Abstract
An unlabeled probe assay relies on a double-stranded DNA-binding dye to detect and verify target based on amplicon and probe melting. During the development and application of unlabeled probe assays, aberrant melting peaks are sometimes observed that may interfere with assay interpretation. In this report, we investigated the origin of aberrant melting profiles observed in an unlabeled probe assay for exon 10 of the RET gene. It was determined that incomplete 3′ blocking of the unlabeled probe allowed polymerase-mediated probe extension resulting in extension products that generated the aberrant melting profiles. This report further examined the blocking ability of the 3′ modifications C3 spacer, amino-modified C6, phosphate, inverted dT, and single 3′ nucleotide mismatches in unlabeled probe experiments. Although no 3′ blocking modifications in these experiments were 100% effective, the amino-modified C6, inverted dT, and C3 spacer provided the best blocking efficiencies (1% or less unblocked), phosphate was not as effective of a block (up to 2% unblocked), and single nucleotide mismatches should be avoided as a 3′ blocking modification.
Unlabeled probe and amplicon melting analysis have been used for detecting single nucleotide polymorphisms and small deletions in the human RET, cystic fibrosis, and factor V genes, among others.1,2,3,4 The method uses a 3′ blocked unlabeled probe in combination with the double-stranded (ds)DNA-binding dye LCGreen Plus (Idaho Technology, Salt Lake City, UT) for detection and confirmation of target.5,6 Unlabeled probes possess no fluorescent moiety; rather, they produce a signature melting profile when used with a dsDNA-binding dye by annealing with single-stranded (ss)DNA produced by asymmetric polymerase chain reaction (PCR). Unlike other detection methods that only interrogate the region of complementarity covered by the probe, an unlabeled probe system can detect polymorphisms outside the probe region by analysis of dsDNA amplicon melting.7,8
The high sensitivity and low specificity of a dsDNA-binding dye requires unambiguous melting profiles to accurately identify insertions/deletions or polymorphisms. During unlabeled probe optimization, the majority of aberrant melting peaks are primer dimers or nonspecific amplicons, which can be reduced or eliminated by altering annealing temperatures or by redesigning primer sequences. Once optimized, an unlabeled probe assay produces two melting peaks: the melting of the dsDNA amplicon and the melting of the probe off the ssDNA product derived by asymmetric PCR. During the course of implementing a previously optimized unlabeled probe assay, we have occasionally observed aberrant melting profiles when using older probes or when replacing a proven probe with a newly synthesized probe of identical sequence. Removing the unlabeled probe from these PCR reactions eliminated these aberrant melting profiles, suggesting that the quality of the probe is important for optimal results. It was hypothesized that the aberrant melting profiles might be caused by probe degradation, or probe extension by DNA polymerase because of incomplete 3′ blocking.
In the current study, aberrant melting profiles observed in an unlabeled probe assay specific for exon 10 of the tyrosine kinase receptor gene RET were investigated. Mutations in exon 10 lead to multiple endocrine neoplasia type-2 or familial medullary thyroid carcinomas.9,10 This report determined the identity of aberrant amplification products and melting profiles, and assessed the effectiveness of common 3′ blocking modifications [phosphate, C3 spacer, amino-C6, dideoxynucleotides, and inverted dT (InvT)], used to prevent DNA polymerase extension.11
Materials and Methods
Human Genomic DNA Sample
A previously characterized wild-type human genomic DNA sample (hgDNA) was used for these experiments. The clinical sample used was residual and deidentified following the Health Insurance Portability and Accountability Act of 1996 and was used in accordance with University of Utah Institutional Review Board protocol number 7275, which covers research conducted by ARUP Laboratories.
Primers and Probes
All oligonucleotides were manufactured by Integrated DNA Technologies (Coralville, IA). The sequences are summarized in Table 1. ReadyMade primer M13 Forward-20 was used for sequencing of unknown amplification products. Primers used for real-time amplification include 10F, 10R, and probe-OH. Oligonucleotide probes used for unlabeled probe experiments were designed with two internal mutations, represented in Table 1 as underlined As, to differentiate mutant probe extension. The probes possessed the following 3′ blocking modifications: a single nucleotide mismatch (probe-mis), a phosphate (probe-phos), a C3 spacer (probe-C3), an amino-modified C6 (probe-C6), and an inverted dT (probe-InvT). All oligonucleotides were desalted with no postsynthesis purification except for ReadyMade primer, which was polyacrylamide gel electrophoresis purified.
Table 1.
Oligonucleotides Used for Sequencing, Real-Time PCR Amplification, and Detection
| Name | Sequence 5′ to 3′ | 3′ modification |
|---|---|---|
| 10F | 5′-GGGCAGCATTGTTGGGGGAC-3′ | None |
| 10R | 5′-TGGTGGTCCCGGCCGCCA-3′ | None |
| M13 for −20 | 5′-GTAAAACGACGGCCAGT-3′ | None |
| Probe-OH | 5′-GGAGAAGTACTTCTACGAGCCCGAAGACATC-3′ | None |
| Probe-mis | 5′-GGAGAAGTACTTCTACGAGCCCGAAGACATC G-3′ | G mismatch |
| Probe-phos | 5′-GGAGAAGTACTTCTACGAGCCCGAAGACATC-3′ | Phosphate |
| Probe-C3 | 5′-GGAGAAGTACTTCTACGAGCCCGAAGACATC-3′ | C3 spacer |
| Probe-C6 | 5′-GGAGAAGTACTTCTACGAGCCCGAAGACATC-3′ | C6 amino |
| Probe-InvT | 5′-GGAGAAGTACTTCTACGAGCCCGAAGACATC-3′ | Reverse T |
Unmodified oligonucleotides were used as primers and oligonucleotides with 3′ modifications were used as probes. The underlined As in the probe sequences represent mismatches in the probe sequence.
All probes used for blocking efficiency experiments were resuspended on day 0 in RNase/DNase-free water (Quality Biological, Gaithersburg, MD) and adjusted to a 200-μmol/L concentration based on OD260 measurements. Each primer was split into two 150-μl aliquots and processed by adding 150 μl of water or 150 μl of 2× TE (20 mmol/L Tris-Cl, pH 8.0, and 2 mmol/L ethylenediaminetetraacetic acid) to obtain a final concentration of 100 μmol/L in water or 1× TE. Oligonucleotides were further split into 20-μl aliquots for subsequent experiments.
PCR Reactions and Melting Analysis
PCR reactions were performed in a LightCycler (Roche Diagnostics, Indianapolis, IN). Each reaction consisted of 1× Roche FastStart DNA hybridization mix (includes dNTPs, dUTP, and 1 mmol/L MgCl2), 0.028 μmol/L forward primer 10F, 0.25 μmol/L reverse primer 10R, 0.5 μmol/L probe, 1× LCGreen Plus, an additional 2 mmol/L MgCl2, 10 U/ml uracil DNA glycosylase (Roche), and ∼30 ng (∼14 zmol) of hgDNA in a 10-μl reaction volume. Cycling conditions were performed using the following protocol at a transition rate of 20°C per second: (55°C (10:00) + 95°C (10:00) + [95°C (0:01) + 62°C (0:01) + 72°C (0:10)] × 55 cycles + 95°C (0:00) + 40°C (0:20) + 50°C → 95°C at 0.5°C per second + 40°C (0:00)]), where (0:00) is equal to (minutes:seconds). Melting analysis after amplification was also evaluated using a high-resolution melting analysis platform (HR-1; Idaho Technology, Salt Lake City, UT) with the following melting protocol: 4°C (≥10:00) + 60°C → 95°C at 0.3°C per second.
Cloning of PCR Products
PCR products were amplified using the previously described cycling conditions with Roche FastStart DNA polymerase without dUTP. The PCR products were ethanol-precipitated, run on a 4% agarose gel, bands excised, and gel-purified using a QIAQuick spin gel purification kit (Qiagen, Valencia, CA). The gel-purified bands were cloned into pCRII TOPO TA (Invitrogen, Carlsbad, CA) by adding 4 μl of gel-purified amplicon to 1 μl of pCRII TOPO TA plasmid and 1 μl of 6× salt solution (1.2 mol/L NaCl and 0.06 mol/L MgCl2) and allowed to incubate at room temperature for 30 minutes. The cloning solution was transformed into chemically competent TOP10 cells (Invitrogen), following the manufacturer’s protocol, plated onto LB-AMP agar plates (50 μg/ml ampicillin), and incubated at 37°C overnight. Single colonies were picked from plates and grown up in 50 μg/ml LB-AMP overnight. Plasmid DNA was isolated using a Qiagen QIAprep spin miniprep kit before sequencing.
Estimation of Blocking Efficiency
The probe blocked with an amino-C6 modification showed limited detectable extension by real-time PCR and was used as a theoretical 100% blocked primer in primer-mixing experiments. Ratios of unblocked probe-OH to probe-C6 were used in real-time PCR reactions to estimate percent extension of all blocked probes using the previously described reaction conditions. Percentages of probe-OH to probe-C6 tested were 1, 2, 3, 4, 5, 10, 25, 50, and 75% (0.500 μmol/L combined total probe concentration). The percentage of blocked probe was estimated by measuring band pixel density using Gel Doc Quantity One software (Bio-Rad, Hercules, CA) and/or by interpolation using HR-1 software.
Freeze-Thaw Experiments
Oligonucleotides resuspended in water or TE were tested for blocking stability after multiple freeze-thaw cycles. Baseline blocking efficiency using real-time PCR and HR-1 melting analysis was performed and analyzed using probe-OH to probe-C6 as previously described. Precautions were taken to ensure all oligos were subjected to the same environment conditions and processed in parallel. A freeze-thaw cycle consisted of placing oligonucleotides at −80°C for 10 minutes and transferring to a water bath at 22°C for 5 minutes. Once an aliquot was used for a real-time experiment, it was excluded from the remaining experiments unless otherwise specified. Oligonucleotides were frozen and removed for each probe (water and 1× TE) and tested for blocking efficiency after 1, 20, 30, 40, and 50 freeze-thaw cycles.
Time Course Experiments
Baseline blocking efficiency using real-time PCR and HR-1 melting analysis was performed and analyzed using probe-OH to probe-C6 as previously described. Oligonucleotides were placed at −20°C, 4°C, 22°C (room temperature), and 37°C for a time period of 1, 2, and 8 weeks. Blocking stability was estimated by mixing experiments for each time point as measured by real-time PCR and HR-1.
Results
Unlabeled Probe Melting Peak Analysis
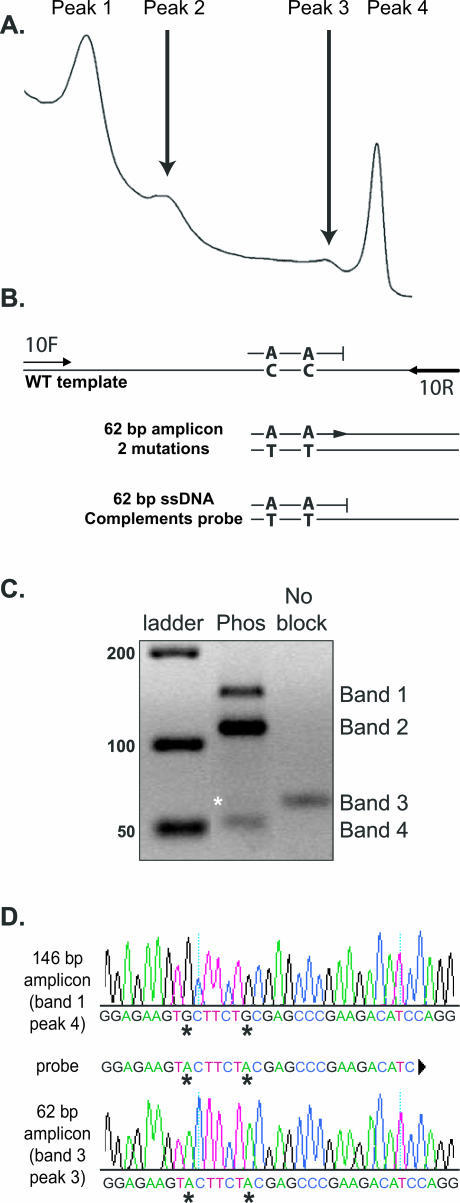
The melting profile for the unlabeled probe assay for exon 10 of the RET gene should yield only two melting peaks (Figure 1A), one corresponding to the unlabeled probe melting off the ssDNA generated during asymmetric PCR (peak 1) and the other resulting from dsDNA amplicon melting (peak 4). Peaks 2 and 3 are observed when the unlabeled probe is incompletely blocked. For these experiments, probe-phos—a probe with a 3′ phosphate block and two mismatches—was chosen to visualize peaks 2 and 3. If a fraction of probe-phos is unblocked, it will act as a forward primer in conjunction with primer 10R, creating a 62-bp amplicon demonstrated by peak 3. Peak 2 is observed when probe-phos melts off the ssDNA generated by unblocked probe-phos and 10R primers (Figure 1B). The probe-phos and mutated ssDNA bottom strand have 100% complementarity, which results in the higher melting temperature seen in peak 2 compared with peak 1. The hgDNA sample was cloned and confirmed to be wild type at the locus of interest (GenBank no. AJ243297). The HR melting profiles were identical between the hgDNA sample and the cloned plasmid, suggesting template complexity did not contribute to aberrant melting peaks. Experiments with additional templates, including other RET exons and herpes simplex virus, also displayed high-resolution melting anomalies and additional agarose gel bands when a poorly blocked unlabeled probe was used (S.D. and R.M., unpublished data).
Figure 1.
Unlabeled probe melting analysis of exon 10 of the RET gene. A: High-resolution melting profile observed with 3′ phosphate blocked unlabeled probe (probe-phos). B: Diagram of amplification products expected using primers 10F, 10R, and incompletely blocked probe-phos. C: Unlabeled probe reaction visualized on a 4% agarose gel. Products include 146-bp dsDNA using primers 10F and 10R (band 1), 146-nucleotide ssDNA (band 2), 62-bp amplicon derived from extension of unblocked probe-phos and 10R (band 3), and 62-nucleotide ssDNA (band 4). Lane Phos used probe-phos and is represented by the melting profile shown in A. Lane No block was run with probe-OH and 10R primers to demonstrate size differences between the 62-nucleotide ssDNA and 62-bp dsDNA amplicon (asterisk). The asterisk denotes the 62-bp probe-phos and 10R amplicon. D: Sequence of cloned unlabeled probe products. Sequence of 146-bp wild-type amplicon 10F and 10R (band 1, C) and the 62-bp probe-phos and 10R-derived amplicon (band 3, C). The amplification products were isolated from the same unlabeled probe reaction, demonstrating that the 62-bp amplicon was derived by extension of unblocked probe-phos.
The identities of each peak were further examined using primer-specific amplification and/or TOPO cloning. Peaks 1, 2, and 3 were detected only when unlabeled probe was present in the asymmetric real-time PCR. Removal of the probe results in the full-length amplicon melting peak only (peak 4). Melting signatures for peaks 3 and 4 (Figure 1A) were confirmed using probe-OH and 10R (peak 3) and 10F and 10R (peak 4). The probe-phos unlabeled probe and the probe-OH and 10R reactions were recovered and run on a 4% agarose gel (Figure 1C). All three bands in the Phos lane were gel purified and processed for cloning. The lower band (retrospectively designated band 4) did not migrate as expected. The 62-bp amplicon from probe-OH and 10R migrated at 62 bp (lane no block), but the expected 62-bp product from the probe-phos and 10R migrated at ∼55 bp (Figure 1C). We hypothesized that the 62-bp dsDNA band was not visible because the asymmetric, full-length ssDNA band migrated faster than the dsDNA band and had greater intensity than the full-length dsDNA (bands 1 and 2). Based on asymmetric primer mixing experiments with 10F and 10R, and probe-OH and 10R, it was shown that band 1 was the expected full-length 146 bp, band 2 was the 146-nucleotide ssDNA, band 3 was the expected 62 bp, and band 4 was the probe-derived ssDNA (Supplemental Figure 1A at http://jmd.amjpathol.org/). To obtain enough sample for cloning using probe-phos, a large-scale PCR reaction was performed, ethanol precipitated, examined on a gel, and gel extracted (Supplemental Figure 1B at http://jmd.amjpathol.org/). Only two bands could be cloned and sequenced: the 146-bp amplicon (band 1, primers 10F and 10R) and the 62-bp amplicon (band 3, probe-phos and 10R). The 146-bp amplicon was confirmed by sequence as wild type, and the 62-bp amplicon was verified to possess two mutations, confirming that the 62 nucleotide amplification product was derived from probe-phos (Figure 1D). Bands 2 and 4, the presumed ssDNA from the asymmetric amplification, could not be cloned or sequenced using primer 10R but could be digested by mung bean nuclease (Supplemental Figure 1C at http://jmd.amjpathol.org/).
Evaluation of 3′ Blocking Modifications
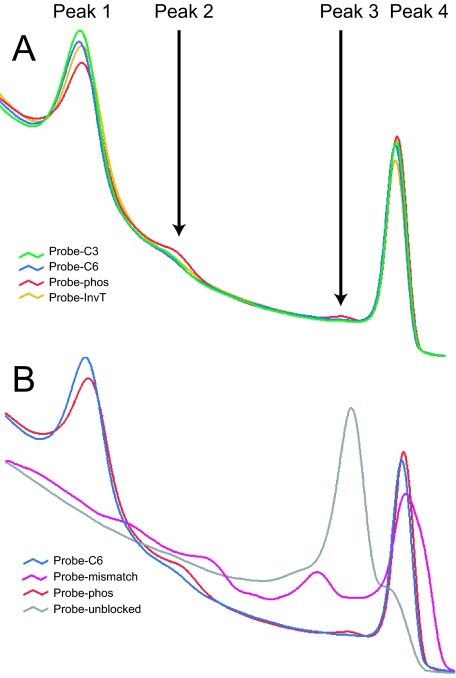
The C3, C6, phosphate, InvT, and single nucleotide mismatch-blocked oligonucleotides were tested on day 0 to establish baseline melting profiles and blocking efficiency. Melting analysis showed that C3-, C6-, and InvT-blocked probes provide the best inhibition of DNA polymerase extension, whereas probe-phos extended at a higher rate, represented by peaks 2 and 3 (Figure 2A). Both the 3′ single nucleotide mismatch and the unblocked probe extended readily (Figure 2B). All probes, regardless of 3′ block, exhibited some degree of extension as assessed by high-resolution melting analysis. There was no difference between oligonucleotides resuspended in water or TE on day 0.
Figure 2.
Day 0 high-resolution melting analysis of blocked probes. A: Probes blocked with C3, C6, and InvT provide the least amount of DNA polymerase extension on day 0, as shown by the heights of peaks 2 and 3. Probe-phos extension is higher as a new oligonucleotide and shown in red for comparison. B: Probe-mis, phos, C6, and probe-OH are shown for comparison. Probe-mis displays aberrant melting profiles and cannot be used for melting analysis.
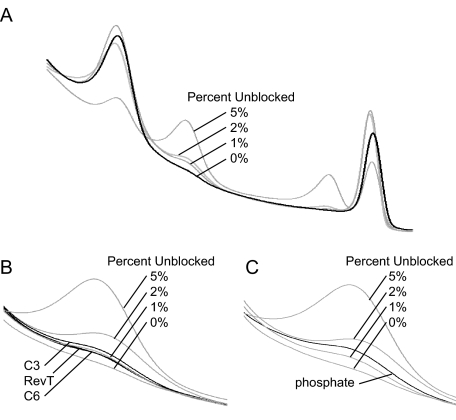
Blocking stability was estimated by mixing experiments using day 0 probe-C6 as a theoretical 100% blocked probe mixed with different percentages of the unblocked probe-OH, as described in Materials and Methods. Data for 50 freeze-thaw cycles are shown in Figure 3. The C6 block displays the least amount of extension (∼0.5%) followed by InvT, C3 (both at ∼0.75%), and phosphate at ∼1.5% (Figure 3, B and C). Experiments to determine whether oligonucleotides kept for 8 weeks at −20°C, 4°C, 22°C, and 37°C affected blocking efficiency also display the same general trend (data not shown). After 8 weeks, blocking efficiency of oligonucleotides resuspended in TE is minimally changed compared with day 0, regardless of temperature. In both the freeze-thaw and time-course experiments, oligonucleotides resuspended in water seemed to lose blocking efficiency at a higher rate (less than 0.20% based on mixing experiments) than oligonucleotides resuspended in TE (data not shown). Based on this data as well as other probe sets not reported, the C6 modification seems to possess the best overall blocking efficiency and 3′ stability compared with C3 and InvT.
Figure 3.
Estimation of blocking stability after 50 freeze-thaw cycles. A: Complete melting profile of unblocked to blocked probes representing 0, 1, 2, and 5% unblocked. B: Melting profiles of C3, RevT, and C6 compared with unblocked percentage. The best blocking stability is amino C6, followed by InvT, and C3. C: Melting profile of phosphate compared with unblocked percentages shows that phosphate is the least stable of all blocks tested.
Discussion
Assessment of Amplification Products
Known dsDNA products were generated using full-length amplicon primers 10F and 10R and the 62-bp probe-derived amplicon, probe-OH and 10R. The dsDNA melting signatures using HR-1 melting analysis and the agarose gel banding patterns were defined for each amplicon (peaks 3 and 4 and bands 1 and 3, respectively). With the probe-OH and 10R melting signature confirmed, experiments with probe-phos and 10R suggested that peak 3 was produced by an incompletely blocked probe. The presumed probe-derived 62-bp product was gel purified, cloned, sequenced, and shown to possess two mismatches introduced by probe-phos, in which the full-length amplicon from the same reaction had no mutations. Melting profiles for peaks 1 and 2 represent melting of the probe off ssDNA generated during asymmetric amplification.
Single-stranded DNA molecules cannot be detected using LCGreen Plus and HR-1 melting analysis. As a result, differing asymmetric primer ratios were used to monitor the shift of ssDNA to dsDNA on an agarose gel. Bands 2 and 4 were observed only when asymmetric ratios greater than 1:5 were used in the unlabeled probe reactions. Bands 2 and 4 could not be cloned or sequenced using primer 10R but could be digested by mung bean nuclease, supporting the hypothesis that the bands were single-stranded amplification products.
Blocking Efficiency and Stability
Mixing experiments with probe-OH and probe-C6 were used to estimate the amount of unblocked probe for each modification. By mixing, less than 1% of unblocked probe leads to amplification and generation of aberrant melting profiles. A previous report using a fluorescence resonance energy transfer-based system noted the lower quality and higher instability of the 3′ phosphate block compared with a C3 block.12 We observed polymerase extension using both the phosphate- and C3-blocked oligonucleotides and wanted to determine whether there was a block more suitable for unlabeled probe assays. We evaluated multiple lots of phosphate-blocked probes in unlabeled probe experiments and each has displayed varying day 0 blocking efficiencies—from less than 1% to almost 2% unblocked based on estimates (data not shown). Day 0 melting profiles for C3-, C6-, and InvT-blocked probes show the best inhibition of DNA polymerase extension, whereas probe-phos routinely extends at a higher rate. Other hot-start DNA polymerases, such as Platinum Taq (Invitrogen), have been used for unlabeled probe assays and display similar melting profiles, suggesting that extension of blocked probes is not FastStart-specific (data not shown). Both the single 3′ single nucleotide mismatch and unblocked probe extend readily.
The stability of the different blocks in water and TE was examined by multiple freeze-thaw cycles and time course experiments. Only after 50 freeze-thaw cycles was a difference in stability between the C6, InvT, and C3 blocks detectable. Probes resuspended in water were marginally less stable than their TE counterparts. Similar results were observed with probes kept at −20°C, 4°C, 22°C, and 37°C for 8 weeks. There was no difference in DNA polymerase extension for C6, InvT, C3, and phosphate between day 0 and week 8—even with probes kept at 37°C. Amino C6, InvT, and C3 all display excellent initial blocking quality. Freeze-thaw stability shows that amino C6 and InvT seem the most stable, whereas C3 and phosphate degrade at a higher rate. However, C3 after 50 freeze-thaw cycles will prevent DNA polymerase extension better than a new phosphate-blocked oligonucleotide.
Probes with single nucleotide 3′ mismatches routinely extend but display a depressed crossing threshold (CT), suggesting that the addition of a mismatch affects amplification efficiency (data not shown). Because FastStart DNA polymerase has no detectable 3′ exonuclease activity (Roche Package Insert. FastStart Taq DNA Polymerase, December ed. Mannheim, Germany, 2005, p 20),13 the exact mechanism of extension remains unclear. This observation raises concerns about assays that require single-nucleotide mismatches such as genotype-specific amplification in multiplex PCR reactions.14,15 Limiting the total number of PCR cycles, using locked nucleic acids, or using two mismatches at positions 3′-0 and 3′-2 could prove useful for reducing unwanted amplification based on 3′ mismatches.16,17,18 To avoid unwanted amplification off the probe in an unlabeled probe assay, a single nucleotide 3′ mismatch should not be used for a 3′ block.
Blocked oligonucleotides are required for many types of experiments, including unlabeled probe assays, fluorescence resonance energy transfer analysis, single-stranded tailing, and the prevention of 3′ exonuclease activity, among others.19,20,21,22 One advantage of unlabeled probes is that they are less expensive than fluorescently labeled probes. For example, the cost of an unlabeled probe with a C6 amino modifier is only 5 to 10% that of an adjacent hybridization probe pair used in fluorescence resonance energy transfer melting analysis. Unlabeled probes are especially useful in assay development, because the low cost of the probes allows a researcher to order multiple oligonucleotides for the cost of one fluorescence resonance energy transfer probe pair. However, when established assays are considered, the cost of LCGreen Plus (required with unlabeled probes) makes the running costs of both assays similar. The blocks used for these experiments were chosen based on affordability, compared with other 3′ blocks, and universal availability. Other 3′ modifications that have distinct functions other than blocking, such as a fluorophore, quencher, or dsDNA-binding enhancer, were not examined.
These experiments defined the products associated with aberrant melting peaks, proposed solutions to eliminate and/or minimize unwanted melting profiles, and established which blocks performed the best in an unlabeled probe assay as new oligonucleotides. Further experimentation defined the minimum amount of probe extension needed to produce aberrant melting profiles and examined the stability of the given oligonucleotides in both TE and water. The blocked oligonucleotides used for these experiments were examined by high-resolution melting analysis and defined to possess extension based on detectable melting profiles.
To minimize aberrant melting profiles in an unlabeled probe assay, amino C6 and InvT are the best oligonucleotide blocks to prevent DNA polymerase extension, followed by C3 (based on stability experiments), and finally phosphate (based on initial oligonucleotide quality and stability). The use of a 3′ mismatch, commonly used for allele-specific PCR, should be avoided to prevent unwanted polymerase extension. Although the exact mechanism of blocking stability was not examined, the initial quality of the oligonucleotide seems to be more important than routine handling as defined by multiple freeze-thaws and time-course experiments. General handling and storage guidelines supplied with oligonucleotides, such as resuspension in TE, should be followed to help prevent long-term stability issues.
Supplementary Material
Acknowledgments
We thank Maria Erali for comments regarding the manuscript.
Footnotes
S.D. and R.L.M. contributed equally to this article.
Supplemental material for this article can be found on http://jmd.amjpathol.org/.
Aspects of high-resolution melting analysis are licensed from the University of Utah to Idaho Technology. C.T.W. holds equity interest in Idaho Technology.
References
- Graham R, Liew M, Meadows C, Lyon E, Wittwer CT. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295–1298. doi: 10.1373/clinchem.2005.051516. [DOI] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Highsmith WE, Holtegaard LM, Wittwer CT. Mutation scanning of the RET protooncogene using high-resolution melting analysis. Clin Chem. 2006;52:138–141. doi: 10.1373/clinchem.2005.052951. [DOI] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Wittwer CT. Masking selected sequence variation by incorporating mismatches into melting analysis probes. Hum Mutat. 2006;27:269–278. doi: 10.1002/humu.20290. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang L, Palais R, Pryor R, Wittwer CT. High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin Chem. 2005;51:1770–1777. doi: 10.1373/clinchem.2005.054924. [DOI] [PubMed] [Google Scholar]
- Zhou L, Myers AN, Vandersteen JG, Wang L, Wittwer CT. Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin Chem. 2004;50:1328–1335. doi: 10.1373/clinchem.2004.034322. [DOI] [PubMed] [Google Scholar]
- Liew M, Nelson L, Margraf R, Mitchell S, Erali M, Mao R, Lyon E, Wittwer C. Genotyping of human platelet antigens 1 to 6 and 15 by high-resolution amplicon melting and conventional hybridization probes. J Mol Diagn. 2006;8:97–104. doi: 10.2353/jmoldx.2006.050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Eng C. RET proto-oncogene in the development of human cancer. J Clin Oncol. 1999;17:380–393. doi: 10.1200/JCO.1999.17.1.380. [DOI] [PubMed] [Google Scholar]
- Santoro M, Melillo RM, Carlomagno F, Vecchio G, Fusco A. RET: normal and abnormal functions. Endocrinology. 2004;145:5448–5451. doi: 10.1210/en.2004-0922. [DOI] [PubMed] [Google Scholar]
- Ortigão JF, Rosch H, Selter H, Frohlich A, Lorenz A, Montenarh M, Seliger H. Antisense effect of oligodeoxynucleotides with inverted terminal internucleotidic linkages: a minimal modification protecting against nucleolytic degradation. Antisense Res Dev. 1992;2:129–146. doi: 10.1089/ard.1992.2.129. [DOI] [PubMed] [Google Scholar]
- Cradic KW, Wells JE, Allen L, Kruckeberg KE, Singh RJ, Grebe SK. Substitution of 3′-phosphate cap with a carbon-based blocker reduces the possibility of fluorescence resonance energy transfer probe failure in real-time PCR assays. Clin Chem. 2004;50:1080–1082. doi: 10.1373/clinchem.2004.033183. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li K, Pardinas JR, Sommer SS, Yao KT. Proofreading genotyping assays mediated by high fidelity exo+ DNA polymerases. Trends Biotechnol. 2005;23:92–96. doi: 10.1016/j.tibtech.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- Landegren U, Nilsson M, Kwok PY. Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res. 1998;8:769–776. doi: 10.1101/gr.8.8.769. [DOI] [PubMed] [Google Scholar]
- Latorra D, Campbell K, Wolter A, Hurley JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- Di Giusto DA, King GC. Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: implications for genotyping assays. Nucleic Acids Res. 2004;32:1–8. doi: 10.1093/nar/gnh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp AC, Pinsonneault JK, Cooke G, Sadee W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques. 2003;34:1068–1072. doi: 10.2144/03345dd03. [DOI] [PubMed] [Google Scholar]
- Behlke MA, Dames SA, McDonald WH, Gould KL, Devor EJ, Walder JA. Use of high specific activity StarFire oligonucleotide probes to visualize low-abundance pre-mRNA splicing intermediates in S. pombe. Biotechniques. 2000;29:892–897. doi: 10.2144/00294pf01. [DOI] [PubMed] [Google Scholar]
- Zendegui JG, Vasquez KM, Tinsley JH, Kessler DJ, Hogan ME. In vivo stability and kinetics of absorption and disposition of 3′ phosphopropyl amine oligonucleotides. Nucleic Acids Res. 1992;20:307–314. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper HB, Reed MW, Cox T, Virosco JS, Adams AD, Gall AA, Scholler JK, Meyer RB., Jr Facile preparation of nuclease resistant 3′ modified oligodeoxynucleotides. Nucleic Acids Res. 1993;21:145–150. doi: 10.1093/nar/21.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JP, Kent K, Bird J, Fishback J, Froehler B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991;19:747–750. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.