Abstract
The LRR receptor serine/threonine kinases are a major eukaryotic receptor family, for which the central regulatory mechanism of endosomal trafficking remains largely unadressed. We show that the steroid receptor BRI1 localizes to both plasma membrane and early endosomal compartments, even when observed at low, endogenous expression levels, and that its localization and turnover are independent of ligand. However, increasing endosomal localization of BRI1 enhances activation of the pathway and genomic responses. Our data indicate distinct signaling and trafficking mechanisms within this receptor class and show that the use of endosomes as signaling compartments is an unexpectedly broad phenomenon in eukaryotes.
Keywords: Brassinosteroid, endosomes, plant receptor kinase, Arabidopsis, receptor trafficking
Receptor endocytosis has long been viewed as a mechanism to inactivate receptors and down-regulate signaling. This concept was challenged more than a decade ago, initially based on the observation that many activated receptors show considerable accumulation in endosomes (Baass et al. 1995). It has now become apparent that key signaling components are localized exclusively to endosomes and that endocytosis is required to bring them into contact with their activated receptors, thereby allowing signaling to take place (Vieira et al. 1996; Wunderlich et al. 2001; Panopoulou et al. 2002). Such a requirement for endosomal localization appears to be widespread in metazoans, as it has been observed in diverse receptor families, such as RTKs, TGF-β-Rs, and GPCRs (DeFea et al. 2000; Wunderlich et al. 2001; Panopoulou et al. 2002; Shenoy et al. 2006). On the other hand, endocytosis of the Ste2p pheromone receptor in yeast appears only necessary for termination of the signal, but not for its transduction per se (Hicke et al. 1998). Thus, it is unclear whether endosomes as signaling platforms are specific to animal cells or whether they represent a more general feature of receptor signaling in eukaryotes.
As the other big branch of multicellular life, plants have independently evolved a different set of receptors and show considerable divergence in endosomal structures and trafficking components (Shiu and Bleecker 2001; Geldner and Jurgens 2006). The >200 LRR receptor Ser–Thr kinases (unrelated to TGF-β receptors) represent the predominant receptor family in the plant kingdom. Although the vast majority of them are orphan receptors (Morillo and Tax 2006), the steroid receptor BRI1 has been intensively studied and provides a good model for plant receptor kinase activation (Vert et al. 2005).
BRI1 is the receptor for brassinosteroids (BRs), which are crucial growth-promoting hormones in plants. Knock-out of BRI1 leads to extremely dwarfed and entirely BR-insensitive plants (Clouse et al. 1996; Li and Chory 1997; Kinoshita et al. 2005). Binding of brassinolide (BL), the most active BR, to BRI1’s extracellular domain activates the BRI1 kinase, releases the inhibitory BKI1 protein from the plasma membrane and increases it affinity for BAK1, a second LRR kinase (Wang et al. 2001, 2005; Nam and Li 2002; Wang and Chory 2006). This leads to dephosphorylation and consequent dimerization and DNA binding of the nuclear-localized BES1/BZR1 transcription factors, which control the genomic BR response (Yin et al. 2005; Vert and Chory 2006). This dephosphorylation is thought to be caused by a combination of inactivation of the GSK3/Shaggy kinase BIN2 (Vert and Chory 2006) and the phosphatase activity of BSU1 (Mora-Garcia et al. 2004). It is currently unknown how receptor activation leads to BIN2 inactivation, and the immediate relevant downstream targets of the activated receptor complex remain to be identified (Belkhadir and Chory 2006). Nonetheless, the known components provide a straightforward and specific read-out of receptor activity.
BRI1 has been shown to localize to the plasma membrane and endosomal structures, and co-overexpression of BRI1 together with the BAK1 coreceptor in protoplasts leads to increased internal accumulation of BRI1. Based on this, it was proposed that BAK1 regulates BRI1 trafficking (Russinova et al. 2004). However, an effect of BAK1 on BRI1 localization could not be demonstrated in intact plants, and the inherent limitations of the protoplast system precluded attempts to functionally connect endocytosis to signaling of BRI1.
Results and Discussion
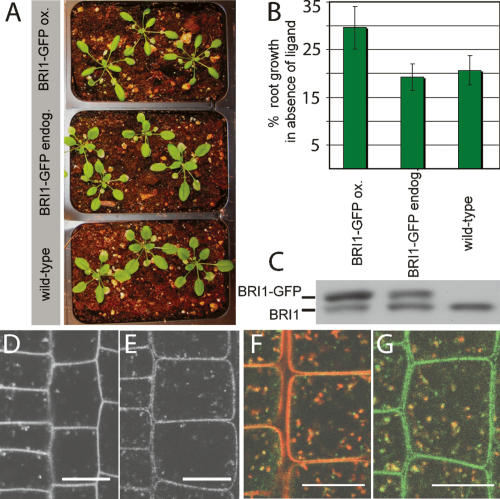
BRI1 is expressed and required in root meristems, a stem cell tissue that is accessible to live imaging without dissection. This provides the rare opportunity to address the connection between receptor localization and signaling in an intact organ. Previously, BRI1 localization to endosomal compartments has been observed exclusively in a BRI1-GFP-overexpressing line that shows phenotypes of enhanced BR response (Fig. 1A), displaying increased root elongation in the absence of ligand, indicative of inappropriate, partially constitutive receptor activity (Fig. 1B). Since we were concerned that the observed endosomal localization was an artifact of overexpression, we constructed a new GFP fusion and selected a line with endogenous expression levels (Fig. 1C). We ascertained that this line had no overexpression phenotypes and rescued a null mutant (Fig. 1A,B; Supplementary Table 1). We detected a fairly weak but specific GFP signal in root meristem cells (Fig. 1D,E; Supplementary Fig. 1), both in the plasma membrane and highly mobile, pleiomorphic intracellular structures (Supplementary Movie 1). These BRI1-positive structures are at least in part early endosomes because they colocalize with the endocytic tracer FM4-64 after 5–10 min of uptake (Fig. 1F). Additionally, they display partial colocalization with VHA-a1-RFP (Fig. 1G), a marker for TGN/early endosomes in plants (Dettmer et al. 2006).
Figure 1.
Endogenously expressed BRI1-GFP localizes to endosomes. (A) Representative pictures of rosette stage Arabidopsis grown under identical conditions. The BRI1-GFP line expressing at endogenous levels (endog.) is indistinguishable from wild type, whereas the overexpressing line (ox.) shows the reported overexpression phenotypes of narrow leaf blades and elongated, twisting petioles (leaf stalks). (B) Roots were depleted of endogenous BRs by growth on 5 μM brassinazole for 3 d. Primary root growth was assessed after three more days (n = 10 per line). Percent growth relative to untreated control is shown. The line expressing BRI1-GFP at endogenous levels is inhibited to wild-type levels, whereas the overexpressing line (ox.) shows less inhibition of root growth in the absence of ligand. (C) Immunoblot of the same lines detected with α-BRI1 antibodies. Intensity of the BRI1-GFP (top) band is much stronger than the band of endogenous BRI1 (bottom) in the overexpressing line, but both bands show the same intensity in middle lane. (D,E) Subcellular localization and levels of BRI1-GFP in root meristem epidermal cells. (D) BRI1-GFP-overexpressing line. (E) BRI1-GFP endogenous expresser. (F) BRI1-GFP (green) partially colocalizes with the endocytic tracer FM4-64 (red) after 5–10 min uptake. (G) BRI1-GFP (green) also colocalizes partially with VHA-a1-RFP (red). See Supplementary Figure 1 for individual channels of overlays. Bars, 10 μm.
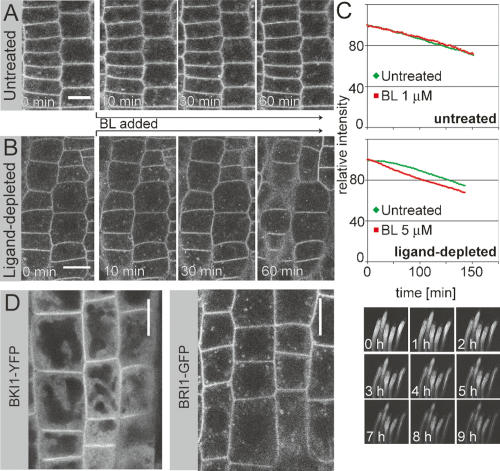
Root meristematic cells are thought to continuously produce and perceive BRs as a crucial signal for continued cell division and elongation. To test whether the endosomal BRI1 in this growing organ represented activated receptors targeted to the endosome for either degradation or recycling, we applied exogenous BL and sampled over time. We did not observe any changes in levels or localization of BRI1-GFP, even at saturating concentrations of ligand (Fig. 2A; Supplementary Movie 2).
Figure 2.
Ligand-independent localization and turnover of BRI1. (A) Seedling roots were treated with saturating amounts of ligand (BL, 1 μM), after recording of first frame (0 min) and were observed at 1-min intervals. BRI1-GFP localization remains unaltered. (B) As in A, but roots were depleted of endogenous BR by growing on 5-μM brassinazole for 3 d. BRI1-GFP localization remains unchanged. (C) In vivo pulse-chase analysis of BRI1-YFP. Transgenic lines driving BRI1-YFP under a heat-shock-inducible promoter were used to measure the half-life of BRI1 in vivo and exclusively in root meristematic cells by quantitative confocal microscopy. YFP signal intensity peaked 4 h after heat shock, when mRNA levels were already back to basal levels (not shown). Turnover was recorded as the reduction in YFP intensity relative to peak expression. Data points were recorded every 2 min. Half-life of BRI1 was determined to be ∼5 h and this turnover rate appeared to be entirely independent of ligand, as much in untreated roots (left) as in ligand-depleted roots (right) (grown as in B). (Below) Picture row shows an example of signal recordings for five untreated control root meristems. (D, left) BKI-YFP displays clear PM accumulation over cytosolic background, whereas no comparable signal can be observed in endosomal compartments. (Right) BRI1-GFP grown under identical conditions. Bars: A,B, 10 μm.
It is possible that root meristem cells have close to maximal levels of activated receptors, precluding the detection of ligand-dependent localization changes when adding exogenous BL. Therefore, we depleted roots of endogenous BRs by growing them for 3 d on 5 μM brassinazole, a specific inhibitor of BR biosynthesis (Sekimata et al. 2001). To our surprise, neither endosomal localization nor levels of BRI1-GFP were affected by this treatment (Fig. 2B, left). Further, the levels and localization of BRI1-GFP remained unaltered even when high amounts of BL were added back to these ligand-devoid roots, which should induce a rapid shift toward completely ligand-occupied receptors (Fig. 2B; Supplementary Movie 3). We conclude that BRI1 localizes to endosomes independently of its activation state.
Activation of signaling pathways can lead to a relevant increase in protein flux between compartments, without necessarily shifting their steady-state accumulation in one or the other direction (Ando et al. 2004). In the case of BRI1, an increase in endocytosis and degradation could be offset by increased synthesis and secretion, without causing observable changes in overall localization or levels of BRI1-GFP. Therefore, we decided to directly measure BRI1 turnover rates in planta. To do so, we placed BRI1-YFP under the control of a heat-shock promoter, generating lines that allow pulsed expression of the tagged BRI1 by a 20–30-min induction at 37°C (Supplementary Movie 4). In combination with quantitative confocal microscopy, this allowed for a pulse-chase analysis exclusively in intact root meristems (Fig. 2C; Supplementary Fig. 2; Supplementary Movie 5). We determined the half-life of BRI1-YFP to be ∼5 h. This relative stability suggests that BRI1 either recycles during its lifetime or that only a minor fraction of BRI1 engages in endocytic transport to the vacuole with a majority remaining immobile. Again, the measured degradation rate of BRI1 was unaffected by addition of BL either to untreated or ligand-depleted roots (Fig. 2C). Based on these results, it is unlikely that BRs cause any difference in BRI1 intracellular transport, and we conclude that BRI1 trafficking is constitutive.
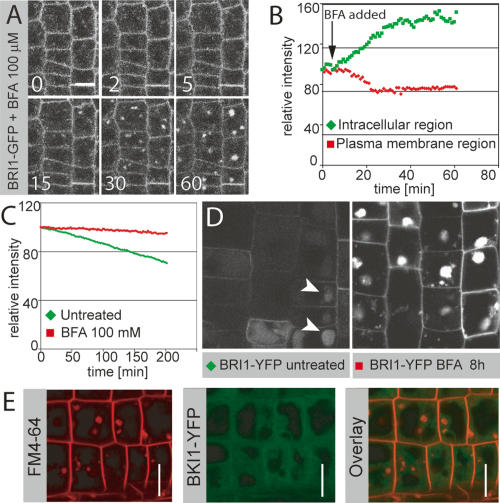
The activity status of BRI1 cannot be monitored directly at subcellular resolution. However, in untreated root cells a significant amount of plasma-membrane-localized BRI1 is apparently in an inactive state, as judged by the presence of the inhibitory BKI1 protein, which is released by BRI1-dependent phosphorylation after BR stimulation (Wang and Chory 2006). We did not observe any BKI1 accumulation in endosomal structures, suggesting that the endosomal BRI1 may represent an active receptor pool (Fig. 2D). To test this idea, we attempted to specifically manipulate the subcellular localization of BRI1 and to monitor the consequences on signaling activity. A number of known chemical inhibitors of trafficking were tested (data not shown), but only Brefeldin A (BFA) turned out to be sufficiently specific for our purpose. BFA is a widely used inhibitor of endosomal trafficking in plants (Geldner 2004). BFA had the expected effects on BRI1 localization, although we observed a fairly strong persistence of BRI1 signals at the plasma membrane. Nonetheless, we quantified plasma membrane versus intracellular signal in time-lapse images and showed that BFA induced a shift of BRI1 localization into endosomal compartments (Fig. 3A,B; Supplementary Movie 6). We also showed by pulse-chase analysis that BFA blocks translocation of BRI1 from earlier to late endosomal compartments and vacuoles, thereby interfering with BRI1-YFP degradation (Fig. 3C,D). This block in vacuolar transport is visualized, because BFA-treated cells did not accumulate residual YFP signals in vacuoles some hours after the expression pulse (Fig. 3D). Again, we did not observe any BKI1 accumulation in these BFA-induced endosomal aggregates, suggesting that the BRI1 therein could be part of active receptor complexes (Fig. 3E). For summary cartoon of BFA effects on BRI1 trafficking, see Supplementary Figure 3.
Figure 3.
BFA increases endosomal localization of BRI1. (A) Time-lapse analysis of BFA-induced shift of BRI1 from the plasma membrane to endosomal compartments. (B) Quantification of time lapse in A. BFA induces a rapid drop in plasma membrane signal with a concomitant increase in intracellular fluorescence. A new steady state is apparently reached after ∼30 min. (C) Quantification of the effect of BFA on BRI1-YFP degradation in a pulse-chase experiment. BFA leads to a nearly complete block of BRI1 turnover. (D) BRI1-YFP localization in seedling root meristems 8 h after heat-shock-induced peak expression, with (left) or without (right) BFA treatment, taken under identical settings. BFA leads to strong accumulation of BRI1-YFP in endosomal aggregates, whereas only a weak and predominantly vacuolar BRI1-YFP signal is observed in the untreated control (arrowheads). (E) FM4-64 (red, left) and BKI1-YFP (green, middle) after 50 μM BFA treatment for 30 min. (Right) Overlay. Note the absence of BKI1 signal in BFA compartments. Bars: A,D,E, 10 μm.
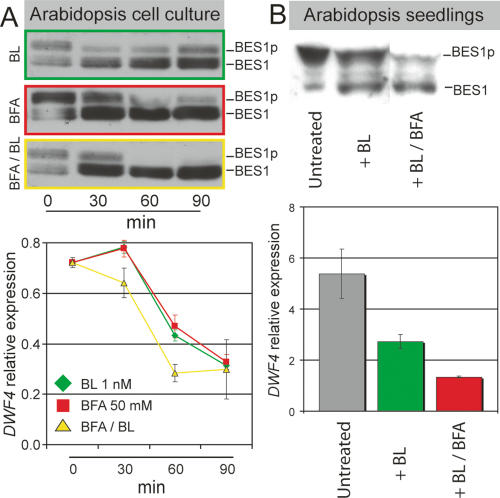
We then investigated if increasing BRI1 endosomal accumulation could affect signaling activity. We first used a highly BR-sensitive cell suspension culture that shows BR-induced dephosphorylation of the BES1 transcription factor when treated with scant amounts of BL (1 nM vs. 10 nM in intact seedlings). Dephosphorylation of BES1 and its close family member, BZR1, leads to reduced expression of the BR early-response gene DWF4 (Fig. 4A). Remarkably, BFA on its own was able to induce strong dephosphorylation of BES1 and to cause suppression of downstream genes over a similar time course and to a similar extent as exogenously applied BL (Fig. 4A). Cotreatment of BFA together with exogenous BL further enhanced activation of the pathway (Fig. 4A). The cell culture results were corroborated using intact seedlings overexpressing BRI1-GFP. We observed a small, but consistent, enhancement of BR signaling when seedlings exposed to low amounts of BL were cotreated with BFA (Fig. 4B).
Figure 4.
BRI1 signaling is induced by BFA treatment. (A) A highly sensitive Arabidopsis green cell suspension line shows dephosphorylation of BES1 (green, above) and suppression of early-response gene, DWF4 (green, below) already at 1 nM BL for 30 min. (Middle) BFA treatment alone leads to an efficient activation of BES1 (red) and suppression early-response genes. External BL enhances the BFA effect on BES1 (yellow, above) and DWF4 (yellow, below). (B) Enhancement of BR signaling by BFA can also be observed in BRI1-overexpressing seedlings, when treating with low amounts of BL (10 nM) for 4 h, which leads to only half-maximal dephosporylation of BES1 (above) and a twofold reduction in DWF4 levels (green, below). Adding BFA (100 μM) after 30 min of BL pretreatment significantly enhances BES1 activation (above) and DWF4 suppression (red, below).
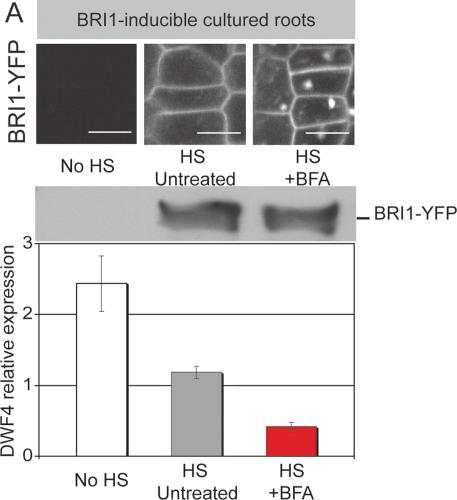
The fairly mild enhancements of BR signaling in intact seedlings compared with cell cultures could be explained by problems of uptake and activity of BFA into the different BR-responsive organ and tissue types contained in whole seedlings. We therefore attempted to establish a more homogenous system where the same tissues could be simultaneously analyzed for the effect of BFA on BRI1 signaling and localization. We generated a bri1-null mutant line that contained heat-shock-inducible BRI1-YFP as the sole source of BRI1 activity. Since bri1-null mutants cannot be propagated as homozygous individuals, we established a cultured root system, where large amounts of root meristems were continuously formed from pre-existing roots. We then induced the expression of BRI1, which consequently led to induction of BRI1 signaling (Fig. 5). BFA had the expected effect on BRI1 localization (Fig. 5, top panel), without increasing the actual amount of BRI1 over the time scale of the treatment (Fig. 5, middle panel). Nonetheless, BFA was able to enhance BRI1-dependent signaling (Fig. 5, bottom panel), again suggesting that BRI1 signals preferentially from endosomes.
Figure 5.
Induced BRI-YFP expression in cultured roots. (A) BFA effects can also be observed in intact organs using a bri1-null mutant root culture containing heat-shock-inducible BRI1-YFP. In this system, the effects of BFA on localization and signaling can be simultaneously assessed. Top panel shows BRI-YFP signals, 8 h after a 30-min heat-shock or control treatment. BFA treatment (100 μM) started 4 h after heat shock. Middle panel shows BRI1-YFP levels as detected on immunoblot with α-BRI1 after same treatments as above. Bottom panel shows heat-shock-induced suppression of DWF4 (gray) and its further reduction by BFA treatment (red). Bar, 10 μm.
Our findings highlight the relevance of subcellular compartmentation of signaling components for the regulation of plant receptor kinase pathways. We show that the endosomal and plasma membrane pool of BRI1 represent two distinct subpopulations, since it is only the latter that is complexed with an inhibitory scaffold protein. This subcellular partitioning appears to be functionally relevant, since increasing the ratio of endosomal to plasma membrane-localized BRI1 significantly enhances signaling activity. One prediction from our findings would be that the as yet unidentified direct downstream targets of the activated receptor complex may localize preferentially to endosomal compartments (Supplementary Fig. 4). This is not unlike the situation for the unrelated TGF-β receptor in animals, which also has been shown to traffic in a ligand-independent fashion, but nonetheless transduces signals from endosomes (Panopoulou et al. 2002). However, this contrasts to the recently demonstrated ligand-dependent trafficking of the related LRR-receptor kinase FLS2 (Robatzek et al. 2006).
Together, this now provides us with two plant receptor models that display opposite trafficking behavior. Interestingly, this difference in trafficking matches an underlying difference in the biology of these two receptors. Whereas FLS2 is a pathogen receptor evolved for rare but acute signaling, BRI1 is thought to perceive a more or less continuous signal, whose relevance lies in its modulation more than in acute presence or absence.
A number of interesting speculations have been put forward to explain why endosomal signaling might have evolved in animals. It was proposed that signaling from numerous motile endosomes might overcome problems of diffusion-based signaling cascades in large cells and that timing and specificity of signaling events can be better controlled in endosomes (Miaczynska et al. 2004). Our data now indicate that endosomes in multicellular plants also act as signaling compartments, in spite of their independent sets of receptors and a differently organized endosomal system. We propose that a common, fundamental problem might have independently driven the development of signaling endosomes. Both multicellular plants and animals show vast increases of surface-localized receptor families. In such a situation, the available plasma membrane surface could become a limiting factor and trafficking of activated receptors to endosomes a means to increase the effective surface area available for signaling. This would restrict the dwelling time of receptors at the plasma membrane to ligand binding and activation, while compartmentalizing the longer-lasting downstream signaling events to the much less restricted inner surfaces of endosomal compartments.
Materials and methods
Plant material and growth conditions
Arabidopsis Columbia seedlings were grown vertically for 4–6 d with 24 h light at 22°C on 0.5× LS (Linsmayer-Skoog plant growth mixture) agar plates. Green cell suspension cultures were originally obtained from Ruishuang Geng (Ohio State University, Columbus, OH) and grown as described by Kim et al. (2003). Suspension-grown root meristem cultures were obtained according to an online protocol at http://www.bio.net/bionet/mm/arab-gen/1992-September/000708.html.
Constructs and generation of transgenic lines
A BRI1-GFP construct was cloned into a pGREEN II vector derivative containing nos-promoter-driven Basta resistance. It contains 1689 base pairs (bp) upstream of the BRI1 translational start codon and 629 bp downstream from the stop codon. The EGFP sequence was inserted at the C terminus as described by Friedrichsen et al. (2000). The contruct was transformed into wild type, and a line with close to identical expression levels was selected. This line therefore still contains twice the amount of overall BRI1 product (endogenous BRI1 plus BRI1-GFP). The promoter of the HS∷BRI1-YFP construct was described by Knox et al. (2003). EYFP was fused to the C terminus as above.
Confocal microscopy and image quantification
Confocal microscopy was done with a Leica SP/2 inverted microscope. Live imaging of roots was done by placing root tips on an agar block that fit into silicon chambers with a coverslip bottom (Grace Biolabs). Specimens were either sealed with a slide (long-term observation) or overlaid with sufficient liquid media (short-term observations). Image analysis was done with the Leica SP/2 software package and the ImageJ bundle provided by the Wright Cell Imaging facility.
Hormone and inhibitor treatments
BL was obtained from CIDtech Research, Inc., and Brefeldin A was obtained from Sigma. For long-term treatments, hormones or drugs were dissolved into agar blocks onto which roots were placed (see above). Short-term observations were done by mixing the hormone/drug into a liquid drop overlaying the agar-embedded root tips (see above) while continuously scanning.
Heat-shock pulse-chase analysis
For quantification of the BRI1 degradation rate, seedlings of homozygous HS∷BRI1-YFP lines were transferred into in 200 μL of liquid 0.5× LS in a 2-mL cap and heat-shocked in a 37°C water bath for 20–30 min. Seedlings were then placed back onto an agar plate for 4.5 h, before being mounted for observation (see above). For initial experiments, entire agar plates with seedlings were sealed and left floating in the water bath for at least 45 min. Turnover rates were determined by measuring signal intensities in five regions of interest along the root meristem, as depicted in Supplementary Figure 2, of at least five roots per experiment.
Acknowledgments
We thank Y. Belkhadir, M. Chen, and S. Salvadi-Goldstein for critical reading of the manuscript; Y. Yin for providing BES1 antibody; T. Dabi for maintenance of cell cultures; and F. Schöffl for the heat-shock promoter fragment. The work was supported by initial funding from the European Molecular Biology Organisation (EMBO) and continued funding from a Human Frontier Science Project (HFSP) long-term fellowship to N.G., by the Howard Hughes Medical Institute, and by USDA and NSF grants to J.C.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1561307
References
- Ando R., Mizuno H., Miyawaki A., Mizuno H., Miyawaki A., Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- Baass P.C., Di Guglielmo G.M., Authier F., Posner B.I., Bergeron J.J., Di Guglielmo G.M., Authier F., Posner B.I., Bergeron J.J., Authier F., Posner B.I., Bergeron J.J., Posner B.I., Bergeron J.J., Bergeron J.J. Compartmentalized signal transduction by receptor tyrosine kinases. Trends Cell Biol. 1995;5:465–470. doi: 10.1016/s0962-8924(00)89116-3. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Chory J., Chory J. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science. 2006;314:1410–1411. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C., Langford M., McMorris T.C., McMorris T.C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFea K.A., Zalevsky J., Thoma M.S., Dery O., Mullins R.D., Bunnett N.W., Zalevsky J., Thoma M.S., Dery O., Mullins R.D., Bunnett N.W., Thoma M.S., Dery O., Mullins R.D., Bunnett N.W., Dery O., Mullins R.D., Bunnett N.W., Mullins R.D., Bunnett N.W., Bunnett N.W. β-Arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K., Stierhof Y.D., Schumacher K., Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A., Li J., Hunter T., Chory J., Joazeiro C.A., Li J., Hunter T., Chory J., Li J., Hunter T., Chory J., Hunter T., Chory J., Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N. The plant endosomal system—Its structure and role in signal transduction and plant development. Planta. 2004;219:547–560. doi: 10.1007/s00425-004-1302-x. [DOI] [PubMed] [Google Scholar]
- Geldner N., Jurgens G., Jurgens G. Endocytosis in signalling and development. Curr. Opin. Plant Biol. 2006;9:589–594. doi: 10.1016/j.pbi.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Hicke L., Zanolari B., Riezman H., Zanolari B., Riezman H., Riezman H. Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Geng R., Somers D.E., Geng R., Somers D.E., Somers D.E. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. 2003;100:4933–4938. doi: 10.1073/pnas.0736949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Cano-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J., Cano-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J., Hiranuma S., Fujioka S., Yoshida S., Chory J., Fujioka S., Yoshida S., Chory J., Yoshida S., Chory J., Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Knox K., Grierson C.S., Leyser O., Grierson C.S., Leyser O., Leyser O. AXR3 and SHY2 interact to regulate root hair development. Development. 2003;130:5769–5777. doi: 10.1242/dev.00659. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J., Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Miaczynska M., Pelkmans L., Zerial M., Pelkmans L., Zerial M., Zerial M. Not just a sink: Endosomes in control of signal transduction. Curr. Opin. Cell Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Mora-Garcia S., Vert G., Yin Y., Cano-Delgado A., Cheong H., Chory J., Vert G., Yin Y., Cano-Delgado A., Cheong H., Chory J., Yin Y., Cano-Delgado A., Cheong H., Chory J., Cano-Delgado A., Cheong H., Chory J., Cheong H., Chory J., Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes & Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo S.A., Tax F.E., Tax F.E. Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 2006;9:460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Nam K.H., Li J., Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Panopoulou E., Gillooly D.J., Wrana J.L., Zerial M., Stenmark H., Murphy C., Fotsis T., Gillooly D.J., Wrana J.L., Zerial M., Stenmark H., Murphy C., Fotsis T., Wrana J.L., Zerial M., Stenmark H., Murphy C., Fotsis T., Zerial M., Stenmark H., Murphy C., Fotsis T., Stenmark H., Murphy C., Fotsis T., Murphy C., Fotsis T., Fotsis T. Early endosomal regulation of Smad dependent signaling in endothelial cells. J. Biol. Chem. 2002;277:18046–18052. doi: 10.1074/jbc.M107983200. [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T., Chinchilla D., Boller T., Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes & Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Cano-Delgado A., Yin Y., Chory J., de Vries S.C., Borst J.W., Kwaaitaal M., Cano-Delgado A., Yin Y., Chory J., de Vries S.C., Kwaaitaal M., Cano-Delgado A., Yin Y., Chory J., de Vries S.C., Cano-Delgado A., Yin Y., Chory J., de Vries S.C., Yin Y., Chory J., de Vries S.C., Chory J., de Vries S.C., de Vries S.C. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata K., Kimura T., Kaneko I., Nakano T., Yoneyama K., Takeuchi Y., Yoshida S., Asami T., Kimura T., Kaneko I., Nakano T., Yoneyama K., Takeuchi Y., Yoshida S., Asami T., Kaneko I., Nakano T., Yoneyama K., Takeuchi Y., Yoshida S., Asami T., Nakano T., Yoneyama K., Takeuchi Y., Yoshida S., Asami T., Yoneyama K., Takeuchi Y., Yoshida S., Asami T., Takeuchi Y., Yoshida S., Asami T., Yoshida S., Asami T., Asami T. A specific brassinosteroid biosynthesis inhibitor, Brz2001: Evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta. 2001;213:716–721. doi: 10.1007/s004250100546. [DOI] [PubMed] [Google Scholar]
- Shenoy S.K., Drake M.T., Nelson C.D., Houtz D.A., Xiao K., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Drake M.T., Nelson C.D., Houtz D.A., Xiao K., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Nelson C.D., Houtz D.A., Xiao K., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Houtz D.A., Xiao K., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Xiao K., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R.J., Premont R.T., Lichtarge O., Lefkowitz R.J., Lichtarge O., Lefkowitz R.J., Lefkowitz R.J. β-Arrestin-dependent, G protein-independent ERK1/2 activation by the β2 adrenergic receptor. J. Biol. Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B., Bleecker A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Chory J., Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Vert G., Nemhauser J.L., Geldner N., Hong F., Chory J., Nemhauser J.L., Geldner N., Hong F., Chory J., Geldner N., Hong F., Chory J., Hong F., Chory J., Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- Vieira A.V., Lamaze C., Schmid S.L., Lamaze C., Schmid S.L., Schmid S.L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- Wang X., Chory J., Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J., Seto H., Fujioka S., Yoshida S., Chory J., Fujioka S., Yoshida S., Chory J., Yoshida S., Chory J., Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D., Asami T., Yoshida S., Huber S.C., Clouse S.D., Yoshida S., Huber S.C., Clouse S.D., Huber S.C., Clouse S.D., Clouse S.D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich W., Fialka I., Teis D., Alpi A., Pfeifer A., Parton R.G., Lottspeich F., Huber L.A., Fialka I., Teis D., Alpi A., Pfeifer A., Parton R.G., Lottspeich F., Huber L.A., Teis D., Alpi A., Pfeifer A., Parton R.G., Lottspeich F., Huber L.A., Alpi A., Pfeifer A., Parton R.G., Lottspeich F., Huber L.A., Pfeifer A., Parton R.G., Lottspeich F., Huber L.A., Parton R.G., Lottspeich F., Huber L.A., Lottspeich F., Huber L.A., Huber L.A. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 2001;152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J., Tao Y., Yoshida S., Asami T., Chory J., Yoshida S., Asami T., Chory J., Asami T., Chory J., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]