Abstract
Amplicon melting is a closed-tube method for genotyping that does not require probes, real-time analysis, or allele-specific polymerase chain reaction. However, correct differentiation of homozygous mutant and wild-type samples by melting temperature (Tm) requires high-resolution melting and closely controlled reaction conditions. When three different DNA extraction methods were used to isolate DNA from whole blood, amplicon Tm differences of 0.03 to 0.39°C attributable to the extractions were observed. To correct for solution chemistry differences between samples, complementary unlabeled oligonucleotides were included as internal temperature controls to shift and scale the temperature axis of derivative melting plots. This adjustment was applied to a duplex amplicon melting assay for the methylenetetrahydrofolate reductase variants 1298A>C and 677C>T. High- and low-temperature controls bracketing the amplicon melting region decreased the Tm SD within homozygous genotypes by 47 to 82%. The amplicon melting assay was 100% concordant to an adjacent hybridization probe (HybProbe) melting assay when temperature controls were included, whereas a 3% error rate was observed without temperature correction. In conclusion, internal temperature controls increase the accuracy of genotyping by high-resolution amplicon melting and should also improve results on lower resolution instruments.
Amplicon melting analysis is a simple closed-tube genotyping method that uses a saturating DNA binding dye instead of fluorescently labeled primers or probes.1 High-resolution melting analysis can detect single base changes and other variations in single or multiplex polymerase chain reaction (PCR).2 Wild-type and homozygous mutant samples typically have sharp, symmetric melting transitions, whereas heterozygous samples have more complex, gradual melting curves. Homozygous sequence changes result in characteristic shifts in melting temperature (Tm).2,3,4,5 In contrast, heterozygous samples are identified by melting peak shape and width and not by Tm. Correct identification of sample genotype by amplicon melting requires standardization of reaction conditions to achieve reproducible, characteristic melting profiles. Reaction conditions can vary between lots of PCR reagents, including different buffers introduced by the DNA isolation method. Ionic strength, in particular, significantly affects Tm.6,7,8,9,10
The current study introduces the use of one or more internal controls for temperature calibration between reactions. Complimentary, unlabeled oligonucleotides that do not interfere with the PCR were designed so that they melt outside the temperature region of PCR product melting. Any buffer differences that affect duplex Tms affects both the amplicon and the internal temperature controls, allowing subsequent temperature correction of melting profiles. As a genotyping target, the 1298A>C and 677C>T variants of the methylenetetrahydrofolate reductase (MTHFR) gene were used. A single-color duplex amplicon melting assay (with and without internal temperature correction) was compared with a duplex multicolor HybProbe melting assay.
Materials and Methods
DNA Extraction and Study Design
Sixty whole blood samples were submitted to ARUP for clinical evaluation of MTHFR (1298A>C and 677C>T) genotype with K3 ethylenediaminetetraacetic acid, sodium-heparin, or citrate-phosphate-dextrose anticoagulation. Samples were blinded and deidentified according to a global ARUP protocol under institutional review board no. 7275. DNA was extracted with the Roche MagNA Pure LC system (Roche, Indianapolis, IN), resulting in concentrations of 20 to 40 ng/μl by absorbance at A260. All samples were genotyped by the duplex multicolor HybProbe assay. Thirty-seven of these samples were selected by genotype and the duplex amplicon melting assay also performed. These samples were blinded and genotypes determined with no temperature correction, only low-temperature correction, only high-temperature correction, and both low- and high-temperature correction. A single-tailed F-test was used to assess the variance in the Tms of homozygous genotypes. (Excel; Microsoft, Redmond, WA).
DNA was extracted from an additional 10 whole blood samples by three methods: the Roche MagNA Pure LC system (Roche), the Puregene DNA blood kit (Gentra Systems Inc., Minneapolis, MN), and the Qiagen QIAamp DNA blood mini kit (Qiagen Inc., Valencia, CA). Extracted QIAamp and MagNA Pure DNA samples were 20 to 40 ng/μl in concentration and used without dilution, whereas Puregene samples (initially at 160 to 500 ng/μl) were diluted with water to 40 ng/μl. The duplex amplicon melting assay was performed, and the effect of the extraction method on Tm was evaluated by paired t-tests. The average difference in Tm was also calculated between each extraction method, both before and after correction with low and high internal temperature controls.
MTHFR (1298A>C and 677C>T) HybProbe Genotyping Assay
A duplex, two-color HybProbe assay11,12 for the 677C>T and 1298A>C13 variants of the MTHFR gene (GenBank AY338232) was used to establish MTHFR genotypes. The MTHFR 1298A>C variation was interrogated by amplifying a 108-bp fragment with primers 5′-GAGGAGCTGCTGAAGATGTGG-3′ (forward) and 5′-CACTTTGTGACCATTCCGGTTTG-3′ (reverse), using probes 5′-GAGCTGACCAGTGAAGCAAGT-3′-FITC and LCRed 705–5′-CTTTGAAGTCTTTGTTCTTTACCTCTCGGG-C3–3′. The underlined base indicates the position of the variation, and C3 is a C3 spacer used to prevent the 3′ end of the oligonucleotide from extending.14 The MTHFR 677C>T variation was interrogated by amplifying a 94-bp fragment with primers 5′-CAACCCCGAAGCAGGGAG-3′ (forward) and 5′-GCCTCAAAGAAAAGCTGCGTG-3′ (reverse), using probes 5′-AAGCACTTGAAGGAGAAGGTGTCT-3′-FITC and LCRed 640–5′-CGGGAGCCGATTTCA-C3–3′. Probes containing the C3 spacer were acquired from Idaho Technology (Salt Lake City, UT). All other oligonucleotides were acquired from Integrated DNA Technologies, Inc. (Coralville, IA).
PCR was performed in 12-μl volumes with 1× LightCycler DNA Master HybProbes (Roche), 0.08 μmol/L of the both forward primers, 0.41 μmol/L of the 1298A>C reverse primer, 0.16 μmol/L of the 677C>T reverse primer, 0.16 μmol/L of each probe, 2.8 mmol/L MgCl2 (including 1 mmol/L MgCl2 contributed by the LightCycler Master solution), 0.1 U/reaction AmpErase UNG (Perkin-Elmer, Foster City, CA), and 2 μl of extracted DNA. PCR was done in a LightCycler (Roche) with an initial hold at 50°C for 1 minute followed by a hold at 94°C for 1 minute, and 40 cycles of 94°C for 0 seconds, 62°C for 20 seconds (with fluorescence acquisition), and 72°C for 0 seconds. The temperature transition rate between 62 and 72°C was 1°C/second; all other rates were programmed at 20°C/second. After PCR, the samples were denatured for 30 seconds at 95°C, followed by 30-second holds at 70, 60, and 35°C. Melting curve data were gathered by continuous fluorescence acquisition from 35 to 85°C with a transition rate of 0.1°C/second. Genotyping was based on negative first derivative melting curves and comparison of unknowns to genotyped controls.
Internal Temperature Controls
Complementary oligonucleotides that varied in G/C content and length (Table 1) were obtained from Integrated DNA Technologies, Inc. For some oligonucleotides, Tms were further decreased by deletions or increased with locked nucleic acids on one strand but not the compliment.15 Theoretical Tms were calculated using IDT SciTools OligoAnalyzer 3.0 software (Integrated DNA Technologies, Inc.). The internal temperature controls were blocked from extending during PCR by incorporating a phosphate group on the 3′ end of each oligonucleotide.
Table 1.
Sequence and Predicted/Observed Tms of Internal Temperature Controls
| Name | Predicted Tm (°C) | Observed Tm (°C) | |
|---|---|---|---|
| High-temperature sequences* | |||
| 50 bp | G C G G T C A G T C G G C C T A G C G G T A G C C A G C T G C G G C A C T G C G T G A C G C T C A G | 87.0 | 88.5 |
| 50 bp(4LNA)† | … …A … … . .A… … … … … … …A … … . A … … . | 90.2 | 90.8 |
| 50 bp(8LNA)† | G … . . A … . . C … … . . A … . . C … … A … … . A … … G | 92.4 | 92.6 |
| 40 bp | … … … … … … … … … … … … . .G … . . | 87.1 | 87.7 |
| 30 bp | … . G . G T C G . C . G G C T G . C A G A G G C . G C . | 88.4 | 87.3 |
| Low-temperature sequences* | |||
| 50 bp | A T C G T G A T T T C T A T A G T T A T C T A A G T A G T T G G C A T T A A T A A T T T C A T T T T | 68.5 | 68.5 |
| 40 bp | … … … … … … … … … … . C T . . A . T A . | 69.0 | 69.2 |
| 35 bp(del)‡ | … … . .–––… … … … … C … A T A T A | nd§ | 62.9 |
| 35 bp | … … … … … … … … . . C … A T A T A | 68.3 | 69.0 |
| 30 bp | … … . . C … . G … … … … C … | 68.3 | 68.5 |
Duplex controls consist of the listed oligonucleotide and its complement. All sequences are shown 5′ to 3′ and both sequence and complement are blocked at the 3′ end with a phosphate.
Bases in bold type indicate that locked nucleic acids are present in the indicated strand, but not in the complement.
Dashes indicate a deletion in the listed sequence but not in the complement strand.
Not determined because thermodynamic parameters are not available.
MTHFR (1298A>C and 677C>T) Amplicon Melting Assay
The duplex amplicon melting assay included an 80-bp fragment for genotyping the 1298A>C variant amplified by primers 5′-GGGAGGAGCTGACCAGTGAAG-3′ and 5′-CACTTTGTGACCATTCCGGTTTGGTTCTCC-3′ and a 120-bp fragment for genotyping the 677C>T variant amplified by primers 5′-GAAGCAGGGAGCTTTGAGGCTGACCTG-3′ and 5′-TGCCTTCACAAAGCGGAAGAATGTGTCAGC-3′. PCR was performed in 10-μl volumes with 1× LightCycler FastStart DNA Master HybProbes (Roche), 1.0 μmol/L each of the 1298 primers, 0.5 μmol/L each of the 677 primers, 0.2 μmol/L of the 50-bp high and low internal temperature controls (Table 1), 3.5 mmol/L MgCl2 (including 1 mmol/L MgCl2 contributed by the LightCycler Master solution), 0.01 U/reaction heat-labile uracil-DNA glycosylase (Roche), 1× LCGreen Plus (Idaho Technology), and 2 μl of extracted DNA. PCR was done on a LightCycler (Roche) with an initial hold at 95°C for 10 minutes, followed by 40 cycles of 95°C for 1 second, 60°C for 0 seconds, and 72°C for 2 seconds with fluorescence acquisition. All heating and cooling steps during PCR were done with ramp rates programmed at 20°C/second. After PCR, samples were prepared for melting analysis by rapid cooling in the LightCycler from 95 to 40°C.
High-resolution melting analysis was performed on the HR-1 instrument (Idaho Technology). Melting curves were generated using continuous fluorescence acquisition from 60 to 95°C with a temperature transition rate of 0.2°C/second. After fluorescence normalization, the exponential background was removed16 and derivative melting curves displayed. Heterozygotes were easily identified by melting peak width and shape. Homozygotes were genotyped by Tm (melting peak maxima) either before or after internal temperature control correction. Temperature correction of derivative melting curves was performed using custom software developed in LabView (National Instruments, Austin, TX). The Tms of the control peaks were first identified and then aligned by shifting and scaling the temperature axis. Scaling was performed by linear expansion or compression when two temperature controls were used.
Results
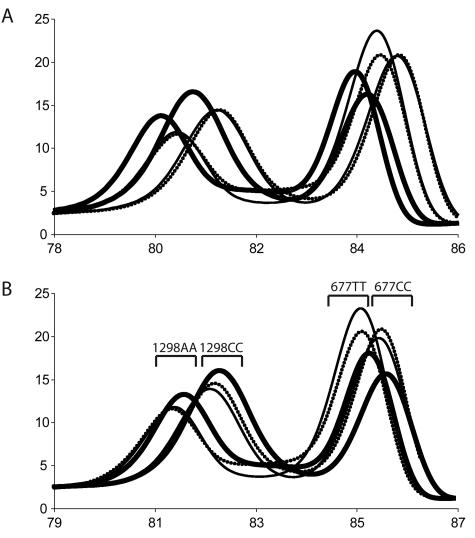
To demonstrate that common DNA extraction methods can affect PCR product Tm, DNA was isolated from 10 whole blood samples by MagNA Pure, QIAamp, and Puregene DNA methods. Using a duplex amplicon genotyping assay for the MTHFR variants 1298A>C and 677C>T, high-resolution melting curves of PCR products were obtained and Tms calculated. Although significantly different (P = 0.028), QIAamp and Puregene Tms differed on average by only 0.03°C, whereas MagNA Pure Tms differed from QIAamp by 0.36°C (P = 7.3 × 10−15) and from Puregene by 0.39°C (P = 8.8 × 10−14).
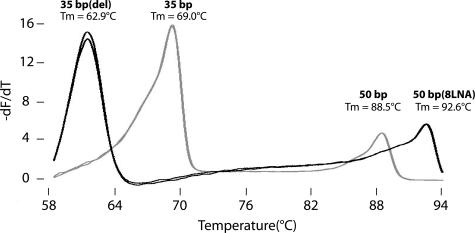
Ten sets of complimentary oligonucleotides were synthesized as potential internal temperature controls for high-resolution melting analysis (Table 1). Melting profiles of four of these duplexes under PCR conditions are shown in Figure 1. A 3-bp deletion in one strand of the 35-bp duplex lowered the Tm 6.1°C compared with a perfectly matched hybrid, whereas locked nucleic acids increased Tm in a 50-bp duplex by 0.51 to 0.58°C for every locked nucleic acid incorporated. After attempted MTHFR duplex PCR in the absence of template, only the expected products were observed on derivative melting plots without visible primer dimers. Nearest neighbor stability predictions and observed Tms (Table 1) were very close with almost no systematic bias (ΔTm = −0.03°C) and a SD for ΔTm of 0.69°C. In subsequent studies, the 50-bp high- (without locked nucleic acids) and low-temperature controls were used. When applied to the different DNA extraction methods, temperature correction reduced the Tm differences by 33 to 50% (the ΔTm of QIAmp versus Puregene was 0.02°C; MagNA Pure versus QIAmp, 0.18°C; and MagNA Pure versus Puregene, 0.21°C). Representative derivative melting plots of the different extraction methods are shown in Figure 2.
Figure 1.
Derivative melting curves of selected internal temperature controls (Table 1). The 35-bp low-temperature control was combined with the 50-bp high-temperature control (gray curve). The black curve included the 35-bp low-temperature control with a 3-bp deletion on one strand and the 50-bp high-temperature control with eight locked nucleic acid bases on one strand. Both mixtures were melted under PCR conditions in duplicate.
Figure 2.
Derivative melting plots of MTHFR samples extracted using different methods. Samples were extracted in parallel using the MagNA Pure LC system (thick black line), QIAamp DNA blood mini kit (thin black line), and the Puregene DNA blood kit (dotted black line). The sample genotypes shown were 1298AA/677CC and 1298CC/677TT. A: Derivative melting profiles shown without temperature correction. B: Derivative melting profiles shown with temperature correction. The brackets represent Tm ranges expected for the homozygous MTHFR genotypes.
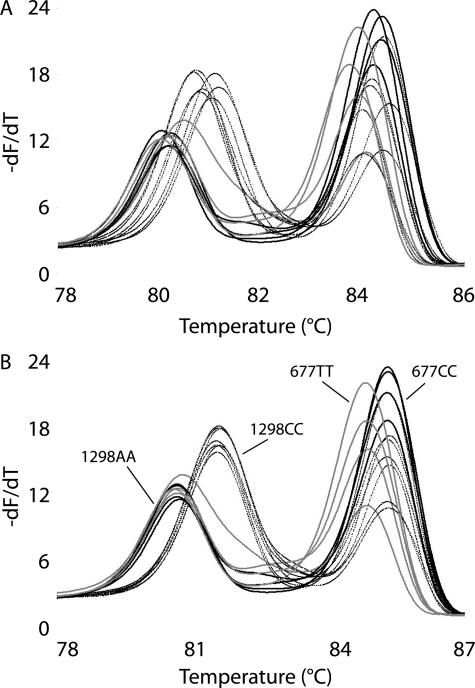
Internal temperature controls also reduced temperature variation when the DNA extraction method remained the same. Figure 3 shows derivative amplicon melting data for selected samples with MagNA Pure isolated DNA, before and after temperature correction. Samples heterozygous at either locus are easily identified and are not shown for clarity. The high- and low-temperature controls are also off scale in Figure 3 to magnify the amplicon melting region. Before temperature correction, it is difficult to visually genotype the homozygous samples because of temperature variation (Figure 3A). Table 2 shows the means and SDs of each homozygous Tm with and without correction using the high- and/or low-temperature controls. Before temperature correction, the Tm standard deviations were 0.16 to 0.18°C, decreasing to 0.04 to 0.14°C with one temperature control, and 0.03 to 0.10°C with two temperature controls. When only one temperature control was used, the most significant variance reductions were observed at temperatures closest to the control Tm.
Figure 3.
Duplex MTHFR derivative melting plots for genotyping the 1298A>C and 677C>T variants. Genotypes shown are 1298AA/677CC (thick black line), 1298CC/677CC (thin black line), and 1298AA/677TT (gray line). A: Without temperature correction. B: With temperature correction using high- and low-temperature controls (not shown).
Table 2.
Effect of Internal Temperature Controls on the Apparent Tm of MTHFR Homozygous Genotypes
| Temperature controls |
MTHFR 1298A>C
|
MTHFR 677C>T
|
|||
|---|---|---|---|---|---|
| AA (n = 7) | CC (n = 7) | TT (n = 4) | CC (n = 10) | ||
| None | Mean Tm (°C) | 80.20 | 80.96 | 83.97 | 84.41 |
| SD (°C) | 0.16 | 0.17 | 0.19 | 0.18 | |
| Low | Mean Tm (°C) | 79.94 | 80.73 | 83.73 | 84.15 |
| SD (°C) | 0.07 | 0.08 | 0.14 | 0.11 | |
| P value* | 4.4 × 10−8 | 0.016 | 0.2 | 0.048 | |
| High | Mean Tm (°C) | 80.83 | 81.63 | 84.68 | 85.01 |
| SD (°C) | 0.10 | 0.06 | 0.10 | 0.04 | |
| P value* | 0.07 | 4.3 × 10−3 | 0.066 | 1.3 × 10−7 | |
| Low and high | Mean Tm (°C) | 80.69 | 81.50 | 84.60 | 84.95 |
| SD (°C) | 0.06 | 0.03 | 0.10 | 0.03 | |
| P value* | 1.1 × 10−3 | 1.3 × 10−4 | 0.073 | 4.1 × 10−9 | |
Single-tailed F-test against the "no Tm control″ group.
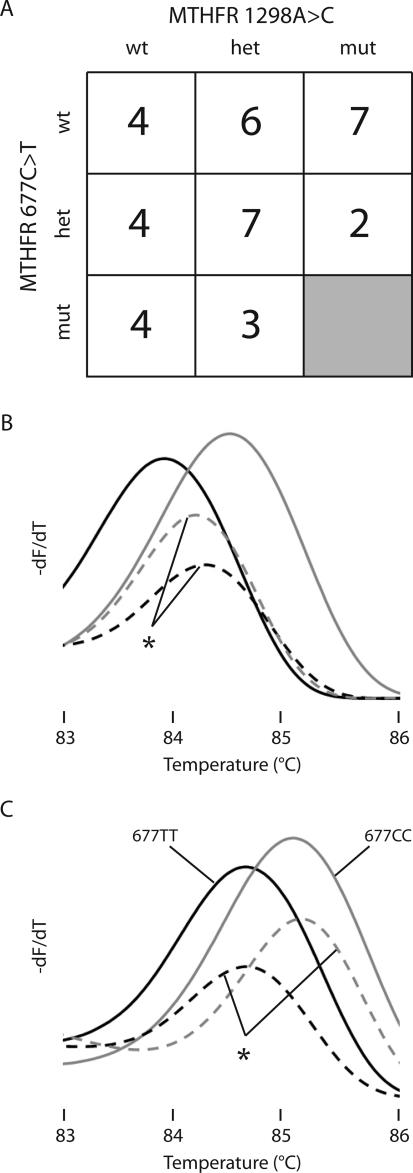
To assess further the utility of incorporating internal temperature controls, a blinded study of 37 selected samples was performed by duplex amplicon melting. The genotype distribution of the selected samples is shown in Figure 4A. All 37 MTHFR 1298A>C genotypes were correctly assigned whether or not temperature correction was performed. All but two of the MTHFR 677C>T genotypes were also correctly assigned without temperature correction—one TT genotype was misread as CC and one CC genotype was misread as TT. Derivative melting profiles of the two discordant samples, before and after temperature correction, are shown in Figure 4, B and C. When the discordant curves were adjusted by temperature correction, both were correctly genotyped.
Figure 4.
Genotype distribution and derivative melting plots for 677C>T MTHFR genotyping. Thirty-seven selected DNA samples were blinded and genotyped by the duplex amplicon melting assay. A: Genotype distribution determined by HybProbe analysis. The MTHFR 1298CC/677TT genotype was not available and is presumed lethal.13,23 B: Two controls and the two discordant samples are shown without temperature correction. C: The same samples are shown after temperature correction. The solid black line shows 677TT and the solid gray line shows 677CC genotypes. The dotted black and gray lines marked with the asterisk indicate the samples that required temperature correction for correct genotyping.
Discussion
Amplicon melting analysis1,2 is a simple, cost-effective alternative to other closed-tube genotyping approaches that require probes.17,18,19,20 Heterozygotes are easily identified by a change in shape of the melting curve.3 Homozygous variants are more difficult to detect and may produce only small differences in Tm that are best detected on high-resolution melting instruments.21 When the ΔTm is very small, genotyping accuracy depends on the temperature resolution of the instrument and any solution chemistry differences between samples. Instrument resolution concerns are most severe on heating block instruments with 96 or 384 wells.21 Variations in the PCR buffer or ionic strength between samples can also compromise the reproducibility of Tm measurements, as evidenced by differences dependent on the template DNA extraction method.
Internal temperature controls, comprised of complementary oligonucleotides, can partly correct resolution limitations imposed by the instrument and/or variable solution chemistry. This was demonstrated with a duplex amplicon melting assay (MTHFR 1298A>C and 677C>T) requiring fine temperature resolution for correct genotyping. Although some genotyping errors were made without temperature correction, genotyping was 100% concordant to a HybProbe assay when temperature correction was applied. Although one temperature control significantly decreased Tm variance, particularly at temperatures nearer the control, two temperature controls bracketing the amplicon melting region gave the best results. Internal temperature controls can potentially enable the use of rare archived DNA samples, in which the buffer chemistry is unknown, in amplicon melting assays. A uniform sample extraction method becomes less important when temperature correction can be applied to melting curves that vary because of solution chemistry differences.
High-resolution melting analysis without probes has the potential benefit of new sequence variant discovery during amplicon melting.22 The ability to discriminate new variants depends on temperature resolution, which can be improved by including internal temperature controls. This study was performed on the HR-1 instrument, which has the highest reported resolution of any DNA melting instrument.21 Use of internal temperature controls on lower resolution instruments was not investigated. However, genotyping by amplicon melting should be improved with internal temperature controls on any instrument.
Unlabeled oligonucleotides make convenient internal temperature controls that use the same saturating DNA dyes needed for amplicon melting. Control Tms can be adjusted over a wide range by varying the GC content and length and by including deletions or LNAs. Another option is to use melting controls that are covalently labeled with a dye of a second color for use in multicolor instruments. Such controls could be designed to melt at the same temperature as the amplicon without interference. Internal temperature controls decrease Tm variations attributable to the instrument or solution chemistry. However, they will not control for Tm variation secondary to the concentration of amplified DNA. Luckily, this variation is minor,10 and the PCR plateau tends to equalize any difference in starting DNA concentration.
Acknowledgments
We thank David Pattison for his assistance in preparing the blinded study, Nora Arias for the deidentification of clinical samples, Lisa Collins for illustration design assistance, and the Advanced Technology Group for their valued input on this project.
Footnotes
Supported by the ARUP Institute of Clinical and Experimental Pathology.
Aspects of high-resolution melting are licensed by the University of Utah to Idaho Technology and from Idaho Technology to Roche Applied Systems. C.T.W. holds equity interest in Idaho Technology.
References
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen., Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Graham R, Liew M, Meadows C, Lyon E, Wittwer CT. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295–1298. doi: 10.1373/clinchem.2005.051516. [DOI] [PubMed] [Google Scholar]
- Palais RA, Liew MA, Wittwer CT. Quantitative heteroduplex analysis for single nucleotide polymorphism genotyping. Anal Biochem. 2005;346:167–175. doi: 10.1016/j.ab.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- Owczarzy R, You Y, Moreira BG, Manthey JA, Huang L, Behlke MA, Walder JA. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004;43:3537–3554. doi: 10.1021/bi034621r. [DOI] [PubMed] [Google Scholar]
- Rouzina I, Bloomfield VA. Heat capacity effects on the melting of DNA. 2. Analysis of nearest-neighbor base pair effects. Biophys J. 1999;77:3252–3255. doi: 10.1016/S0006-3495(99)77156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzina I, Bloomfield VA. Heat capacity effects on the melting of DNA. 1. General aspects. Biophys J. 1999;77:3242–3251. doi: 10.1016/S0006-3495(99)77155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AR, Meehan T. The effect of sodium ion concentration on intrastrand base-pairing in single-stranded DNA. Nucleic Acids Res. 1994;22:3147–3150. doi: 10.1093/nar/22.15.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry CN, Vandersteen JG, Reed GH, Pryor RJ, Chen J, Wittwer CT. Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin Chem. 2003;49:396–406. doi: 10.1373/49.3.396. [DOI] [PubMed] [Google Scholar]
- Bernard PS, Lay MJ, Wittwer CT. Integrated amplification and detection of the C677T point mutation in the methylenetetrahydrofolate reductase gene by fluorescence resonance energy transfer and probe melting curves. Anal Biochem. 1998;255:101–107. doi: 10.1006/abio.1997.2427. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Herrmann MG, Gundry CN, Elenitoba-Johnson KS. Real-time multiplex PCR assays. Methods. 2001;25:430–442. doi: 10.1006/meth.2001.1265. [DOI] [PubMed] [Google Scholar]
- van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradic KW, Wells JE, Allen L, Kruckeberg KE, Singh RJ, Grebe SK. Substitution of 3′-phosphate cap with a carbon-based blocker reduces the possibility of fluorescence resonance energy transfer probe failure in real-time PCR assays. Clin Chem. 2004;50:1080–1082. doi: 10.1373/clinchem.2004.033183. [DOI] [PubMed] [Google Scholar]
- Chou LS, Meadows C, Wittwer CT, Lyon E. Unlabeled oligonucleotide probes modified with locked nucleic acids for improved mismatch discrimination in genotyping by melting analysis. Biotechniques. 2005;39:644–648. doi: 10.2144/000112050. [DOI] [PubMed] [Google Scholar]
- Erali M, Palais R, Wittwer CT. SNP genotyping by unlabeled probe melting analysis. Seitz O, Marx A, editors. Totowa: Humana Press; Methods in Molecular Biology. 2006 doi: 10.1007/978-1-60327-040-3_14. [DOI] [PubMed] [Google Scholar]
- Bernard PS, Ajioka RS, Kushner JP, Wittwer CT. Homogeneous multiplex genotyping of hemochromatosis mutations with fluorescent hybridization probes. Am J Pathol. 1998;153:1055–1061. doi: 10.1016/s0002-9440(10)65650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- Zhou L, Myers AN, Vandersteen JG, Wang L, Wittwer CT. Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin Chem. 2004;50:1328–1335. doi: 10.1373/clinchem.2004.034322. [DOI] [PubMed] [Google Scholar]
- Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem. 2006;52:494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- von Ahsen N. Two for typing: homogeneous combined single-nucleotide polymorphism scanning and genotyping. Clin Chem. 2005;51:1761–1762. doi: 10.1373/clinchem.2005.057729. [DOI] [PubMed] [Google Scholar]
- Volcik KA, Blanton SH, Northrup H. Examinations of methylenetetrahydrofolate reductase C677T and A1298C mutations—and in utero viability. Am J Hum Genet. 2001;69:1150–1153. doi: 10.1086/324066. [DOI] [PMC free article] [PubMed] [Google Scholar]