Abstract
Many metazoan gene transcripts exhibit neuron-specific splicing patterns, but the developmental control of these splicing events is poorly understood. We show that the splicing of a large group of exons is reprogrammed during neuronal development by a switch in expression between two highly similar polypyrimidine tract-binding proteins, PTB and nPTB (neural PTB). PTB is a well-studied regulator of alternative splicing, but nPTB is a closely related paralog whose functional relationship to PTB is unknown. In the brain, nPTB protein is specifically expressed in post-mitotic neurons, whereas PTB is restricted to neuronal precursor cells (NPC), glia, and other nonneuronal cells. Interestingly, nPTB mRNA transcripts are found in NPCs and other nonneuronal cells, but in these cells nPTB protein expression is repressed. This repression is due in part to PTB-induced alternative splicing of nPTB mRNA, leading to nonsense-mediated decay (NMD). However, we find that even properly spliced mRNA fails to express nPTB protein when PTB is present, indicating contributions from additional post-transcriptional mechanisms. The PTB-controlled repression of nPTB results in a mutually exclusive pattern of expression in the brain, where the loss of PTB in maturing neurons allows the synthesis of nPTB in these cells. To examine the consequences of this switch, we used splicing-sensitive microarrays to identify different sets of exons regulated by PTB, nPTB, or both proteins. During neuronal differentiation, the splicing of these exon sets is altered as predicted from the observed changes in PTB and nPTB expression. These data show that the post-transcriptional switch from PTB to nPTB controls a widespread alternative splicing program during neuronal development.
Keywords: Alternative splicing, neuronal development, nonsense-mediated decay, polypyrimidine tract-binding proteins, splicing microarray, ultraconserved element
Alternative pre-mRNA splicing is a common mechanism for diversifying genetic output in metazoan organisms (Black 2003; Matlin et al. 2005). Alternative choices in exons and splice sites can create substantial changes in the encoded protein and its activity. Changes in splicing can also affect downstream regulatory processes such as nonsense-mediated decay (NMD), and thus direct additional levels of post-transcriptional gene regulation (Lewis et al. 2003; Lejeune and Maquat 2005; Hughes 2006). Transcripts exhibiting multiple splicing patterns are especially prevalent in the mammalian nervous system, where alternative splicing affects important processes such as axon guidance, synaptogenesis, and the regulation of membrane physiology (Black and Grabowski 2003; Lipscombe 2005; Ule and Darnell 2006). The choice of splicing pattern within a transcript is generally controlled by RNA-binding proteins that bind to the pre-mRNA to enhance or silence particular splicing events (Black 2003; Matlin et al. 2005). Some splicing regulators are expressed in a tissue-specific manner and have been shown to influence the alternative splicing of particular gene transcripts. However, little is known about how the expression of these factors is regulated during development, their roles in defining the differentiated states of a cell, or even whether changes in their expression influence large or small numbers of target genes.
One well-studied splicing regulator is the polypyrimidine tract-binding protein (PTB), which has been shown to repress the splicing of several exons (Wagner and Garcia-Blanco 2001; Spellman and Smith 2006). PTB binds to CU-rich regulatory elements near PTB-repressed exons and alters assembly of the spliceosome at adjacent splice sites (Oberstrass et al. 2005). In some cases, PTB may simply block access of the spliceosome to splice sites (Lin and Patton 1995; Singh et al. 1995; Amir-Ahmady et al. 2005). Alternatively, the PTB bound to the pre-mRNA can prevent spliceosome assembly subsequent to initial splice site recognition (Izquierdo et al. 2005; Sharma et al. 2005; Spellman and Smith 2006). Finally, differences in the cofactors required to repress different exons indicate that the mechanism of PTB-dependent exon silencing may vary from transcript to transcript (Gromak et al. 2003; Rideau et al. 2006). PTB is found in many tissues, but its expression is relatively low in brain and muscle, and many PTB-repressed exons are induced to splice in these tissues (Ashiya and Grabowski 1997; Spellman et al. 2005; Boutz et al. 2007).
In the mammalian brain, a PTB paralog called neural PTB (nPTB, also called brPTB and PTBP2) is also expressed. nPTB is highly homologous to PTB, but is encoded by a separate gene and is more restricted in its expression; in addition to the brain, nPTB is prevalent in testis and is also found in several other tissues (Kikuchi et al. 2000; Markovtsov et al. 2000; Polydorides et al. 2000; Lillevali et al. 2001). PTB and nPTB are remarkably similar in structure across all four of their RNA recognition motifs (RRMs). Except for one lysine-to-arginine and one phenylalanine-to-tyrosine substitution, nPTB has the same residues at positions of RNA contact as PTB. The two proteins bind to the same sequence elements, although with some differences in affinity (Markovtsov et al. 2000; Oberstrass et al. 2005). Nevertheless, nPTB has different splicing regulatory properties. It is a significantly weaker repressor than PTB of several neuron-specific exons and is known to bind additional neuronally regulated transcripts (Ashiya and Grabowski 1997; Chan and Black 1997; Irwin et al. 1997; Markovtsov et al. 2000; Underwood et al. 2005). nPTB also interacts with the neuron-specific splicing regulator Nova 1 (Polydorides et al. 2000). How nPTB and PTB differ in activity and thus serve distinct biological roles is not understood.
NMD is a form of post-transcriptional gene regulation that can be coupled to alternative splicing. The exon junction complex (EJC) is a specific assembly of proteins that is deposited on each exon–exon junction of an mRNA during splicing and removed during the first, or “pioneer,” round of translation (Ishigaki et al. 2001; Singh and Lykke-Andersen 2003; Conti and Izaurralde 2005; Maquat 2005). If translation terminates more than ∼50 nucleotides (nt) upstream of an EJC, the EJC can recruit essential NMD factors, including Upf1/Rent1, that stimulate degradation of the mRNA (Lykke-Andersen et al. 2000; Mendell et al. 2002). The NMD pathway serves to prevent the translation of mutant or aberrantly spliced mRNAs that would produce a truncated protein due to the presence of a premature termination codon (PTC). NMD can also function to control the expression from a gene that is alternatively spliced to introduce a PTC (Lewis et al. 2003; Lejeune and Maquat 2005; Pan et al. 2006). This splicing-coupled NMD is seen in the PTB transcript. The PTB gene contains a 34-nt exon (exon 11) that is normally included in the mRNA to encode full-length PTB protein. If exon 11 is skipped during splicing of the mRNA, the reading frame is shifted to introduce a PTC that targets the transcript to the NMD pathway (Wollerton et al. 2004). Interestingly, PTB protein itself represses exon 11, creating a negative feedback loop that allows PTB to control its own synthesis. Similar autoregulatory loops, where a protein controls its own expression through the coupling of alternative splicing to NMD, have been described for multiple other splicing regulators (Chabot et al. 1997; Jumaa and Nielsen 1997; Sureau et al. 2001; Stoilov et al. 2004; Dredge et al. 2005; Kumar and Lopez 2005; Lareau et al. 2007; Ni et al. 2007).
In this study, we examined the regulation of PTB and nPTB expression in the brain. We found that PTB and nPTB protein expression are mutually exclusive, with PTB restricted to nonneuronal lineages and nPTB found only in post-mitotic neurons. Interestingly, the presence of PTB strongly represses nPTB protein expression. Because of this post-transcriptional repression, the down-regulation of PTB in differentiating neurons is sufficient to induce nPTB expression, and this switch triggers substantial changes in the neuronal alternative splicing program.
Results
In the brain, nPTB expression is restricted to post-mitotic neurons and PTB is restricted to nonneuronal cells
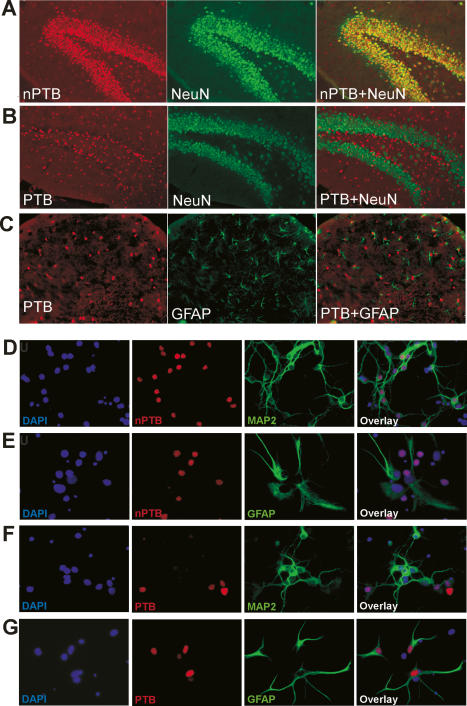
nPTB protein was previously detected in the brain as well as in cell lines of neuronal origin (Ashiya and Grabowski 1997; Chan and Black 1997; Irwin et al. 1997; Markovtsov et al. 2000; Polydorides et al. 2000). To examine which cells within the adult brain expressed each protein, we performed immunofluorescence on mouse brain sections, using antibodies that recognize only PTB (PTB-NT) or nPTB (nPTB-IS2) (Supplementary Fig. 1). At the cellular level, the divergent expression of PTB and nPTB is quite striking. As seen by costaining with the neuronal marker NeuN, nPTB is highly specific to neurons. In the dentate gyrus (DG) region of the hippocampus, nearly all of the neurons express nPTB and none of the nonneuronal cells do (Fig. 1A). nPTB was similarly neuron specific in the cortex and cerebellum (data not shown). In contrast, PTB was absent from NeuN-positive neurons (Fig. 1B). Most PTB-positive nuclei are associated with GFAP-positive processes (Fig. 1C), indicating they are astroglia. PTB is also found in cells negative for both NeuN and GFAP, most notably in the meninges, in the vascular endothelium, and in the subventricular zone of the cortex (data not shown). In the adult brain, cells that express both proteins were very rare.
Figure 1.
nPTB protein is expressed in post-mitotic neurons in the mouse brain. (A–C) Immunofluorescence images of coronal adult brain sections. (A) Immunostaining of nPTB (red, left panel) and NeuN (green, middle panel) in the DG of the hippocampus. (Right panel) Cells expressing nPTB are also positive for NeuN (overlay), indicating that they are neurons. (B) Overlay image of PTB (red) and NeuN (green) immunostaining shows that most PTB-expressing cells in the DG are negative for NeuN. (C) Double-staining for PTB (red) and GFAP (green) in the region proximal to the DG shows most PTB-expressing cells to be GFAP-positive astrocytes. (D–G) Immunofluorescence images of dissociated cerebellar cell cultures after 6 d in vitro. nPTB (D,E) or PTB (F,G) are stained in red, as indicated. The same fields are stained for the neuronal marker MAP2 (D,F) or the glial marker GFAP (E,G) (both green), and with the DNA stain DAPI (blue). The right-most panels are an overlay of the three colors.
Similarly divergent expression was seen in postnatal cultures of dissociated cells from the Hippocampus (P1–P2) (data not shown) and the cerebellum (P3–P5) (Fig. 1D–G). In cerebellar cultures, nPTB is found in differentiated MAP2-positive neurons (Fig. 1D), but is absent from GFAP-expressing astrocytes (Fig. 1E). PTB shows the reverse pattern with expression in astrocytes but not in neurons (Fig. 1F,G). In immature cerebellar cultures, some neurons expressed both proteins (data not shown).
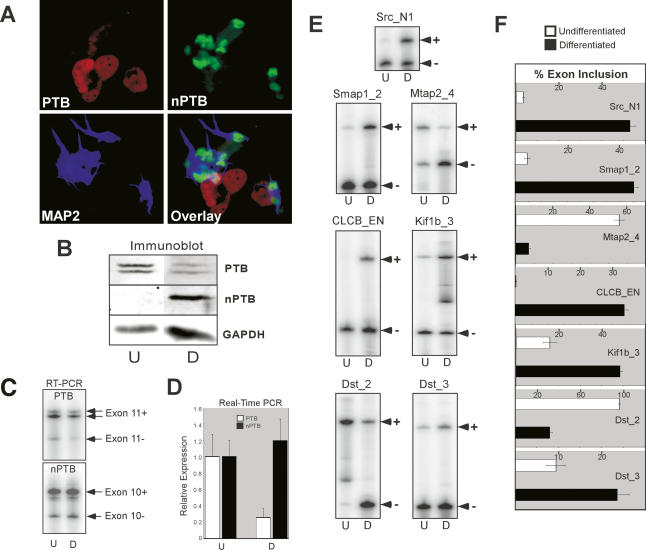
To examine nPTB expression in a model of neuronal differentiation, we used P19 mouse embryonal carcinoma cells, which differentiate into post-mitotic neurons after treatment with retinoic acid (RA). Thirteen days after RA treatment, >80% of the cells exhibited neuronal morphology with small cell bodies and long fasciculated processes, while the remaining cells were flat, adherent, and resembled fibroblasts or other cell types (data not shown). By immunofluorescence, undifferentiated cells predominantly express PTB, but after differentiation the neurons have switched to nPTB expression exclusively (Fig. 2A; data not shown). In contrast, the remaining nonneuronal cells in the culture express only PTB (Fig. 2A). This switch can also be seen by immunoblot, with nPTB protein being strongly induced during differentiation and PTB protein decreasing (Fig. 2B). The decrease in PTB protein is paralleled by a decrease in PTB mRNA, as measured by RT–PCR and by quantitative real-time PCR (Fig. 2C,D). Interestingly, the nPTB mRNA level and splicing pattern remained relatively constant with differentiation (Fig. 2C,D), indicating that a post-transcriptional mechanism(s) controls the induction of nPTB expression.
Figure 2.
Developing neurons switch from PTB to nPTB expression concomitant with changes in alternative splicing. (A) Immunofluorescence of differentiated P19 cells 13 d after RA treatment. The field of cells is stained for PTB (red), nPTB (green), and the neuronal marker MAP2 (blue). An overlay of the three colors is shown in the bottom right panel. (B) Protein lysates from undifferentiated (U) and 13-d differentiated (D) P19 cells were immunoblotted for PTB and nPTB, with GAPDH as a loading control. Note that PTB appears as a doublet due to the presence of alternative splicing of exon 9. (C) Semiquantitative RT–PCR of PTB and nPTB alternative splice variants. PTB exon 11-included (Exon 11+) and exon 11-skipped (Exon 11−) and nPTB exon 10-included (Exon 10+) and exon 10-skipped (Exon 10−) splice variants are indicated. Note an additional band in the PTB Exon 11+ lane is due to a second alternative splice at a different location. (D) Real-time PCR quantification of PTB and nPTB mRNA measured relative to β-actin mRNA. (E) Alternative splicing of PTB/nPTB-regulated exons assayed by RT–PCR. The exon IDs and the RT–PCR products corresponding to exon-included (+) and exon-skipped (−) forms are indicated. (F) Quantification of exon inclusion in E. The percent exon inclusion is graphed for differentiated (black bars) and undifferentiated (white bars) cells. The error bars in D and F indicate the standard error among three separate experiments.
To assess the functional consequence of the change from PTB to nPTB, we assayed a subset of PTB target exons by RT–PCR to determine whether they also switch in splicing in the differentiated P19 cells. As seen in Figure 2, E and F, all of the target transcripts tested exhibited changes in exon inclusion upon P19 differentiation. These included previously identified PTB targets (e.g., Src N1, ClCB EN), as well as new targets identified by a splicing microarray (described below); among them are exons that increase in inclusion, and exons that decrease in inclusion.
Skipping exon 10 leads to NMD of the nPTB transcript
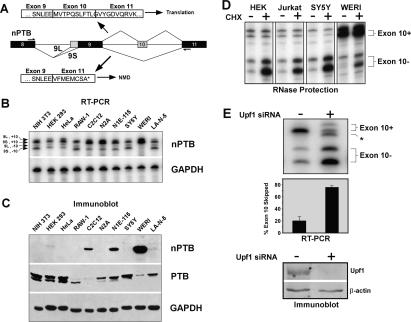
The presence of nPTB mRNA in undifferentiated P19 cells, which contained little nPTB protein, suggested a post-transcriptional mechanism was controlling nPTB expression. Similar to the homologous PTB exon 11, nPTB exon 10 inclusion is required to produce the full-length nPTB, while exon 10 skipping would shift the reading frame and introduce a PTC (Fig. 3A; Wollerton et al. 2004). To examine the role of exon 10 in affecting nPTB expression, we assayed a variety of cell lines for the presence of the different nPTB spliced isoforms (diagrammed in Fig. 3A). Most of the cell lines tested contained significant amounts of nPTB mRNA, both including and skipping exon 10 (Fig. 3B). However, like undifferentiated P19 cells, many of these cells express PTB, but little or no nPTB protein (Fig. 3C).
Figure 3.
nPTB exon 10-skipped splice variants are degraded by NMD. (A) nPTB gene structure in the region containing exons 8–11. Constitutive exons are shown as black boxes, and alternative exons are shown as gray boxes. Alternative splicing patterns are indicated by lines connecting the exons. Use of one of two alternative 3′ splice sites upstream of exon 9 results in the 9L or 9S variants, as labeled. Exon 10 is either skipped (−) or included (+). The combination of the two alternative splicing patterns results in four possible variants: 9L/10+, 9S/10+, 9L/10−, and 9S/10−. RT–PCR primers are shown as arrows above exon 8 and below exon 11. White boxes indicate the predicted amino acid sequence encoded by each splice variant with exon boundaries shown. A PTC (*) is generated in the exon 10-skipped variant. (B) Splicing of nPTB exons 9 and 10 in diverse cell lines. RT–PCR generates four specific bands corresponding to the splice variants indicated on the left. GAPDH amplification indicates equal amounts of template were used in each reaction. (C) Immunoblotting of protein samples collected from the same cells as in B for nPTB and PTB. GAPDH was used as a loading control for total protein. (D) Cultured cells were incubated with (CHX+) or without (CHX−) cycloheximide for 6 h prior to harvesting total RNA. Splice variants were assayed by RNase protection. The two exon 10-containing (Exon 10+) variants and the two exon 10-skipped (Exon 10−) variants are indicated. (E) N1E-115 cells were transfected with an siRNA targeting Upf1 (+) or a control siRNA (−) and were incubated for 48 h prior to harvesting total RNA and assay by RT–PCR. An unknown product band is indicated (*). Quantification of the exon 10-skipped variant relative to the total is graphed below. Error bars indicate the standard error among three separate experiments. (Bottom panel) Immunoblot of Upf1 protein after treatment with the Upf1 siRNA (+) or the control siRNA (−), using β-actin as a loading control.
The ongoing production of nPTB mRNA could be much higher than observed from the steady-state mRNA levels if exon 10-skipped mRNA were targeted by the NMD pathway. NMD requires active translation, and NMD-targeted transcripts accumulate when translation is blocked (Belgrader et al. 1993; Menon and Neufeld 1994). To examine whether mRNA was being lost to NMD, we treated cells with the translational inhibitor cycloheximide and measured the abundance of each splice variant by RNase protection. Indeed, we observed a significant increase in the two exon 10-skipped products after cycloheximide treatment of a variety of cell lines (Fig. 3D). The level of mRNA that includes exon 10 did not change significantly upon cycloheximide treatment, regardless of the initial ratio of the different splice variants. The accumulation of exon 10-skipped mRNA was observed in all cells tested. These included the retinoblastoma cell line WERI-1, the embryonic kidney cell line HEK293, the neuroblastoma SY5Y, and the T-cell line Jurkat (Fig. 3D), as well as the other cell lines HeLa, HepG2, N10, and LA-N-5 (data not shown).
To determine directly whether the exon 10-skipped isoforms were degraded by the NMD pathway, we used a small interfering RNA (siRNA) to knock down the essential NMD pathway component Upf1/Rent1 (Mendell et al. 2002). In N1E-115 cells, knockdown of the Upf1 protein led to a fourfold increase in the exon 10-skipped splice variants of nPTB (Fig. 3E). The observed effect was similar to that of cycloheximide, except that changes were also seen in the exon 10-included isoforms. These changes were due specifically to Upf1 knockdown, as we obtained similar results with a short-hairpin RNA (shRNA) targeting a different region of Upf1, whereas siRNAs targeting an unrelated RNA-binding protein (CUGBP1) had no effect (data not shown). Thus, nPTB mRNA is expressed in many cells, but a majority of these transcripts are missing exon 10 and are degraded by the NMD pathway. Moreover, many cells that produce the full-length exon 10-plus mRNA still fail to produce a significant amount nPTB protein, indicating that additional post-transcriptional mechanisms are blocking nPTB expression.
PTB regulates both the splicing and protein expression of nPTB
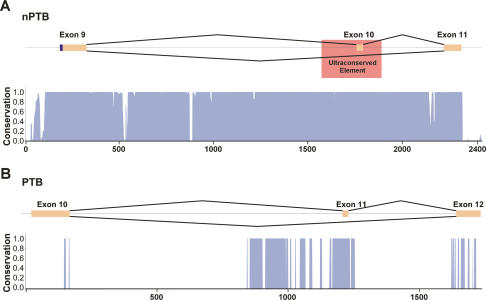
nPTB exon 10 is located within a 2-kb region exhibiting 85% identity among mammals and 70% identity between mammals and birds (Fig. 4A; Rahman et al. 2004). The degree of conservation in this region is highly unusual for a noncoding sequence. In contrast, the region surrounding the homologous PTB exon 11 shows conservation typical of a regulated exon, having conserved blocks of sequence larger than those seen in unregulated introns, but not the long stretches of interspecies identity found surrounding nPTB exon 10 (Fig. 4B; Sorek and Ast 2003; Yeo et al. 2005; Sugnet et al. 2006). A portion of this highly conserved region is an invariant sequence previously described as an “ultraconserved element” (Bejerano et al. 2004). This is one of the most conserved regions in mammalian genomes, consisting of 297 contiguous nucleotides with 100% identity in five mammalian species. The functions of ultraconserved elements are mostly unknown, although a number of them are seen to encompass other alternative exons (Bejerano et al. 2004; Lareau et al. 2007; Ni et al. 2007).
Figure 4.
nPTB exon 10 is within an ultraconserved region. (A) Schematic showing the genomic region of nPTB exons 9–11, with exons shown in orange, and introns shown as black lines. The location of the previously defined ultraconserved element is indicated by a red box. A histogram displaying the degree of conservation of this region among 17 vertebrate species is shown below in blue, as determined by the UCSC genome browser (http://genome.ucsc.edu). A score of 1 indicates 100% identity among all species at that nucleotide position. A distance scale in nucleotides is shown below the histograms. (B) The homologous region of PTB, exons 10–12, is shown with the same annotation as in A.
To examine whether the ultraconserved sequence was important for controlling splicing, genomic fragments containing exon 10 and varying portions of its flanking introns (denoted as A–C) were inserted between two constitutive exons in a splicing reporter plasmid (Fig. 5A). These constructs were transfected into HEK293 cells and the splicing of nPTB exon 10 was assayed by RT–PCR (Fig. 5B, lane 1). The intron upstream of exon 10 contains two unusually distant putative branchpoints ∼400 nt upstream of the splice acceptor. Construct A lacks these natural branchpoints but contains a branchpoint sequence from the reporter gene intron. Transcripts from this plasmid spliced exon 10 poorly under all conditions (Fig. 5B, top panel). Construct B, containing an upstream intron sequence that includes a potential distal branchpoint, showed more efficient exon 10 splicing (Fig. 5B, middle panel). This was further enhanced by the addition of a downstream intron sequence in construct C (Fig. 5B, bottom panel), indicating the presence of enhancer elements within the downstream intron.
Figure 5.
PTB represses nPTB expression. (A) Schematic of the nPTB exon 10 genomic region, with solid bars below indicating the fragments used for minigene constructs. Construct A contains 108 nt of upstream intron sequence and uses the branchpoint from the constitutive globin intron. Constructs B and C contain 418 nt of the upstream nPTB intron sequence that includes a putative distal branchpoint (indicated by arrowhead). Constructs A and C contain 83 and 179 nt, respectively, of downstream nPTB intron sequence. (B) Minigene constructs A, B, and C carrying nPTB exon 10 were cotransfected with shRNA vectors into HEK293 cells, and splicing was assayed by RT–PCR. Exon 10 is shown as a gray box, nPTB intron sequence is shown as a solid line, globin intron sequence is shown as a broken line, and constitutive exons are shown as black boxes. (C) RNAi against PTB or both PTB and nPTB was induced using shRNA-expression vectors, with empty vector serving as a negative control. Western blots were probed for PTB, nPTB, and GAPDH as a loading control. (D) Different cell lines (indicated above) were transfected with PTB (+) and control (−) siRNAs. (Top panel) RNA samples were collected and assayed by RT–PCR for the nPTB exon 9 and 10 region or for GAPDH. (Bottom panels) Protein lysates were probed for PTB, nPTB, and GAPDH as a loading control.
To examine the dependence of the exon on PTB, we knocked down PTB by RNA interference (RNAi) in the HEK293 cells, which normally express only PTB. Strikingly, endogenous nPTB expression is strongly up-regulated upon PTB knockdown (Fig. 5C, lane 2; see also below). An shRNA targeting nPTB eliminated this expression when combined with the PTB shRNA (Fig. 5C, lane 3). The nPTB shRNA had no effect when expressed on its own in these cells (data not shown). Notably, the splicing of exon 10 in constructs B and C was substantially enhanced by the knockdown of PTB (Fig. 5B, lane 2). The double knockdown of PTB and nPTB had a still stronger effect (Fig. 5B, lane 3), indicating that nPTB controls the splicing of its own message. These results demonstrate that the ultraconserved introns flanking nPTB exon 10 contain sequences required for its regulated splicing, and confirm the roles of PTB and nPTB as splicing repressors of this exon, similar to PTB exon 11.
We were particularly interested in the increase in nPTB protein expression seen upon PTB knockdown (Fig. 5B, lane 2). To examine the splicing and protein expression of endogenous nPTB in cells depleted of PTB, we performed RT–PCR and Western blot assays on a variety of cell lines after transfection of a PTB siRNA (Fig. 5D). Knockdown of PTB resulted in a loss of exon 10-skipped variants in all cell lines tested (Fig. 5D, top panel, lanes 2,4,6). This produced a 20%–70% increase in the amount of exon 10-included mRNA, depending on the cell line (Fig. 5D, top panels; data not shown). The increase in exon 10 inclusion after PTB knockdown can be reversed by stable expression of either PTB or its homolog Rod1 (Supplementary Fig. 2). These data indicate that PTB or its homologs are essential for the formation of the NMD-sensitive spliced form of the nPTB transcript.
As seen before, nPTB protein is strongly induced in all cells in which PTB is eliminated (Fig. 5D, bottom panel, lanes 2,4,6). Interestingly, the increase in nPTB protein is often significantly greater than can be accounted for by the increase in exon 10-included mRNA. This is most apparent in the N2A neuroblastoma, where the mRNA-encoding full-length nPTB protein increases 2.5-fold upon PTB knockdown, but nPTB protein increases >14-fold (Fig. 5D; data not shown). Thus, besides affecting splicing, PTB represses nPTB protein expression by additional mechanisms, possibly by inhibiting nPTB mRNA translation.
Neuronal progenitor cells (NPCs) express PTB
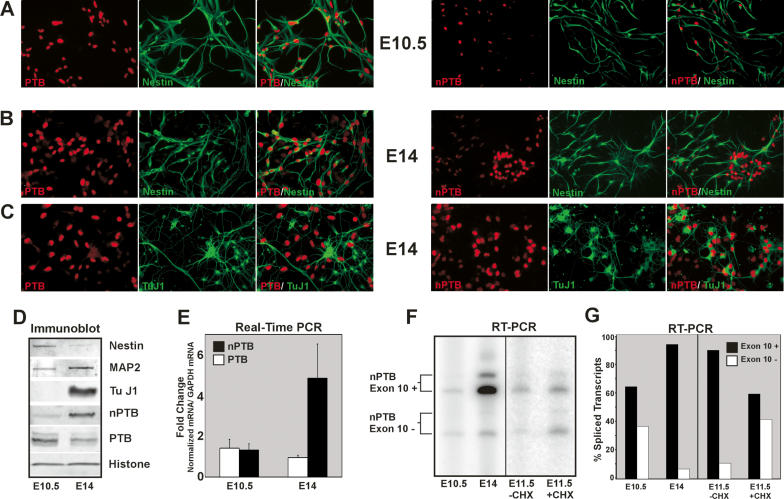
Despite the presence of nPTB mRNA in many cells, nPTB protein is largely suppressed by PTB by both NMD and non-NMD mechanisms. Given this strong cross-regulation, the loss of PTB expression during neuronal maturation should contribute to the induction of nPTB seen in these cells. To assess this, we examined the expression of these two proteins in cultures of NPCs that give rise to neurons and glia (Ahlemeyer and Baumgart-Vogt 2005; Feng et al. 2005). These cells can be identified by their expression of the marker Nestin (Cattaneo and McKay 1990; Reynolds and Weiss 1992). Immunofluorescent staining of cells isolated from embryonid day 10.5 (E10.5) brains indicated that they are strongly positive for Nestin and for PTB but show only weak nPTB expression. At this day of isolation very few of the cells are positive for the neuronal marker TuJ1 (Fig. 6A; data not shown). When isolated from E14 embryos, the cultures still contain these nestin-positive, PTB-high, nPTB-low cells (Fig. 6B). However, the neurogenesis during this period gives rise to additional TuJ1-positive differentiating neurons with robust nPTB expression and little PTB (Fig. 6C). In summary, the NPCs express PTB, but little nPTB. If these cells differentiate into Astrocytes, they maintain this PTB expression (Fig. 1; data not shown). However, during the differentiation of these cells into neurons, PTB expression is lost and nPTB is induced.
Figure 6.
nPTB replaces PTB as NPCs mature into neurons. (A–C) Immunofluorescence of E10.5 (A) and E14 (B,C) NPCs in culture. Individual staining for PTB (left panels, red) or nPTB (right panels, red), Nestin (A,B, green), or TuJ1 (C, green) are shown along with overlays of the two-color staining for each field. Signal gain for the nPTB staining in the right panel of A has been increased to show the fainter nPTB signal. (D) Immunoblot of lysates prepared from E10.5 and E14 cultures, probed for the NCP marker Nestin, the neuronal markers MAP2 and TuJ1, nPTB, PTB, and Histone H3 as a loading control. (E) Real-time PCR of cDNAs prepared from E10.5 and E14 cell lysates. PTB (white bars) and nPTB (black bars) mRNA levels were quantified relative to GAPDH mRNA, and the fold change in each mRNA from E10.5 to E14 is graphed. (F) RT–PCR of the nPTB alternatively spliced region. Exon 10-included (Exon 10+) and exon 10-skipped (Exon 10−) splice variants are indicated for E10.5 and E14 cells, and E11.5 cells incubated for 6 h without (−CHX) or with (+CHX) cycloheximide prior to harvesting. (G) Quantification of splice variants shown in F. The percentage of total spliced transcripts in which exon 10 is included (+, black bars) or skipped (−, white bars) is graphed.
This induction of nPTB protein and loss of PTB during embryonic neurogenesis is also seen by immunoblot (Fig. 6D). At the mRNA level, nPTB is strongly up-regulated between E10.5 and E14 as the number of neurons in the cultures increases (Fig. 6C,D). There is a particular increase in the expression of the nPTB mRNA isoforms that include exon 10 (Fig. 6C,D). To determine whether exon 10-skipped nPTB transcripts are expressed in NPCs, but are subject to NMD, we treated E11.5 cells with cycloheximide and assayed the splicing of exon 10 by RT–PCR (Fig. 6C,D). Compared with the untreated control cells, cycloheximide-treated NPCs exhibited a threefold increase in exon 10-skipped transcripts, indicating that a substantial portion of the nPTB mRNA produced in these cells is lost to NMD. Thus, the loss of PTB during neuronal differentiation should be sufficient to switch the expressed nPTB transcripts to exon 10 inclusion and allow expression of nPTB protein. There may also be a transcriptional induction of nPTB during this process.
PTB and nPTB regulate different exon sets
The sharp difference in the expression patterns of PTB and nPTB, and their cross-regulation, indicate that the two proteins have distinct physiological functions, despite their similar sequence and RNA-binding properties. To understand the consequences of the PTB-to-nPTB switch in neurons, and to look more broadly at the splicing changes controlled by these proteins, we applied a microarray-based alternative splicing assay (Srinivasan et al. 2005). These arrays used oligonucleotide probes that hybridize to exon–exon junctions similar in design to systems described previously (Clark et al. 2002; Johnson et al. 2003; Le et al. 2004; Pan et al. 2004; Blanchette et al. 2005; Srinivasan et al. 2005; Ule et al. 2005; Sugnet et al. 2006). The arrays probed the splicing of ∼1300 exons that were selected for their likely functional significance (see Materials and Methods).
To identify splicing events dependent on these proteins, we examined N2A mouse neuroblastoma cells in which the expression of PTB, nPTB, or both proteins (double knockdown) was reduced or eliminated by RNAi. N2A cells exhibit highly efficient shRNA-induced RNAi and express PTB and a small amount of nPTB (Supplementary Fig. 3). Arrays were cohybridized with cDNA from shRNA-treated cells and control-treated cells. After scanning, the Cy5/Cy3 ratios for the oligonucleotide probes were extracted, normalized, and analyzed (see the Supplemental Material).
Exons showing significant changes in inclusion between the knockdown and the control were ranked by False Discovery Rate (FDR) value (see Materials and Methods). From these ranked lists, RT–PCR was performed on 72–77 exons for each knockdown to validate the changes determined by microarray. Of these exons subjected to secondary tests, 93% were confirmed by RT–PCR for the PTB knockdown, 80% for the nPTB knockdown, and 96% for the double knockdown. The higher rate of false positives in the nPTB knockdown experiments is presumably due to the lower level of nPTB in these cells resulting in a lower magnitude of observable change. Supplementary Table 1 lists the results of the microarray analysis and RT–PCR verification. Knocking down other splicing factors was found to alter different exon groups (data not shown), indicating that the changes in splicing observed here are specific to PTB/nPTB knockdown and are not due to general affects of the RNAi machinery.
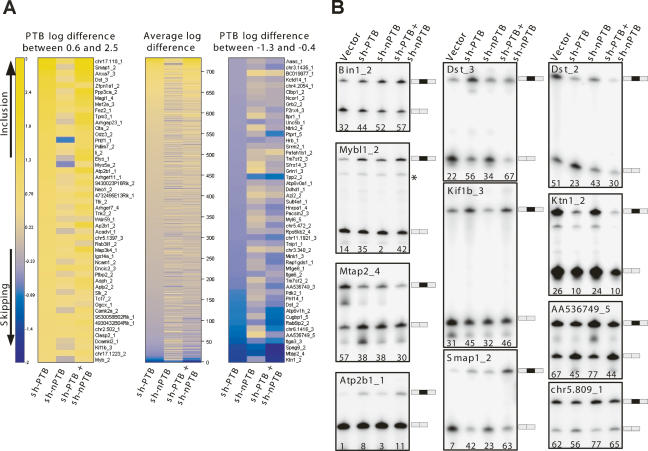
The splicing changes measured for each knockdown are presented as a heat map in Figure 7A, listed from increased inclusion (yellow) to increased skipping (blue) for the log2 change in the PTB knockdown relative to the control (mock knockdown). This spectrum is aligned with the corresponding values for each exon in the nPTB and double knockdowns. The 50 highest and 50 lowest values are expanded with the gene names shown. The RT–PCR validation of some of these exons is shown in Figure 7B. This analysis identified a large number of exons whose splicing is affected by PTB, nPTB, or both proteins. These include known PTB targets (e.g., Tpm) and many new transcripts.
Figure 7.
Microarrays reveal that PTB and nPTB direct distinct alternative splicing programs. Alternative exon microarrays were probed with cDNA from cells after PTB, nPTB, or PTB and nPTB knockdown. (A) Heat maps showing splicing changes ranging from increased inclusion (yellow) to increased skipping (blue). The change for each exon was calculated by subtracting the average log ratio for the skipping probe set from the average log ratio for the inclusion probe set. The exons were sorted by this log ratio difference for the PTB knockdown, with the scale on the far left. This spectrum is aligned with values for the nPTB and double knockdowns. The middle panel shows the average log difference for all exons giving a detectable signal in the assay. The top 50 exons (log difference from +0.6 to +2.5) and the bottom 50 exons (log difference from −1.3 to −0.4) are expanded to the left and right respectively. (B) Examples of RT–PCR verification of exons identified in the microarray experiments. RT–PCR was performed using primers in the flanking constitutive exons. The exon ID is shown on the top of each panel with the percent exon inclusion shown at the bottom. The included and skipped forms are indicated on the right.
As seen in Figure 7A, it is clear that exons affected by PTB knockdown can behave quite differently upon nPTB or double knockdown, and that PTB and nPTB are regulating different but overlapping target sets. These exons can be grouped according to their responsiveness to the different proteins. One group of exons, such as that in Kinesin Intermediate Filament (Kif1b_3), increases in splicing upon PTB knockdown, is unaffected by nPTB knockdown, and shows no additional increase in the double knockdown (Fig. 7B). These exons are presumably repressed by PTB but not by nPTB. Another group of exons, such as found in Bin1_2 (Bridging Integrator 1), Dst_3 (Dystonin), and Smap1_2 (Stromal membrane-associated protein 1), shows increased inclusion in each single knockdown, and a larger increase when both PTB and nPTB are eliminated (Fig. 7B). This behavior defines a group of exons that are repressed by both proteins. However within this group, there is variability in the relative effect of PTB and nPTB. For Dst_3, the double knockdown shows only a modest increase from the PTB knockdown alone, indicating a relatively weak effect from nPTB. For Smap1_2, the alternative exon is included at substantially higher levels in the double knockdown than with either single shRNA treatment, indicating a strong effect from both proteins. For Bin1_2, the nPTB knockdown gives a larger effect than PTB and is only modestly enhanced in the double knockdown. This exon appears more strongly repressed by nPTB. Exons that behave like Kif1b_3 and Dst_3 are the two most common types. We found relatively few exons that show stronger derepression upon nPTB knockdown than upon PTB knockdown (like Bin1_2). It may be that such exons are less common, but the N2A cells express less nPTB than PTB, prior to PTB knockdown. This could lower the magnitude of the changes observed after nPTB knockdown, making them more difficult to identify. In sum, the array data identify a large number of exons that are repressed by PTB, nPTB, or both proteins.
Exons repressed by PTB and/or nPTB respond to the knockdown of these proteins with increased inclusion. In contrast, exons such as Dst_2 (Dystonin), Mtap2_4 (Microtubule-Associated Protein 2), and Ktn1_2 (Kinectin 1) sharply increase in skipping upon single or double knockdown. These exons and others behave as if one or both PTB proteins act positively to promote their inclusion. Interestingly, within the same Dst transcript one exon is positively regulated by PTB, while another exon appears to be repressed by it. There is also a third alternative exon in Dst that is unaffected by either knockdown (data not shown). Finally, a small group of exons, including Rip3 (Rho-Interacting Protein 3; AA536749_5) and Exoc1 (Sec3; chr5.809_1), shows opposite directions of regulation by the two proteins (Fig. 7B). In these cases, the knockdown of PTB leads to a decrease in exon inclusion, while the nPTB knockdown increases the inclusion of the exon.
Exons showing different dependencies on PTB and nPTB would be predicted to change in splicing during neuronal differentiation. Many of these exons were confirmed to change during P19 differentiation by RT–PCR (Fig. 2). To confirm the splicing changes in the PTB target exons on a larger scale, we compared RNA from differentiated cells with that from undifferentiated P19 on the microarrays. These results are listed in Supplementary Table 2. Multiple RNA-binding proteins change in expression upon P19 differentiation and, not surprisingly, many exons are altered in their splicing. For the transcripts expressed in both P19 and N2A cells, 83% of the exons (54 of 65) altered by PTB knockdown show the equivalent change during P19 differentiation (Supplementary Fig. 4). As seen with the RT–PCR of individual exons, these included both exons that increase and decrease in inclusion. Random resampling tests indicate that the probability of observing by chance 54 exons from the N2A experiments overlapping with the 202 exons that change upon P19 differentiation is close to 0 (Supplementary Fig. 4). This concordance between the splicing changes seen in response to PTB knockdown and the changes upon P19 differentiation indicates that the reprogramming of alternative splicing of many exons during neuronal development is primarily dependent on the switch from PTB to nPTB expression. From these data, we estimate that ∼25% of the exons on the array that change during P19 differentiation are PTB dependent (54 out of 202), with the rest presumably affected by other splicing regulators.
Discussion
A switch in splicing regulation during neuronal differentiation
The genetic programs that direct neuronal and glial differentiation are known to involve cascades of transcriptional regulators, epigenetic maintenance of chromatin states, and post-transcriptional regulation by microRNAs (Sun et al. 2001, 2003; Bertrand et al. 2002; Ballas et al. 2005; Fan et al. 2005; Johnston et al. 2005; Conaco et al. 2006). We find that, during the differentiation of a cell into a post-mitotic neuron, a novel genetic switch takes place in which the splicing regulator PTB is replaced with its close relative, nPTB. This switch in RNA-binding proteins reprograms the splicing of a large number of alternative exons to be specifically included or excluded in neuronal mRNA. Thus, the splicing alteration directed by the PTB/nPTB switch adds a new layer of genetic change determining the function of a post-mitotic neuron.
Gene ontology analyses of the PTB and nPTB target gene sets show enrichment for genes affecting cytoskeletal rearrangement (e.g., Mtap2_4, Dst_2), and vesicular or protein transport (e.g., Kinesin, Dynein) (data not shown). These functions are essential to remodeling cell morphology during neurite outgrowth, the development of connectivity, and synaptogenesis—processes that are roughly coincident with the switch in splicing regulators (Dalpe et al. 1998; Zhao et al. 2001; Hirokawa and Takemura 2004; Dehmelt and Halpain 2005). However, exons controlled by PTB/nPTB are diverse and additional analyses will be needed to place them in common cellular pathways.
PTB-controlled exons whose function is characterized testify to the wide-ranging consequences of the splicing switch. The transcription factor MEF2 controls a large number of genes that play important roles in neuronal function and survival (Mao et al. 1999; Heidenreich and Linseman 2004; Shalizi and Bonni 2005). The MEF2 gene contains an alternative exon β whose inclusion greatly stimulates the protein’s ability to activate transcription (Zhu et al. 2005). We found that the β exon is strongly repressed by PTB and not nPTB in N2A cells and is induced during P19 differentiation into neurons (data not shown). Thus, the switch from PTB to nPTB influences the transcriptional program in neurons through its effect on this regulator.
A different function is affected by PTB regulation of Rab6IP2 splicing. Rab6IP2 protein (also called ERC1) is involved in membrane trafficking and is expressed in two spliced isoforms. ERC1a is ubiquitous, but inclusion of a neuron-specific exon generates ERC1b that interacts with specialized RIM proteins to modulate neurotransmitter release at the synapse (Wang et al. 2002). We found that the ERC1b-specific exon is repressed by PTB in nonneuronal cells. The knockout of PTB expression leads to altered splicing and the synthesis of the synaptic form, ERC1b. Thus, the loss of PTB-dependent repression during neuronal differentiation is needed for the synthesis of components required for presynaptic function.
It will be important to examine the kinetics of the PTB/nPTB switch and to understand whether it develops with stages at which both proteins are expressed. Initial experiments indicate that committed neuroblasts express both proteins, but this will need additional analysis (Q. Li, unpubl.). Interestingly, both PTB and nPTB levels in the brain decrease with age after birth, indicating that another transition in splicing regulation occurs during postnatal development (data not shown).
Ensembles of coregulated Exons
Using a newly designed oligonucleotide microarray, we identified sets of exons regulated by PTB, nPTB, or both proteins. The microarray data make clear that exons can exhibit a wide range of dependencies on PTB and nPTB, with some exons repressed by one or the other protein and some repressed by both, as well as some exons that are affected positively by one or both proteins. The statistical analysis of these exons is ongoing, but the enrichment of CU-rich elements in the introns flanking these exons indicates that the majority are likely direct targets of the protein (data not shown). These elements are predicted PTB-binding sites and their enrichment in these regions is a known feature of exons repressed by the protein (Ashiya and Grabowski 1997; Amir-Ahmady et al. 2005). Although the majority of these exons have the hallmarks of direct PTB/nPTB targets, one cannot rule out that indirect effects from other proteins also occur on individual exons. Indeed, several other RNA-binding proteins seem to be PTB targets (see Supplementary Table 1). From the number of exons affected and the variety of their responses, it is clear that the change from PTB to nPTB alters the splicing of a complex ensemble of exon targets.
Besides identifying cellular functions subject to PTB/nPTB control, the identification of PTB- and nPTB-responsive exon sets opens up interesting avenues of investigation regarding their mechanisms of action. One application will be to define the sequence requirements for an exon to show regulation by one or both of these proteins. For PTB, these sequences have been defined biochemically on model exons (Wagner and Garcia-Blanco 2001; Amir-Ahmady et al. 2005; Spellman et al. 2005). However, the arrangement and sequence of PTB-binding sites can vary substantially from exon to exon, and it is not clear what allows an exon to be repressed by one protein and not the other. Is there a specific arrangement of binding sites needed? Are specific cofactors needed? Are exons that are repressed by both PTB and nPTB specific to tissues that are low in both proteins, such as muscle? A number of muscle-specific exons have been shown to be repressed by both proteins (Boutz et al. 2007; R. Spellman and C. Smith, pers. comm.) Answers to these mechanistic questions will help us understand how the PTB proteins interact with the splicing apparatus, as well as predict which exon groups will be spliced in particular tissues and conditions.
Of particular interest are the exons that show apparent positive regulation by PTB or nPTB. These have not been previously described and the mechanism by which PTB could activate splicing is not clear. For other splicing regulators, the position of a binding site relative to the regulated exon can determine whether the binding factor will positively or negatively affect its splicing (Kanopka et al. 1996; Hui et al. 2005; Ibrahim el et al. 2005; Ule et al. 2006). It will be very interesting to compare the PTB/nPTB-binding sites adjacent to the exons that show positive regulation by PTB or nPTB with those that are repressed by one or both proteins. This approach proved very effective for exons regulated by the Nova proteins (Ule et al. 2006).
Mechanisms of post-transcriptional gene regulation
The post-transcriptional repression of nPTB by PTB allows for mutually exclusive expression of these two proteins between the neuronal and astrocytic lineages. The down-regulation of PTB in neurons should be sufficient to induce nPTB expression. The mechanism directing the loss of PTB is not yet clear, and is presumably directed by other genetic programs controlling neuronal maturation. We found that nPTB is targeted by muscle-specific microRNAs during myotube maturation, thereby limiting nPTB expression to immature myoblasts—the converse of what is seen in neurons (Boutz et al. 2007). In muscle, both PTB proteins are lost during development, but in neurons expression switches from one protein to the other. Interestingly, PTB is a predicted target for miR-124, a neuron-specific microRNA (Lim et al. 2005). The induction of miR-124 during neuronal differentiation could contribute to the loss of PTB protein seen during this process, but this will require further study.
An intriguing feature of the nPTB gene is its extreme conservation within the region surrounding exon 10. One role of this nPTB ultraconserved element is to control alternative exon inclusion, with portions of the conserved region having enhancing effects on exon 10, and other regions allowing splicing repression by PTB. This regulation provides a functional role for the sequence, but it does not indicate why this sequence would be even more highly conserved than those surrounding other alternative exons. There are other ultraconserved elements surrounding exons in ASF/SF2, hnRNP H, Nova 1, Tra2β, and more than a dozen other potential splicing regulators (Bejerano et al. 2004). One function of these exons is to allow autoregulation by the encoded protein, as demonstrated for PTB, SRp20, Nova, and other proteins (Jumaa and Nielsen 1997; Wollerton et al. 2004; Dredge et al. 2005; Kumar and Lopez 2005; Lareau et al. 2007; Ni et al. 2007). For nPTB exon 10, there is similar autoregulation by nPTB itself. However, there is a larger effect from PTB that allows for cross-regulation of one protein by the other and the mutually exclusive pattern of expression seen in neural cell lineages. Thus, besides acting as an autoregulatory mechanism, splicing-mediated NMD takes part in the larger regulatory pathways of RNA-binding proteins, similar to the classical cascade of splicing regulation in the Drosophila sex determination pathway (Black 2003). In fact, although not all are defined as ultraconserved, many other splicing regulatory proteins show evidence in the EST database for alternative splicing-mediated NMD (P. Stoilov, unpubl.). In addition to these RNA-binding proteins, some of the other PTB target exons identified in our analysis are also predicted to mediate NMD. Thus, the PTB/nPTB splicing regulatory pathway does not just create modified protein products, but can also control overall product levels.
It is important to note that the cellular effects of the PTB-to-nPTB switch are likely not limited to changes in splicing. PTB is known to affect the translation of several cell cycle and apoptotic regulators, as well as having other effects in the cytoplasm (Pilipenko et al. 2001; Mitchell et al. 2003; Knoch et al. 2004; Tillmar and Welsh 2004; Cho et al. 2005). It will be particularly interesting to examine the “post-splicing” effects of PTB on the nPTB transcript itself. Most cells make observable amounts of exon 10 plus nPTB mRNA but do not appear to produce protein, unless PTB is removed by RNAi. Thus, besides NMD, there are further PTB-dependent mechanisms maintaining the repression of nPTB. This additional layer of regulation could occur through effects on mRNA transport or protein stability. However, the implication of PTB in controlling the translation of other mRNAs makes this an appealing model. It will be important to examine these mechanisms if we are to understand how the switch from PTB to nPTB is controlled during development.
Materials and methods
Tissue culture
Cell lines were grown following standard tissue culture procedure, with guidelines provided by the American Type Culture Collection (http://www.atcc.org). P19 cells were cultured and differentiated according to previously published protocols (Yao et al. 1995). Cortical neural precursors were cultured according to Feng et al. (2005) with minor modifications (Ahlemeyer and Baumgart-Vogt 2005; Feng et al. 2005). Briefly, cortices from E10.5 or E14 mouse embryos were dissected in Hank’s balanced salt solution (HBSS). E10.5 cortices were dissociated mechanically by fire-polished glass pipette; E14 cortices were incubated in 0.1% trypsin in DMEM/F12 (Invitrogen), washed with DMEM/F12 containing 5% fetal bovine serum (FBS), and dissociated mechanically. Dissociated cells were plated on poly-ornithine-coated (10 μg/mL) and fibronectin-coated (10 μg/mL) dishes with serum-free DMEM/F12 medium supplemented with B27 (Invitrogen) and penicillin/streptomycin (50 U/mL and 50 μg/mL, respectively). Cultures were fed with 10 ng/mL basic fibroblast growth factor (bFGF) (Peprotech) at the time of plating. Cerebellar culture was carried out according to Ahlemeyer and Baumgart-Vogt (2005). Cerebelli were dissected out from P3–P5 mice in Neurobasal medium (Invitrogen) containing 0.02% BSA and were digested at 37°C in 0.25% trypsin in HBSS. The cerebellar tissue sample was washed three times with ice-cold HBSS with EDTA and 0.05% DNase solution (0.05% DNase plus 12 mM MgSO4 in HBSS) and dissociated mechanically with a flame-polished pasture pipette. Dissociated cells were plated onto poly-L-lysine-coated (10 mg/mL) dishes in Neurobasal medium with B27 supplement, 25 mM KCl, 2 mM glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin. All cultures were either fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 20 min at room temperature for immunocytochemistry or harvested for RNA isolation or Western blot analysis of proteins.
Western blotting
Western blots and preparation of tissue culture cell lysates were performed as previously described (Boutz et al. 2007). Adult mouse tissues obtained by standard dissection were lysed in RIPA buffer and sonicated to homogenize samples using a W-385 Ultrasonic Processor (Heat Systems) at 10 Hz. After a 30-min incubation at 4°C, samples were frozen at −20°C. PAGE and Immunoblotting were performed as with tissue culture cell lysates. Antibodies were used at the following dilutions: α-GAPDH (1:200,000; Research Diagnostics, Inc.), α-nPTB IS2 (1:500) (Sharma et al. 2005), α-PTB/nPTB antibody cPTB (1:2500) (Amir-Ahmady et al. 2005), α-PTB antibody PTB-NT (1:3000) (Markovtsov et al. 2000), α-TuJ1 (1:2000; Covance), α-Nestin (1:250; Chemicon), α-Histone H3 (1:1000; AbCam), α-Upf1/RENT1 (1:1000; Bethyl), α-MAP2 (1:250; Chemicon), α-β-actin (1:2000; Novus), α-U1 70k (1:2000) (Sharma et al. 2005), and HRP-conjugated secondary antibodies (1:5000; Amersham Pharmacia) or ECL Plex Cy5-conjugated goat α-mouse and goat α-rabbit secondary antibodies (1:2500; GE Healthcare).
Immunohistochemistry/immunocytochemistry
Adult mice were perfused transcardially with ice-cold PBS, followed by ice-cold 4% PFA/PBS. The brains were removed, post-fixed in 4% PFA/PBS overnight, cryoprotected in 20% sucrose-PBS, frozen in 4-methyl-butane, and stored at −80°C until use. Ten-micron sections were cut on a cryostat and collected onto Superfrost plus slides (Fisher) at −80°C until use. Sections were thawed, post-fixed in 10% formalin for 10 min, rinsed twice in PBS, permeablized with 0.5% Triton in PBS for 10 min, and incubated in 1% normal goat serum and 2 mg/mL BSA in PBS for 1 h. Sections were incubated at room temperature overnight with primary antibodies and rinsed in PBS with 0.1% Triton (PBST) before incubation with Alexa-conjugated secondary antibodies for 2 h at room temperature. Slides were rinsed in PBST and mounted with a Prolong Gold AntiFade mounting medium containing nuclear stain DAPI (Molecular Probes). In all cases, controls with no primary antibody yielded no labeling. Similar procedures were used for immunofluorescence staining of cortical precursors and cerebellar cells, except that they are fixed with 4% PFA/PBS for 20 min and incubated with primary antibody for 2 h. Primary antibodies were used at the following concentrations: α-nPTB IS2 (1:200), α-PTB antibody PTB-NT (1:500), α-GFAP (1:1000; Chemicon), α-NeuN (1:100; Chemicon), α-Nestin (1:200), α-TuJ1 (1:250), and α-MAP2 (1:500). Secondary antibodies, Alexa488 anti-mouse IgG and Alexa568 anti-rabbit IgG (Molecular Probes), were used at 1:1000 dilutions.
RT–PCR
Total RNA was collected from adherent tissue culture cells using Trizol (Invitrogen) according to the manufacturer’s instructions. Adult mouse tissue RNA was harvested using Trizol followed by homogenization with a Tissue Tearor (Biospec). RT–PCR was performed as previously described (Boutz et al. 2007). Sequences of the primers used for PCR are listed in the Supplemental Material.
siRNA synthesis
The anti-PTB siRNA was described previously (Amir-Ahmady et al. 2005). siRNAs were constructed by T7 polymerase transcription as described previously (Amir-Ahmady et al. 2005; Boutz et al. 2007). Template oligonucleotides used to construct siRNAs are listed in the Supplemental Material. siRNA transfections were performed in 24-well plates, with 100,000 N1E, 250,000 HEK293, or 120,000 N2A cells per well. One microgram of pUC18 plasmid was used as a carrier, and 2 μL of Lipofectamine 2000 (Invitrogen). Twenty-six nanomolar siRNA was used in each transfection. All transfections were performed as previously described (Underwood et al. 2005).
Short hairpin design and RNAi
Hairpins were designed to target the 3′ untranslated regions (UTRs) of PTB and nPTB. A single-nucleotide mismatch was introduced close to the 3′ end of the sense strand of the hairpin duplex to destabilize the end and ensure that the antisense strand would be selected in the RISC complex. The hairpins were constructed by annealing two synthetic oligonucleotides and filling the ends with Klenow DNA polymerase. The duplexes were then digested with BsrGI and cloned behind the H1 promoter of pBlsH1 (a kind gift from M. Gencheva and R.J. Lin, City of Hope, Duarte, CA). The oligos used to make the hairpins are listed in the Supplemental Material. For transfection of shRNA constructs, exponentially growing N2A cells were trypsinized and resuspended in growth medium (DMEM 10% FBS) at 2.5 × 105 cells per milliliter. DNA/Lipofectamine 2000 (Invitrogen) complexes were assembled according to the manufacturer’s protocol using 20 μg of DNA and 50 μL of Lipofectamine 2000. Cells (2.5 × 106) were added to the complexes and were incubated in suspension and mixed occasionally to prevent cells from sticking to the tube walls and forming clumps. After 4 h the cells were collected by centrifugation, resuspended in 30 mL of growth medium, and plated in 30-cm culture dishes. After 3 d the cells were collected and the knockdown was tested by Western blot.
RNase protection
Thirty micrograms of total RNA were coprecipitated with 100,000 counts per minute (cpm) of gel-purified, single-stranded RNA probe. The pellet was resuspended in hybridization buffer (400 mM NaCl, 15 mM PIPES at pH 6.5, 1 mM EDTA, 80% formamide) and denatured for 10 min at 85°C. The hybridization was carried out overnight at 50°C. The hybridization mix was then diluted with 300 μL of RNase solution (300 μM NaCl, 10 mM Tris-HCl at pH 7.5, 5 mM EDTA). One microliter of RNase T1/RNase A mix (Ambion) was then added to the mix and it was incubated at 37°C for 1 h. The digestion products were purified by extraction with phenol/chloroform, followed by ethanol precipitation. They were dissolved in formamide gel loading buffer and resolved by electrophoresis on 8% denaturing polyacrylamide gels.
Real-time PCR
Real-time PCR was performed on cDNAs reverse-transcribed as explained in the RT–PCR section. All real-time PCRs were performed as described previously (Boutz et al. 2007). Primers are listed in the Supplemental Material.
Microarray design
Oligonucleotide and microarray design have been described previously (Clark et al. 2002; Srinivasan et al. 2005). To chose exons whose regulation was likely to be functionally significant, we used exons from five data sets: (1) exons identified from the literature that exhibit neuron-specific or muscle-specific regulation, or have partially characterized mechanisms of regulation; (2) exons derived from genes coding for known splicing factors or protein kinases involved in splicing where EST, phylogenetic, and other evidence supports a role for these exons in regulating protein expression by coupling to NMD (similar to PTB exon 11); (3) exons in genes associated with cancer and tumor development, whose splicing is thought to have important functional effects on the encoded proteins; (4) exons derived from the MAASE database of alternative splicing (Zheng et al. 2005) (these exons affect proteins that have well-described functions in the Swiss-prot database); (5) a subset of the exons identified by Sugnet et al. (2004) as showing conserved regulation between human and mouse. A detailed description of the microarray construction, probing, and data analysis is provided in the Supplemental Material.
Acknowledgments
We particularly thank Chris Smith for multiple helpful discussions and prior communication of unpublished data. We thank Abigail Silver and members of the Black laboratory, as well as one of our reviewers for helpful advice. Special thanks go to Shalini Sharma for affinity purification of the IS2 antibody, and to Jian Feng and Guoping Fan for help with the NPC culture. This work was supported by NIH grants RO1 GM49662 and R24 GM070857 to D.L.B., RO1040478 and R24 GM070857 to M.A., and a Warsaw Family Fellowship at UCLA to P.L.B. D.L.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1558107
References
- Ahlemeyer B., Baumgart-Vogt E., Baumgart-Vogt E. Optimized protocols for the simultaneous preparation of primary neuronal cultures of the neocortex, hippocampus and cerebellum from individual newborn (P0.5) C57Bl/6J mice. J. Neurosci. Methods. 2005;149:110–120. doi: 10.1016/j.jneumeth.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Amir-Ahmady B., Boutz P.L., Markovtsov V., Phillips M.L., Black D.L., Boutz P.L., Markovtsov V., Phillips M.L., Black D.L., Markovtsov V., Phillips M.L., Black D.L., Phillips M.L., Black D.L., Black D.L. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiya M., Grabowski P.J., Grabowski P.J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: Evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G., Grunseich C., Lu D.D., Speh J.C., Mandel G., Lu D.D., Speh J.C., Mandel G., Speh J.C., Mandel G., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Maquat L.E., Cheng J., Maquat L.E., Maquat L.E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl. Acad. Sci. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Castro D.S., Guillemot F., Castro D.S., Guillemot F., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Black D.L., Grabowski P.J., Grabowski P.J. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Green R.E., Brenner S.E., Rio D.C., Green R.E., Brenner S.E., Rio D.C., Brenner S.E., Rio D.C., Rio D.C. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes & Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P.L., Chawla G., Stoilov P., Black D.L., Chawla G., Stoilov P., Black D.L., Stoilov P., Black D.L., Black D.L. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes & Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo E., McKay R., McKay R. Proliferation and differentiation of neuronal stem cells regulated by nerve growth factor. Nature. 1990;347:762–765. doi: 10.1038/347762a0. [DOI] [PubMed] [Google Scholar]
- Chabot B., Blanchette M., Lapierre I., La Branche H., Blanchette M., Lapierre I., La Branche H., Lapierre I., La Branche H., La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.C., Black D.L., Black D.L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Kim J.H., Back S.H., Jang S.K., Kim J.H., Back S.H., Jang S.K., Back S.H., Jang S.K., Jang S.K. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T.A., Sugnet C.W., Ares M., Sugnet C.W., Ares M., Ares M. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.J., Mandel G., Otto S., Han J.J., Mandel G., Han J.J., Mandel G., Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E., Izaurralde E., Izaurralde E. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Dalpe G., Leclerc N., Vallee A., Messer A., Mathieu M., De Repentigny Y., Kothary R., Leclerc N., Vallee A., Messer A., Mathieu M., De Repentigny Y., Kothary R., Vallee A., Messer A., Mathieu M., De Repentigny Y., Kothary R., Messer A., Mathieu M., De Repentigny Y., Kothary R., Mathieu M., De Repentigny Y., Kothary R., De Repentigny Y., Kothary R., Kothary R. Dystonin is essential for maintaining neuronal cytoskeleton organization. Mol. Cell. Neurosci. 1998;10:243–257. doi: 10.1006/mcne.1997.0660. [DOI] [PubMed] [Google Scholar]
- Dehmelt L., Halpain S., Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge B.K., Stefani G., Engelhard C.C., Darnell R.B., Stefani G., Engelhard C.C., Darnell R.B., Engelhard C.C., Darnell R.B., Darnell R.B. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Martinowich K., Chin M.H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., Martinowich K., Chin M.H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., Chin M.H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., Hattori D., Ge W., Shen Y., Wu H., Ge W., Shen Y., Wu H., Shen Y., Wu H., Wu H., et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Feng J., Chang H., Li E., Fan G., Chang H., Li E., Fan G., Li E., Fan G., Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Gromak N., Rideau A., Southby J., Scadden A.D., Gooding C., Huttelmaier S., Singer R.H., Smith C.W., Rideau A., Southby J., Scadden A.D., Gooding C., Huttelmaier S., Singer R.H., Smith C.W., Southby J., Scadden A.D., Gooding C., Huttelmaier S., Singer R.H., Smith C.W., Scadden A.D., Gooding C., Huttelmaier S., Singer R.H., Smith C.W., Gooding C., Huttelmaier S., Singer R.H., Smith C.W., Huttelmaier S., Singer R.H., Smith C.W., Singer R.H., Smith C.W., Smith C.W. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich K.A., Linseman D.A., Linseman D.A. Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Mol. Neurobiol. 2004;29:155–166. doi: 10.1385/MN:29:2:155. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R., Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr. Opin. Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hughes T.A. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Hui J., Hung L.H., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Hung L.H., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Neumuller N., Reither G., Haas S.A., Bindereif A., Reither G., Haas S.A., Bindereif A., Haas S.A., Bindereif A., Bindereif A. Intronic CA-repeat and CA-rich elements: A new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim el C., Schaal T.D., Hertel K.J., Reed R., Maniatis T., Schaal T.D., Hertel K.J., Reed R., Maniatis T., Hertel K.J., Reed R., Maniatis T., Reed R., Maniatis T., Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin N., Baekelandt V., Goritchenko L., Benowitz L.I., Baekelandt V., Goritchenko L., Benowitz L.I., Goritchenko L., Benowitz L.I., Benowitz L.I. Identification of two proteins that bind to a pyrimidine-rich sequence in the 3′-untranslated region of GAP-43 mRNA. Nucleic Acids Res. 1997;25:1281–1288. doi: 10.1093/nar/25.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L.E., Li X., Serin G., Maquat L.E., Serin G., Maquat L.E., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Izquierdo J.M., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Castelo R., Guigo R., Bilbao D., Valcarcel J., Guigo R., Bilbao D., Valcarcel J., Bilbao D., Valcarcel J., Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Schadt E.E., Stoughton R., Shoemaker D.D., Stoughton R., Shoemaker D.D., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Chang S., Etchberger J.F., Ortiz C.O., Hobert O., Chang S., Etchberger J.F., Ortiz C.O., Hobert O., Etchberger J.F., Ortiz C.O., Hobert O., Ortiz C.O., Hobert O., Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H., Nielsen P.J., Nielsen P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A., Muhlemann O., Akusjarvi G., Muhlemann O., Akusjarvi G., Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- Kikuchi T., Ichikawa M., Arai J., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Ichikawa M., Arai J., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Arai J., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Fu L., Higuchi K., Yoshimura N., Higuchi K., Yoshimura N., Yoshimura N. Molecular cloning and characterization of a new neuron-specific homologue of rat polypyrimidine tract binding protein. J. Biochem. 2000;128:811–821. doi: 10.1093/oxfordjournals.jbchem.a022819. [DOI] [PubMed] [Google Scholar]
- Knoch K.P., Bergert H., Borgonovo B., Saeger H.D., Altkruger A., Verkade P., Solimena M., Bergert H., Borgonovo B., Saeger H.D., Altkruger A., Verkade P., Solimena M., Borgonovo B., Saeger H.D., Altkruger A., Verkade P., Solimena M., Saeger H.D., Altkruger A., Verkade P., Solimena M., Altkruger A., Verkade P., Solimena M., Verkade P., Solimena M., Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- Kumar S., Lopez A.J., Lopez A.J. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 2005;24:2646–2655. doi: 10.1038/sj.emboj.7600723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E., Inada M., Green R.E., Wengrod J.C., Brenner S.E., Green R.E., Wengrod J.C., Brenner S.E., Wengrod J.C., Brenner S.E., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Le K., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Wang Q., Xu Q., Nelson S.F., Lee C., Xu Q., Nelson S.F., Lee C., Nelson S.F., Lee C., Lee C.2004Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data Nucleic Acids Res. 32e180 . 10.1093/nar/gnh173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Maquat L.E., Maquat L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Green R.E., Brenner S.E., Green R.E., Brenner S.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillevali K., Kulla A., Ord T., Kulla A., Ord T., Ord T. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech. Dev. 2001;101:217–220. doi: 10.1016/s0925-4773(00)00566-9. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin C.H., Patton J.G., Patton J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.D., Steitz J.A., Shu M.D., Steitz J.A., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Mao Z., Bonni A., Xia F., Nadal-Vicens M., Greenberg M.E., Bonni A., Xia F., Nadal-Vicens M., Greenberg M.E., Xia F., Nadal-Vicens M., Greenberg M.E., Nadal-Vicens M., Greenberg M.E., Greenberg M.E. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- Markovtsov V., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L., Goldman J.A., Turck C.W., Chou M.Y., Black D.L., Turck C.W., Chou M.Y., Black D.L., Chou M.Y., Black D.L., Black D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A.J., Clark F., Smith C.W., Clark F., Smith C.W., Smith C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys C.M., Dietz H.C., ap Rhys C.M., Dietz H.C., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Menon K.P., Neufeld E.F., Neufeld E.F. Evidence for degradation of mRNA encoding α-L-iduronidase in Hurler fibroblasts with premature termination alleles. Cell. Mol. Biol. (Noisy-le-grand) 1994;40:999–1005. [PubMed] [Google Scholar]
- Mitchell S.A., Spriggs K.A., Coldwell M.J., Jackson R.J., Willis A.E., Spriggs K.A., Coldwell M.J., Jackson R.J., Willis A.E., Coldwell M.J., Jackson R.J., Willis A.E., Jackson R.J., Willis A.E., Willis A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Shiue L., Clark T.A., Blume J.E., Ares M., Clark T.A., Blume J.E., Ares M., Blume J.E., Ares M., Ares M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Amir-Ahmady B., Pitsch S., Black D.L., Pitsch S., Black D.L., Black D.L., et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Babak T., Siu H., Hughes T.R., Morris Q.D., Siu H., Hughes T.R., Morris Q.D., Hughes T.R., Morris Q.D., Morris Q.D. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Maquat L.E., Frey B.J., Blencowe B.J., Frey B.J., Blencowe B.J., Blencowe B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Viktorova E.G., Guest S.T., Agol V.I., Roos R.P., Viktorova E.G., Guest S.T., Agol V.I., Roos R.P., Guest S.T., Agol V.I., Roos R.P., Agol V.I., Roos R.P., Roos R.P. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001;20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydorides A.D., Okano H.J., Yang Y.Y., Stefani G., Darnell R.B., Okano H.J., Yang Y.Y., Stefani G., Darnell R.B., Yang Y.Y., Stefani G., Darnell R.B., Stefani G., Darnell R.B., Darnell R.B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman L., Bliskovski V., Kaye F.J., Zajac-Kaye M., Bliskovski V., Kaye F.J., Zajac-Kaye M., Kaye F.J., Zajac-Kaye M., Zajac-Kaye M. Evolutionary conservation of a 2-kb intronic sequence flanking a tissue-specific alternative exon in the PTBP2 gene. Genomics. 2004;83:76–84. doi: 10.1016/s0888-7543(03)00207-6. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Rideau A.P., Gooding C., Simpson P.J., Monie T.P., Lorenz M., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W., Gooding C., Simpson P.J., Monie T.P., Lorenz M., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W., Simpson P.J., Monie T.P., Lorenz M., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W., Monie T.P., Lorenz M., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W., Lorenz M., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W., Singer R.H., Matthews S., Curry S., Smith C.W., Matthews S., Curry S., Smith C.W., Curry S., Smith C.W., Smith C.W. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Shalizi A.K., Bonni A., Bonni A. Brawn for brains: The role of MEF2 proteins in the developing nervous system. Curr. Top. Dev. Biol. 2005;69:239–266. doi: 10.1016/S0070-2153(05)69009-6. [DOI] [PubMed] [Google Scholar]
- Sharma S., Falick A.M., Black D.L., Falick A.M., Black D.L., Black D.L. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Lykke-Andersen J., Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcarcel J., Green M.R., Valcarcel J., Green M.R., Green M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Sorek R., Ast G., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R., Smith C.W., Smith C.W. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Spellman R., Rideau A., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., Rideau A., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., Grellscheid S.N., Southby J., Wollerton M., Smith C.W., Southby J., Wollerton M., Smith C.W., Wollerton M., Smith C.W., Smith C.W. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Srinivasan K., Shiue L., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Shiue L., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Bryant J., Smith M., Rommelfanger C., Smith M., Rommelfanger C., Rommelfanger C., et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Stoilov P., Daoud R., Nayler O., Stamm S., Daoud R., Nayler O., Stamm S., Nayler O., Stamm S., Stamm S. Human tra2-β1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- Sugnet C.W., Kent W.J., Ares M., Haussler D., Kent W.J., Ares M., Haussler D., Ares M., Haussler D., Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004;9:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- Sugnet C.W., Srinivasan K., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Srinivasan K., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Williams A., Kulp D., Blume J.E., Haussler D., Kulp D., Blume J.E., Haussler D., Blume J.E., Haussler D., Haussler D., et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vicens M., Misono S., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E., Nadal-Vicens M., Misono S., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E., Misono S., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E., Zubiaga A., Hua X., Fan G., Greenberg M.E., Hua X., Fan G., Greenberg M.E., Fan G., Greenberg M.E., Greenberg M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Sun Y.E., Martinowich K., Ge W., Martinowich K., Ge W., Ge W. Making and repairing the mammalian brain—Signaling toward neurogenesis and gliogenesis. Semin. Cell Dev. Biol. 2003;14:161–168. doi: 10.1016/s1084-9521(03)00007-7. [DOI] [PubMed] [Google Scholar]
- Sureau A., Gattoni R., Dooghe Y., Stevenin J., Soret J., Gattoni R., Dooghe Y., Stevenin J., Soret J., Dooghe Y., Stevenin J., Soret J., Stevenin J., Soret J., Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmar L., Welsh N., Welsh N. Glucose-induced binding of the polypyrimidine tract-binding protein (PTB) to the 3′-untranslated region of the insulin mRNA (ins-PRS) is inhibited by rapamycin. Mol. Cell. Biochem. 2004;260:85–90. doi: 10.1023/b:mcbi.0000026059.56089.e4. [DOI] [PubMed] [Google Scholar]
- Ule J., Darnell R.B., Darnell R.B. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Wang H., Clark T., Fraser C., Ruggiu M., Clark T., Fraser C., Ruggiu M., Fraser C., Ruggiu M., Ruggiu M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Gaasterland T., Blencowe B.J., Darnell R.B., Blencowe B.J., Darnell R.B., Darnell R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Underwood J.G., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L., Dougherty J.D., Stoilov P., Black D.L., Stoilov P., Black D.L., Black D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu X., Biederer T., Sudhof T.C., Liu X., Biederer T., Sudhof T.C., Biederer T., Sudhof T.C., Sudhof T.C. A family of RIM-binding proteins regulated by alternative splicing: Implications for the genesis of synaptic active zones. Proc. Natl. Acad. Sci. 2002;99:14464–14469. doi: 10.1073/pnas.182532999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Garcia-Blanco M.A., Smith C.W., Smith C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Yao M., Bain G., Gottlieb D.I., Bain G., Gottlieb D.I., Gottlieb D.I. Neuronal differentiation of P19 embryonal carcinoma cells in defined media. J. Neurosci. Res. 1995;41:792–804. doi: 10.1002/jnr.490410610. [DOI] [PubMed] [Google Scholar]
- Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B., Van Nostrand E., Holste D., Poggio T., Burge C.B., Holste D., Poggio T., Burge C.B., Poggio T., Burge C.B., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Takita J., Tanaka Y., Setou M., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Takita J., Tanaka Y., Setou M., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Tanaka Y., Setou M., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Setou M., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Yang H.W., Terada S., Nakata T., Takei Y., Terada S., Nakata T., Takei Y., Nakata T., Takei Y., Takei Y., et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- Zheng C.L., Kwon Y.S., Li H.R., Zhang K., Coutinho-Mansfield G., Yang C., Nair T.M., Gribskov M., Fu X.D., Kwon Y.S., Li H.R., Zhang K., Coutinho-Mansfield G., Yang C., Nair T.M., Gribskov M., Fu X.D., Li H.R., Zhang K., Coutinho-Mansfield G., Yang C., Nair T.M., Gribskov M., Fu X.D., Zhang K., Coutinho-Mansfield G., Yang C., Nair T.M., Gribskov M., Fu X.D., Coutinho-Mansfield G., Yang C., Nair T.M., Gribskov M., Fu X.D., Yang C., Nair T.M., Gribskov M., Fu X.D., Nair T.M., Gribskov M., Fu X.D., Gribskov M., Fu X.D., Fu X.D. MAASE: An alternative splicing database designed for supporting splicing microarray applications. RNA. 2005;11:1767–1776. doi: 10.1261/rna.2650905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Ramachandran B., Gulick T., Ramachandran B., Gulick T., Gulick T. Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J. Biol. Chem. 2005;280:28749–28760. doi: 10.1074/jbc.M502491200. [DOI] [PubMed] [Google Scholar]