Abstract
Many elongation factors in eukaryotes promote gene expression by increasing the processivity of RNA polymerase II (Pol II). However, the stability of RNA Pol II elongation complexes suggests that such complexes are not inherently prone to prematurely terminating transcription, particularly at physiological nucleotide concentrations. We show that the termination factor, Pcf11, causes premature termination on an HIV provirus. The transcription that occurs when Pcf11 is depleted from cells or an extract is no longer sensitive to 6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), a compound that causes premature termination. Hence, Pcf11 can act as a negative elongation factor to repress RNA Pol II gene expression in eukaryotic cells.
Keywords: RNA polymerase II, transcription termination, HIV, gene expression
Transcription elongation can be defined as the process by which RNA polymerase traverses a gene while synthesizing RNA. For protein-encoding genes in eukaryotic cells, a plethora of proteins have been identified that either stimulate or inhibit elongation by RNA polymerase II (Pol II) (Sims et al. 2004; Saunders et al. 2006). Many proteins influence elongation by directly associating with Pol II, while others act indirectly by modifying the chromatin structure. Under conditions that inhibit elongation, Pol II is prone to premature termination, which is often described as a reduction in processivity. For example, treatment of cells with the transcription inhibitor 6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) causes production of short transcripts both in vitro and in vivo, suggesting that DRB reduces the processivity of the Pol II by promoting premature termination (Fraser et al. 1979; Tamm et al. 1980; Marciniak and Sharp 1991). Mutations in several elongation factors in yeast were found to have little impact on the rate of elongation; instead, the processivity of Pol II was diminished (Mason and Struhl 2005). These and other studies of Pol II elongation imply the existence of forces that cause Pol II to prematurely dissociate and terminate transcription. Yet there is little evidence that an elongation complex containing only Pol II and nucleic acids is intrinsically unstable as the complexes remain intact following treatment with high salt, detergents, and even proteases. Moreover, at physiological nucleotide concentrations, Pol II typically transcribes to the end of a DNA template (Izban and Luse 1991).
Only two proteins are known that have the capacity to dissociate Pol II elongation complexes from the DNA template. TTF2 dissociates the elongation complex in an ATP-dependent manner (Jiang et al. 2004). The protein is concentrated in the cytoplasm through out most of the cell cycle and associates with chromosomes during M phase to dissociate elongation complexes during chromosome condensation. There is no evidence that TTF2 affects transcription outside of M phase of the cell cycle in vivo. The second protein is Pcf11. Pcf11 is one of a collection of proteins involved in 3′ end processing of mRNA and transcription termination of protein-encoding genes (Buratowski 2005; Rosonina et al. 2006). It is the only protein in this collection that has been demonstrated to dissociate transcriptionally engaged Pol II from DNA; thus, Pcf11 could play a pivotal role in termination (Zhang et al. 2005; Zhang and Gilmour 2006).
Chromatin immunoprecipitation (ChIP) analyses show that Pcf11 associates with the promoter region of some genes, although it is most concentrated at the 3′ end (Licatalosi et al. 2002; Kim et al. 2004; Zhang and Gilmour 2006). Here we investigate the possibility that Pcf11 can act to inhibit transcription in vivo by causing premature termination. We chose the HIV provirus as a model because studies have demonstrated that inefficient transcription of the provirus is associated with premature termination (Kao et al. 1987; Laspia et al. 1989; Cullen 1990, 1991; Feinberg et al. 1991; Marciniak and Sharp 1991). We provide evidence that Pcf11 represses transcription of the provirus by causing premature termination.
Results and Discussion
Depletion of Pcf11 with small interfering RNA (siRNA) results in increased virus production
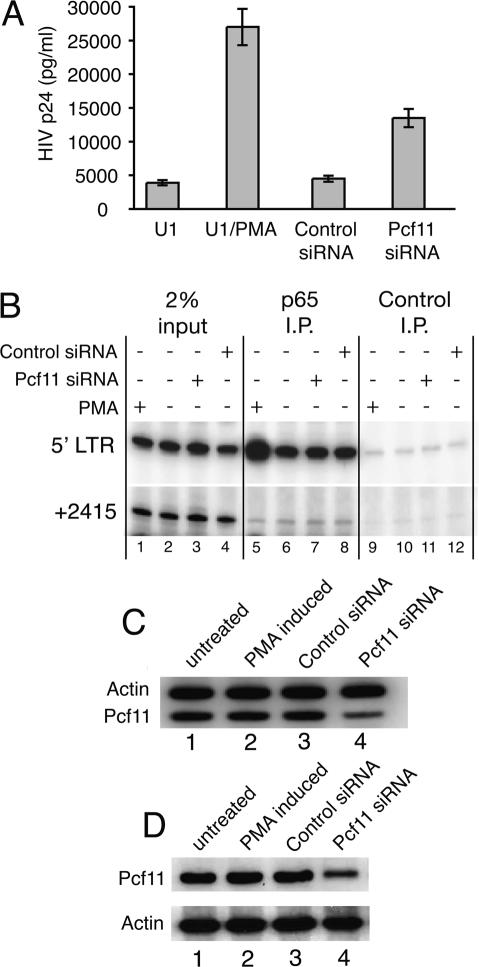
U1 cells contain two transcriptionally repressed copies of the HIV provirus (Verdin et al. 1993). Upon treatment with the phorbol ester PMA, provirus transcription and replication were increased sevenfold as measured by levels of HIV p24 in the culture media (Fig. 1A). This agrees with previous studies, and suggests that transcription of the provirus in our untreated cells was subject to repression (Folks et al. 1987, 1988). When we depleted Pcf11 from U1 cells using siRNA, we observed a threefold increase in the level of HIV p24 (Fig. 1A).
Figure 1.
Depletion of Pcf11 increases HIV replication in latently infected U1 cells. (A) U1 cells were either untreated or treated with 10 ng/mL PMA, control siRNA, or Pcf11 siRNA. Relative levels of virus production were determined by measuring viral protein p24 in the culture medium using an ELISA assay. (B) ChIP analysis for the p65 subunit of NF-κB. Cells were treated as described by the matrix above the lanes. (C) RT–PCR analysis of Pcf11 and actin mRNA shows that Pcf11 mRNA is diminished in cells treated with Pcf11 siRNA. (D) Western blot analysis of Pcf11 and actin protein in crude nuclear preparations shows that Pcf11 protein is diminished in cells treated with Pcf11 siRNA. Pcf11 migrated slightly slower than a 175-kDa marker protein on 8% SDS-PAGE.
Induction of HIV provirus transcription with PMA involves NF-κB, and ChIP showed that the level of NF-κB associating with the provirus increased fourfold (Fig. 1B, cf. lanes 5 and 6). In contrast, depletion of Pcf11 did not alter the level of the p65 subunit of NF-κB associated with the provirus (Fig. 1B, cf. lanes 7 and 8). Therefore, induction of HIV transcription and replication did not appear to be a result of ectopic activation of U1 cells by siRNA but rather a direct consequence of depleting Pcf11. RT–PCR and Western blot analysis indicated that the Pcf11 siRNA was indeed depleting Pcf11 message and protein (Fig. 1C,D).
ChIP analysis detects Pcf11 in the promoter region of the HIV provirus
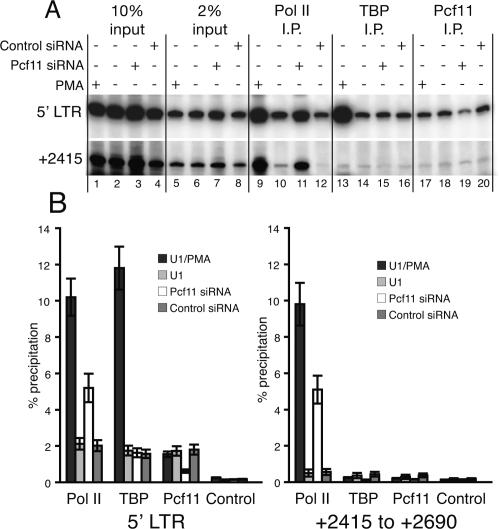
If Pcf11 was mediating premature termination, we predicted that it might be located at the HIV promoter within the 5′ long terminal repeat (LTR). ChIP analyses revealed significantly more Pcf11 in the HIV promoter region than in a region ∼2.4 kb downstream (Fig. 2A [lane 18], B). Moreover, depletion of Pcf11 with siRNA reduced the level of Pcf11 associated with the promoter region (Fig. 2A [cf. lanes 19 and 20], B). These results indicate that the increased expression of virus caused by the Pcf11 siRNA could be a direct consequence of reducing the level of Pcf11 associated with the promoter region of the provirus.
Figure 2.
Depletion of Pcf11 increases the density of Pol II downstream from the promoter region. Interactions of Pol II, TBP, and Pcf11 with the provirus were determined by ChIP. (A) Representative data for one ChIP experiment. Immunoprecipitated DNA, 10% input DNA, and 2% input DNA were subjected to PCR amplification for the regions corresponding to the promoter region in the 5′ LTR (+1 to +248) or a downstream region (+2415 to +2690). (B) Quantification of ChIP data from three independent experiments. The control samples recovered with nonspecific antibody are shown in Figure 1B.
ChIP analysis reveals that depletion of Pcf11 increases the processivity of Pol II
To investigate the mechanism by which depletion of Pcf11 increased HIV expression, we used ChIP to monitor the interactions of Pol II and TBP. TBP was monitored to determine if treatments increased the transcriptional potential of the LTR by promoting binding of TBP (Raha et al. 2005). TBP and Pol II were clearly present at the promoter in untreated U1 cells (Fig. 2A [lanes 10,14], B). This agrees with previous studies showing that HIV transcription is controlled at least in part by regulating Pol II after it associates with the HIV promoter (Cullen 1991). Treatment of U1 cells with PMA increased binding of TBP and Pol II by fivefold, indicating that binding of TBP is in part limiting HIV transcription in U1 cells prior to PMA treatment. In contrast, depleting Pcf11 increased Pol II binding by 2.5-fold in the promoter region but had no effect on TBP. Therefore, the increase in transcription seen upon depletion of Pcf11 is not likely due to an increase in the number of initiation complexes.
Depletion of Pcf11 could be increasing the processivity of Pol II, thus leading to an increase in virus production. Measurement of the association of Pol II with a region 2.4 kb downstream from the transcription start site supports this conclusion. Minimal Pol II could be detected at the downstream region in untreated cells; there was ∼10-fold more Pol II at the promoter than the region 2.4 kb downstream (Fig. 2A [lanes 10,12], B). This indicates that those Pol II molecules associated with the promoter region rarely transcribe to the distal region in uninduced cells. In contrast, 2.5-fold and fivefold more Pol II was associated with these downstream sequences upon Pcf11 depletion with siRNA or upon PMA induction, respectively (Fig. 2A [lanes 9,11], B). Furthermore, similar amounts of Pol II were detected at the 5′ LTR and the 2.4-kb region, indicating that the polymerases were more processive than in untreated or siRNA control treated cells.
It is interesting to note that we observed no change in the level of Pcf11 associated with the promoter region following PMA induction (Fig. 2A, lanes 17,18). However, if we consider the increase in Pol II observed in the promoter region, then the ratio of Pcf11 to Pol II decreases fivefold. Hence, a significant fraction of the Pol II molecules initiating transcription in the presence of PMA appear not to interact with Pcf11. This is consistent with the hypothesis that transcriptional activation inhibits the association of Pcf11 with the elongation complex.
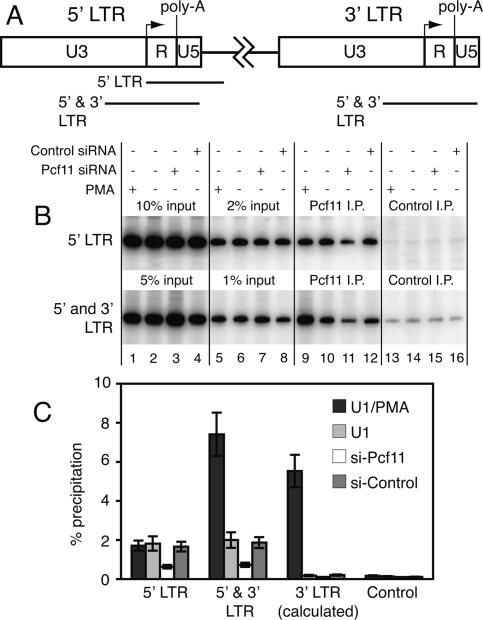
To further investigate the association of Pcf11 with the provirus, we monitored the interaction of Pcf11 with the 3′ LTR using ChIP. This analysis is complicated by the fact that the 5′ and 3′ LTRs of the provirus have the same sequence. Consequently, we PCR-amplified a region that corresponded to both the 5′ and 3′ LTR and compared the results with PCR amplification of the 5′ LTR alone; the latter was achieved by having one PCR primer hybridize to a site just downstream from the 5′ LTR. Amplification of both LTRs from Pcf11 ChIP DNA resulted in ∼2.5-fold more signal for cells induced with PMA than uninduced or siRNA-control treated cells (Fig. 3B [cf. lanes 9 and 10 and 12], C). As expected, the signal for the cells treated with Pcf11 siRNA was significantly less than the control samples (Fig. 3B, lane 11). Amplification of only the 5′ LTR from the Pcf11 ChIP DNA resulted in similar signals for the PMA-induced and uninduced U1 cells (Fig. 3B [cf. lanes 9 and 10 and 12], C). Since amplification of both LTRs yielded a 2.5-fold stronger signal for PMA-induced cells than uninduced cells while amplification of only the 5′ LTR yielded similar signals, we conclude that PMA induction results in increased recruitment of Pcf11 to the 3′ LTR. Presumably the Pcf11 that associates with the 3′ LTR participates in 3′ end formation and transcription termination. However, since depletion of Pcf11 increases virus production, Pcf11 is either not essential or the level of depletion is insufficient to interfere with processes involving the 3′ LTR. For example, transcription termination mediated by the Xrn2 RNA exonuclease could provide an alternative pathway for termination (West et al. 2004).
Figure 3.
HIV induction by PMA results in recruitment of Pcf11 to the 3′ LTR. (A) Schematic showing the 5′ and 3′ LTRs, the location of PCR-amplified regions corresponding to the 5′ LTR and both the 5′ and 3′ LTRs, the transcription start site (arrow), potential polyadenylation sites. (B) Representative data for one ChIP experiment. Immunoprecipitated DNA and the indicated percentages of input DNA were subjected to PCR amplification of the 5′ LTR alone or both the 5′ and 3′ LTRs. The amount of input for amplification of both the 5′ and 3′ LTRs was half that of the input for the 5′ LTR. (C) Quantification of ChIP data from three independent experiments. The signals for the 3′ LTR were calculated by subtracting the intensity of the signals for the 5′ LTR alone from the signals for the sum of the 5′ and 3′ LTRs.
Transcription in cells caused by depletion of Pcf11 is insensitive to the elongation inhibitor DRB
If Pcf11 induces premature termination, then transcription caused by depletion of Pcf11 might no longer be sensitive to DRB, a transcriptional inhibitor that causes premature termination (Fraser et al. 1979; Tamm et al. 1980; Marciniak and Sharp 1991). DRB sensitivity has been ascribed to two factors, NELF and DSIF, but neither of these factors has been shown to cause Pol II to dissociate from DNA; they merely slow the rate of elongation (Wada et al. 1998; Yamaguchi et al. 1999).
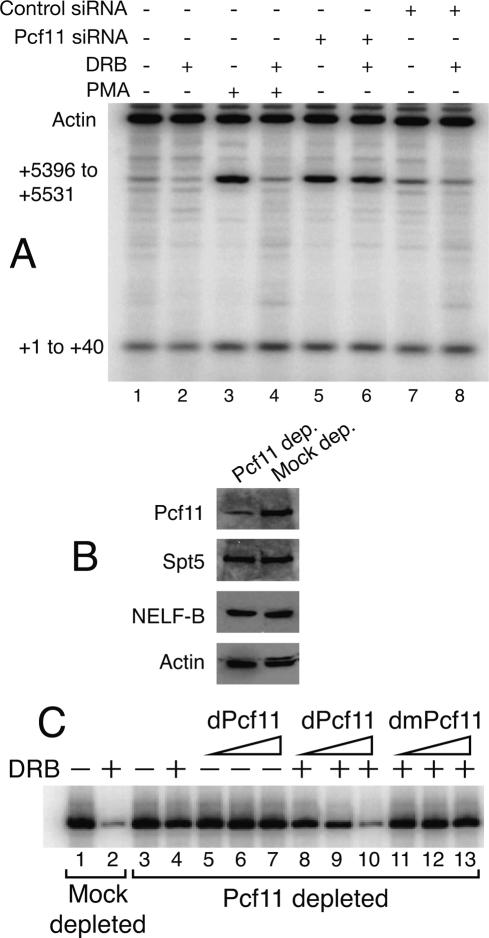
We tested if Pcf11 might be involved in conferring DRB sensitivity by comparing the DRB sensitivity of transcription in cells induced by PMA and by siRNA-mediated depletion of Pcf11. Transcription of the first 40 nucleotides (nt) of the provirus and of a region 5.3 kb downstream from the transcription start were monitored by RT–PCR. The level of transcript derived from the first 40 nt of the provirus was unchanged by any of our treatments (Fig. 4, all lanes, +1 to +40). This was expected because transcription of this region occurs even under uninduced conditions, and stable short transcripts accumulate in the cytoplasm of these cells (Toohey and Jones 1989; Kessler and Mathews 1992). PMA treatment caused a significant increase in transcription of the distal region consistent with an increase in the processivity of Pol II (Fig. 4, cf. lanes 1 and 3, +5396 to +5531). In accord with its action as an elongation inhibitor, DRB repressed PMA-dependent transcription of the distal region (Fig. 4, cf. lanes 3 and 4). In contrast, transcription induced by depletion of Pcf11 was insensitive to DRB (Fig. 4, cf. lanes 5 and 6). We note that similar results were obtained with DRB concentrations of 5 and 20 μM (data not shown). These low concentrations of DRB were chosen to reduce inhibition of transcription of cellular genes (Mancebo et al. 1997).
Figure 4.
Depletion of Pcf11 from cells or cell extracts renders transcription insensitive to DRB. (A) RT–PCR analysis of transcripts in U1 cells. Double-stranded RNA was transfected into U1 cells using an Amaxa Nucleofector II. Approximately 2 million cells (2 mL of cells treated with siRNA or not) were placed in a culture dish. Twelve hours post-transfection (or not), cells were incubated with or without 5 μM DRB. After 12 h, cells were stimulated with 2 ng/mL PMA or not for 12 h. Total RNA was isolated, and transcripts spanning +1 to +40 and +5396 to +5531 of HIV RNA and actin were detected by RT–PCR. (B) Western blot analysis of Pcf11-immunodepleted and mock-depleted HeLa nuclear extracts. Spt5 and NELF-F B are subunits of DSIF and NELF, respectively. (C) In vitro transcription of the 5′ LTR in Pcf11-immunodepleted (lanes 3–13) or mock-depleted (lanes 1,2) HeLa nuclear extracts. DRB at 50 μM was present in those samples marked with plus signs. dPcf11 and dmPcf11, which were isolated from Escherichia coli, encompass amino acids 1–283. The amounts of exogenously added Pcf11 correspond to 0.1 μg, 0.5 μg, and 1 μg.
Pcf11 contributes to the DRB sensitivity of transcription in vitro
To determine if Pcf11 contributes to DRB sensitivity in vitro, we performed Pcf11 immunodepletion and Pcf11 add-back experiments with HeLa nuclear extracts. Western blot analysis shows that Drosophila Pcf11 antibody selectively depletes Pcf11 from the extract (Fig. 4B). Spt5 and NELF-B are subunits of DSIF and NELF, respectively, and neither is depleted by the Pcf11 antibody.
Transcription reactions were performed on an HIV template spanning the region from −415 to +214 with mock-depleted and Pcf11-depleted nuclear extracts. DRB at 50 μM dramatically inhibited transcription in the mock-depleted extract but had little effect on the Pcf11-depleted extract (Fig. 4C, lanes 1–4). To attribute the loss of DRB sensitivity to depletion of Pcf11, we tested if addition of purified Pcf11 to the extract would restore DRB sensitivity. Human Pcf11 is not well characterized (de Vries et al. 2000). Hence, we added back a 283-amino-acid piece of Drosophila Pcf11 that had been previously shown to dismantle an elongation complex (Zhang and Gilmour 2006). Addition of dPcf11 to the Pcf11-depleted extract had no effect on transcription in the absence of DRB (Fig. 4C, lanes 5–7). In contrast, transcription by the Pcf11-depeleted extract became sensitive to DRB upon addition of the Drosophila Pcf11 (Fig. 4, lanes 8–10). We tested if the ability of Pcf11 to restore DRB sensitivity correlated with the capacity of Pcf11 to dismantle elongation complexes by determining if a mutant would restore DRB sensitivity. Mutation of amino acids 75, 76, and 77 inactivates the dismantling activity of Pcf11 (Zhang and Gilmour 2006). Addition of this mutant to the Pcf11-depleted extract failed to restore DRB sensitivity (Fig. 4, lanes 11–13), suggesting that the ability of Pcf11 to dismantle the elongation complex was involved in conferring DRB sensitivity.
Conclusion
The processivity of Pol II is recognized as an important parameter in regulating transcription (Mason and Struhl 2005). Pol II with poor processivity is by definition a form of the enzyme that is prone to premature termination. However, the molecular basis for premature termination is unknown. Results presented here identify the termination factor, Pcf11, as a factor that causes premature termination and negatively regulates gene expression. Since HIV expression increases when Pcf11 is depleted, premature termination plays a significant role in maintaining the transcriptional latency of the provirus.
The discovery that Pcf11 is involved in causing DRB sensitivity provides a possible explanation of how DRB causes premature termination. Biochemical analyses have resulted in the following model for DRB sensitivity (Peterlin and Price 2006). Transcription elongation by Pol II is inhibited by the combination of two proteins, NELF and DSIF, which associate with the elongation complex. The kinase, P-TEFb, overcomes this inhibition by phosphorylating one or more members of the trio of Pol II, NELF, and DSIF (Renner et al. 2001). P-TEFb is inhibited by DRB, thus allowing DRB to interfere with P-TEFb’s function in overcoming the inhibition by NELF and DSIF. This model, however, lacks an explanation for premature termination because NELF and DSIF have only been shown to slow elongation. Based on our data, we propose that Pcf11 causes premature termination by acting on Pol II elongation complexes that have paused as a consequence of their association with NELF and DSIF. Previously, we obtained evidence that Pcf11 acts only on paused elongation complexes (Zhang and Gilmour 2006). Recently, we determined that NELF causes Pol II to pause in the promoter proximal region of the HIV provirus (Zhang et al. 2007). We posit that the pause induced by DSIF and NELF allows Pcf11 to dismantle the elongation complex.
Since DRB inhibits transcription of a wide spectrum of genes in human cells (Lam et al. 2001), it is possible that Pcf11 could be a significant component in the regulation of gene expression. Indeed, we predict that any interaction that causes Pol II to pause could render the elongation complex susceptible to Pcf11. This might explain why depleting Pcf11 caused an increase in HIV transcription in cells but not in the cell extract (Fig. 4, cf. A and C). The DNA template in cells is packaged into chromatin, which could slow elongation and render Pol II susceptible to Pcf11. Hence, in cells, chromatin structure could act in addition to repression by NELF and DSIF. In contrast, the DNA templates in our in vitro transcription reactions were unlikely to be packaged into chromatin. Consequently, NELF and DSIF might be primarily responsible for rendering the Pol II susceptible to Pcf11 in these cell-free reactions. A possible role for the plethora of positive elongation factors is to counteract the premature termination activity of Pcf11.
Significant new insight into transcription regulatory mechanism might be uncovered by investigating parameters that control the activity of Pcf11. A hint of such regulation is provided by our observation that the ratio of Pcf11 to Pol II is substantially greater at the 5′ LTR of the inactive provirus than the 5′ LTR of the PMA-induced provirus. Thus, part of the activation mechanism could involve steps that prevent Pcf11 from associating with the elongation complex.
Materials and methods
Antibodies
Anti-p65 (A, sc-109), anti-Pol II (N-20, sc-899), and anti-Spt5 (sc-28678) were obtained from Santa Cruz Biotechnology, Inc. Mouse anti-β-actin was obtained from Sigma-Aldrich. Anti-NELF-B antibody was obtained from Rong Li (University of Texas Health Science Center at San Antonio, San Antonio, TX) (Aiyar et al. 2004). Rabbit anti-Pcf11 antiserum was previously described (Zhang and Gilmour 2006).
Depletion of Pcf11 from U1 cells with siRNA
siRNA against Pcf11 (siGENOME SMARTpool reagent M-015381-00-0005) and control siRNA were obtained from Dharmacon. siRNA (350 nM) was transfected into 1 × 106 ∼ 2 × 106 U1 cells using the Amaxa Nucleofector II. Cells were recovered in RPMI media supplemented with 10% fetal bovine serum. Twenty-four hours post-transfection, fresh RPMI was added and cells were harvested at 48 h post-transfection.
ChIP
ChIP analyses were performed as previously described (Zhang et al. 2007). The following primers were used to amplify different regions of the HIV-1 transcription unit: 5′ LTR, +1 to +248 (GGGTCTCTCTGGT TAGA and AGTCCTGCGTCGAGAGAG), HIV +2415 to +2690 (GTAA CAGTACTGGATGTGGGTGATG and CTGCCCTATTTCTAAGTCA GATCC), and 5′ and 3′ LTR (ACTGCTGACATCGAGCTTGC and CCACACTGACTAAAAGGGTCTG).
RT–PCR
RT–PCR was performed as previously described (Zhang et al. 2007). The following primers were used to amplify different regions of the HIV-1 gene: initiated short transcripts, +1 to +40 (GGGTCTCTCTGGTTAGA and AGAGCTCCCAGGCTCA); elongated transcripts, +5396 to +5531 (GACTAGAGCCCTGGAAGCA and GCTTCTTCCTGCCATAGGAG). β-Actin was detected with the primers GTCGACAACGGCTCCG GC and GGTGTGGTGCCAGATTTTCT. Ten microliters of each PCR reaction were run on a 9% nondenaturing acrylamide gel. Gels were fixed in 10% acetic acid for 20 min, dried, and then analyzed with a PhosphorImager (Molecular Dynamics). The intensity of each band was quantified by volume analysis using ImageQuant software.
Western blot analysis for Pcf11 in U1 cells
Nuclei were prepared by lysing 2 × 106 cells with 1% Nonidet P-40 in buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF) and collecting crude nuclei by centrifugation in a microfuge at 10,000 rpm. Nuclei were resuspended in 50 μL of buffer C (20 mM HEPES at pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF). An equal volume of nuclear suspension was added to 2× SDS loading buffer (100 mM Tris-HCl at pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mM DTT) and run on an 8% SDS–polyacrylamide gel. After transfer of proteins to the nitrocellulose membrane, anti-Pcf11 polyclonal antiserum was used to probe for Pcf11, or mouse anti-β-actin Ab was used to detect β-actin. HRP-conjugated goat anti-rabbit IgG and goat anti-mouse IgG (Sigma-Aldrich) were used as secondary antibodies. Proteins were detected using the ECL Plus Western blotting detection system (Amersham Biosciences).
Immunodepletion of Pcf11 from HeLa nuclear extracts and in vitro transcription
Pcf11 antibody sepharose was made by incubating 100 μL of Pcf11 antiserum with 50 μL of protein A-sepharose followed by covalent coupling with dimethyl pimelimidate (Harlow and Lane 1999). Control antibody sepharose was made from preimmune antiserum. To reduce nonspecific binding during depletions, the antibody sepharose was first incubated for 4 h with 100 μL of nuclear extract generated from U1 cells (Cook et al. 2003). Antibody-sepharose was then washed extensively with 0.1 M KCl/HEMG (100 mM KCl, 25 mM HEPES at pH 7.6, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 mM DTT), followed by two washes with 0.1 M glycine (pH 2.5) and extensive washing with 0.1 M KCl/HEMG. Antibody-sepharose was stored at 4°C.
HeLa cell nuclear extracts were obtained from Promega (HeLaScribe Nuclear Extract in vitro Transcription System). For mock and Pcf11 depletion, 40 μL of nuclear extract were incubated for 1.5 h with 20 μL of antibody-sepharose at 4°C. Supernatants were collected with a Handee Spin Cup (Pierce) and then incubated with a fresh 20-μL portion of antibody-sepharose. Depleted extracts were stored at −80°C. The DNA template used for in vitro transcription consisted of the HIV 5′ LTR region spanning from −450 to +214 that had been PCR-amplified with the primers TGGAAGGGCTAATTCACTCCC and TTCGCTTTCAGGTCCCT GTTC.
Transcription reactions were performed as instructed by the HeLaScribe Nuclear Extract in vitro Transcription System (Promega) with minor modifications. First assembled was a 22-μL solution containing 200 ng of HIV DNA template, 7 μL of HeLa Nuclear Extract 1× Transcription Buffer (Promega), 4 μL of HeLa Nuclear Extract (Pcf11- or mock-depleted), 1.5 μL of 50 mM MgCl2, 10 U of RNasin, and 0.5 mM DTT. Where indicated, 1 μL of dPcf11 1–238 or dmPcf11 1–283 (freshly diluted in TE) at 0.1 μg/μL, 0.5 μg/μL, or 1.0 μg/μL was then added. The mixture was incubated at 37°C for 10 min to allow formation of preinitiation complexes. Following preinitiation complex formation, 0.5 μL of 2.5 mM DRB (in ethanol) or ethanol was added. Transcription was started by adding 3 μL of the following nucleotide mix: 1.0 μL of 10 mM ATP, 10 mM UTP, 10 mM GTP, 0.4 mM CTP, and 2.0 μL of [32P] CTP (6000 Ci/mmol, 10 mCi/mL). Transcription was allowed to occur at 37°C for 30 min. Transcription was stopped by adding 175 μL of HeLa Extract Stop Solution (Promega), and RNA was isolated as directed by the manufacturer. RNA was analyzed on a 6% polyacrylamide gel containing 8 M urea.
Acknowledgments
We thank Rong Li (University of Texas Health Science Center at San Antonio) for providing NELF-B antibody. This project is supported by Pennsylvania State University Tobacco Formula Funds and NIH grants AI62467 (to A.J.H.) and GM47477 (to D.S.G.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1542707
References
- Aiyar S.E., Sun J.L., Blair A.L., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Sun J.L., Blair A.L., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Blair A.L., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., Mukherjee A., Ren D.M., Handa H., Ren D.M., Handa H., Handa H., et al. Attenuation of estrogen receptor α-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes & Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Cook J.A., Albacker L., August A., Henderson A.J., Albacker L., August A., Henderson A.J., August A., Henderson A.J., Henderson A.J. CD28-dependent HIV-1 transcription is associated with Vav, Rac, and NF-κ B activation. J. Biol. Chem. 2003;278:35812–35818. doi: 10.1074/jbc.M302878200. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. The HIV-1 Tat protein: An RNA sequence-specific processivity factor. Cell. 1990;63:655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. Regulation of HIV-1 gene expression. FASEB J. 1991;5:2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- de Vries H., Ruegsegger U., Hubner W., Friedlein A., Langen H., Keller W., Ruegsegger U., Hubner W., Friedlein A., Langen H., Keller W., Hubner W., Friedlein A., Langen H., Keller W., Friedlein A., Langen H., Keller W., Langen H., Keller W., Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M.B., Baltimore D., Frankel A.D., Baltimore D., Frankel A.D., Frankel A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks T.M., Justement J., Kinter A., Dinarello C.A., Fauci A.S., Justement J., Kinter A., Dinarello C.A., Fauci A.S., Kinter A., Dinarello C.A., Fauci A.S., Dinarello C.A., Fauci A.S., Fauci A.S. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- Folks T.M., Justement J., Kinter A., Schnittman S., Orenstein J., Poli G., Fauci A.S., Justement J., Kinter A., Schnittman S., Orenstein J., Poli G., Fauci A.S., Kinter A., Schnittman S., Orenstein J., Poli G., Fauci A.S., Schnittman S., Orenstein J., Poli G., Fauci A.S., Orenstein J., Poli G., Fauci A.S., Poli G., Fauci A.S., Fauci A.S. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J. Immunol. 1988;140:1117–1122. [PubMed] [Google Scholar]
- Fraser N.W., Sehgal P.B., Darnell J.E., Sehgal P.B., Darnell J.E., Darnell J.E. Multiple discrete sites for premature RNA chain termination late in adenovirus-2 infection: Enhancement by 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole. Proc. Natl. Acad. Sci. 1979;76:2571–2575. doi: 10.1073/pnas.76.6.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D., Lane D. Using antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- Izban M.G., Luse D.S., Luse D.S. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes & Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liu M., Spencer C.A., Price D.H., Liu M., Spencer C.A., Price D.H., Spencer C.A., Price D.H., Price D.H. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol. Cell. 2004;14:375–385. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- Kao S.Y., Calman A.F., Luciw P.A., Peterlin B.M., Calman A.F., Luciw P.A., Peterlin B.M., Luciw P.A., Peterlin B.M., Peterlin B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kessler M., Mathews M.B., Mathews M.B. Premature termination and processing of human immunodeficiency virus type 1-promoted transcripts. J. Virol. 1992;66:4488–4496. doi: 10.1128/jvi.66.7.4488-4496.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ahn S.H., Krogan N.J., Greenblatt J.F., Buratowski S., Ahn S.H., Krogan N.J., Greenblatt J.F., Buratowski S., Krogan N.J., Greenblatt J.F., Buratowski S., Greenblatt J.F., Buratowski S., Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam L.T., Pickeral O.K., Peng A.C., Rosenwald A., Hurt E.M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Pickeral O.K., Peng A.C., Rosenwald A., Hurt E.M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Peng A.C., Rosenwald A., Hurt E.M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Rosenwald A., Hurt E.M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Hurt E.M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., Zhao H., Davis R.E., Sathyamoorthy M., Davis R.E., Sathyamoorthy M., Sathyamoorthy M., et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001 doi: 10.1186/gb-2001-2-10-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspia M.F., Rice A.P., Mathews M.B., Rice A.P., Mathews M.B., Mathews M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Cilli K., McNeil J.B., Bentley D.L., McNeil J.B., Bentley D.L., Bentley D.L. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Mancebo H.S., Lee G., Flygare J., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Lee G., Flygare J., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Flygare J., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Zhu Y., Peng J., Blau C., Hazuda D., Price D., Peng J., Blau C., Hazuda D., Price D., Blau C., Hazuda D., Price D., Hazuda D., Price D., Price D., et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes & Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R.A., Sharp P.A., Sharp P.A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.B., Struhl K., Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Raha T., Cheng S.W., Green M.R., Cheng S.W., Green M.R., Green M.R. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner D.B., Yamaguchi Y., Wada T., Handa H., Price D.H., Yamaguchi Y., Wada T., Handa H., Price D.H., Wada T., Handa H., Price D.H., Handa H., Price D.H., Price D.H. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- Rosonina E., Kaneko S., Manley J.L., Kaneko S., Manley J.L., Manley J.L. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Saunders A., Core L.J., Lis J.T., Core L.J., Lis J.T., Lis J.T. Breaking barriers to transcription elongation. Nat. Rev. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Sims R.J., Belotserkovskaya R., Reinberg D., Belotserkovskaya R., Reinberg D., Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Tamm I., Kikuchi T., Darnell J.E., Salditt-Georgieff M., Kikuchi T., Darnell J.E., Salditt-Georgieff M., Darnell J.E., Salditt-Georgieff M., Salditt-Georgieff M. Short capped hnRNA precursor chains in HeLa cells: Continued synthesis in the presence of 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole. Biochemistry. 1980;19:2743–2748. doi: 10.1021/bi00553a032. [DOI] [PubMed] [Google Scholar]
- Toohey M.G., Jones K.A., Jones K.A. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes & Dev. 1989;3:265–282. doi: 10.1101/gad.3.3.265. [DOI] [PubMed] [Google Scholar]
- Verdin E., Paras P., Van Lint C., Paras P., Van Lint C., Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., Sugimoto S., Yano K., Hartzog G.A., Winston F., Yano K., Hartzog G.A., Winston F., Hartzog G.A., Winston F., Winston F., et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J., Gromak N., Proudfoot N.J., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H., Furuya A., Sugimoto S., Hasegawa J., Handa H., Sugimoto S., Hasegawa J., Handa H., Hasegawa J., Handa H., Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Gilmour D.S., Gilmour D.S. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol. Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fu J., Gilmour D.S., Fu J., Gilmour D.S., Gilmour D.S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Klatt A., Gilmour D.S., Henderson A.J., Klatt A., Gilmour D.S., Henderson A.J., Gilmour D.S., Henderson A.J., Henderson A.J. J. Biol. Chem. 2007. Negative elongation factor, NELF, represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. [DOI] [PubMed] [Google Scholar]