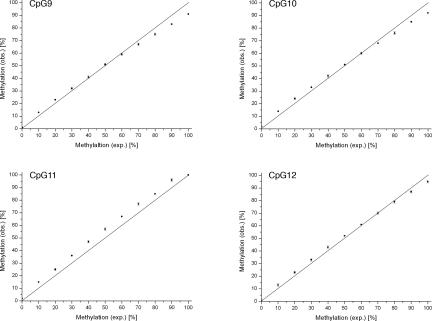
To assess measurement accuracy and linearity at the interrogated CpG positions, we performed a titration experiment. We prepared dilutions corresponding to well-defined DNA methylation levels by mixing PCR products of single clones with known methylation pattern (fully unmethylated versus methylated at CpGs 9 to 12). Before mixing, these PCR products were quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Oxfordshire, UK) and equilibrated. The mixtures were performed in triplicates with a final volume of 40 μl and subjected to pyrosequencing, resulting in three data points for each dilution. The ratio of unmethylated PCR product to methylated PCR product was increased from 100:0 to 0:100 in 10 equidistant steps (Figure 9). Linear regression analysis was performed to assess linearity.
Figure 9.
Titration curves obtained for each individual CpG position interrogated by the pyrosequencing assay. The SD of each data point is shown by a vertical error bar. The straight line reflects the theoretical curve. The titration experiment was performed as described in Materials and Methods. exp., expected; obs., observed.
Statistical Analysis
Statistical analysis was performed using the SPSS statistics package (SPSS for Windows; SPSS Inc., Chicago, IL). Hierarchical clustering was based on the methylation averages and standard deviations of all CpG positions 1 to 25, calculated over all sequenced clones. Between-groups average linkage was used with squared Euclidean distance as interval measure. Logistic regression models were calculated with the WEKA package27 using default parameters. Classification accuracy was estimated using leave-one-out cross-validation, ie, by repeatedly training a logistic regression model on 12 of 13 cases and testing it on the one remaining case.
Results
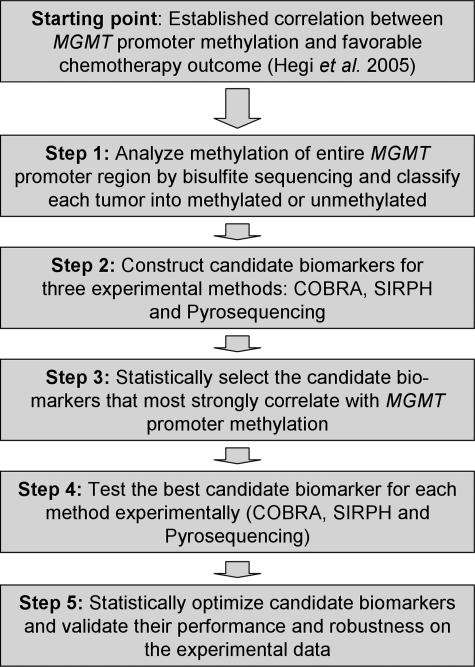
We optimized the analysis of MGMT promoter methylation as an epigenetic biomarker of chemotherapy resistance, using a five-step process (Figure 1). First, DNA was extracted from 22 snap-frozen primary glioblastoma samples as well as from three snap-frozen normal brain controls and subjected to bisulfite sequencing. Based on full methylation patterns for an extensive region covering the transcription start site, the first exon and parts of the first intron of the MGMT gene, tumors were clustered into methylated and unmethylated cases. Second, we determined all CpG positions in this region that can be readily analyzed by COBRA, SIRPH, or pyrosequencing, giving rise to candidate biomarkers of MGMT promoter methylation. Third, each candidate biomarker was statistically evaluated on the full methylation profiles, and for each method optimal marker candidates were selected based on their correlation with overall MGMT promoter methylation. Fourth, all selected markers were experimentally tested on a subset of 14 of 22 tumor samples. Fifth, we statistically optimized each biomarker and assessed its accuracy and robustness.
Figure 1.
Strategy for the optimization of MGMT promoter methylation detection as an epigenetic biomarker for chemotherapy resistance.
Methylation Patterns of the MGMT Promoter Region in Glioblastomas
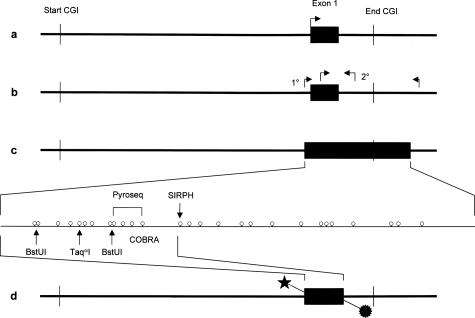
We screened the genomic sequence corresponding to the promoter region and the first exon of the MGMT gene for CpG islands, using the CpG-Plot28 web service (http://www.ebi.ac.uk/emboss/cpgplot/) with default parameters but with a window size of 200 and a step size of 10 (Figure 2a). Next we designed nondiscriminating bisulfite-specific primers (ie, primers that do not contain CpGs because these are subject to methylation-dependent bisulfite conversion) to amplify the region of interest from bisulfite-modified DNA. The amplicon includes all CpG positions analyzed via MSP by Hegi and colleagues,13 who used a nested MSP approach24 with increased sensitivity toward methylated DNA compared with the original single step protocol9 (Figure 2b). Our PCR product comprises 266 bp of DNA sequence and contains 27 CpG positions (Figure 2c). CpGs 1 to 12 span the exon 1, CpGs 13 to 20 are intronic inside the predicted CpG island, and the CpGs 21 to 27 are also intronic but outside the CpG island. The MSP used by Hegi and colleagues13 cover CpG positions 5 to 9 and 13 to 16 (Figure 2b).
Figure 2.
a: General map of the CpG island (CGI) spanning the promoter region and the first exon of the MGMT gene. b: Primer positions for the nested PCR approach used by Hegi et al.13 The second step (2°) used a methylation-sensitive primer established by Esteller et al.9 c: CpG map of the PCR product used for bisulfite sequencing of single clones, SIRPH, and pyrosequencing (biotin label not shown). d: Location of the double-labeled PCR product used for the COBRA assay.
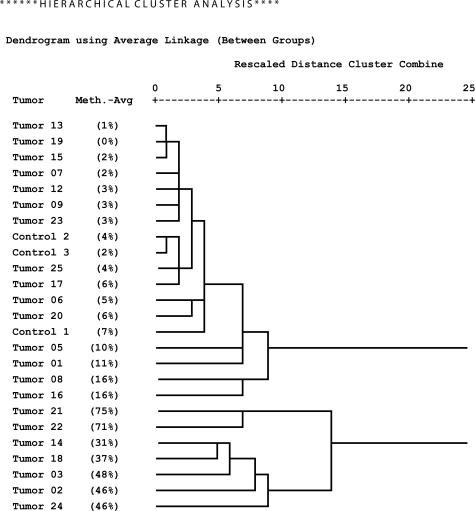
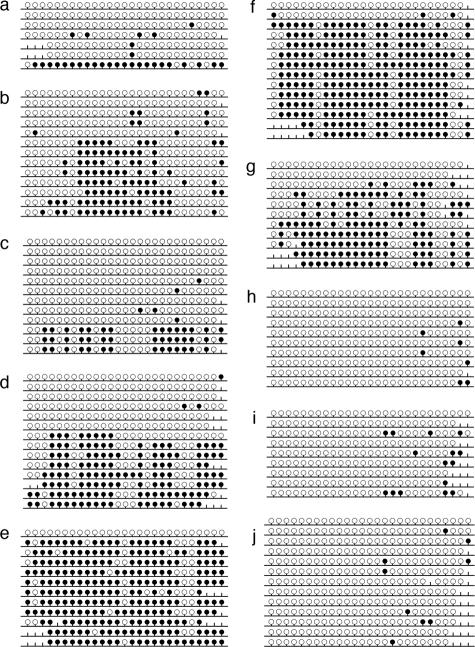
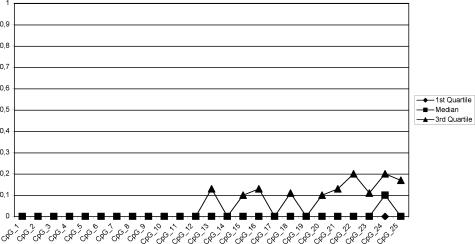
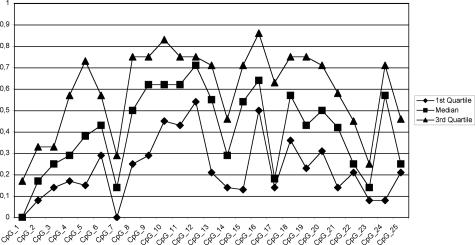
Bisulfite sequencing of DNA from 22 glioblastomas and three normal brain controls as described in Materials and Methods resulted in reliable data for CpGs 1 to 25 (98% average conversion rate, 1.7% missing values), on which we focused our further analyses. We calculated methylation profiles for each sample, averaging over individual clones. Hierarchical clustering of the methylation profiles provides strong evidence for two distinct tumor subclasses (Figure 3). The first subclass consists of tumor samples 01, 05, 06, 07 08, 09, 12, 13, 15, 16, 17, 19, 20, 23, and 25, as well as the primarily unmethylated normal brain controls 1 to 3. The second group contains tumor samples 02, 03, 14, 18, 21, 22, and 24, which show significant methylation levels. Tumors 08 and 16, members of the first subclass, are best described as borderline cases. They exhibit few heavily methylated alleles among a large number of unmethylated alleles (see below; Figure 6, a and c, respectively). Therefore they deviate from the other tumor samples in the first subclass, which only exhibited sporadic methylation (eg, tumor 13, Figure 6h; below) which is also observed among the control samples (eg, normal brain 3, Figure 6j; below). This behavior is also apparent from the dendrogram where they are localized in between the clearly unmethylated and the clearly methylated samples. Based on these peculiarities and the fact that higher variation is present in the methylated subclass, we decided to regard these two intermediate cases as belonging to the methylated subclass for the purpose of methylation profile calculation. Consequently, tumor samples 01, 05, 06, 07, 09, 12, 13, 15, 17, 19, 20, 23, and 25 are classified as unmethylated samples and the corresponding methylation profile of this group (excluding the control samples) is shown in Figure 4, whereas samples 02, 03, 08, 14, 16, 18, 21, 22, and 24 are classified as methylated samples (Figure 5). Figure 5 reveals that the median of the methylation level constantly increases from CpG 1 to 12 with values rising from 0 to 71% (exception, CpG 7). Beyond CpG 13 the methylation pattern becomes irregular and exhibits highly variable medians.
Figure 3.
Hierarchical clustering of samples by their MGMT promoter methylation profiles (methylation averages and standard deviations over all individual clones at CpG positions 1 to 25).
Figure 6.
Methylation pattern obtained by bisulfite sequencing of single clones. a: Sample 08; b: sample 14; c: sample 16; d: sample 18; e: sample 21; f: sample 22; g: sample 24; h: sample 13; i: sample 20; and j: control sample 3. Filled circles correspond to methylated CpG positions, open circles correspond to unmethylated CpG positions, and the vertical lines without a circle correspond to CpG positions not determined in the sequence. The diagrams were generated with BiQ Analyzer software.25
Figure 4.
Methylation profile (methylation averages over all individual clones) of the low-methylation tumor subclass.
Figure 5.
Methylation profile (methylation averages over all individual clones) of the high-methylation tumor subclass.
Construction of Candidate Markers for COBRA, SIRPH, and Pyrosequencing
CpGs at positions 3 to 13 represent stable indicators of the overall methylation level (all but CpG 7 correlate with the overall methylation level with Pearson correlation coefficients greater than 0.85). We therefore focused on this region and determined for each CpG whether it can be readily analyzed by at least one of three bisulfite-based assays used in this study. For COBRA we used a combination of two restriction endonucleases, TaqαI and BstUI, and could therefore assess DNA methylation at positions CpGs 1 and 2 simultaneously (methylation median in methylated samples, 0 and 17%), at CpG 5 (methylation median in methylated samples, 38%), and at CpGs 8 and 9 simultaneously (methylation median in methylated samples, 50 and 62%). By SIRPH, only position CpG 13 (methylation median in methylated samples, 55%) could be targeted. Pyrosequencing enabled us to assess CpGs 9 to 12 at the same time (methylation medians in methylated samples in the range of 62 to 71%).
Statistical Evaluation of Candidate Markers
It is important to note that the different CpG sites, and therefore the corresponding marker candidates have different powers of predicting MGMT methylation, depending on their degree of correlation with the overall promoter methylation (as determined by bisulfite sequencing). We pursued two routes to score the predictiveness of all marker candidates for the overall state of MGMT promoter methylation. First, based on the bisulfite sequencing data of all 22 tumor samples, we calculated correlations between tumor promoter methylation subclass and each marker score or combination of marker scores that is experimentally feasible. Second, for the subset 14 tumor samples, we experimentally reanalyzed all positions with the respective method and again calculated correlations between marker scores and the overall promoter methylation subclass as determined by bisulfite sequencing. Because little is known about the quantitative relationship between DNA methylation levels and gene silencing, we expect markers to reflect overall methylation states both in absolute terms (which can be measured by Pearson’s correlation coefficient) and in relative terms (which can be measured by the rank-based Spearman correlation coefficient). Furthermore, to enable comparison between markers that make use of different numbers of CpG positions, we focused our comparison on the mean methylation per marker candidate and did not perform any marker optimization at this stage. Most marker candidates showed strong correlation with overall methylation state (Table 1). For experimental validation we selected the candidate biomarker with highest correlation and strongest support from each method, namely CO7 for COBRA (five CpG positions), SI01 for SIRPH (one CpG position), and Py15 for pyrosequencing (four CpG positions). Experimental validation was performed on 14 of the 22 glioblastoma multiforme, comprising eight unmethylated and six methylated tumor samples. Two of these are highly methylated (samples 21 and 22), three are moderately methylated (samples 14, 18, and 24), and one is a borderline case (sample 16). The methylation patterns obtained by bisulfite sequencing of single clones are shown in Figure 6.
Table 1.
Analysis of Biomarker Candidates
| Experimental method | Marker ID | Analyzed CpG positions | Correlation for score calculated from bisulfite data
|
Correlation for score calculated from experimental evaluation
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pearson’s r | Performance | Spearman’s rho | Performance | Pearson’s r | Performance | Spearman’s rho | Performance | |||
| COBRA
|
CO1
|
1 to 2 (comb.)
|
0.696
|
−
|
0.554
|
−
|
0.433
|
− | 0.670 | − |
| CO2 | 8 to 9 (comb.) | 0.887 | − | 0.804 | − | 0.938 | + | 0.879 | + | |
| CO3 | 5 | 0.928 | − | 0.857 | − | 0.933 | + | 0.856 | + | |
| CO4 | 1 to 2 (comb.), 8 to 9 (comb.) | 0.927 | − | 0.881 | Ø | 0.916 | + | 0.875 | + | |
| CO5 | 1 to 2 (comb.), 5 | 0.937 | Ø | 0.869 | Ø | 0.926 | + | 0.857 | + | |
| CO6 | 8 to 9 (comb.), 5 | 0.949 | Ø | 0.908 | + | 0.947 | + | 0.856 | + | |
| CO7* | 1 to 2 (comb.), 8 to 9 (comb.), 5 | 0.961 | + | 0.929 | + | 0.942 | + | 0.856 | + | |
| SIRPH | SI01* | 13 | 0.955 | Ø | 0.888 | Ø | 0.908 | Ø | 0.844 | Ø |
| Pyrosequencing | Py01 | 9 | 0.877 | − | 0.734 | − | 0.901 | Ø | 0.838 | Ø |
| Py02 | 10 | 0.894 | − | 0.817 | − | 0.886 | Ø | 0.792 | Ø | |
| Py03 | 11 | 0.962 | + | 0.898 | + | 0.867 | − | 0.740 | − | |
| Py04 | 12 | 0.959 | Ø | 0.910 | + | 0.872 | − | 0.825 | Ø | |
| Py05 | 9, 10 | 0.950 | Ø | 0.868 | Ø | 0.903 | Ø | 0.792 | Ø | |
| Py06 | 9, 11 | 0.935 | Ø | 0.858 | − | 0.886 | − | 0.774 | − | |
| Py07 | 9, 12 | 0.952 | Ø | 0.867 | Ø | 0.888 | Ø | 0.825 | Ø | |
| Py08 | 10, 11 | 0.954 | Ø | 0.886 | Ø | 0.895 | Ø | 0.748 | − | |
| Py09 | 10, 12 | 0.943 | Ø | 0.888 | Ø | 0.887 | Ø | 0.796 | Ø | |
| Py10 | 11, 12 | 0.967 | + | 0.898 | + | 0.873 | − | 0.758 | − | |
| Py11 | 9, 10, 11 | 0.958 | Ø | 0.872 | Ø | 0.898 | Ø | 0.787 | Ø | |
| Py12 | 9, 10, 12 | 0.962 | + | 0.874 | Ø | 0.895 | Ø | 0.792 | Ø | |
| Py13 | 9, 11, 12 | 0.957 | Ø | 0.863 | Ø | 0.883 | − | 0.793 | Ø | |
| Py14 | 10, 11, 12 | 0.961 | + | 0.887 | Ø | 0.888 | Ø | 0.769 | − | |
| Py15* | 9, 10, 11, 12 | 0.964 | + | 0.873 | Ø | 0.892 | Ø | 0.784 | Ø | |
| Minimum | 0.696 | 0.554 | 0.433 | 0.670 | ||||||
| 1st quartile | 0.932 | 0.861 | 0.886 | 0.779 | ||||||
| Median | 0.952 | 0.873 | 0.895 | 0.793 | ||||||
| 3rd quartile | 0.960 | 0.888 | 0.912 | 0.850 | ||||||
| Maximum | 0.967 | 0.929 | 0.947 | 0.879 | ||||||
This table shows the correlation between the overall methylation of the MGMT promoter region on the one hand and the candidate biomarker scores (before optimization) on the other hand. In columns four to seven, scores are calculated from the bisulfite sequencing data for all tumor samples, whereas in the four rightmost columns the scores are based on the experimental values of the candidate biomarkers on tumor samples 12 to 25. All correlation coefficients but one (0.433) are significantly different from zero (P < 0.01 for each individual test). In the Performance columns, a − indicates that the correlation is among the bottom 25% of the column, a + indicates that it is among the top 25%, and a Ø indicates that it falls in between. The asterisks in the second column from the left highlight the candidate biomarkers that were selected for each experimental method. Comb, combined.
COBRA
For fragment analysis we chose a system based on the restriction endonucleases TaqαI and BstUI with optimized reaction conditions (see Materials and Methods). Cleavage sites for both enzymes are shown in Figure 2d. TaqαI has the recognition site 5′-TCGA-3′, whereas BstUI cuts the site 5′-CGCG-3′. The results obtained by analysis with TaqαI are quantitative because the analysis recognizes only a single CpG position. The values for BstUI are semiquantitative because both neighboring CpG positions have to be left unconverted by bisulfite treatment to permit digestion. TaqαI cuts the appropriate pattern for CpG 5, whereas BstUI cuts the appropriate pattern for CpG 1/2 and CpG 8/9.
To increase the sensitivity of the assay and to facilitate quantification, we used a two-step PCR approach, which resulted in generation of a small PCR product double labeled with fluorescent dyes (Figure 2d). The first step was performed using primers containing M13 tails at their 5′-end. In the second round, M13 primers labeled with 6-FAM (forward) and JOE (reverse) were used to increase the detection limit of the PCR product and to increase the number of amplicons. The resulting PCR product consists of 136 bp (97 bp representing bisulfite- converted DNA) and comprises CpG positions 1 to 12. Measures were highly reproducible as demonstrated by small standard deviations (Table 2). For the unmethylated tumor samples and for the normal brain controls, the TaqαI site showed low levels of methylation (<10%), but no evidence of methylation was detectable by BstUI. For the tumor samples classified as methylated evidence of significant methylation was detected for both restriction enzymes. However, in few cases (samples 16, 21, 22, and 24) the quantitative results obtained by TaqαI differed considerably from the values obtained by bisulfite sequencing of single clones.
Table 2.
Experimental Data
| Sample | Method | CpG 01 (%) | CpG 02 (%) | CpG 05 (%) | CpG 08 (%) | CpG 09 (%) | CpG 10 (%) | CpG 11 (%) | CpG 12 (%) | CpG 13 (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Tu12 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 11 ± 31 | 11 ± 31 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 2 ± 3 | 0 ± 0 | 0 ± 0 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 3 ± 2 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0 ± 0 | |||||||||
| Tu13 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 2 ± 3 | 4 ± 3 | 4 ± 3 | 5 ± 3 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 5 ± 3 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.08 ± 0.01 | |||||||||
| Tu14 | Bis-Seq. | 0 ± 0 | 8 ± 27 | 15 ± 36 | 54 ± 50 | 62 ± 49 | 62 ± 49 | 62 ± 49 | 54 ± 50 | 46 ± 50 |
| Pyroseq. | 56 ± 4 | 57 ± 7 | 59 ± 3 | 62 ± 3 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 27 ± 6 | 25 ± 3 | 25 ± 3 | |||||
| SIRPH | Q = 0.23 ± 0.05 | |||||||||
| Tu15 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 2 ± 3 | 3 ± 4 | 3 ± 4 | 2 ± 4 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 7 ± 3 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.02 ± 0.03 | |||||||||
| Tu16 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 25 ± 43 | 25 ± 43 | 0 ± 0 | 25 ± 43 | 25 ± 43 | 0 ± 0 |
| Pyroseq. | 23 ± 3 | 19 ± 1 | 21 ± 4 | 26 ± 3 | ||||||
| COBRA | 12 ± 3 | 12 ± 3 | 18 ± 6 | 17 ± 1 | 17 ± 1 | |||||
| SIRPH | Q = 0.27 ± 0.02 | |||||||||
| Tu17 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 13 ± 33 | 0 ± 0 | 0 ± 0 | 13 ± 33 |
| Pyroseq. | 2 ± 4 | 3 ± 4 | 3 ± 5 | 2 ± 4 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 4 ± 2 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.03 ± 0.01 | |||||||||
| Tu18 | Bis-Seq. | 15 ± 36 | 15 ± 36 | 57 ± 50 | 50 ± 50 | 57 ± 50 | 57 ± 50 | 57 ± 50 | 57 ± 50 | 21 ± 41 |
| Pyroseq. | 51 ± 1 | 49 ± 1 | 54 ± 3 | 49 ± 1 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 40 ± 7 | 31 ± 6 | 31 ± 6 | |||||
| SIRPH | Q = 0.12 ± 0.04 | |||||||||
| Tu19 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 2 ± 4 | 6 ± 9 | 0 ± 0 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 3 ± 3 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.03 ± 0.02 | |||||||||
| Tu20 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 2 ± 3 | 7 ± 3 | 5 ± 3 | 7 ± 3 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 6 ± 3 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.12 ± 0.01 | |||||||||
| Tu21 | Bis-Seq. | 60 ± 49 | 40 ± 49 | 82 ± 39 | 92 ± 28 | 83 ± 37 | 83 ± 37 | 92 ± 28 | 75 ± 43 | 83 ± 37 |
| Pyroseq. | 59 ± 8 | 53 ± 9 | 60 ± 10 | 57 ± 8 | ||||||
| COBRA | 7 ± 2 | 7 ± 2 | 47 ± 8 | 47 ± 7 | 47 ± 7 | |||||
| SIRPH | Q = 0.46 ± 0.03 | |||||||||
| Tu22 | Bis-Seq. | 17 ± 37 | 67 ± 47 | 85 ± 36 | 86 ± 35 | 86 ± 35 | 86 ± 35 | 86 ± 35 | 86 ± 35 | 83 ± 37 |
| Pyroseq. | 53 ± 2 | 50 ± 1 | 57 ± 4 | 50 ± 5 | ||||||
| COBRA | 3 ± 1 | 3 ± 1 | 37 ± 4 | 30 ± 3 | 30 ± 3 | |||||
| SIRPH | Q = 0.43 ± 0.01 | |||||||||
| Tu23 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 11 ± 31 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 4 ± 2 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.04 ± 0.01 | |||||||||
| Tu24 | Bis-Seq. | 0 ± 0 | 33 ± 47 | 73 ± 45 | 36 ± 48 | 64 ± 48 | 46 ± 50 | 73 ± 45 | 73 ± 45 | 55 ± 50 |
| Pyroseq. | 64 ± 3 | 35 ± 7 | 86 ± 4 | 64 ± 4 | ||||||
| COBRA | 3 ± 0 | 3 ± 0 | 44 ± 5 | 22 ± 2 | 22 ± 2 | |||||
| SIRPH | Q = 0.44 ± 0.03 | |||||||||
| Tu25 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 10 ± 30 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 5 ± 4 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.03 ± 0.01 | |||||||||
| NB01 | Bis-Seq. | 10 ± 30 | 0 ± 0 | 10 ± 30 | 10 ± 30 | 20 ± 40 | 10 ± 30 | 20 ± 40 | 10 ± 30 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 5 ± 4 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.03 ± 0.01 | |||||||||
| NB02 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 8 ± 28 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 3 ± 2 | 0 ± 0 | 3 ± 3 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 4 ± 2 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.06 ± 0.00 | |||||||||
| NB03 | Bis-Seq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Pyroseq. | 0 ± 0 | 0 ± 0 | 0 ± 0 | 3 ± 2 | ||||||
| COBRA | 0 ± 0 | 0 ± 0 | 3 ± 3 | 0 ± 0 | 0 ± 0 | |||||
| SIRPH | Q = 0.05 ± 0.00 |
Experimental data obtained by bisulfite sequencing of single clones (Bis-Seq.) for selected CpG positions, pyrosequencing (Pyroseq.), COBRA, and SIRPH.
The values are given by mean ± SD. The values for the SIRPH assay (CpG 13) are given as unit-free quotients Q (see Materials and Methods).
SIRPH
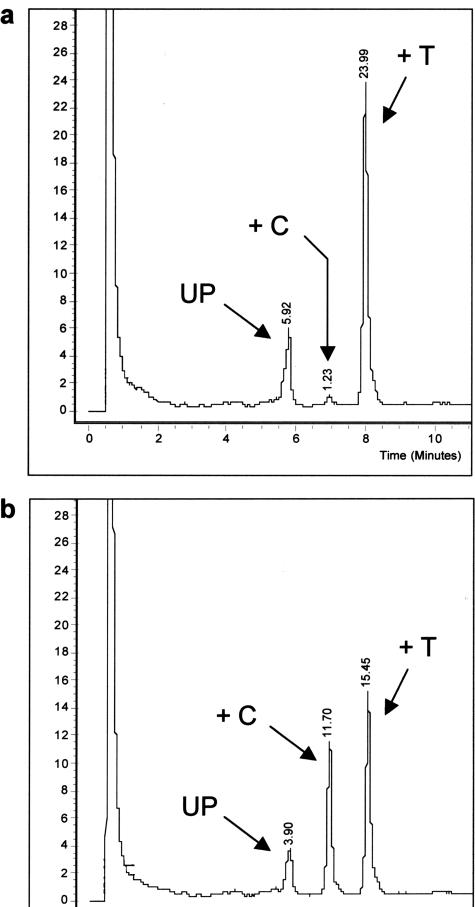
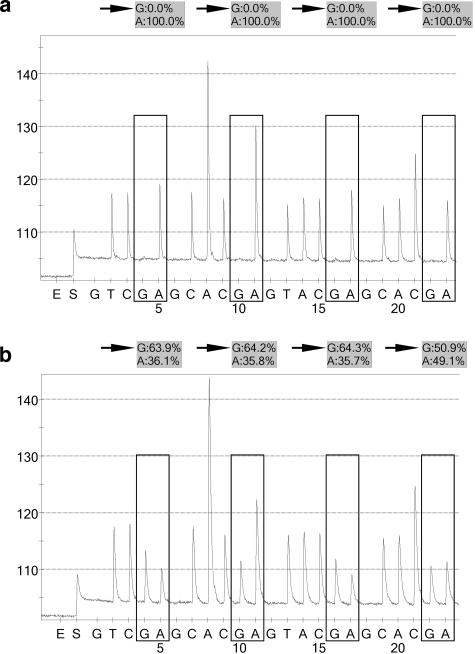
The SIRPH approach is based on a primer extension reaction (SNuPE), in which the primer is located directly in front of the CpG position to scan. Extension is performed by the appropriate dideoxynucleotides, and separation of the extended primer products used denaturing HPLC. The CpG position 13 (Figure 2c) is separated by 19 bases from CpG 12 and thus a SNuPE primer can be easily placed in front of CpG 13. A second reason to choose this position for this assay is its median value of methylation, which is relatively high (55%) and therefore suitable for a predictive methylation marker. Although we observed that this position is not methylated or less methylated in samples 16 (Figure 6c) and 18 (Figure 6d), care should thus be taken when using classification results based on only one CpG position. We decided to run the SIRPH assay qualitatively. A qualitative SIRPH assay avoids the necessity of calibration curves and comparative determination of the methylation level, by calculating quotients from signals measured in a single run.
The normal brain controls and unmethylated tumor samples showed either no or a very faint signal for the ddCTP elongated primer (methylated position), whereas a high signal was observed for ddTTP (unmethylated position) (Figure 7a). The methylated samples showed a significantly higher signal for the ddCTP extended primer (Figure 7b). Each generated PCR product showed high reproducibility for both values, whereas the values between two different PCR products sometimes differed. Strong variations were observed for the methylated tumor samples 14 and 18. Samples 18 and 20 showed a ratio of 0.12, which lies in between the quotients observed for the other unmethylated samples and those for methylated samples (Table 2). This may also explain the relatively high background noise observed for sample 20 using pyrosequencing (Table 2) or bisulfite sequencing of single clones (Figure 6i), whereas the quotient obtained for sample 18 was a result of the differing values measured for each PCR product (data not shown), reflecting the low-methylation level for CpG 13 in this sample.
Figure 7.
Typical chromatograms obtained in the SIRPH assay for an unmethylated sample (a: sample 15) and for a methylated sample (b: sample 21). UP, signal of the unextended primer; +C and +T, signal of the ddCTP and ddTTP extended primer, respectively. The signal seen in the beginning of the chromatogram is attributable to loading the sample on the column.
Pyrosequencing
The region analyzed by pyrosequencing includes CpGs 9 to 12 (Figure 2c), which are individually assayed. The basic principle of pyrosequencing is a primer extension reaction as for the SIRPH assay. However, while SIRPH measures the end product, pyrosequencing can accurately quantify the act of incorporation of different nucleotides. The unmethylated tumor samples as well as the control samples showed ratios less than 10% at all positions, with small SD (Table 2). Only sample 20 exhibited a slight increase in background noise of 5 to 7% for three of four CpGs. This tendency was also observed for the SIRPH assay and for bisulfite sequencing (Figure 6i), indicating that this was not attributable to a technical artifact. The values for the methylated tumor samples 14, 16, 18, and 24 are consistent with the results obtained by bisulfite sequencing. However, samples 21 and 22 demonstrated considerably lower methylation levels when compared with bisulfite sequencing data. Sample 21 exhibited differences in the range of 18 to 32% and relatively high standard deviations (8 to 10%), whereas sample 22 showed differences between 29 and 36% with small SDs (1 to 5%). Nevertheless, the general tendency of both results is consistent with high-methylation levels in all cases. Typical pyrograms obtained by this method for one unmethylated and one methylated sample are shown in Figure 8.
Figure 8.
Typical pyrograms obtained for an unmethylated sample (a: sample 25) and for a methylated sample (b: sample 14). Each box represents one of four CpG positions interrogated by the pyrosequencing assay, starting on the left side with CpG 12 because of reverse sequencing the upper strand of the PCR product. As a consequence, a C/TpG position appears as a CpG/A. The incorporation of the base guanine in this context represents the methylated fraction (arrows), and incorporation of adenine represents the unmethylated fraction.
To test accuracy and linearity for each individual CpG position, appropriate PCR templates were analyzed as serial mixtures of methylated and unmethylated products, with 10% methylation increments (Figure 9). For each titration curve, linearity was observed, and the measured data points were in good agreement with the theoretical curve (straight line). The arrangement of the data points given for CpG 9, CpG 10, and CpG 12 shared the same shape. Some degree of underestimation was observed for the completely methylated PCR product (91, 92, and 95%, respectively). The titration curve for CpG 11 showed minor but consistent overestimation in the range of 5 to 7% in between the methylation level of 10 to 90% but reflected accurately the values expected for the completely unmethylated and the fully methylated PCR product.
Statistical Validation of Candidate Biomarker Accuracy and Robustness
The experimental analyses reported in the previous section indicate that all candidate biomarkers can help discriminate between tumor samples with methylated MGMT promoters and those with unmethylated MGMT promoters. However, to select the optimal biomarker, additional statistics are required. To be able to compare the accuracy of the most promising candidate biomarker of each method (COBRA, CO7; SIRPH, SI01; pyrosequencing, Py15), we performed leave-one-out cross-validation on the experimental data, using a logistic regression model for classification (tumor 16 was excluded because it showed intermediate behavior for all technologies and may therefore constitute an outlier). For both the CO7 and the Py15 candidate biomarker, logistic regression led to correct classification of all 13 tumor samples (100% test set accuracy as determined by leave-one-out cross-validation). For the SI01 candidate biomarker, 12 of 13 tumor samples were classified correctly (92% test set accuracy). This result is consistent with our observation that all marker candidates performed well, but indicates that the SIRPH marker is less robust than the other two markers.
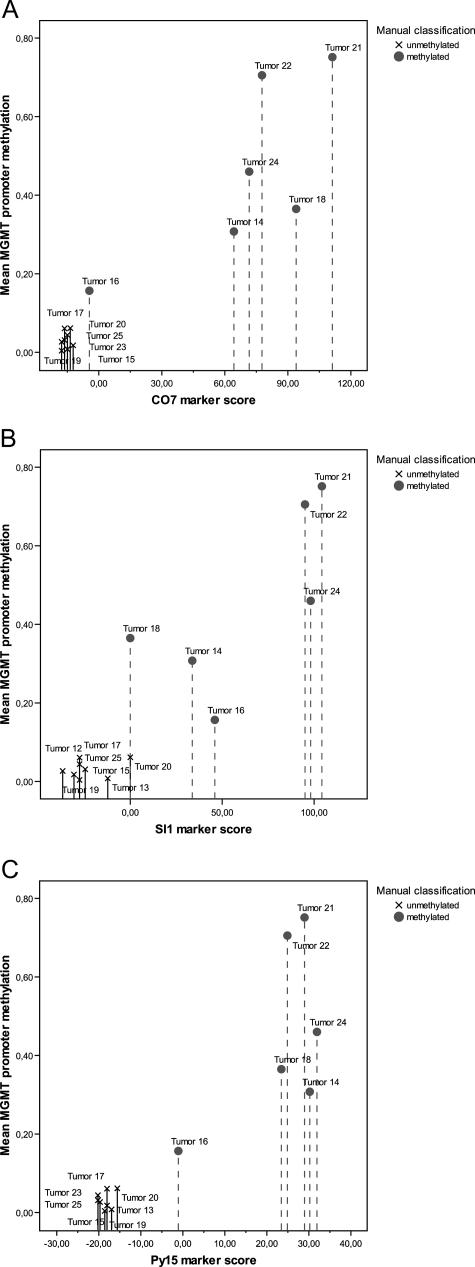
Next, we used logistic regression on the full validation dataset (again excluding tumor 16) to calculate optimal separation between unmethylated and methylated cases. This gave rise to the following three classification formulae: COBRA: ScoreCO7 = −339.385 · CpG1/2 + 196.192 · CpG8/9 + 137.296 · CpG5 − 21.803; SIRPH: ScoreSI01 = 306.601 · CpG13 − 36.792; and pyrosequencing: ScorePy15 = 21.330 · CpG9 + 24.806 · CpG10 + 18.637 · CpG11 + 21.503 · CpG12 − 20.197. The CpG variables refer to the measured methylation score at each position, and overall positive scores predict the presence of significant promoter methylation whereas negative scores predict absence of promoter methylation (for COBRA, we observed an unexpected negative coefficient at CpG position 1/2, indicating that a high-methylation level at this position was not a good predictor of a high overall methylation level). Application of these formulae to new tumor samples is straightforward: experimentally determine the values for the CpG variables, plug these into the formula, calculate the overall score, and compare this value to thresholds that we derived (see next section). To select thresholds that distinguish between clearly unmethylated cases, clearly methylated cases, and borderline cases, we reapplied the classification formulae to the full validation dataset, now including the borderline case 16.
For CO7 (Figure 10a), we observed that scores less than −15 were highly indicative of overall absence of MGMT promoter methylation, whereas scores greater than 60 were consistently associated with the presence of promoter methylation. Tumor 16 fell between these two thresholds, indicating that no clear conclusion is possible in the region between −15 and 60. For SI01 (Figure 10b), tumors with scores less than −10 should be classified as unmethylated and tumors with scores greater than 80 can safely be regarded as methylated. However, SI01 gives rise to a large interval in which no clear conclusion is possible, a high degree of variance among the two subclasses. For Py15 (Figure 10c), this uncertainty interval is substantially smaller, attributable to lower score variance among subclasses. Scores less than −10 provide strong evidence of an unmethylated MGMT promoter, whereas scores greater than 10 indicate substantial MGMT promoter methylation. Figure 10 visually confirmed our previous observation that candidate marker Py15 used for pyrosequencing is superior to both CO7 and SI01, and that CO7 is superior to SI01.
Figure 10.
Marker scores for all three methods plotted against overall methylation level. The CO7 marker (a) is based on the analysis of CpGs 1, 2, 5, 8, and 9. The SI01 marker (b) is based on the analysis of CpG 13, and the Py15 marker (c) is based on the analysis of the CpG positions 9, 10, 11, and 12.
Because the Py15 biomarker incorporates four CpG positions, it is relatively tolerant toward errors. Simulation showed that flipping any one position to zero or to one (which could happen because of incomplete bisulfite conversion or rare C-T-SNPs at the analyzed CpG dinucleotide) can only convert a clearly methylated or a clearly unmethylated sample into a borderline case (or vice versa), but cannot, for none of the samples we analyzed, convert a clearly methylated case into a clearly unmethylated case (or vice versa). Therefore, we conclude that the Py15 marker and, to a lesser extent, the CO7 marker as described here provide the necessary statistical robustness to cope with borderline cases.
Applicability of the Pyrosequencing Assay on FFPE Samples
To test the applicability and reproducibility of the pyrosequencing method on FFPE clinical specimens, we compared the results from the snap-frozen samples with matched FFPE tissues (age, 8 years). Genomic DNA was available from 10 of the 14 tumor samples. For two tissues, no FFPE material was left in the archives, whereas for the other two samples the amount of extracted DNA was not sufficient for further processing. After bisulfite conversion and subsequent PCR, we obtained 8 of 10 PCR products applicable for pyrosequencing. For seven samples (87%) the pyrograms could be analyzed, whereas one sample (13%) failed. The results are summarized in Table 3. A comparison of the pyrograms obtained from matched frozen and FFPE tissues showed little differences. However, tumor 18 exhibited higher methylation levels for the snap-frozen sample at CpG positions 9 and 12, which may be attributable to slight heterogeneity of MGMT promoter methylation patterns in spatially separated regions of the same tumor. In all cases, the overall methylation score (Table 3, right column) led to the same molecular diagnostic decision. Further investigations of samples aged between several weeks and up to 3 years (n = 4) showed that all of them (100%) could be analyzed by the pyrosequencing assay (data not shown).
Table 3.
Applicability of the Pyrosequencing Assay on FFPE Samples
| Sample | Source | CpG 09 (%) | CpG 10 (%) | CpG 11 (%) | CpG 12 (%) | Score (calculated) |
|---|---|---|---|---|---|---|
| Tu15 | Snap-frozen | 2 ± 3 | 3 ± 4 | 3 ± 4 | 2 ± 4 | −18 |
| FFPE | 0 ± 0 | 7 ± 9 | 0 ± 0 | 9 ± 11 | −17 | |
| Tu16 | Snap-frozen | 23 ± 3 | 19 ± 1 | 21 ± 4 | 26 ± 3 | −1 |
| FFPE | 31 ± 8 | 26 ± 7 | 28 ± 6 | 33 ± 4 | 5 | |
| Tu17 | Snap-frozen | 2 ± 4 | 3 ± 4 | 3 ± 5 | 2 ± 4 | −18 |
| FFPE | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | −20 | |
| Tu18 | Snap-frozen | 51 ± 1 | 49 ± 1 | 54 ± 3 | 49 ± 1 | 23 |
| FFPE | 0 ± 0 | 57 ± 15 | 59 ± 24 | 21 ± 20 | 9 | |
| Tu20 | Snap-frozen | 2 ± 3 | 7 ± 3 | 5 ± 3 | 7 ± 3 | −16 |
| FFPE | 8 ± 11 | 8 ± 12 | 12 ± 18 | 10 ± 15 | −12 | |
| Tu22 | Snap-frozen | 53 ± 2 | 50 ± 1 | 57 ± 4 | 50 ± 5 | 25 |
| FFPE | 42 ± 11 | 40 ± 6 | 46 ± 9 | 37 ± 7 | 15 | |
| Tu23 | Snap-frozen | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | −20 |
| FFPE | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | −20 |
Comparison of the experimental data obtained by pyrosequencing from matched snap-frozen and FFPE tissues, respectively. The experimental values are given as mean ± SD.
Discussion
MSP is an important bisulfite-based method for analyzing the DNA methylation status with high sensitivity. However, its application in clinical settings is constrained by several problems. First, mosaic methylation patterns with variable grades of methylation at the primer position can lead to both false-positive and false-negative results. This mispriming risk increases further when high numbers of PCR cycles or nested primers are used,16 as is typically necessary for processing DNA from FFPE samples. Second, MSP reliability strongly depends on the use of reproducible amounts of high-quality DNA, which is difficult to guarantee in clinical settings. Third, MSP cannot provide reliable quantitative information because band strength depends on multiple factors that cannot be fully controlled (eg, PCR performance and concentration of bisulfite-treated DNA). Taking these problems into account, it is not surprising that Hegi and colleagues13 reported significant difficulties when using MSP on FFPE tumor samples (for only 67% of these samples the MGMT methylation status could be determined by MSP). Further difficulties mentioned in recent studies29,30 substantiate the conclusion that MSP analysis of MGMT promoter methylation is unsuitable as a robust and widely applicable biomarker for clinical settings. Taking the importance of MGMT as a predictive marker into account, alternative experimental methods have to be established for reliable analysis of the MGMT methylation status:
1) Bisulfite sequencing of single clones is currently regarded as the gold standard for the analysis of DNA methylation profiles because it can provide both single bp resolution and quantitative methylation information (by analyzing multiple clones). This method is widely used in biomedical research, but it is too complex, time-consuming, and expensive for routine application in clinical settings. For these reasons, bisulfite sequencing was used to determine the most appropriate CpG positions for an MGMT promoter methylation biomarker to assess, but not for the construction of the biomarker itself.
2) The restriction-based COBRA method is typically used when the methylation profile is known, and the most informative CpG positions within an amplicon have been identified. With the help of restriction endonucleases that recognize DNA methylation at specific sites, the informative positions can be analyzed. The recognition of the restriction site and the number of cutting sites are the most limiting factors of this technology because in many cases there is no enzyme available that fulfills the demands for analyzing the position(s) of interest. Furthermore, the reaction conditions for a suitable enzyme must be carefully established to guarantee complete digestion while minimizing side effects. When an appropriate enzyme or enzyme combination is available, COBRA provides a cost-effective assay for obtaining quantitative or semiquantitative information of the methylation level for the screened CpG sites. We established one COBRA assay for the analysis of MGMT promoter methylation, which provided robust distinction between methylated and unmethylated cases. This assay is suitable for both high-quality and low-quality DNA material for four reasons. First, the amplified region spans 12 CpG positions with a total length of only 97 bp. Second, we applied a pseudo-nested PCR approach to enrich the yield of amplicons without saturating the PCR reaction. Third, the use of two restriction endonucleases TaqαI and BstUI provides additional robustness when conditions for one enzyme are less optimal. Fourth, TaqαI here functions as a bisulfite conversion-specific enzyme, ie, the recognition site TCGA is only present when the first base of the cleavage site, in the genomic DNA a cytosine residue, is converted by the bisulfite treatment and appears finally as thymine.
3) The SIRPH method requires designing a primer of at least six bases with no overlapping CpGs adjacent to the relevant CpG position. Therefore, it is only applicable to a subset of CpG positions and is particularly restricted in the context of CpG islands. Furthermore, the use of more than one primer requires considerable optimization to obtain sufficient resolution in the denaturing HPLC chromatogram. We established one SIRPH assay for the analysis of MGMT promoter methylation, which did not perform as well as the COBRA and pyrosequencing assays that we developed. We therefore do not recommend the use of the SIRPH assay as a clinical biomarker of MGMT promoter methylation.
4) Pyrosequencing is a sequencing-based method which is capable of analyzing several CpG positions simultaneously (up to 30 bp amplicon length). It generates quantitative results for each analyzed CpG position individually and enables rapid parallel processing of a large number of samples. By careful design of the extension primer, which is also the step that limits the applicability of pyrosequencing to a (relatively large) subset of CpG positions, it is possible to minimize the risk of assaying DNA that was not fully converted during bisulfite treatment. We established one pyrosequencing assay for the analysis of MGMT promoter methylation, which assays four CpG positions. Plotting the score of the optimized pyrosequencing marker (Py15) against overall MGMT promoter methylation (Figure 10c) shows that this marker provides excellent separation between methylated and unmethylated cases (tumor sample 16 falls between both groups, consistent with the observation that it exhibited borderline characteristics throughout this analysis). Overall, Py15 is the most accurate and most robust of all MGMT methylation markers that we explored in this study and is therefore recommended for use in clinical settings. However, because pyrosequencers are not yet widely available, we can also recommend the COBRA-based CO7 marker as the second-best method to assess MGMT promoter methylation. Nevertheless, further validation of both assays in a larger number of samples is still needed.
The main focus of our work was to translate the important epigenetic marker MGMT into a robust and easy-to-use clinical diagnosis tool that can be applied to DNA extracted from clinical samples. The marker’s robustness was confirmed on FFPE specimens, which makes it possible to investigate MGMT promoter methylation of archival tissues in a cost-efficient and accurate way. Finally, this study also contributes to the methodology of translational medicine by describing and prototyping a generally applicable workflow for the optimization of an epigenetic marker for clinical application. We conclude that the described pyrosequencing assay is suitable for clinical applications and allows accurate and sensitive identification of MGMT promoter methylation to support therapeutic decision-making.
Acknowledgments
We thank Ulrike Milde and Ortrud Schmidt for excellent technical assistance and Ulrich Klatt for photographic work.
Footnotes
Supported by the Nationales Genomforschungsnetz (grant NGFN-2, N3KR-S04T01), the Brain Tumor Network, and SMP Epigenetics (grant NGFN-2, PEG-S04T02).
References
- Ludlum DB. DNA alkylation by the haloethylnitrosoureas: nature of modifications produced and their enzymatic repair or removal. Mutat Res. 1990;233:117–126. doi: 10.1016/0027-5107(90)90156-x. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Dolan ME, Moschel RC. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- Wibley JEA, Pegg AE, Moody PCE. Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28:393–401. doi: 10.1093/nar/28.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P, Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- Margison GP, Povey AC, Kaina B, Santibáñez Koref MF. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2003;4:625–635. doi: 10.1093/carcin/bgg005. [DOI] [PubMed] [Google Scholar]
- Citron M, Decker R, Chen S, Schneider S, Graver M, Kleynerman L, Kahn LB, White A, Schoenhaus M, Yarosh D. O6-Methylguanine-DNA methyltransferase in human normal brain and tumor tissue from brain, lung and ovary. Cancer Res. 1991;51:4131–4134. [PubMed] [Google Scholar]
- Silber JR, Mueller BA, Ewers TG, Berger MS. Comparison of O6-methylguanine-DNA methyltransferase activity in brain tumors and adjacent normal brain. Cancer Res. 1993;53:3416–3420. [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, García-Lopez JL, Piquer J, Safont M-J, Balaña C, Sanchez-Cespedes M, García-Villanueva M, Arribas L, Esteller M. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JEC, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Roos WP, Batista LF, Naumann SC, Wick W, Weller M, Menck CF, Kaina B. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26:186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks S, Lentjes MHFM, Hellebrekers DMEI, de Bruïne AP, Herman JG, van Engeland M. Methylation-specific PCR unraveled. Cell Oncol. 2004;26:291–299. doi: 10.1155/2004/370301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Herbiniaux U, Walter J, Oldenburg J. A rapid, quantitative, non-radioactive bisulfite-SNuPE-IP RP HPLC assay for methylation analysis at specific CpG sites. Nucleic Acids Res. 2002;30:e25. doi: 10.1093/nar/30.6.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nürnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Webster KC. Lyon: Oxford University Press,; Pathology and genetics of tumours of the nervous system. World Health Organization Classification of Tumours, (ed 2) 2000 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor: Cold Spring Harbor Laboratory Press,; 1989 [Google Scholar]
- Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky S. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulphite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, Fuchs CS. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques with Java Implementations, ed xxv. Morgan Kaufmann: San Francisco,; 2000:p 371. [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Akufo-Tetteh EK, Risk JM, Field JK, Liloglou T. Methylation enrichment pyrosequencing: combining the specificity of MSP with validation by pyrosequencing. Nucleic Acids Res. 2006;34:e78. doi: 10.1093/nar/gkl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes AA, Tosoni A, Cavallo G, Reni M, Franceschi E, Bonaldi L, Bertorelle R, Gardiman M, Ghimenton C, Iuzzolino P, Pession A, Blatt V, Ermani M. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J Clin Oncol. 2006;24:4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]