Abstract
Currently used clinical and histopathological parameters imprecisely define the risk of distant recurrence in breast cancer, underscoring the need for more informative prognostic markers. In the present fluorescence in situ hybridization study of archived surgical specimens, we derived an algorithm for computing a prognostic index (PI) from DNA copy numbers of three genomic regions (CYP24, PDCD6IP, and BIRC5) for estrogen/progesterone receptor-positive (ER/PR+) cancers and a distinct PI (based on NR1D1, SMARCE1, and BIRC5) for estrogen/progesterone receptor-negative (ER/PR−) cancers. Among independent test cases stratified by PI, recurrence rates were significantly higher among high-risk patients than low-risk patients for both ER/PR+ (odds ratio = 9.52, 95% confidence interval >2.12, P = 0.0024) and ER/PR− (odds ratio = 12.3, 95% confidence interval >1.45, P = 0.0188) cancers. Among the entire population, recurrences were significantly more prevalent for cases with PI above the medians for both ER/PR+ (Fisher’s exact, P = 1.19 × 10−5) and ER/PR− (P = 0.0025) patients and for the node-negative subsets (ER/PR+ node-negative, P = 0.042 and ER/PR− node-negative, P = 0.039). In conclusion, these markers perform well in comparison with other criteria for recurrence risk assessment and can be used with routinely formalin-fixed, paraffin-embedded surgical specimens.
The risk of distant recurrence in patients with invasive breast carcinoma is imperfectly predicted by factors such as age at diagnosis, clinical and pathological stage, tumor cell nuclear grade, and estrogen/progesterone receptor (ER/PR) status. Approximately 30% of breast cancer patients with early-stage disease and no detectable axillary lymph node involvement eventually will have a distant metastasis, whereas 30% of patients at higher risk according to conventional parameters would not have a recurrence if treated with local therapy only.1,2 Difficulty in accurately identifying patients destined for a recurrence complicates decisions regarding the use of adjuvant chemotherapy.3
Analyses of a variety of tumor cell molecular markers may be useful in the assessment of patients with breast carcinomas. The presence of estrogen and progesterone receptors is associated with less aggressive tumors and response to hormone therapy. Acquisition of extra copies (amplification) of the HER2gene in tumors correlates with poorer outcome4 and is an indication for treatment with the immunotherapeutic drug trastuzumab.5 Methods for global quantitation of mRNAs via microarray analysis have made it possible to develop gene expression profiles of breast carcinomas useful in predicting outcome. A 70-gene expression profile was identified that correlates with good outcome in relatively young women with early-stage lymph node-negative cancers,6 and another 76-gene expression profile predictive of survival in pre- or postmenopausal women with small tumors has also been described previously.7 An expression panel of 21 genes was used to generate a recurrence risk algorithm for women with ER/PR+ lymph node-negative breast cancers who had been treated with tamoxifen8, and a prognostic marker based on the expression ratio of two genes has been identified from gene expression profiling of tamoxifen-treated early-stage cancers.9 Such expression profiling holds promise as an approach for developing prognostic markers. However, the markers have been validated in a limited spectrum of disease presentations, namely early-stage disease and ER/PR+ cases. Furthermore, quantification of multiple mRNA levels in tumors is expensive and technically demanding and is not readily available in a routine clinical setting.
Chromosomal aberrations associated with breast cancer have been studied using comparative genomic hybridization assays,10,11,12,13 and associations between poor prognosis and amplification (increased copy number) of specific loci, such as the HER2locus on chromosome 1714 and the CYP24locus on chromosome 20,15,16,17 have been identified. Higher overall numbers of chromosome aberrations have been correlated with a greater risk of recurrence,18 and preliminary evidence for global patterns of genomic amplifications and deletions correlated with recurrence have been described previously.19,20 An analysis of a series of breast tumors, combining gene expression and comparative genomic hybridization data, indicated that there is a good correlation between genome amplification/deletion and gene expression for some, but not all, genes.21
We applied a concurrent data-mining technique to publicly available microarray-based gene expression6 and comparative genomic hybridization21,22 data to identify chromosomal regions containing genes characterized by DNA copy numbers that are coordinated with expression levels and are prognostic for breast cancer recurrence or prognostic for recurrence irrespective of gene expression levels.23 In this study, 17 such chromosomal regions (Table 1) were examined further to identify subsets with prognostic significance. A supplemental description of this data-mining technique is available online at http://jmd.amjpathol.org.
Table 1.
Bacterial Artificial Chromosome Clones Used as Probes for FISH Assays
| BAC name | Genetic locus | GenBank accession number | Chromosome location | Clone size (nucleotides) |
|---|---|---|---|---|
| RP11-499K24 | AL080059 | AL080059 | 8q22.1 | 165,883 |
| RP11-159A16 | STK3 | NM_006281 | 8q22.2 | 212,805 |
| RP11-486B24 | EXT1 | NM_000127 | 8q24.11 | 180,390 |
| RP11-367C15 | RAD21 | NM_006265 | 8q24.11 | 192,954 |
| RP11-529C24 | ANZA11 | NM_001157 | 10q22.3 | 187,470 |
| RP11-354M24 | FANCA | NM_000135 | 16q24.3 | 163,945 |
| RP11-610D13 | ZNF144 | NM_007144 | 17q12 | 218,801 |
| RP11-372J02 | SMARCE1 | NM_003079 | 17q21.2 | 174,341 |
| RP11-141D15 | BIRC5 | NM_001168 | 17q25.3 | 177,622 |
| RP11-683H06 | PDCD6IP | NM_013374 | 3p23 | 153,782 |
| RP11-563O04 | GRB7 | NM_005310 | 17q12 | 151,040 |
| RP11-689B15 | MLN64 | NM_006804 | 17q12 | 170,163 |
| RP11-092B18 | CYP24 | NM_000782 | 20q13.2 | 170,508 |
| RP11-067F19 | IMPA1 | NM_005536 | 8q21.13 | 194,492 |
| RP11-737K14 | HEPSIN | NM_002125 | 19q13.12 | 184,710 |
| RP11-278E15 | NR1D1 | X72631 | 17q21.1 | 161,993 |
| RP11-299H03 | ZNF207 | NM_003457 | 17q11.2 | 171,427 |
The specific chromosomal position of the bacterial artificial chromosome (BAC) clones used in this study can be identified within databases residing at http://genome.ucsc.edu (accessed November 3, 2006).24
We determined the DNA copy number for each of these genomic regions via fluorescent in situ hybridization (FISH) analysis of archived biopsy and resection specimens isolated from an independent group of women with stage I to III invasive breast carcinomas and known clinical outcomes. This allowed us to evaluate the prognostic significance of tumor copy number for each of the 17 identified chromosomal regions. For discovery of the most sensitive prognostic marker panels, we analyzed the correlation between recurrence in subset combinations of the 17 regions. Two trios of chromosomal markers applicable to ER/PR-positive or -negative tumors were identified. Prognostic indices that predict risk of distant recurrence were calculated, enabling the classification of women into low-, medium-, and high-risk categories.
Materials and Methods
Patients
This study was approved by the University of New Mexico Institutional Review Board. Medical records of all patients diagnosed with breast cancer at the University of New Mexico Hospital between 1986 and 1999 were examined, and all cases presenting with stage I, II, or III invasive ductal carcinoma were reviewed further. Patients were excluded from the study if their clinical or pathology records were inadequate (see below), if their archived specimens were inadequate for FISH, if they experienced an isolated local recurrence (to the breast or chest wall), or if they received neoadjuvant therapy and their pretreatment specimen was not available for FISH. A minimum of 4 years of follow-up was required unless a recurrence occurred sooner.
Clinicopathological data and archived slides were reviewed for all patients and verified on fresh hematoxylin and eosin-stained thin sections. Data collected for the study included method of biopsy or surgical excision, pathological and/or clinical node status, tumor size, overall stage, nuclear grade, margin status, ER/PR status, treatment history, and clinical outcome. Where necessary, fresh sections were prepared from the archived tumors and re-examined for surgical margins and nuclear grade. When retested, ER/PR status was assessed for all tumors with newly prepared unstained sections from archived biopsies. ER/PR testing was performed in a Clinical Laboratory Improvements Amendment-licensed, College of American Pathologists-accredited laboratory (TriCore Reference Laboratories, Albuquerque, NM) using a standard Ventana immunohistochemistry system with a threshold for positivity of 1% of the cells examined. Tumors were considered to be ER/PR-positive when they were positive for ER or PR. All ER/PR results were reviewed by the central study pathologist. HER2status, not generally obtained when these patients were initially diagnosed, was determined by FISH analysis in the same laboratory used for the ER/PR studies. HER2 was considered to be amplified when the HER2-to-CEP17ratio was greater than or equal to 2.0. Study endpoints were defined as recurrence (distant metastasis or death from breast cancer, regardless of length of follow-up) or nonrecurrence (disease free throughout follow-up). Mean follow-up was 8.9 years, and more than 96% of the patients were followed for at least 5 years.
Fluorescence in Situ Hybridization Assays
Genomic probes (Table 1) were selected from the 32K Re-Array library of bacterial artificial chromosomes.24 Each bacterial artificial chromosome contained the coding sequence of only one of the 17 genes selected for study. Bacterial artificial chromosome identities were confirmed with sequence and size analysis of polymerase chain reaction-amplified exons or 3′-untranslated regions and via hybridization to banded metaphase chromosomes. Bacterial artificial chromosomes were fluorescently labeled with either Spectrum Green or Spectrum Orange (Vysis, Inc., Downers Grove, IL) using a standard random priming reaction25 and were generally hybridized as pairs to 4-μm de-paraffinized protease sections prepared from formalin-fixed paraffin-embedded tissue. Labeled probes were coprecipitated with a 100-fold excess of human Cot-1 DNA and resuspended in hybridization buffer (50% formamide, 2× standard saline citrate, 10% dextran sulfate, and 0.01% Tween 20). Slides were co-denatured with labeled probes at 73°C for 6 minutes and incubated at 37°C for 16 to 20 hours. Hybridized slides were then washed in 2× standard saline citrate containing 0.3% Igepal (Sigma, Inc., St. Louis, MO) at 73°C for 2 minutes and counterstained with 4,6-diamidino-2-phenylindole.
A Metasystems Metafer 4 image analysis workstation (MetaSystems GmbH, Altlussheim, Germany) was used for image capture and signal analysis. FISH signals were counted in at least 100 cells and in at least three 40× fields for each probe. Signals from stromal and inflammatory cells were excluded from the analysis. Stromal cells and lymphocytes served as controls for probes: We did not observe probe signals greater than 2 in these cells. A few specimens (<5%) yielded smaller numbers of analyzable cells. Signal counts were collected in a “tiling” pattern to minimize the effects of nonuniform distribution of nuclei in thin sections.26 Raw data (DNA copy number per tile) were normalized to copy number per nuclear equivalent volume (NEV).27 Explicit thresholds for gene amplification were not set; instead, we related copy numbers per NEV for each gene as set forth by the formulas in Results. Personnel involved in the FISH data collection were blinded to probe identities and patient outcomes. Manual counting of multiple microscopic fields concurrent with automated signal counting revealed that <1% of cases demonstrated significant heterogeneity of signal counts from field to field.
Derivation of the Prognostic Index
We developed an algorithm for ranking combinations of the 17 markers, based on their ability to categorize samples into two or more risk groups. The algorithm employs a linear combination of the log (copy numbers) with coefficients computed from a logistic regression analysis. We term the sum of this linear combination the PI. From the value of the prognostic index, the samples were categorized into risk groups. A fitness function, based on the actual risk difference between assigned low- and high-risk groups, was used in a comprehensive search to rank marker combinations and identify significant prognostic combinations.
The data were analyzed separately according to ER/PR status. In the search phase, only a “training” subset (50% of cases for ER/PR+; 25% for ER/PR−) of the data was used. The remaining samples, blinded as to recurrence, were withheld as the test set. ER/PR+ cases were divided 50%/50% between training and test sets; the smaller number of ER/PR− cases were partitioned 25%/75% between training and test sets to obtain a reasonably sized test set. We binned the training data PI values into approximately equal thirds to produce risk category cutoff values and then evaluated the risk of recurrence for each of these categories using the withheld test data. Prognostic markers identified in the ER/PR+ and ER/PR− cases were further tested on the subset of cases that were lymph-node negative (ER/PR+ N0; or ER/PR− N0). Negative and positive predictive values of each of the best combinations were calculated on the basis of patients assigned to the low-risk and high-risk categories.
Statistical Analysis
One-sided Fisher’s exact tests assessed for increased incidence of distant recurrence with ER/PR status, node status (N+), tumor size (T >1), overall stage (>I), nuclear grade (>1), age (>50 years), or PI (above the median). Furthermore, Wilcoxon’s rank-sum tests evaluated differences between the recurrent and nonrecurrent groups with respect to the individual chromosome regions or the PI. Survival curves were derived from the time of surgery to the time of recurrence or death if from cancer or to the time the patient was censored, and the odds ratios (OR) comparing the recurrence rates of the low- and high-risk groups were evaluated using Fisher’s exact test. Relative risks were calculated from the probabilities of recurrence. Statistical analysis was done using R, version 2.1.1.R (Development Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2005. http://www.R-project.org). Cox proportionate hazard regression analysis was performed to study covariates.
Results
Patients
A total of 229 patients were available to the study after examination of 723 patient records and fluorescence in situ hybridization studies. Of the 723 cases reviewed, 57.4% did not meet the clinicopathological inclusion criteria, 0.7% were excluded due to local recurrence in the absence of eventual distant metastasis, 4% were excluded due to neoadjuvant therapy, and 6% were lost due to hybridization failure despite meeting the inclusion criteria. Patient characteristics are summarized in Table 2. The average age at diagnosis was 54.4 years; 72% had no recurrence; 41% presented with stage I disease, 41% with stage II, and 19% with stage III; 21% had nuclear grade 1 tumors, 53% had grade 2, and 25% had grade 3; 57% were node-negative, 27% were N1, 9% were N2, and 7% were N3; and 71% were classified ER/PR+. With respect to treatment, 28% received hormone therapy, 22% received chemotherapy, 16% received both chemotherapy and hormone treatment, and 34% received local therapy only. Patients were non-Hispanic Caucasians (60%), Hispanic (30%), African American (1%), Asian (2%), or Native American (3%), or had unknown or mixed ethnicity (4%). We found no significant difference in the rate of recurrence in study patients with HER2+ and HER2− tumors, for both ER/PR+ tumors (P = 0.28, Fisher’s exact) and ER/PR− tumors (P = 0.43, Fisher’s exact).
Table 2.
Clinicopathological Characteristics of Invasive Cancers in Study Population
| N | Age
|
ER/PR receptor (4 N/A)
|
Overall stage (0 N/A)
|
Tumor size (1 N/A)
|
Node status (0 N/A)
|
Nuclear grade (12 N/A)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 229 (total) | Average (years) | <50 years (N = 92) | >50 years (N = 137) | ER/PR+ | ER/PR− | I | II and III | T1 | T >1 | N0 | N >0 | I | II to III | |
| No recurrence | 165 | 56.5 | 56 | 109 | 121 | 41 | 83 | 82 | 109 | 56 | 113 | 52 | 40 | 117 |
| Recurrence | 64 | 50.9 | 36 | 28 | 42 | 21 | 10 | 54 | 20 | 43 | 18 | 46 | 8 | 52 |
Patients are stratified by recurrence status, age, ER/PR status, overall stage, tumor size, lymph node status, and nuclear stage.
N/A, data not available.
Amplification of Genomic Markers in Tumors
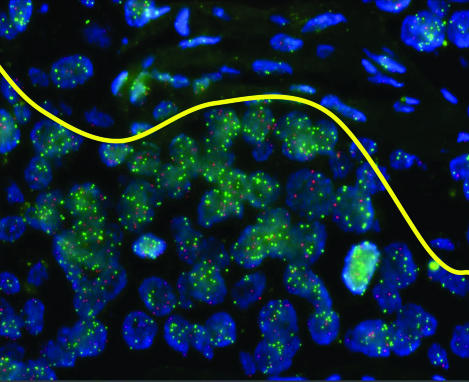
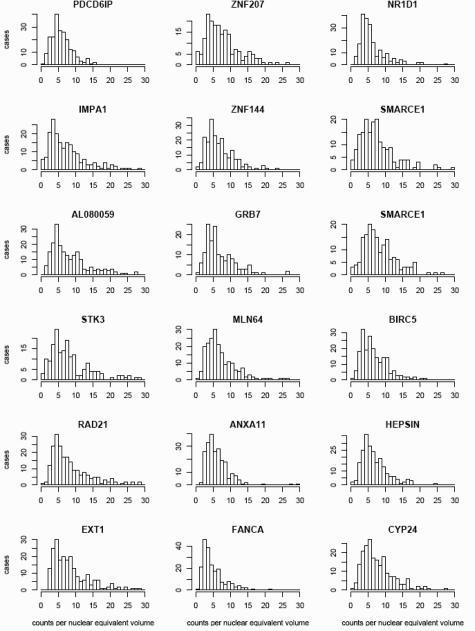
Figure 1 is a representative FISH image of a specimen hybridized with one of the probe pairs. The average DNA copy numbers per NEV for all 17 probes among all 229 specimens are summarized as a set of histograms (Figure 2). The mean copy numbers among all cases ranged from 5.0 to 9.0, and the median copy numbers ranged from 3.9 to 7.3 (SD range, 3.3 to 6.6), indicating a substantial variation in the degree of amplification in the specimen set at all these chromosomal regions. For most of the probes, the number of DNA copies per NEV peaks at four or five in 20 to 40 cases and increases to as many as 20 to 30 copies in some cases. FANCA, the probe with the lowest level of amplification peaking at three copies per NEV in more than 40 cells, maps to a chromosome region (16q) seldom amplified in breast cancer.
Figure 1.
Dual hybridization of the probes SMARCE1 (Spectrum Green) and AL080059 (Spectrum Orange) to an archived breast carcinoma tissue section is shown. The yellow line represents “virtual microdissection” of the image, so that only tumor cells (below the line) are used in assessment of genomic marker status, excluding the mostly stromal cells above the line.
Figure 2.
The individual histograms display the number of cases (y axis) containing the average DNA copy number per nuclear equivalent volume indicated by each bar (x axis) for each probe. Data for all 229 specimens are included. Two histograms are shown for SMARCE1, because hybridization for this probe was performed in duplicate.
Derivation of Prognostic Index
We evaluated the patterns of genomic aberrations of the 17 chromosomal markers, in subsets, to identify combinations that correlate with patient outcome. In women with ER/PR-positive tumors, the amplification pattern of three chromosome regions, at CYP24, PDCP6IP, and BIRC5, formed the best predictor of recurrence. The PI for this combination is given by PI = 0.183[log(CNBIRC5)] + 0.128[log(CNCYP24)] − 0.173[log(CNPDCD6IP)], where CN is copy number.
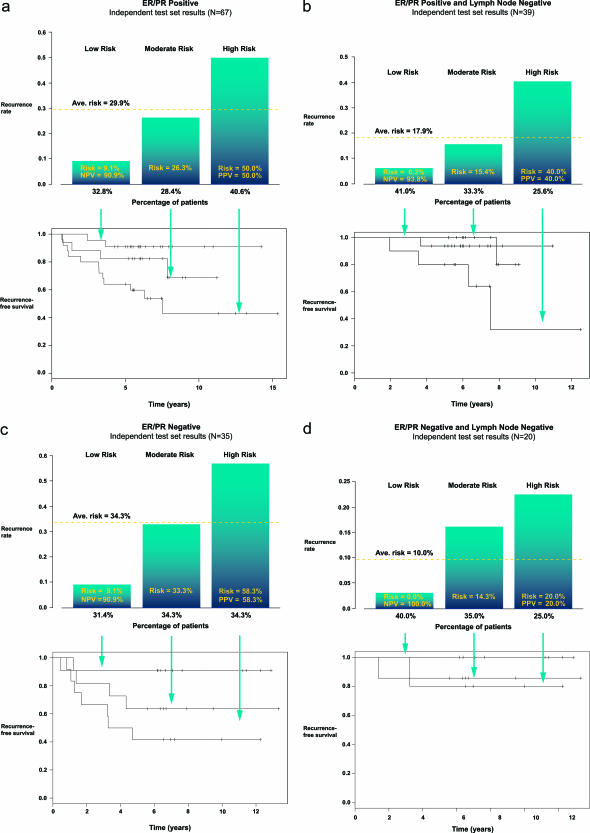
The division of the training set patients, based on the PI calculations, into approximately equal categories of low, moderate, and high risk and the test set data using the same risk category cutoff values, is summarized in Table 3. The relationship between risk categories, with PI ranges of <0.248, 0.248 to 0.328, and >0.328, respectively, and rates of recurrence for the ER/PR+ test set are shown in Figure 3a. The overall recurrence rate for the ER/PR+ test set is 29.9%. The rates were 9% for the low-risk and 50% for the high-risk categories, respectively. Risk for the intermediate category decreased slightly from the test set average to 26%. The difference in rates between the low-risk and the high-risk categories was significant [OR = 9.52, 95% confidence interval (95% CI) >2.12, P = 0.0024], with relative risks for each category of 0.305 and 1.68, respectively (Table 4). Negative predictive value (NPV) and positive predictive value (PPV) for the ER/PR+ algorithm were 91 and 50%, respectively. Performance of the ER/PR+ algorithm on all three risk categories of the ER/PR+ test set patients is shown by the survival curves (bottom panels in Figure 3, a and b).
Table 3.
Recurrence in Training and Test Set Cases with Low, Moderate, and High Prognostic Indices
| Training sets
|
Test sets
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All ER/PR+ (N = 58)
|
All ER/PR− (N = 13)
|
All ER/PR+ (N = 67)
|
All ER/PR− (N = 35)
|
|||||||||
| Low PI | Mod PI | High PI | Low PI | Mod PI | High PI | Low PI | Mod PI | High PI | Low PI | Mod PI | High PI | |
| No recurrence | 17 | 16 | 7 | 4 | 3 | 2 | 20 | 14 | 13 | 10 | 8 | 5 |
| Recurrence | 2 | 3 | 13 | 0 | 1 | 3 | 2 | 5 | 13 | 1 | 4 | 7 |
|
|
ER/PR+ N0 (N = 31)
|
ER/PR− N0 (N = 5)
|
ER/PR+ N0 (N = 39)
|
ER/PR− N0 (N = 20)
|
||||||||
| Low PI | Mod PI | High PI | Low PI | Mod PI | High PI | Low PI | Mod PI | High PI | Low PI | Mod PI | High PI | |
| No recurrence | 11 | 12 | 5 | 1 | 2 | 0 | 15 | 11 | 6 | 8 | 6 | 4 |
| Recurrence | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 4 | 0 | 1 | 1 |
The study population, divided according to ER/PR status, was further divided into training and test sets for ER/PR+ and ER/PR− cases. The PI algorithms, identified using training set data, were used to calculate the PI on both the training and test patients, and each case was assigned to a predicted risk category. The numbers of recurrent and nonrecurrent cases in each risk category are shown. Risk category assignments and actual outcome data are summarized for the node negative subsets of the training and test sets in the lower portion of the table.
Figure 3.
The prognostic index algorithm derived from the ER/PR+ and ER/PR− training sets was used to calculate PI for the patients in the test sets. Patients were placed into risk categories according to their PI, with PI increasing on the x axes and rates of recurrence on the y axes. The percentages of patients assigned to each risk category are shown below the bins in the top panels. The average risk of recurrence for each group is shown as a bar across all three risk categories, and the rates of recurrence for each risk group are shown within each bin. NPV and PPV for the low-risk and high-risk groups are also shown within each bin. Survival curves for each risk category are shown in the bottom panels. a: ER/PR+ test set (OR = 9.52, 95% CI >2.12, P = 0.0024). b: ER/PR+ N0 test set (OR = 9.05, 95% CI >0.958, P = 0.0549). c: ER/PR− test set (OR = 12.3, 95% CI >1.45, P = 0.0188). d: ER/PR− N0 test set (OR = infinity, 95% CI >0.0842, P = 0.385).
Table 4.
Relative Risk of Recurrence for Patients with Low and High Genomic Marker Prognostic Indices
| All ER/PR+ | ER/PR+ N0 | All ER/PR− | ER/PR− N0 | |
|---|---|---|---|---|
| Low-risk PI | 0.305 | 0.348 | 0.265 | 0 |
| High-risk PI | 1.68 | 2.22 | 1.70 | 2.0 |
Relative risk of assigned category to group risk (test sets). Risks were calculated for each of the four categories of patients (all ER/PR+, ER/PR+ N0, all ER/PR−, and ER/PR− N0) using the test set data only. Relative risk for each risk category was calculated based on the risk of recurrence in each risk category in comparison with the average rate of recurrence for that patient group. Data are summarized for both lymph node-positive and -negative cases. Note that prognostic indices were calculated for ER/PR+ and ER/PR− cases only when raw data (signals per NEV) were available for a case for all three chromosome region markers; thus, the number of cases available for this analysis is less than shown in Table 2.
The overall recurrence rate in the ER/PR+ N0 subgroup was less than 15%, and rates for the low- and high-risk categories were 6.3 and 40%, respectively (Figure 3b). The difference in recurrence rates between the risk groups was marginally significant (OR = 9.05, 95% CI >0.958, P = 0.0549). The recurrence rate in the intermediate risk category remained relatively unchanged (15.4% recurrence rate), with relative risks of 0.348 and 2.22, respectively (Table 4). The NPV and PPV for the ER/PR+ algorithm in the ER/PR+ N0 group are 93.8 and 40.0%, respectively. Performance of the algorithms on all three risk categories are illustrated by the survival curves (bottom panels in Figure 3, a and b).
The same approach was taken for discovery of a predictive marker among the ER/PR− cases. The amplification pattern of three chromosome regions, at NR1D1, SMARCE1, and BIRC5, formed the most predictive algorithm for risk among the ER/PR− group. The prognostic index for this combination is given by PI = 0.310 +.311[log(CNNR1D1) − 0.155[log(CNSMARCE1)] − 0.112[log(CNBIRC5)].
The distribution of the training and test sets of ER/PR− patients into three risk categories based on their calculated PIs and their actual rates of recurrence are summarized in Table 3. Performance of this algorithm among the ER/PR− test set is illustrated in Figure 3c, with PI ranges of <0.140, 0.140 to 0.329, and >0.329 for the low-, moderate-, and high-risk categories, respectively. The overall recurrence rate in the ER/PR− group was 34.3%, and the recurrence rates for the low-, moderate-, and high-risk patients were 9.1, 33.3, and 58.3%, respectively. The difference in recurrence rates between the low-risk and the high-risk categories was significant (OR = 12.3, 95% CI >1.45, P = 0.0188), with relative risks for each category of 0.265 and 1.70, respectively (Table 4). The NPV and PPV of the ER/PR− algorithm were 91 and 58%, respectively. Performance of the ER/PR− algorithm is shown by the survival curves (bottom panel).
The N0 cases of the ER/PR− test set are shown in Figure 3d, and although the number of patients in this subset was small, the results were consistent with the trend demonstrated by the ER/PR+ and ER/PR− groups and the ER/PR+ N0 groups. The recurrence rates among the low-risk and high-risk groups were 0 and 20%, respectively. The odds ratio for the difference in recurrence rates could not be computed because no low-risk cases had a recurrence (OR = infinity, 95% CI >0.0842, P = 0.385). The low- and high-risk categories had relative risk of 0 and 2.0, respectively, and the NPV and PPV for the algorithm were 100 and 20%, respectively. Survival curves for the four categories are illustrated in Figure 3d (bottom panel).
PI as an Independent Predictor of Recurrence
Among the 229 women with breast cancer (Table 5), clinicopathological features associated with recurrence included age (younger than 50 years at diagnosis), positive node status, tumor size > T1, overall stage > I, and nuclear grade >1 (one-sided Fisher’s Exact tests). ER/PR status was not found to be significantly associated with recurrence. DNA copy numbers of 11 of the 17 chromosome regions differed significantly with recurrence (P < 0.05; Table 6). When stratified by hormone receptor status, the associations between clinicopathological traits and recurrence were similar between the ER/PR+ cases and the entire population. However, among the ER/PR− patients, tumor size was marginally significant, but neither age nor overall stage was significantly associated with recurrence, and no association between nuclear grade and recurrence could be calculated because all of the ER/PR− grade 1 cases experienced a recurrence (Table 5). Recurrences were significantly more prevalent for cases with PI above the median for both ER/PR+ (Fisher’s exact, P = 1.19 × 10−5) and ER/PR− (P = 0.0025) patients and for the node-negative subsets (ER/PR+ N0, P = 0.042 and ER/PR− N0, P = 0.039). The PI was significantly greater among recurring than nonrecurring cases (ER/PR+ Wilcoxon’s, P = 2.67 × 10−13; ER/PR−, P = 0.000303).
Table 5.
Association between Clinicopathological Features and Recurrence
| ER/PR+ and ER/PR− cases | ER/PR+ cases | ER/PR− cases | |
|---|---|---|---|
| Lymph node-negative and node-positive cases | |||
| N | 173 | 125 | 48 |
| Age <50 years | |||
| OR (95% CI) | 2.49 (>1.05) | 3.61 (>1.72) | 0.776 (>0.236) |
| P | 0.00173 | 0.00132 | 0.768 |
| Node status >0 | |||
| OR (95% CI) | 5.31 (>3.00) | 6.12 (>2.81) | 5.50 (>1.55) |
| P | 6.02 × 10−8 | 1.04 × 10−5 | 0.00887 |
| T >1 | |||
| OR (95% CI) | 4.16 (>2.38) | 6.10 (>2.81) | 3.56 (>1.06) |
| P | 2.9 × 10−6 | 1.15 × 10−5 | 0.0407 |
| Stage > I | |||
| OR (95% CI) | 5.43 (>2.80) | 6.47 (>2.69) | 3.29 (>0.857) |
| P | 6.64 × 10−7 | 2.79 × 10−5 | 0.0806 |
| Nuclear grade >1 | |||
| OR (95% CI) | 2.21 (>1.05) | 3.65 (>1.30) | Not applicable |
| P | 0.0371 | 0.0142 | Not applicable |
| ER/PR-negative | |||
| OR (95% CI) | 1.57 (>0.880) | Not applicable | Not applicable |
| P | 0.105 | Not applicable | Not applicable |
| Prognostic index | |||
| OR (95% CI) | 6.34 (>2.81) | 7.88 (>2.04) | |
| P
|
Not applicable
|
1.19 × 10−5
|
0.00250
|
| Lymph node-negative cases | |||
| N | 95 | 70 | 25 |
| Age <50 years | |||
| OR (95% CI) | 1.07 (>0.369) | 1.54 (>0.304) | 0.180 (>0.00604) |
| P | 0.550 | 0.410 | 0.983 |
| T >1 | |||
| OR (95% CI) | 2.51 (>0.948) | 3.90 (>0.966) | 0.554 (>0.0187) |
| P | 0.0613 | 0.0551 | 0.856 |
| Stage > I | |||
| OR (95% CI) | 2.20 (>0.814) | 2.62 (>0.604) | 0.677 (>0.0226) |
| P | 0.104 | 0.161 | 0.812 |
| Nuclear grade >1 | |||
| OR (95% CI) | 0.898 (>0.308) | 1.05 (>0.231) | All recurred |
| P | 0.693 | 0.636 | All recurred |
| ER/PR-negative | |||
| OR (95% CI) | 2.27 (>0.840) | Not applicable | Not applicable |
| P | 0.0932 | Not applicable | Not applicable |
| Prognostic index | |||
| OR (95% CI) | 4.79 (>1.06) | Infinity (>1.13) | |
| P | Not applicable | 0.0420 | 0.0391 |
One-sided Fisher’s exact test was used to calculate the associations between age, node status, tumor size, overall stage, nuclear grade, ER/PR status, and the prognostic index for the entire study population and subsets of the population stratified by ER/PR and node status. Data are summarized lymph node-negative cases only. Note that prognostic indices were calculated for ER/PR+ and ER/PR− cases only when raw data (signals per NEV) were available for a case for all three chromosome region markers; thus, the number of cases available for this analysis is less than shown in Table 2.
Table 6.
Association Between Copy Number of Individual Genomic Regions and Distant Recurrence
| Genomic region | P (Wilcoxon)* |
|---|---|
| CYP24 | 0.001 |
| EXT1 | 0.002 |
| NR1D1 | 0.003 |
| MLN64 | 0.003 |
| FANCA | 0.004 |
| BIRC5 | 0.007 |
| ZNF144 | 0.008 |
| RAD21 | 0.012 |
| GRB7 | 0.016 |
| HEPSIN | 0.02 |
| ZNF207 | 0.03 |
| STK3 | 0.051 |
| IMPA1 | 0.062 |
| AL080059 | 0.073 |
| SMARCE1 | 0.075 |
| ANXA11 | 0.086 |
| PDCD6IP | 0.526 |
Associations between risk of distant recurrence and DNA copy number determined by FISH for each chromosome region marker are shown (Wilcoxon’s rank sum test). The cutoff for a significant association (α) was set at 0.05.
Among the node-negative group, there was no association between recurrence and age, stage, tumor size, or nuclear grade, in either the ER/PR-positive or -negative groups (Table 5). However, PIs were significantly associated with recurrence in the hormone receptor-positive and -negative and node-negative subsets of patients (Table 5; ER/PR+ N0, Fisher’s exact, P = 0.0420; ER/PR− N0, P = 0.0391), and PI was significantly greater among recurring node-negative cases than node-negative cases without a recurrence (ER/PR+ N0, Wilcoxon’s, P = 1.10 × 10−9; ER/PR− N0, P = 0.0441).
Cox proportional hazard regressions were performed to determine whether the prognostic index provides information beyond known prognostic factors. Using backward elimination to remove insignificant covariates from an initial model containing age, stage, nuclear grade, tumor size, and categorical PI, we found PI to be a significant independent predictor of recurrence in both the ER/PR+ and ER/PR− subgroups. For the ER/PR+ group, age (P = 0.0057), nuclear grade (P = 0.025), tumor size (P = 0.0014), and categorical PI (P = 0.0003) remained significant. For the ER/PR− group, only categorical PI (P = 0.0082) remained a significant predictor of recurrence.
Discussion
We have described two sets of genomic markers for which relative copy numbers in breast cancers have prognostic significance in women with stage I to III invasive ductal carcinoma: a CYP24/PDCD6IP/BIRC5trio of markers for ER/PR-positive cancers and a NR1D1/SMARCE1/BIRC5trio for ER/PR-negative cancers. Prognostic indices, calculated from algorithms based on DNA copy numbers of the two trios of genomic markers, are significant independent predictors of distant recurrence of ductal carcinomas within 5 years of surgery for women with either ER/PR-positive or -negative cancers. Each of the algorithms has a NPV of 91% for their test sets. In the lymph node-negative subsets of test cases, the ER/PR+ marker has an NPV of 93.8%, and the ER/PR− marker has an NPV of 100%.
We have compared the performance of these algorithms to the National Institutes of Health and St. Gallens guidelines for treatment and risk assessment. According to the National Institutes of Health guidelines,28 96% of the women in this study would be advised to receive chemotherapy and/or hormone therapy, and the St. Gallens criteria29 would classify 87% of them at greater than minimal risk. The algorithms described here correctly identified 91% of the recurrent patients as moderate or high risk and correctly identified 43% of the nonrecurrent patients as low risk. These results outperform the National Institutes of Health and St. Gallens classifications.
Odds ratios for the algorithms described here are 9.52 (95% CI >2.12, P = 0.0024) and 12.3 (95% CI >1.45, P = 0.0188) for the ER/PR+ and ER/PR− test sets, respectively. Previously published prognosis signatures confer similar odds ratios for distant metastasis within 5 years: 15 (95% CI 4 to 56, P = 4.1 × 10−6)30 and 11.9 (95% CI 4.04 to 35.1, P < 0.0001) for the ER/PR+ and ER/PR− test sets, respectively.7 However, it should be noted that the genomic markers described here were identified from a more heterogeneous patient population than the expression-based prognosis signatures, with respect to stage of disease at presentation, clinicopathological features, and treatment histories.
Our approach to discovery of genomic markers predictive of breast cancer recurrence is distinct from the strategies of other groups. First, we correlated DNA copy number data and gene expression data to identify genes with prognostic expression levels that ultimately could be assayed by DNA copy number measurement, and we included chromosome regions for which copy numbers are correlated with prognosis irrespective of gene expression levels.23 Second, we evaluated the prognostic value of genomic regions in combinations rather than building marker sets from single genes rank ordered by significance. An advantage to this approach is that chromosome region redundancy in the best prognostic combination is avoided and instead emphasizes the pattern of the chromosome regions that the solution comprises. These points are illustrated by considering that PDCD6IP, individually, is not significantly correlated with recurrence (Table 6). However, when considered together with CYP24and BIRC5in the ER/PR+ cancers, PDCD6IP adds prognostic information.
The genomic regions with prognostic significance that we identified in these studies do not entirely coincide with those found to be aberrant in other studies. For example, the long arm of chromosome 1 is amplified in a large proportion of breast cancers (Table 1), but we did not identify genes mapping to that region in which copy number predicted recurrence. Amplification of the HER2 locus on chromosome 17q21 is known to be correlated with poor prognosis. However, HER2was not identified as one of our initial 17 candidate genomic regions. Two other genes in the same region of chromosome 17, GRB7 and MLN64, were found to be significant markers of recurrence risk.
Although the genomic marker sets that we identified would not necessarily be expected to include genes with known roles in breast cancer pathogenesis, several of the genomic markers include genes known to be involved in carcinogenesis. For example, CYP24encodes a hydroxylase that participates in vitamin D catabolism, is known to be amplified and overexpressed in breast cancers, and may act by abrogating the growth regulatory activity of vitamin D.17 The overexpression of PDCD6IP restores contact inhibition, promotes detachment-induced apoptosis, and reduces tumorigenicity in nude mice.31,32 Overexpression of BIRC5, an inhibitor of apoptosis, is correlated with poor prognosis in many types of cancer.33 BIRC5is positively correlated with increasing PI in the ER/PR+ patients, when considered in the context of CYP24and PDCD6IP, yet negatively correlated with increasing PI in the ER/PR− patients, when considered together with NR1D1and SMARCE1, again illustrating the importance of the pattern of the chromosome regions that the solution comprises. NR1D1, a steroid hormone receptor transcription factor, was recently shown to be amplified in breast cancer34 and coexpressed with HER2,35 whereas SMARCE1, a component of the SWI/SNF chromatin-remodeling complex that appears to participate in regulation of estrogen-responsive genes, has not been directly implicated in carcinogenesis of receptor-negative tumors.
Assessing copy numbers of each of the genomic markers via FISH can be performed on surgical specimens routinely formalin fixed and paraffin embedded in a pathology laboratory. Because these genomic markers were identified in a heterogeneous population of women, with respect to age and stage of breast carcinoma presentation, these results should be validated in additional studies of different patient populations in other institutions.
Supplementary Material
Acknowledgments
We are indebted to the patients whose biopsy and surgical samples and outcome information allowed the studies and analyses described here. We appreciate advice provided regarding statistical analysis from Curtis Hunt and the support of this work by R. Phillip Eaton and Mary F. Lipscomb.
Footnotes
Supplemental material for this article can be found on http://jmd.amjpathol.org.
References
- Early Breast Cancer Trialists’ Collaborative Group Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group Polychemotherapy for early breast cancer: an overview of the randomised trials. Lancet. 1998;352:930–942. [PubMed] [Google Scholar]
- Caldas C, Aparicio SA. The molecular outlook. Nature. 2002;415:484–485. doi: 10.1038/415484a. [DOI] [PubMed] [Google Scholar]
- Carr JA, Havstad S, Zarbo RJ, Divine G, Mackowiak P, Velanovich V. The association of HER-2/neu amplification with breast cancer recurrence. Arch Surg. 2000;135:1469–1474. doi: 10.1001/archsurg.135.12.1469. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wang ZR, Yan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatr G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Lichter P, Fischer K, Joos S, Fink T, Baudis M, Potkul RK, Ohl S, Solinas-Toldo S, Weber R, Stilgenbauer S, Bentz M, Dohner H. Efficacy of current molecular cytogenetic protocols for the diagnosis of chromosome aberrations in tumor specimens. Cytokines Mol Ther. 1996;2:163–169. [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Cai WW, Mao JH, Chow CW, Damani S, Balmain A, Bradley A. Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat Biotechnol. 2002;20:393–396. doi: 10.1038/nbt0402-393. [DOI] [PubMed] [Google Scholar]
- Ravdin PM, Chamness GC. The c-erbB-2 proto-oncogene as a prognostic and predictive marker in breast cancer: a paradigm for the development of other macromolecular markers-a review. Gene. 1995;159:19–27. doi: 10.1016/0378-1119(94)00866-q. [DOI] [PubMed] [Google Scholar]
- Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, Gray JW, Kallioniemi OP, Isola J. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, Spichtin H, Maurer R, Mirlacher M, Kochli O, Zuber M, Dieterich H, Mross F, Wilber K, Simon R, Sauter G. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- Somiari SB, Shriver CD, He J, Parikh K, Jordan R, Hooke J, Hu H, Deyarmin B, Lubert S, Malicki L, Heckman C, Somiari RI. Global search for chromosomal abnormalities in infiltrating ductal carcinoma of the breast using array-comparative genomic hybridization. Cancer Genet Cytogenet. 2004;155:108–118. doi: 10.1016/j.cancergencyto.2004.02.023. [DOI] [PubMed] [Google Scholar]
- Callagy G, Pharoah P, Chin SF, Sangan T, Daigo Y, Jackson L, Caldas C. Identification and validation of prognostic markers in breast cancer with the complementary use of array-CGH and tissue microarrays. J Pathol. 2005;205:388–396. doi: 10.1002/path.1694. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshiran R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein , Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C: Discovery of multiplex genomic markers for predicting breast cancer recurrence (abstract). Presented at the San Antonio Breast Cancer Symposium, 2004 Dec 8–11, San Antonio, TX [Google Scholar]
- Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong PJ. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Lörch T, Piper J, Tomisek J. Tile sampling: a new method for the automated quantitative analysis of samples with high density and its application to Her2 scanning. Altlussheim, Germany: Metasystems, Incorporated,; Proceedings of Quantitative Molecular Cytogenetics. 2002 [Google Scholar]
- Pahlplatz MM, de Wilde PC, Poddighe P, van Dekken H, Vooijs GP, Hanselaar AG. A model for evaluation of in situ hybridization spot-count distributions in tissue sections. Cytometry. 1995;20:193–202. doi: 10.1002/cyto.990200302. [DOI] [PubMed] [Google Scholar]
- Eifel P, Axelson JA, Costa J, Crowley J, Curran WJ, Jr, Deshler A, Fulton S, Hendricks CB, Kemeny M, Kornblith AB, Louis TA, Markman M, Mayer R, Roter D. National Institutes of Health consensus development conference statement: adjuvant therapy for breast cancer, November 1–3, 2000. J Natl Cancer Inst. 2001;93:979–989. doi: 10.1093/jnci/93.13.979. [DOI] [PubMed] [Google Scholar]
- Goldhirsch A, Woos WC, Gelber RD, Coates AS, Thürliman B, Senn J-J. Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. J Clin Oncol. 2003;21:3357–3356. doi: 10.1200/JCO.2003.04.576. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wu Y, Pan S, Che S, He G, Nelman-Gonzalez M, Weil MM, Kuang J. Overexpression of Hp95 induces G1 phase arrest in confluent HeLa cells. Differentiation. 2001;67:139–153. doi: 10.1046/j.1432-0436.2001.670406.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Pan S, Luo W, Lin SH, Kuan J. Hp95 promotes anoikis and inhibits tumorigenicity of HeLa cells. Oncogene. 2002;21:6801–6808. doi: 10.1038/sj.onc.1205849. [DOI] [PubMed] [Google Scholar]
- Altieri DC, Marchisio PC. Survivin apoptosis: an interloper between cell death and cell proliferation in cancer. Lab Invest. 1999;79:1327–1333. [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Dressman MA, Baras A, Malinowski R, Alvis LB, Kwon I, Walz TM, Polymeropoulos MH. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer Res. 2003;63:2194–2199. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.