Abstract
MicroRNAs (miRNAs) are a recently discovered class of endogenous, small, noncoding RNAs that regulate gene expression. Although miRNAs are highly expressed in the heart, their roles in heart diseases are currently unclear. Using microarray analysis designed to detect the majority of mammalian miRNAs identified thus far, we demonstrated that miRNAs are aberrantly expressed in hypertrophic mouse hearts. The time course of the aberrant miRNA expression was further identified in mouse hearts at 7, 14, and 21 days after aortic banding. Nineteen of the most significantly dysregulated miRNAs were further confirmed by Northern blot and/or real-time polymerase chain reaction, in which miR-21 was striking because of its more than fourfold increase when compared with the sham surgical group. Similar aberrant expression of the most up-regulated miRNA, miR-21, was also found in cultured neonatal hypertrophic cardiomyocytes stimulated by angiotensin II or phenylephrine. Modulating miR-21 expression via antisense-mediated depletion (knockdown) had a significant negative effect on cardiomyocyte hypertrophy. The results suggest that miRNAs are involved in cardiac hypertrophy formation. miRNAs might be a new therapeutic target for cardiovascular diseases involving cardiac hypertrophy such as hypertension, ischemic heart disease, valvular diseases, and endocrine disorders.
MicroRNAs (miRNAs) are a recently discovered class of endogenous, small, noncoding RNAs that regulate gene expression.1,2,3 Mature miRNAs are the result of sequential processing of primary transcripts (pri-miRNAs) mediated by two RNase III enzymes, Drosha and Dicer.4 Mature 18- to 24-nucleotides-long miRNAs negatively regulate protein expression of specific mRNA by either translational inhibition or mRNA degradation.5 Currently, more than 400 miRNAs have been cloned and sequenced in humans, and the estimated number of miRNA genes is as high as 1000 in the human genome.6,7 As a group, miRNAs are estimated to regulate 30% of the genes of the human genome.8
Analogous to the first RNA revolution in the 1980s with Zaug and Cech9 discovering the enzymatic activity of RNA, this recent discovery of RNAi and miRNA may represent the second RNA revolution.10 Large scale cDNA sequencing and genome tiling array studies have shown that ∼50% of genomic DNA in humans is transcribed, of which 2% is translated into proteins and the remaining 98% is noncoding RNAs (ncRNAs). The term ncRNA is commonly used for RNA that does not encode a protein, but this does not mean that such RNAs do not contain information or have function.11 Indeed, Zaug and Cech9 first reported the enzymatic activity of RNA in the 1980s. More excitingly, with the finding of RNAi technology,12 two regulatory small ncRNAs were discovered, small interfering RNAs (siRNAs) and miRNAs.1,13,14 siRNAs and miRNAs have a similar mechanism for gene expression regulation; however, they are different from each other.13,14 The chief difference lies in their origins.14,15 siRNAs are produced from long double-stranded (bimolecular) RNAs or long hairpins, often of exogenous origin, and usually target sequences at the same locus or elsewhere in the genome for destruction (gene silencing),16,17 the phenomenon termed RNAi.12 In contrast, miRNAs are endogenous. They are encoded within the genome and come from endogenous short hairpin precursors and usually target sequences at other loci. Therefore, miRNAs are more important because they are endogenous regulators for gene expression.
We are just beginning to understand how this novel class of gene regulators is involved in biological functions. Although only a small number of the hundreds of identified miRNAs have been characterized, a growing body of exciting evidence suggests that miRNAs are important regulators for cell growth, differentiation, and apoptosis.14,18,19 Therefore, miRNAs may be important for normal development and physiology. Consequently, dysregulation of miRNA function may lead to human diseases.20 In this respect, the most exciting research area is the role of miRNAs in cancer, given that cell dedifferentiation, growth, and apoptosis are important cellular events in the development of cancer. Indeed, both basic and clinical studies have demonstrated that miRNAs are aberrantly expressed in diverse cancers.21,22,23,24 miRNAs are currently thought to function as both tumor suppressors and oncogenes.25
Cardiovascular disease has long been the leading cause of death in developed countries, and it is rapidly becoming the number one killer in developing countries.26 Cardiac hypertrophy, the common pathological response to a number of cardiovascular diseases such as hypertension, ischemic heart disease, valvular diseases, and endocrine disorders, is a major determinant of mortality and morbidity in cardiovascular diseases. Although miRNAs are highly expressed in the heart, the roles of these miRNAs in cardiovascular diseases including cardiac hypertrophy are still unclear.20,27 Because cardiac cell growth (hypertrophy) is the key cellular event in the formation of cardiac hypertrophy, we therefore hypothesized that expression of miRNAs in hypertrophic heart may be different from that in normal heart and these aberrantly expressed miRNAs may play important roles in cardiac hypertrophy.
Materials and Methods
Cardiac Hypertrophy Animal Model
To determine the expression changes of miRNAs in hypertrophic hearts, we applied a well-established mouse cardiac hypertrophy model by aortic banding as described.28,29,30 In brief, 12-week-old C576BJ mice were anesthetized with ketamine (80 mg/kg i.p.) and xylazine (5 mg/kg i.p.). Under sterile conditions, aortic binding constriction was performed under a dissecting microscope by ligating the abdominal aorta between the diaphragm and renal arteries with a blunted 27-gauge needle and a 6-0 silk suture. After that, the needle was quickly removed. Age-matched mice with a sham surgical operation will be used as a control group. All protocols were approved by the Institutional Animal Care and Use Committee at the University of Tennessee and were consistent with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 85-23, revised 1985).
Evaluation of Cardiac Hypertrophy in Vivo
Cardiac hypertrophy was evaluated by histopathological analysis of heart size, the ratio of heart weight to body weight (HW:BW), and cardiomyocyte size in heart cross-section.28,29,30 After sacrifice with an overdose of pentobarbital, the hearts were carefully isolated. The HW was measured, and the HW:BW ratio was then calculated. The hearts were then put into 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned. Heart sections cut at the level of the papillary muscle were selected and used to measure the cardiomyocyte size by fluorescein isothiocyanate-labeled wheat germ agglutinin.31 Briefly, the sections were deparaffinized, rehydrated, and incubated for 1 hour at room temperature with fluorescein isothiocyanate-labeled wheat germ agglutinin (1:500 dilution; Vector Laboratories, Inc., Burlingame, CA) to visualize myocyte membranes. The images were taken with a fluorescence microscope, and morphometric analysis was performed with software (Scion Image CMS-800; Scion Corp., Frederick, MD). Mean value of cell size was calculated by using the measurements from 100 cells in an individual mouse.
Cardiac Myocyte Culture and Cell Models for Cardiac Myocyte Hypertrophy
Primary cultures of neonatal rat cardiac ventricular myocytes were performed as described.31,32 In brief, hearts from 1- to 2-day-old Sprague-Dawley rats were removed after hypothermia anesthesia immersion in ice water and placed in ice-cold 1× phosphate-buffered saline solution. After repeated rinsing, the atria were cut off, and the ventricles were minced with scissors. The minced tissue and ventricular cells were dispersed by digestion with collagenase type IV (0.45 mg/ml; Sigma, St. Louis, MO), 0.1% trypsin (Life Technologies, Inc., Grand Island, NY), and 15 μg/ml DNase I (Sigma). Cardiomyocytes (0.33 × 106 cells/ml) were cultured in the cardiac myocyte culture medium containing Dulbecco’s modified Eagle’s medium/F-12 supplemented with 5% horse serum, 4 μg/ml transferrin, 0.7 ng/ml sodium selenite (Life Technologies, Inc.), 2 g/L bovine serum albumin (fraction V), 3 mmol/L pyruvic acid, 15 mmol/L HEPES, 100 μmol/L ascorbic acid, 100 μg/ml ampicillin, 5 μg/ml linoleic acid, 1% penicillin and 1% streptomycin, and 100 μmol/L 5-bromo-2′-deoxyuridine (Sigma), and seeded into six-well plates. Two cell models for cardiac myocyte hypertrophy were applied as described.32,33 The first model is cardiac myocyte hypertrophy stimulated with angiotensin II (Ang II) (1 μm),31 and the second model is hypertrophy stimulated with phenylephrine (PE) (10 μm)32 in the serum-free medium. The stimulation lasted 48 hours.
Evaluation of Cardiac Myocyte Hypertrophy in Vitro
The extent of cardiac myocyte hypertrophy was evaluated by two parameters: [3H]leucine incorporation assay for protein synthesis and cell surface area measurement. Protein synthesis was determined by [3H]leucine incorporation assay.31,32 After 42 hours of treatment with Ang II, PE, or vehicle, [3H]leucine (1.0 μCi/ml) was added into the culture medium, and the cells were further incubated for another 6 hours. After that, cells were washed three times with ice-cold 1× phosphate-buffered saline and incubated with 10% trichloroacetic acid for 1 hour at 4°C to precipitate the proteins. The precipitates were washed twice with 95% ethanol, and then dissolved and scraped in 1 mol/L NaOH. The resulting solution, containing the trichloroacetic acid-insoluble fraction, was neutralized with 1 mol/L HCl, and the radioactivity was counted in a liquid scintillation counter. For cell surface area measurement, cell images were taken with a digital camera (Nikon, Melville, NY) fixed to a microscope (Nikon).31,32 Then, the cardiomyocyte surface area was analyzed with an image processing and analysis system (Scion Image CMS-800). One hundred cells from randomly selected fields in one well were examined, and the average cell surface area was used.
Gene Microarray Analysis for miRNA Expression
miRNAs were isolated from mouse hearts using the mirVana miRNA isolation kit (Ambion, Inc., Austin, TX). miRNA expression profiling was determined by miRNA microarray analysis using the mouse miRNA array probes (Chip ID miMouse7.1 version; LC Science, Houston, TX) that include 233 mature mouse miRNAs.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
miRNA levels were determined by qRT-PCR. Briefly, RNAs from both mouse hearts and neonatal rat cardiac ventricular myocytes were isolated with the mirVana miRNA isolation kit (Ambion, Inc.). qRT-PCR was performed on cDNA generated from 200 ng of total RNA by using the protocol of the mirVana qRT-PCR miRNA detection kit (Ambion, Inc). Amplification and detection of specific products were performed with the ABI Prism 7700 sequence detection system with the cycle profile according to the mirVana qRT-PCR miRNA detection kit. As an internal control, U6 primers were used for RNA template normalization. Fluorescent signals were normalized to an internal reference, and the threshold cycle (Ct) was set within the exponential phase of the PCR. The relative gene expression was calculated by comparing cycle times for each target PCR. The target PCR Ct values were normalized by subtracting the U6 Ct value, which was given as the ΔCt value. The relative expression level between treatments was then calculated using the following equation: relative gene expression = 2− (ΔCt sample − ΔCt control).33
Northern Blot Analysis of miRNA
Ten μg of total RNA from snap-frozen tissues and cardiac myocytes were loaded onto a precast 15% denaturing polyacrylamide gel (Bio-Rad, Hercules, CA). The RNA was then electrophoretically transferred to Bright-Star blotting membranes (Ambion, Inc.). The probe sequences of these miRNAs were shown in Table 1. Probes were end-labeled with [γ-32P]ATP by T4 polynucleotide kinase. Prehybridization and hybridization were performed in Ultrahyb Oligo solution (Ambion, Inc.) containing 106 cpm/ml probes overnight at 37°C. The most stringent wash was with 2× standard saline citrate and 1% sodium dodecyl sulfate at 37°C. For reuse, blots were stripped by boiling and reprobed. U6 was used as a loading control to normalize expression levels.
Table 1.
Probe Sequences for Northern Blot Analysis
| miRNA | Probe sequence |
|---|---|
| miR-21 | 5′- TCAACATCAGTCTGATAAGCTA-3′ |
| miR-27a | 5′-GCGGAACTTAGCCACTGTGAA-3′ |
| miR-27b | 5′-GCAGAACTTAGCCACTGTGAA-3′ |
| miR-29a | 5′-AACCGATTTCAGATGGTGCTA-3′ |
| miR-29c | 5′-ACCGATTTCAAATGGTGCTA-3′ |
| miR-30e | 5′-ACATTTGTAGGAACTGACCT-3′ |
| miR-150 | 5′-CACTGGTACAAGGGTTGGGAGA-3′ |
| Mir-185 | 5′-GAACTGCCTTTCTCTCCA-3′ |
| miR-214 | 5′- CTGCCTGTCTGTGCCTGCTGT-3′ |
| miR-341 | 5′-ACTGACCGACCGACCGATCGA-3′ |
| miR-424 | 5′-TTCAAAACATGAATTGCTGCTG-3′ |
| miR-451 | 5′-AACTCAGTAATGGTAACGGTTT-3′ |
| miR-486 | 5′-AGGACATGACTCGACGGGGCTC-3′ |
| U6 | 5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′ |
Antisense Oligonucleotide for miR-21
Antisense oligonucleotide-mediated miRNA depletion was used to knock down miR-21. The antisense oligonucleotide for miR-21, 2′OMe-miR-21, or miRNA inhibitor, was modified at each nucleotide by an O-methyl moiety at the 2′-ribose position: 5′mUmCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmUmA-3′ (Integrated DNA Technologies, Coralville, IA).34 We also used a control modified antisense oligonucleotide for enhanced green fluorescence protein (EGFP) mRNA (2′OMe-EGFP): 5′-mAmAmGmGmCmAmAmGmCmUmGmAmCmCmCmUmGmAmAmGmU-3′ (Integrated DNA Technologies).34 Both oligos were labeled with a fluorescent dye. Oligo transfection was performed according to an established protocol.35 In brief, cells were transfected using transfection reagent (Qiagen, Valencia, CA) for 6 hours. Transfection complexes were prepared according to the manufacturer’s instructions, and 2′OMe-miR-21 or control oligo 2′OMe-EGFP was added directly to the complexes to a final oligonucleotide concentration of 1, 3, 10, 30, and 100 nmol/L. The transfection medium was replaced 6 hours after transfection by the regular culture medium.
Statistics
All data are presented as mean ± SE. For relative gene expression, the mean value of vehicle control group is defined as 100% or 1. Two-tailed unpaired Student’s t-tests and analysis of variance were used for statistical evaluation of the data. SigmaStat Statistical Analysis Program (San Jose, CA) was used for data analysis. A P value <0.05 was considered significant.
Results
miRNAs Are Highly Expressed in Mouse Heart
Tissue-specific expression is one important characteristic of miRNA expression.19 To study the biological functions of miRNA in cardiac hypertrophy, we first determined the miRNA expression profile in mouse hearts through miRNA microarray analysis. We isolated the total RNA from six normal mouse (C576BJ) hearts, and the miRNA microarray analysis was performed by using mouse miRNA array probes (Chip ID miMouse 7.1 version; LC Science) that include 233 mature mouse miRNAs (Sanger v. 7.0; Hinxton, UK). Overall, 157 miRNAs of 233 arrayed miRNAs were found in normal mouse hearts, and 64 of these were highly expressed (Table 2).
Table 2.
miRNAs Highly Expressed in Normal Mouse Heart
| miR-1 | miR-126-3p | let-7a | miR-30c | miR-26a | let-7f |
| miR-133b | let-7d | miR-23b | miR-30b | miR-23a | let-7b |
| miR-125b | let-7g | miR-29a | miR-24 | miR-451 | miR-30a-5p |
| let-7e | miR-30d | miR-98 | miR-16 | let-7i | miR-145 |
| miR-125a | miR-99a | miR-27a | miR-126-5p | miR-195 | miR-22 |
| miR-143 | miR-29c | miR-191 | miR-185 | miR-30e | miR-486 |
| let-7c | miR-21 | miR-152 | miR-27b | mir-214 | miR-146 |
| miR-133a | miR-15a | miR-341 | miR-100 | miR-29b | miR-103 |
| miR-26b | miR-424 | miR-148a | miR-150 | miR-149 | miR-181a |
Aberrant Expression of miRNAs in Mouse Hypertrophic Hearts
The successful aortic banding was first confirmed before animal sacrifice by pressure gradient across the banding site by using two Millar catheters. The banding group had a pressure gradient at 28 ± 5 mm Hg, whereas no pressure gradient was found in sham controls. The animals were sacrificed at 7, 14, and 21 days after surgery. Every time point had at least six mice in each group. As expected, aortic banding led to a rapid and progressive increase in heart size, whereas no effect on heart size from the sham surgical operation was found during the following 3 weeks. HW and HW:BW in the banded group were increased accordingly (Figure 1A). At the end of 3 weeks, the heart mass had increased by ∼60%. The time course of cardiac hypertrophy-induced aortic banding was consistent with previous reports.28,29,30,36 To evaluate further cardiac hypertrophy in vivo at the cellular level, we measured cardiomyocyte size in heart cross sections by fluorescein isothiocyanate-labeled wheat germ agglutinin staining of cell membranes. As shown in Figure 1B, the left ventricular heart cells were significantly larger in the banded group than those in the sham control group. All of the sham groups had no hypertrophy changes both at organ and cellular levels.
Figure 1.
Aortic banding induces cardiac hypertrophy in mice. A: Quantitative analysis of the ratio of HW (mg) to BW (g) (HW/BW) after aortic banding. B: Quantitative analysis of cardiac myocyte size in heart cross sections. *P < 0.05 compared with sham control.
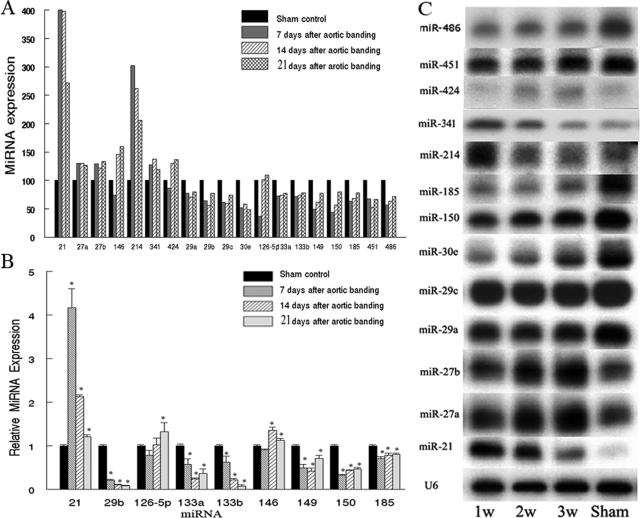
To determine the potential involvement of miRNAs in cardiac hypertrophy, we used microarray analysis to determine miRNA levels in mouse after aortic banding and compared them with levels in age-matched sham control group. Every time point had six mice in each group. We were excited to find that, compared with the sham group, aberrant miRNA expression was a remarkable characteristic in hypertrophic hearts. Seven days after aortic banding, 102 of the 157 heart miRNAs were differentially expressed (P < 0.01); 50 miRNAs were up-regulated, and 52 miRNAs were down-regulated. At 14 days after aortic banding, 69 of the 157 heart miRNAs were differentially expressed (35 up-regulated and 34 down-regulated), whereas at 21 days after aortic banding, 73 of the 157 heart miRNAs were differentially expressed (43 up-regulated and 30 down-regulated). Figure 2A shows the time course changes of miRNAs that are highly expressed in heart and more than 30% dysregulated (down- or up-regulated) after aortic banding. All of the differentially expressed miRNAs at these three time points are listed in the Supplementary Table 1 at http://ajp.amjpathol.org.
Figure 2.
Aberrant expression of miRNAs in mouse hypertrophic hearts. A: Time course changes of miRNAs that are highly expressed in mouse hearts and more than 30% dysregulated (down- or up-regulated) after aortic banding determined by microarray analysis. B: Confirmation of the aberrantly expressed miRNAs after aortic banding by qRT-PCR. C: Confirmation of the aberrantly expressed miRNAs after aortic banding by Northern blot. Note: miRNA levels in sham control hearts are defined as 100. *P < 0.05 compared with those in sham controls.
Confirmation of the Aberrant miRNA Expression in Hypertrophic Hearts by Northern Blot Analysis and/or qRT-PCR
These miRNA genes with expression significantly dysregulated from microarray results were selected for confirmation by Northern blot and/or qRT-PCR. In agreement with the results from microarray analysis, we found that miR-21, -27a, -27b, -146, -214, -341, and -424 were highly up-regulated, whereas miR-29a, -29b, -29c, -30e, -126-5p, -133a, -133b, -149, -150, -185, -451, and -486 were significantly down-regulated after aortic banding (Figure 2, B and C). Remarkably, miR-21 increased more than fourfold compared with the control. Therefore, miR-21 was the selected experimental miRNA using our in vitro cell hypertrophy models. miRNA probes were purchased from Proligo-Sigma (Houston, TX), and their sequences are listed in Table 1. It should be noted that some probes such as miR27a and miR-21b only differ by one base in sequence, so it is likely that Northern analysis might not distinguish between the different isoforms. In that case, Northern blot may be the result of the all of the isoforms of the miRNA. qRT-PCR primers were purchased from Ambion, Inc.
Aberrant miR-21 Expression in Cultured Cardiac Myocytes with Hypertrophy
To determine the potential biological role of these aberrant miRNAs in cardiac hypertrophy, we determined the expression levels of the most up-regulated miR-21 in cultured rat neonatal cardiac myocytes with and without cell hypertrophy. Both Ang II and PE stimulation resulted in the formation of cardiac myocyte hypertrophy. Compared with the in vivo study, a similar mRNA expression pattern was found in PE- and Ang II-stimulated hypertrophic cells (data not shown). Interestingly, miR-21 was the most up-regulated miRNA that was increased more than threefold compared with the control after stimulation with hypertrophic agents (Figures 3 and 4).
Figure 3.
The effect of miR-21 inhibitor 2′OMe-miR-21 on miR-21 expression in cultured cardiac myocytes with hypertrophy. A: Transfection of miR-21 inhibitor 2′OMe-miR-21 (30 nmol/L) and control oligonucleotide (2′OMe-EGFP) (30 nmol/L) labeled with a fluorescent dye (red color) into the cultured cardiac myocytes. B: The effects of miR-21 inhibitor 2′OMe-miR-21 on the expression levels of miR-21 in cultured cardiac myocytes with hypertrophy. Note: Two controls were used in the experiment. The first control was a vehicle control (PBS), and the second control was 2′OMe-EGFP. *P < 0.05 compared with vehicle control.
Figure 4.
The effect of miR-21 inhibitor 2′OMe-miR-21 on cardiac myocyte hypertrophy. A: Inhibition of miR-21 expression by miR-21 inhibitor 2′OMe-miR-21 (30 nmol/L). The mean MiR-21 level in vehicle-treated cells is defined as 1. B: The effect of miR-21 inhibitor 2′OMe-miR-21 on cardiac myocyte size in Ang II- or PE-treated cells. The mean myocyte size in vehicle-treated cells is defined as 1. C: The effect of miR-21 inhibitor 2′OMe-miR-21 on cardiac myocyte protein synthesis in Ang II- or PE-treated cells. The mean value of [3H]leucine incorporation in vehicle-treated cells are defined as 1. D: Representative cell pictures from different treatment groups. *P < 0.05 compared with vehicle control; #P < 0.05 compared with Ang II-treated group; P < 0.05 compared with PE-treated group.
Antisense Oligonucleotide for miR-21 Is Sufficient to Knock Down miR-21 Expression in Cultured Cardiomyocytes
To determine the effect of the up-regulated miR-21 on cell hypertrophy, we applied antisense oligonucleotide-mediated miRNA depletion using 2′OMe-miR-21 to knock down the overexpressed miR-21. We used two controls for this study. The first control was a vehicle control (phosphate-buffered saline, PBS), and the second control was the modified antisense oligonucleotide for EGFP mRNA (2′OMe-EGFP). EGFP gene is a mutant form of green fluorescence protein (GFP) gene. Neither EGFP nor GFP genes are expressed in rat. Thus, 2′OMe-EGFP targeting EGFP mRNA serves as a negative oligonucleotide control.34 As shown in Figure 3A, fluorescent-marked 2′OMe-miR-21 and 2′OMe-EGFP were successfully transfected into the cultured cardiac myocytes. Consistent with the transfection, 2′OMe-miR-21 decreased the miR-21 expression levels (Figure 3A) in a dose-dependent manner, with a significant decrease observed at a concentration of 3 nmol/L and the maximum effect at 100 nmol/L. In contrast, the control oligo, 2′OMe-EGFP, had no effect on miR-21 level, even at the highest concentration (100 nmol/L).
Down-Regulation of the Overexpressed miR-21 Has a Significant Negative Effect on Cardiomyocyte Hypertrophy
According to the dose response of miR-21 inhibitor, we selected 30 nmol/L as our experimental dose to determine the effect of miR-21 inhibition on cardiomyocyte hypertrophy. As shown in Figure 4A, miR-21 inhibitor, 2′OMe-miR-21 decreased miR-21 levels and inhibited myocyte hypertrophy stimulated by either Ang II or PE as shown by the decreased cell size (Figure 4B) and [3H]leucine incorporation (Figure 4C). In contrast, control oligos (2′OMe-EGFP) had no effect on miR-21 expression and cell hypertrophy. Representative cell images from different treatment groups are displayed in Figure 4D.
Discussion
Tissue-specific expression is one important characteristic of miRNA expression. In respect to the cardiovascular system, miRNA expression profile in the heart has been described.19 However, at that time, only a small number of miRNAs were found. In the current study, miRNA expression signature in hearts was further determined using microarray analysis, designed to detect the majority of mammalian miRNAs identified thus far. Indeed, miRNA expression profile in heart is different from that in vessel (our unpublished data). For example, the most abundant miRNAs in heart are miR-1, let-7, miR-133, miR-126-3p, miR-30c, and miR-26a. However, in artery the most abundant miRNAs are miR-145, let-7, miR-125b, miR-125a, miR-23, and miR-143. MiR-1 that is highly expressed in heart is not an abundant miRNA in artery (data not shown). The different expression profiles in different tissues indicate that the physiological functions of miRNAs in different tissues could be different. Identifying these tissue-specific miRNAs and their physiological functions could be important for future studies.
As a novel class of gene regulators, miRNAs play important roles not only in normal development and physiological conditions but also in disease status. In this respect, both basic and clinical studies have demonstrated that miRNAs are aberrantly expressed in diverse cancers.21,22,23,24 miRNAs are currently thought to function as both tumor suppressors and oncogenes.25
Aberrant cardiac cell growth (hypertrophy) is the key cellular event in the formation of cardiac hypertrophy. It is well established that multiple gene are aberrantly expressed in hypertrophic cardiac cells and these aberrantly expressed genes are responsible for the formation of cardiac cell hypertrophy.35 Because miRNAs are endogenous regulators for gene expression, it is reasonable to hypothesize that they may be involved in cardiac hypertrophy. We therefore applied a well-established mouse cardiac hypertrophy model by aortic banding to determine the miRNA expression signature in hypertrophic heart. We are excited to find that multiple aberrant miRNA expression was a remarkable characteristic in hypertrophic heart. The dysregulation and the time course changes of these multiple aberrantly expressed miRNAs match the complex process of cardiac hypertrophy formation in which multiple genes have been dysregulated.37 Determining the effects of these dysregulated miRNAs on cardiac hypertrophy is the prerequisite for this novel research field in cardiovascular diseases.
To determine the potential role of these aberrantly expressed miRNAs in cardiac hypertrophy, we selected an up-regulated miRNA, miR-21, as our first experimental target. Because of the lack of an effective in vivo inhibitor for miR-21 and the lack of availability of miR-21 knockout mice, we thus applied a cardiac myocyte hypertrophy model using the cells from neonatal rat heart. To avoid a drug-specific effect, we have used two different drug-stimulated cardiac cell hypertrophy models. We found that both Ang II and PE are able to induce cardiac myocyte hypertrophy as well as the up-regulation of miR-21 expression. Interestingly, inhibition of miR-21 expression is able to decrease cardiac myocyte hypertrophy stimulated by both Ang II and PE. Therefore, at the cellular level, we have confirmed that miR-21 is involved in cardiac hypertrophy.
There are two limitations in the current study. The detailed gene targets (mRNAs) responsible for miR-21-mediated effects on cardiac hypertrophy were not uncovered by this study. The mRNA targets of miRNAs are very complex as miRNAs are able to bind to their mRNA targets with either perfect or imperfect complementarity. Thus, one miRNA may have multiple mRNA targets. Although we have found that some hypertrophic genes such as ANF, β-MHC, and BNF are decreased by MiR-21 inhibition. However, the decrease is in a nonselective manner (data not shown). Thus, these hypertrophic genes may not be the direct targets for miR-21. Although the detailed mRNA targets responsible for miR-21-mediated effects on cardiac cells are currently unclear, based on the cellular effect of miR-21, we predict that it may target some genes that have anti-growth effects. Although these molecular mechanisms are important for the future studies, identification of the aberrant expression and the cellular effects of these miRNAs as reported in the current study should be the prerequisite for the long-term studies focused on the detailed molecular mechanisms. Another limitation is that we have only demonstrated that cellular effect in cultured cells in vitro. These cellular effects should be further confirmed in vivo with the development of potent specific miRNA inhibitors and miRNA knockout mice. In addition, the roles of other aberrantly expressed miRNAs need to be defined in future studies.
During the writing and submitting of our manuscript, another independent research study of miRNA expression profile in mouse heart hypertrophy has been published.38 Interestingly, they also demonstrated the aberrant expression of miRNAs during heart hypertrophy, in which multiple miRNAs were dysregulated. The results from two independent studies warranted that miRNAs are indeed involved in heart hypertrophy. The differences between our study and the Olson group’s38 study are as follows. First, we have determined the time course changes of miRNA expression during heart hypertrophy, whereas the Olson group has only determined the expression signature at 21 days after aortic banding. Second, we have used microarray, Northern blot, and qRT-PCR to determine the levels of miRNAs, whereas the Olson group has only used microarray and Northern blot. Third, we are focused on the most up-regulated miRNA, miR-21. However, the Olson group is focused on a less up-regulated miRNA, miR-195. We selected miR-21 as our experimental target for the following reasons: 1) it is highly expressed in heart, 2) it is the most up-regulated miRNA during the heart hypertrophy, and 3) it is also overexpressed in cancers.25,39 Thus, miR-21 might be a common regulator for cell growth. Fourth, the Olson group38 has used gain-of-function experiments as their major strategy to determine the potential roles of miRNAs in heart hypertrophy. In contrast, our major strategy is loss-of-function experiments via knocking down the up-regulated miRNAs. In addition, the Olson group38 has used overexpression experiment of miR-195 in vivo, whereas we failed to perform these in vivo studies for miR-21. The reasons for the last difference are as follows: 1) loss-of-function experiments are a good strategy to determine the role of a gene with up-regulation under pathological conditions; 2) there is lack of an established method to efficiently knock down heart miR-21 expression in vivo; and 3) we have failed to up-regulate miR-21 in vitro and in vivo using either miR-21 precursors (Ambion, Inc.) or virus vector-expressing miR-21 precursors. The same result is also shown in the Olson group’s38 reports in that they also failed to overexpress miR-21 using their virus vectors. We hypothesize that the processing of miR-21 precursors into mature miR-21 in normal heart cells may be special, although the posttranscriptional regulation for miR-21 is currently unclear.40 Establishing a method to overexpress miR-21 will be needed for the future study.
In summary, miRNAs are aberrantly expressed in hypertrophic hearts. The miRNA expression signature and antisense-mediated depletion reveal the involvement of miRNAs in cardiac hypertrophy formation. miRNAs may be a new therapeutic target for cardiovascular diseases with cardiac hypertrophy such as hypertension, ischemic heart disease, valvular diseases, and endocrine disorders.
Supplementary Material
Footnotes
Address reprint requests to Chunxiang Zhang, M.D., Ph.D., Cardiovascular Research Laboratory, Vascular Biology Center and Department of Surgery, University of Tennessee Health Science Center, 956 Court Ave., Coleman Building, A331, Memphis, TN 38163. E-mail: czhang1@utmem.edu.
Supported by the National Institutes of Health (grant HL080133), the American Heart Association (grant 0530106N), and the American Diabetes Association (grant 105JF60 to C.Z.).
Y.C., R.J., and J.Y. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Kong Y, Han JH. MicroRNA: biological and computational perspective. Genomics Proteomics Bioinformatics. 2005;3:62–72. doi: 10.1016/S1672-0229(05)03011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Derivation and function of small interfering RNAs and microRNAs. Virus Res. 2004;102:3–9. doi: 10.1016/j.virusres.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkhardt A, Fuchs U, Tuschl T. MicroRNA in chronic lymphocytic leukemia. N Engl J Med. 2006;354:524–525. doi: 10.1056/NEJMc053266. [DOI] [PubMed] [Google Scholar]
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- Van Herck PL, Vrints CJ, Carlier SG. Coronary circulation and interventional cardiology. Ann Biomed Eng. 2005;33:1735–1742. doi: 10.1007/s10439-005-8778-9. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Barbosa ME, Alenina N, Bader M. Induction and analysis of cardiac hypertrophy in transgenic animal models. Methods Mol Med. 2005;112:339–352. doi: 10.1385/1-59259-879-x:339. [DOI] [PubMed] [Google Scholar]
- Zou Y, Hiroi Y, Uozumi H, Takimoto E, Toko H, Zhu W, Kudoh S, Mizukami M, Shimoyama M, Shibasaki F, Nagai R, Yazaki Y, Komuro I. Calcineurin plays a critical role in the development of pressure overload-induced cardiac hypertrophy. Circulation. 2001;104:97–101. doi: 10.1161/01.cir.104.1.97. [DOI] [PubMed] [Google Scholar]
- Zahabi A, Picard S, Fortin N, Reudelhuber TL, Deschepper CF. Expression of constitutively active guanylate cyclase in cardiomyocytes inhibits the hypertrophic effects of isoproterenol and aortic constriction on mouse hearts. J Biol Chem. 2003;278:47694–47699. doi: 10.1074/jbc.M309661200. [DOI] [PubMed] [Google Scholar]
- Hu CM, Chen YH, Chiang MT, Chau LY. Heme oxygenase-1 inhibits angiotensin II-induced cardiac hypertrophy in vitro and in vivo. Circulation. 2004;110:309–316. doi: 10.1161/01.CIR.0000135475.35758.23. [DOI] [PubMed] [Google Scholar]
- Xia Y, Rajapurohitam V, Cook MA, Karmazyn M. Inhibition of phenylephrine induced hypertrophy in rat neonatal cardiomyocytes by the mitochondrial KATP channel opener diazoxide. J Mol Cell Cardiol. 2004;37:1063–1067. doi: 10.1016/j.yjmcc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Li H, Telemaque S, Miller RE, Marsh JD. High glucose inhibits apoptosis induced by serum deprivation in vascular smooth muscle cells via upregulation of Bcl-2 and Bcl-xl. Diabetes. 2005;54:540–545. doi: 10.2337/diabetes.54.2.540. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ha T, Gao X, Kelley J, Williams DL, Browder IW, Kao RL, Li C. NK-κB activation is required for the development of cardiac hypertrophy in vivo. Am J Physiol. 2004;287:H1712–H1720. doi: 10.1152/ajpheart.00124.2004. [DOI] [PubMed] [Google Scholar]
- Dorn GW, II, Hahn HS. Genetic factors in cardiac hypertrophy. Ann NY Acad Sci. 2004;1015:225–237. doi: 10.1196/annals.1302.019. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.