Abstract
Piwi-interacting RNAs (piRNAs) consist of a germline-specific group of small RNAs derived from distinct intergenic loci in the genome. piRNAs function in silencing selfish transposable elements through binding with the PIWI subfamily proteins of Argonautes. Here we show that piRNAs in Drosophila are 2′-O-methylated at their 3′ ends. Loss of Pimet (piRNA methyltransferase), the Drosophila homolog of Arabidopsis HEN1 methyltransferase for microRNAs (miRNAs), results in loss of 2′-O-methylation of fly piRNAs. Recombinant Pimet shows single-stranded small RNA methylation activity in vitro and interacts with the PIWI proteins within Pimet mutant ovary. These results show that Pimet mediates piRNA 2′-O-methylation in Drosophila.
Keywords: piRNA, 2′-O-methylation, HEN1, Pimet, Drosophila
Small, 20- to 30-nucleotide (nt)-long, RNAs trigger gene silencing mechanisms, collectively called RNA silencing, which include cleavage-dependent and -independent mRNA decay, translational repression, DNA methylation, and heterochromatin formation in organisms ranging from fission yeast to humans (Tomari and Zamore 2005; Zaratiegui et al. 2007). RNA silencing-triggering small RNAs are processed from long precursors and are then loaded onto Argonaute proteins (Parker and Barford 2006), guiding the Argonautes to specific sequences within the target RNAs (Tomari and Zamore 2005; Pillai et al. 2007).
Numbers of the Argonaute protein family members, defined by the PAZ and PIWI domains, differ among species (Carmell et al. 2002); for instance, fly has five members (Williams and Rubin 2002) whereas mouse has seven. The Argonautes can be divided into two subgroups, the AGO and PIWI subfamilies, according to sequence similarity. In Drosophila, AGO1 and AGO2 of the AGO subfamily function in the microRNA (miRNA) pathway and RNA interference (RNAi) by binding with miRNAs and short interfering RNAs (siRNAs), respectively (Okamura et al. 2004; Tomari et al. 2004; Miyoshi et al. 2005). Recent studies have revealed that PIWI proteins are specifically associated with a subset of endogenous small RNAs, termed Piwi-interacting RNAs (piRNAs; in flies, these are also called repeat-associated siRNAs, or rasiRNAs), which are distinct from miRNAs in many respects such as size, expression pattern, and even function (O’Donnell and Boeke 2007). Unlike miRNAs, piRNAs are 24–30 nt long, are expressed only in gonads, and function in genome surveillance through association with the PIWI proteins (for review, see Kim 2006; Saito et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2007) by silencing transposable elements that have sufficient potency to invade the genome by inserting themselves into DNA elements.
Recently, a model for piRNA biogenesis in Drosophila was proposed (Brennecke et al. 2007; Gunawardane et al. 2007). In this model, PIWI proteins determine and form 5′ ends of piRNAs, which will then be loaded onto the other PIWI members, by cleaving their target transcripts in a manner depending on the associated piRNA sequence. This process not only continuously generates piRNAs but also keeps destroying target RNAs transcribed from selfish genes. Fish and mammals may have similar mechanisms (Aravin et al. 2007; Houwing et al. 2007). However, many aspects of piRNA biogenesis in Drosophila remain unclear; for example, protein factor(s) determining and forming the 3′ ends of piRNAs are not identified. How nascent piRNAs are loaded onto the other PIWI members also has not been elucidated.
In plants, miRNAs are methylated at the 2′-terminal hydroxyl group on the ribose at the last nucleotide (Yu et al. 2005). A factor responsible for such modification in Arabidopsis is HEN1 (Yu et al. 2005). This is a protein containing a double-stranded RNA (dsRNA)-binding domain at the N terminus and a catalytic domain at the C terminus that transfers methyl groups from S-adenosyl methionine (SAM) to miRNA/miRNA* duplexes. Five consecutive residues (Asp–Phe–Gly–Cys–Gly) residing in the catalytic domain of Arabidopsis HEN1 correspond to the SAM-binding motif. Mutations introduced in the SAM-binding motif abolish the methylation activity. In the hen1 mutant, miRNA are reduced in abundance and when detectable are uridylated, a cause of miRNA destabilization (Li et al. 2005). It can be concluded that HEN1 is the stabilizer of miRNAs in plants. Arabidopsis HEN1 methylates even siRNA duplexes with perfect sequence complementarity (Yang et al. 2006), but single-stranded small RNAs and pre-miRNAs forming a hairpin structure fail to be methylated in vitro by recombinant HEN1 (Yu et al. 2005). It is thus postulated that miRNA methylation by HEN1 occurs between two steps during the miRNA maturation process; namely, after Dicer-mediated excision of miRNA/miRNA* duplexes from the precursors and before the unwinding process of the duplexes.
piRNAs in Drosophila are modified at their 3′ ends, as judged by their resistance to periodate oxidation/β-elimination (Vagin et al. 2006; Gunawardane et al. 2007). In this study, we determined that piRNAs in Drosophila are 2′-O-methylated at their 3′ ends, as in mouse and rat (Houwing et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007). In Drosophila, CG12367 encodes a protein showing similarity at the peptide levels with the methyltransferase catalytic domain of Arabidopsis HEN1. Loss of CG12367 results in loss of piRNA methylation in vivo; thus, we refer to this as Pimet (for piRNA methyltransferase). Indeed, Aub- and Piwi-associated piRNAs isolated from Pimet mutant ovary showed sensitivity to oxidation/β-elimination as oppose to those in wild-type ovary. Recombinant Pimet showed activity in vitro for transferring methyl groups from SAM to single-stranded small RNAs bearing a 2′, 3′-terminal-cis-diol group, but not to 2′-O-methylated small RNAs. Aub-associated piRNAs from Pimet mutant ovary were methylated by recombinant Pimet in vitro; by contrast, AGO1-associated miRNAs did not serve as a substrate for the methylation. Taken together, these results show that Pimet is the factor responsible for piRNA methylation in Drosophila germlines.
Results and Discussion
piRNAs in fly ovary are 2′-O-methylated at their 3′ ends
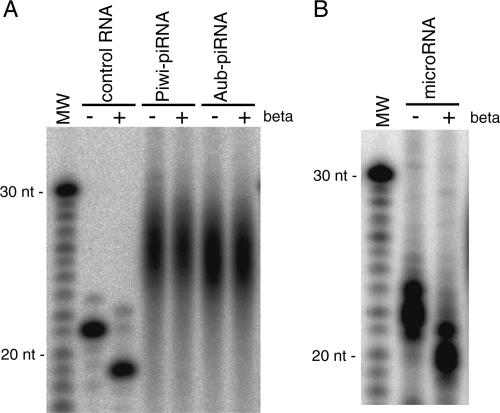
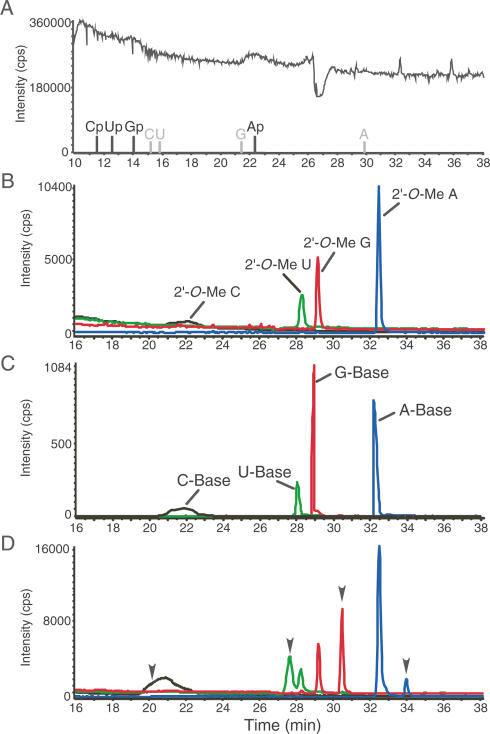
piRNAs in fly ovary are resistant to periodate oxidation/β-elimination treatment, indicating that these piRNAs are modified at their 3′ ends (Vagin et al. 2006; Gunawardane et al. 2007). In this study, similar experiments were performed for piRNAs associated with Aub and Piwi in ovary. In both cases piRNAs did not gain mobility even after periodate oxidation/β-elimination treatment (Fig. 1A), indicating that they are modified at their 3′ ends. Under the same conditions, miRNAs associated with AGO1 increased mobility by nearly 2 nt (Fig. 1B), a feature of RNAs bearing a 2′-, 3′-terminal-cis-diol group (Hutvagner et al. 2001). This is consistent with the fact that animal miRNAs are not modified at their 3′ ends (Elbashir et al. 2001; Hutvagner et al. 2001). piRNAs expressed in mouse and rat have recently been shown to be 2′-O-methylated at their 3′ ends (Houwing et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007). To identify the terminal modification of piRNA in Drosophila, Aub-associated piRNAs were gel-purified and digested by RNase T2. The digest was analyzed by capillary liquid chromatography (LC) and mass spectrometry (MS) (Fig. 2). In this treatment, only 3′-terminal residues are converted to nucleosides (Ohara et al. 2007). As shown in Figure 2B, four kinds of 2′-O-methyl nucleosides were clearly observed as major products. Each proton adduct was further analyzed by MS/MS using collision-induced dissociation (CID). Selected reaction monitoring (SRM) for base-related product ions (Fig. 2C) and mass spectrum (Supplementary Fig. S1) of each 2′-O-methyl nucleoside also confirmed that a methyl group is attached to each ribose portion. To eliminate the possibility that they were 3′-O-methyl nucleosides, RNase T2-digested piRNAs were coinjected with a series of synthetic 3′-O-methyl nucleosides. Each 3′-O-methyl nucleoside eluted at a distinct retention time from each 3′-terminal nucleoside of piRNAs (Fig. 2D). On the other hand, each synthetic 2′-O-methyl nucleoside eluted at the same retention time with its corresponding 3′-terminal nucleoside of piRNAs (data not shown). These results indicate that piRNAs in flies are almost entirely methylated at the 2′-hydroxyl group on the ribose of the last nucleotide.
Figure 1.
piRNAs associated with Aub and Piwi in fly ovary are methylated at their 3′ ends. (A) piRNA associated with Aub and Piwi in fly ovary were subjected to periodate oxidation/β-elimination chemical reactions. In both cases, piRNAs did not gain mobility, indicating that these piRNAs with Aub and Piwi were modified at their 3′ ends. Under these conditions, a synthetic 21-nt small RNA (luc guide siRNA) (Okamura et al. 2004) gained mobility by nearly 2 nt, a feature of RNAs bearing a 2′-, 3′-terminal-cis-diol group at the last nucleotide (Hutvagner et al. 2001). (B) miRNAs associated with AGO1 in fly ovary show that they are not modified at their 3′ ends, in contrast to plant miRNAs (Yu et al. 2005).
Figure 2.
piRNAs associated with Aub in fly ovary are 2′-O-methylated at their 3′ ends. (A) Total ion chromatogram (TIC) of the piRNAs digested by RNase T2. The retention times of the observed 3′-phospho nucleotides (Cp, Up, Gp, and Ap) are indicated. The expected retention times of unmodified nucleosides (C, U, G, and A) are shown in gray. The considerable amount of adenosine observed is due to contamination (data not shown), as in the case of mouse piRNA MS analysis (Ohara et al. 2007). (B) Mass chromatograms for proton adducts of piRNA 3′-terminal nucleosides, 2′-O-Me C (m/z 258; black line), 2′-O-Me U (m/z 259; green line), 2′-O-Me G (m/z 298; red line), and 2′-O-Me A (m/z 282; blue line). (C) Mass chromatograms of SRM for base-related product ions of 2′-O-Me C (m/z 112; black line), 2′-O-Me U (m/z 113; green line), 2′-O-Me G (m/z 152; red line), and 2′-O-Me A (m/z 136; blue line). (D) piRNAs digested by RNase T2 were coinjected with a series of synthetic 3′-O-methyl nucleosides. Mass chromatograms for proton adducts of piRNA 3′-terminal nucleosides and a series of synthetic 3′-O-methyl nucleosides, 2′ or 3′-O-Me C (m/z 258; black line), 2′ or 3′-O-Me U (m/z 259; green line), 2′ or 3′-O-Me G (m/z 298; red line), and 2′ or 3′-O-Me A (m/z 282; blue line). Peaks for 3′-O-methyl nucleosides are indicated by arrowheads.
CG12367 is involved in piRNA methylation in the ovary
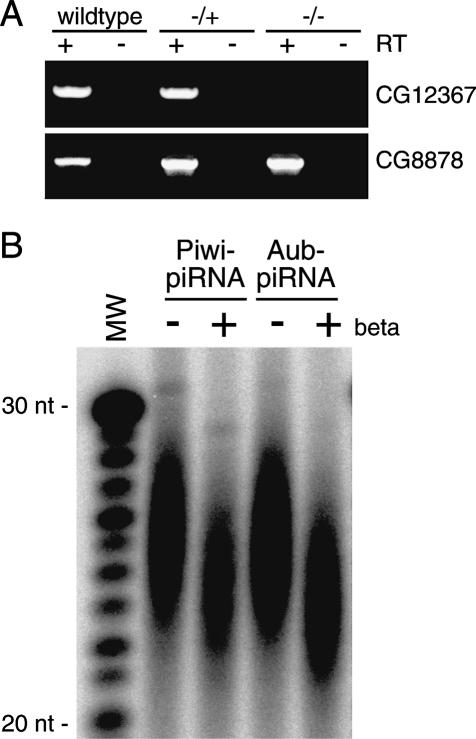
In Arabidopsis, miRNAs are methylated at their 3′ ends by HEN1 while still in a duplex form with miRNA* molecules (Yu et al. 2005). HEN1 is a methyltransferase that contains two characteristic motifs, a dsRNA-binding domain and a catalytic domain necessary for the methyltransferase activity. The Drosophila genome possesses a single gene (CG12367) that encodes a protein showing a strong similarity to the methyltransferase catalytic domain of Arabidopsis HEN1 (Supplementary Fig. S2A). CG12367 lacks the dsRNA-binding domain found in HEN1 (Supplementary Fig. S2A). In fact, many other HEN1 homologs in divergent organisms including nematodes, fish, and mammals also lack dsRNA-binding domains (Supplementary Fig. S2B; Park et al. 2002). To assess possible roles in piRNA methylation, a CG12367 mutant fly (piggyBacf00810) containing a piggyBac insertion within the first intron of CG12367 (Supplementary Fig. S3A) was obtained from the EXELIXIS-Drosophila Stock Collection. The expression levels of CG12367 were checked by RT–PCR. We found that the homozygous piggyBacf00810 mutant does not express CG12367 mRNAs, but expresses transcripts of CG8878, a gene encoded in a region corresponding to the first exon of CG12367 in the reverse orientation (Fig. 3A). piRNAs associated with Aub and Piwi were isolated from CG12367 mutant ovary, and oxidation/β-elimination was performed. Western blotting analysis of the PIWI proteins Aub, Piwi, and AGO3 revealed that their expression levels did not seem to be changed by loss of CG12367 expression (Supplementary Fig. S3B). piRNAs isolated from the mutant ovary were sensitive to β-elimination (Fig. 3B) in contrast to those from wild-type ovary (Fig. 1A). piRNAs from PiggyBacf00810/Df(2R)BSC40 transheterozygote ovaries also showed the sensitivity to the chemical treatment (Supplementary Fig. S3C). These imply that CG12367 is involved in piRNA methylation; thus, being Pimet.
Figure 3.
Involvement of CG12367 in piRNA methylation in fly ovary. (A) RT–PCR shows that the homozygous piggyBacf00810 mutant does not express CG12367 mRNAs, but does express transcripts of CG8878. A schematic drawing of CG12367 (blue) and CG8878 (red) genes in Drosophila is shown in Supplementary Figure S3A. RT–PCR reaction was performed using primers that are indicated in Supplementary Figure S3A. (−/+) PiggyBacf00810/CyO; (−/−) PiggyBacf00810/PiggyBacf00810; (wild type) yellow-white. (B) piRNAs associated with Aub and Piwi in the CG12367 mutant ovary (shown in A) were subjected to oxidation/β-elimination chemical reactions. In both cases, piRNAs gain mobility, indicating that piRNAs associated with Aub and Piwi in the CG12367 mutant are not modified at their 3′ ends.
Recombinant Pimet shows activity to methylate single-stranded small RNAs in vitro
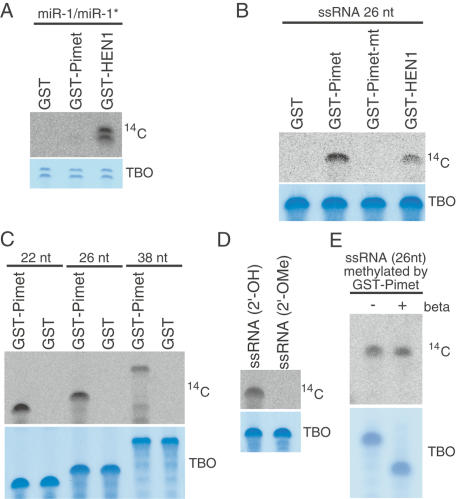
It has been demonstrated that recombinant GST-HEN1 protein produced in Escherichia coli shows activity to transfer methyl groups from SAM to both 3′ ends of miRNA/miRNA* duplex in vitro (Yu et al. 2005; Yang et al. 2006). We produced GST-tagged full-length Pimet recombinant protein in E. coli (Supplementary Fig. S4A) and performed in vitro small RNA methylation assays. GST-HEN1 was employed as a control in the assays. We found that miR-1/miR-1* duplex (0.04 nmol/50 μL) was methylated by GST-HEN1 in the presence of 14C-SAM (Fig. 4A), as reported. By contrast, under the same conditions, GST-Pimet did not methylate the substrate (Fig. 4A). When a 26-nt single-stranded RNA (ssRNA) was the substrate (0.5 nmol/50 μL), even GST-HEN1, which has been reported to fail to methylate ssRNAs (Yu et al. 2005), showed weak activity for ssRNA methylation (Fig. 4B). GST-Pimet methylated ssRNA more efficiently than did GST-HEN1 (Fig. 4B). GST-Pimet mutant (GST-Pimet-mt) (see Supplementary Fig. S4A), in which the residues in the SAM-binding motif were mutated, showed no methylation activity (Fig. 4B). GST-Pimet was able to methylate small ssRNAs ranging from 22 to 38 nt in length (Fig. 4C). It thus seems that Pimet is indifferent to the length of substrates, unlike HEN1 that hardly recognizes 25 nt or longer RNA duplexes as substrates (Yang et al. 2006). 2′-O-methyl modification of 26-nt ssRNAs prevented the methylation (Fig. 4D), suggesting that GST-Pimet transfers methyl groups only onto the 2′-hydroxyl group on the ribose of the far 3′ nucleotide. Similarly, 26-nt ssRNAs phosphorylated at the 3′-hydroxyl group did not serve as substrates (Supplementary Fig. S4B). Pimet seems to require the two hydroxyl groups on the 3′-most nucleotides of substrates, as in the case of HEN1 (Yu et al. 2005). ssRNAs methylated by GST-Pimet show resistance to oxidation/β-elimination treatment (Fig. 4E). These results indicate that Pimet in Drosophila is involved in piRNA methylation.
Figure 4.
GST-Pimet methylates single-stranded small RNAs in vitro. (A) miR-1/miR-1* duplexes (0.04 nmol/50 μL) were incubated with GST, GST-Pimet, and GST-HEN1 in the presence of 14C-labeled SAM. Resultant small RNAs were run on a denaturing acrylamide gel. The top panel (14C; autoradiograph of 14C-labeled RNAs) shows that only GST-HEN1 is able to transfer methyl groups from SAM to miRNA/miRNA* duplexes. The bottom panel shows a staining image (TBO, Toluidine Blue O) of the top gel, which indicates that the same amounts of miRNA/miRNA* duplexes were used in all the lanes. (B, top panel) When single-stranded small RNAs (26 nt) are the substrate for the methylation assay (0.5 nmol/50 μL), GST-Pimet shows stronger activity compared with that of GST-HEN1. The bottom panel shows a staining image of the top gel. (C) ssRNAs (22 nt, 26 nt, and 38 nt) were methylated by GST-Pimet in vitro. GST-Pimet seems to be indifferent to the length of substrates, unlike Arabidopsis HEN1 that hardly recognize 25-nt or longer RNAs as substrates in an in vitro methylation assay (Yang et al. 2006). (D) 2′-O-methyl modification of 26-nt ssRNAs prevents methylation, suggesting that only the 2′-hydroxyl group on the ribose of the far 3′ nucleotide is methylated by Pimet. (E) ssRNAs (26 nt) were methylated by GST-Pimet as in B and subjected to oxidation and β-elimination treatment. Only methylated ssRNAs (not visible in the bottom “TBO” panel) showed resistance (14C) to the chemical treatment.
Recombinant Pimet methylates piRNAs with Aub but not miRNAs with AGO1 from Pimet mutant ovary
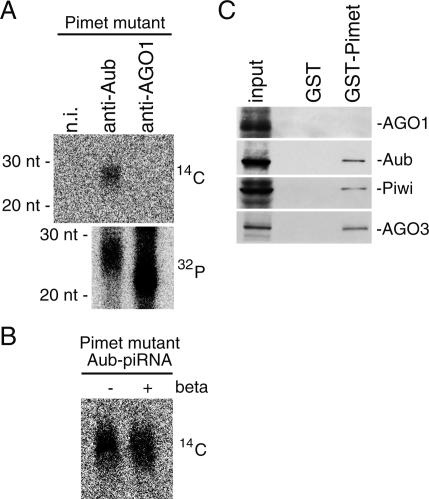
In Pimet mutant ovary, piRNAs associated with Aub and Piwi were not methylated at the 3′ ends (Fig. 3B), most likely due to loss of Pimet expression. We then examined if GST-Pimet is able to methylate these piRNAs associated with the PIWI proteins from Pimet mutant ovary. Aub–piRNA complexes were immunopurified with a specific antibody against Aub (Gunawardane et al. 2007) and subjected to in vitro methylation assays. As a control, miRNAs associated with AGO1 were also obtained through immunoprecipitation using anti-AGO1 (Miyoshi et al. 2005) from ovary lysate. We found that piRNAs were methylated even in a complex form with Aub (Fig. 5A, top panel). piRNA methylated in the assay showed resistance to oxidation and β-elimination treatment (Fig. 5B). Interestingly, miRNAs associated with AGO1 were not methylated, although these miRNAs are single-stranded in a complex form with AGO1 (Miyoshi et al. 2005). Confirmation that the miRNA levels in Figure 5A were several-fold higher than those of piRNAs was provided by phosphorylation of these small RNAs (Fig. 5A, bottom panel). It seems that small RNA methylation by Pimet is largely influenced by the accessibility of the 3′ ends of the substrates to Pimet itself. Structural analysis of Argonaute proteins suggests that the 5′ end of the small guide RNA is anchored in a highly conserved pocket in the PIWI domain, whereas the 3′ end of the small RNA is embedded in the PAZ domain (Ma et al. 2004; Parker and Barford 2006). Taken together, our results suggest that the 3′ ends of Aub-associated piRNAs are not tightly bound to the PAZ domain, but are exposed to the surface of the protein. On the other hand, the 3′ ends of AGO1-associated miRNAs are likely to be embedded in the PAZ domain and therefore are not exposed to the surface of the protein. Alternatively, but not mutually exclusively, it is conceivable that Pimet may interact only with PIWI proteins and not with AGO proteins, thereby methylating only small RNAs associated with PIWI proteins. To test this, we next examined if Pimet associates with PIWI proteins or not. A GST pull-down assay was performed; GST-Pimet was first incubated with Pimet mutant ovary lysate, and after extensive washing the eluates were probed with PIWI protein antibodies. Aub, Piwi, and AGO3 were clearly detected in the bound fraction with GST-Pimet but not with GST itself (Fig. 5C). By contrast, AGO1 was not observed in either lane (Fig. 5C). These results indicated that Pimet is capable of physically interacting with PIWI proteins containing piRNAs that can serve as substrates for Pimet methylation. Addition of RNaseA did not affect the interaction of Pimet with Aub (Supplementary Fig. S4D), suggesting that Pimet is able to associate directly with the PIWI proteins. In Drosophila, piRNA methylation may occur after matured piRNAs are loaded onto PIWI proteins. If so, it clearly differs from the case of miRNA methylation in plants, which likely occurs prior to miRNA loading onto the AGO proteins when miRNAs are still in a duplex form with the complementary miRNA* molecules (Yu et al. 2005).
Figure 5.
(A) Pimet methylates piRNAs associated with Aub obtained from Pimet mutant ovary, but does not methylate miRNAs associated with AGO1. Aub–piRNA complexes were immunopurified from Pimet mutant ovary lysate with anti-Aub antibody (Gunawardane et al. 2007) and subjected to methylation by GST-Pimet. miRNAs associated with AGO1 immunopurified from Pimet mutant ovary lysate were used as a control. Nonimmune IgG (n.i.) was also used as a negative control. (Top panel) piRNAs were even methylated in a complex with PIWI protein (14C). By contrast, miRNAs associated with AGO1 were not methylated, although these miRNAs are single-stranded in a complex form with AGO1 (Miyoshi et al. 2005). (Bottom panel) miRNA levels appearing in A were several-fold higher than those of piRNAs (32P; 5′ end phosphorylation after CIP). (B) Aub–piRNA complexes immunopurified from Pimet ovary lysate were subjected to methylation by GST-Pimet as in A. piRNAs were then isolated and subjected to oxidation and β-elimination treatment. piRNAs methylated in vitro showed resistance to the chemical treatment. (C) Association of Pimet with PIWI proteins. GST-Pimet (Supplementary Fig. S4C) was incubated with the Pimet mutant ovary lysate, and after extensive washing the eluates were probed with antibodies against Aub, Piwi, AGO3, and AGO1. Aub, Piwi, and AGO3 are clearly detected in the bound fraction with GST-Pimet but not with GST. AGO1 was not observed in either lane.
Mutations in Arabidopsis hen1 cause reduced fertility (Chen et al. 2002). Thus, is the piRNA methylation by Pimet crucial in Drosophila? piRNAs function in genome surveillance in germlines in concert with PIWI proteins (Saito et al. 2006; Vagin et al. 2006; Gunawardane et al. 2007). Mutations in aub, piwi, and others like spindle-E (homeless) cause piRNAs not to be accumulated in gonads (Aravin et al. 2004; Savitsky et al. 2006; Vagin et al. 2006), and lead to germ cell malformation and sterility (Gillespie and Berg 1995; Lin and Spradling 1997; Harris and Macdonald 2001). This clearly indicates that piRNAs are necessary for perpetuation of organisms. However, the Pimet mutant fly seems to be viable and fertile. Steady-state levels of piRNAs in the methylation-defective mutant are also similar to those in wild type (data not shown). Expression levels of retrotransposons do not seem to be changed by loss of Pimet expression (data not shown). Thus, the function of 3′ end methylation is currently unknown. Further investigation such as by immunohistochemistry may be required to obtain a more detailed morphology of the mutant. Extensive analyses of the mechanisms underlying piRNA methylation may also provide important clues to more fully elucidating piRNA biogenesis. Aub and AGO3, which determine and form the 5′ end of piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007) in piRNA biogenesis, were shown to be in the protein fraction associated with Pimet (Fig. 4B). Identifying more Pimet-associated proteins may reveal the factors required for formation of the 3′ end of piRNAs.
Materials and methods
Periodate oxidation and β-elimination
AGO1, Aub, and Piwi were immunopurified as described (Saito et al. 2006) from an ovary using anti-AGO1 (Miyoshi et al. 2005), anti-Aub (Gunawardane et al. 2007), and anti-Piwi (Saito et al. 2006) antibodies, respectively. Small RNAs coimmunopurified with AGO1, Aub, and Piwi were isolated from the immunoprecipitates using phenol and phenol: chloroform and were ethanol-precipitated. RNAs isolated were labeled with 32P-γ-ATP after CIP treatment. Ten-thousand counts per minute (cpm) of 32P-labeled RNAs were used per reaction. Periodate oxidation and β-elimination of RNAs were performed as described (Gunawardane et al. 2007). Chemically synthesized luc guide siRNA (Okamura et al. 2004) was used as a control RNA.
Nucleoside analysis of piRNAs by capillary LC nano electrospray ionization (ESI)/MS
piRNAs associated with Aub in wild-type fly ovary were obtained as described in the Periodate Oxidation/β-Elimination section. piRNAs (∼5 pmol) were digested by RNase T2 into 3′-, 5′-diphospho nucleotides, 3′-monophospho nucleotides, and 3′-terminal nucleosides for 3 h at 37°C in 5 μL of reaction mixture containing 10 mM ammonium acetate (pH 5.3) and 1.25 U/mL RNase T2 (GE Healthcare) as described (Ohara et al. 2007; Suzuki et al. 2007). The reaction solution was evaporated and dissolved in 1.5 μL of 50 mM ammonium acetate (pH 5.3). Synthetic 2′ or 3′-O-methyl nucleosides were obtained from ChemGenes Corporation and Sigma-Aldrich. The procedure for LC/MS analysis has been described (Ikeuchi et al. 2006). To analyze a limited quantity of nucleosides, we devised our own system of capillary LC coupled with ESI/MS. A QStar XL Hybrid LC/MS/MS System (Applied Biosystems) equipped with a NanoSpray II nanoelectrospray (Applied Biosystems) source, a Nanovolume Valve (Valco Instruments), and a splitless nano HPLC system DiNa (KYA Technologies) were used to analyze RNase T2-digested piRNAs. The digest (1.5 μL) was loaded onto an ODS capillary column (HiQsil C18H-3, 0.15 × 150 mm; KYA Technologies) directly via a 1-μL loop injection and was chromatographed at a flow rate of 500 nL/min using a linear gradient of 0%–60% of Solvent B (50% acetonitrile containing 5 mM ammonium acetate at pH 5.3)over Solvent A (5 mM ammonium acetate at pH 5.3) in 40 min. The chromatographic eluent was sprayed and ionized from an energized (+2.2-kV) sprayer tip attached to the capillary column. Throughout the separation, ionized molecules were scanned in a positive polarity mode over an m/z range of 108–400. CID of each 2′-O-methylnucleoside was carried out with a collision energy of +25, and scanned in an m/z range of 50–300.
RT–PCR, in vitro small RNA methylation assay, and GST pull-down assay were performed as described in the Supplemental Material.
Acknowledgments
We thank Xuemei Chen for the cDNA of Arabidopsis HEN1 and the miRNA methylation protocol. We also thank Tomoko Mori, Tomoko Nagami, Kazumichi M. Nishida, and Yoshinori Kawamura for technical assistance, and other members of the Siomi laboratory for discussions and comments on the manuscript. K.S. is a post-doctoral fellow of the 21st century COE Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT). This work was supported in part by grants to T.S., M.C.S., and H.S. from MEXT and the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1563607
References
- Aravin A.A., Klenov M.S., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A., Klenov M.S., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A., Bantignies F., Cavalli G., Gvozdev V.A., Cavalli G., Gvozdev V.A., Gvozdev V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J., Girard A., Fejes-Toth K., Hannon G.J., Fejes-Toth K., Hannon G.J., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Kellis M., Sachidanandam R., Hannon G.J., Sachidanandam R., Hannon G.J., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J., Xuan Z., Zhang M.Q., Hannon G.J., Zhang M.Q., Hannon G.J., Hannon G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chen X., Liu J., Cheng Y., Jia D., Liu J., Cheng Y., Jia D., Cheng Y., Jia D., Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D.E., Berg C.A., Berg C.A. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes & Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. A Slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris A.N., Macdonald P.M., Macdonald P.M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Filippov D.V., Blaser H., Raz E., Moens C.B., Blaser H., Raz E., Moens C.B., Raz E., Moens C.B., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., Balint E., Tuschl T., Zamore P.D., Tuschl T., Zamore P.D., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y., Shigi N., Kato J., Nishimura A., Suzuki T., Shigi N., Kato J., Nishimura A., Suzuki T., Kato J., Nishimura A., Suzuki T., Nishimura A., Suzuki T., Suzuki T. Mechanistic insights into sulfur-relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Kim V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes & Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- Kirino Y., Mourelatos Z., Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X., Yang Z., Yu B., Liu J., Chen X., Yu B., Liu J., Chen X., Liu J., Chen X., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Spradling A.C., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Ma J.B., Ye K., Patel D.J., Ye K., Patel D.J., Patel D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C., Tsukumo H., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K.A., Boeke J.D., Boeke J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Miyauchi K., Suzuki T., Suzuki T. The 3′-termini of mouse piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C., Ishizuka A., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X., Li J., Song R., Messing J., Chen X., Song R., Messing J., Chen X., Messing J., Chen X., Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.S., Barford D., Barford D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem. Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Filipowicz W., Bhattacharyya S.N., Filipowicz W., Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M., Kwon D., Georgiev P., Kalmykova A., Gvozdev V., Kwon D., Georgiev P., Kalmykova A., Gvozdev V., Georgiev P., Kalmykova A., Gvozdev V., Kalmykova A., Gvozdev V., Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes & Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Sakaguchi Y., Suzuki T., Sakaguchi Y., Suzuki T., Suzuki T. Mass spectrometric analysis of 3′-terminal nucleosides in non-coding RNAs. Nat. Protoc. 2007 doi: 10.1038/nprot.2007.185. [DOI] [Google Scholar]
- Tomari Y., Zamore P.D., Zamore P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Matranga C., Haley B., Martinez N., Zamore P.D., Matranga C., Haley B., Martinez N., Zamore P.D., Haley B., Martinez N., Zamore P.D., Martinez N., Zamore P.D., Zamore P.D. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Li C., Seitz H., Gvozdev V., Zamore P.D., Seitz H., Gvozdev V., Zamore P.D., Gvozdev V., Zamore P.D., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Williams R.W., Rubin G.M., Rubin G.M. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Ebright Y.W., Yu B., Chen X., Ebright Y.W., Yu B., Chen X., Yu B., Chen X., Chen X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X., Yang M., Padgett R.W., Steward R., Chen X., Padgett R.W., Steward R., Chen X., Steward R., Chen X., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M., Irvine D.V., Martienssen R.A., Irvine D.V., Martienssen R.A., Martienssen R.A. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]