Abstract
Background
Normal preimplantation embryo development encompasses a series of events including first cleavage division, activation of the embryonic genome, compaction and blastocyst formation.
First lineage differentiation starts at the blastocyst stage with the formation of the trophectoderm and the inner cell mass. The main objective of this study was the detection, identification and expression analysis of genes associated with blastocyst formation in order to help us better understand this process. This information could lead to improvements of in vitro embryo production procedures.
Results
A subtractive cDNA library was constructed enriched for transcripts preferentially expressed at the blastocyst stage compared to the 2-cell and 8-cell stage. Sequence information was obtained for 65 randomly selected clones. The RNA expression levels of 12 candidate genes were determined throughout 3 stages of preimplantation embryo development (2-cell, 8-cell and blastocyst) and compared with the RNA expression levels of in vivo "golden standard" embryos using real-time PCR. The RNA expression profiles of 9 (75%) transcripts (KRT18, FN1, MYL6, ATP1B3, FTH1, HINT1, SLC25A5, ATP6V0B, RPL10) were in agreement with the subtractive cDNA cloning approach, whereas for the remaining 3 (25%) (ACTN1, COPE, EEF1A1) the RNA expression level was equal or even higher at the earlier developmental stages compared to the blastocyst stage. Moreover, significant differences in RNA expression levels were observed between in vitro and in vivo produced embryos. By immunofluorescent labelling, the protein expression of KRT18, FN1 and MYL6 was determined throughout bovine preimplantation embryo development and showed the same pattern as the RNA expression analyses.
Conclusion
By subtractive cDNA cloning, candidate genes involved in blastocyst formation were identified. For several candidate genes, important differences in gene expression were observed between in vivo and in vitro produced embryos, reflecting the influence of the in vitro culture system on the embryonic gene expression. Both RNA and protein expression analysis demonstrated that KRT18, FN1 and MYL6 are differentially expressed during preimplantation embryo development and those genes can be considered as markers for bovine blastocyst formation.
Background
The low success rate of in vitro embryo production procedures in cattle, together with early embryonic mortality, lead to the loss of a large number of potential calves, retarded genetic progress and consequently the loss of money and time for the cattle breeding industry. Despite improvements on the culture media and culture conditions only 30–40% of the fertilised oocytes reach the blastocyst stage and it is generally accepted that the quality and developmental competence of in vitro produced (IVP) bovine embryos has failed to keep up with those of their in vivo counterparts [1].
Normal preimplantation embryo development in cattle is characterised by several cleavage divisions of the fertilised egg, activation of the embryonic genome around the 8–16 cell stage, compaction and blastocoel formation leading to the blastocyst. Before the major activation of the embryonic genome, the bovine preimplantation embryo is controlled by maternal genomic information that is accumulated during oogenesis [2]. Both the maternal and embryonic gene expression occurs in a stage- and time-dependent manner [3].
The first important differentiation events take place at the blastocyst stage, resulting in the generation of two distinct cell lineages: the trophectoderm cells (TE) and the inner cell mass (ICM). The ICM gives rise to the embryo, whereas the TE forms the placenta. Since these two lineages are divergent in morphological and biochemical aspects, it is reasonable to hypothesise that many differentiation-related genes are expressed at this stage. Genes being expressed in the blastocyst stage but not in earlier stages are therefore functional candidates for the regulative processes that take place at the onset of differentiation [4]. The identification of novel genes involved in blastocyst formation and the analysis of their expression patterns may help us to understand the mechanisms that control blastocyst formation and may help us to understand why certain embryos do not make it through this stage. As in vitro culture conditions do not fully mimic the in vivo situation and as it has been shown that morphological and physiological differences [5-7] as well as differences in gene expression [8-11] exist between in vivo and in vitro cultured embryos, the RNA expression levels of in vitro produced embryos should be compared with those of in vivo "golden standard" embryos. Changes in transcript abundance between in vitro and in vivo embryos may help to assess the normality of in vitro produced embryos and may help to optimise the in vitro culture conditions.
In previous studies, genes involved in preimplantation embryo development were identified using techniques such as differential display reverse transcription PCR [4,12,13], large scale cDNA library construction [14-16], cDNA microarray [17-19] and suppression subtractive hybridisation (SSH) [4,20-22]. Subtractive cDNA cloning [23] is a powerful tool for the detection of stage-specific transcribed genes. In contrast to e.g. microarray analysis, this PCR-based method can be accomplished without prior knowledge of the genes being expressed, and yields subtracted cDNA pools that are differentially expressed. However, further confirmation of the outcome is required by means of real-time PCR, which is a perfect technique not only to validate the subtractive cDNA cloning results, but also to gather quantitative data concerning the selected transcripts. In the present study subtractive cDNA cloning was used to detect genes that are differentially expressed in the bovine blastocyst compared to genes present in 2-cell and 8-cell stage embryos. The differential RNA expression status of 12 subtracted cDNA clones was validated throughout 3 stages of preimplantation embryo development (2-cell, 8-cell and blastocyst) and compared with the RNA expression levels of their in vivo "golden standard" counterparts using real-time PCR. By immunofluorescent labelling, the protein expression of 3 genes with high differences in RNA expression levels between the developmental stages was examined during preimplantation embryo development to check whether the protein expression patterns were comparable with the RNA expression levels.
Results
Construction and screening of the SSH library
A subtractive cDNA library was constructed for the enrichment of transcripts preferentially expressed in in vitro cultured blastocysts relative to 2-cell and 8-cell embryos.
As bovine embryos contain limited amounts of mRNA and as the subtractive cDNA cloning requires at least 0.5 μg of starting material, a linear amplification step was performed using the SMART PCR cDNA synthesis Kit. By using 27 PCR cycles for linear amplification, the formation of non-specific PCR products due to overcycling was prevented.
The amplification products enriched for blastocyst specific genes were cloned, single clones were randomly picked and single-pass sequenced. Partial sequence information was obtained for 65 clones. Those partial sequences were compared with known sequences in the Genbank by doing a BLAST analysis against the mammal nr database. BLAST searches revealed that 54 sequences were homologous to 36 different known genes, 5 sequences were homologous to genomic sequences and 2 clones had no matches in the database and could not be identified. The 4 remaining clones were false positives, giving similarity with the cloning vector. Among the identified genes, we found 14 genes coding for ribosomal proteins and 10 mitochondrial genes.
The sequence data of the clones were submitted to the NCBI dbEST database [24] and the accession numbers and their putative identities are listed in Table 1.
Table 1.
Overview of the blastocyst specific subtracted cDNA clones compared to known sequences in Genbank
| EST Accession Number | Gene name | Function | Accession Number BlastN | % Identity |
| DQ347577 | B. taurus γ-actin Cytoplasmic 2 | Cytoskeletal protein | NM_001033618 | 99% 443/447 |
| DQ347578 | 100% 237/237 | |||
| DQ347579 | B. taurus actinin, α 1 | Cytoskeletal protein | XM_591685 | 99% 257/258 |
| DQ347580 | B. taurus Fibronectin 1 | Cytoskeletal protein | XM874396 | 95% 457/478 |
| DQ347581 | H. sapiens Microtubule-actin crosslink factor 1 | Cytoskeletal protein | NM_012090 | 88% 425/478 |
| DQ347582 | 89% 317/354 | |||
| DQ347583 | B. taurus Myosin light chain prot. 6 | Cytoskeletal protein | BTMYO217 | 99% 413/417 |
| DQ347584 | B. taurus cytokeratin 18 | Cytoskeletal protein | XM_582930 | 99% 442/443 |
| DQ347585 | 96% 219/228 | |||
| DQ347586 | B. taurus Adenin nucleotide translocator 2 | Transporter activity | AB065433 | 97% 272/279 |
| DQ347587 | 97% 245/259 | |||
| DQ347588 | 99% 300/301 | |||
| DQ347589 | 98% 297/303 | |||
| DQ347590 | B. taurus Ferritin heavy chain polypeptide 1 | Iron storage | NM_174062 | 98% 171/174 |
| DQ347591 | 99% 230/231 | |||
| DQ347592 | AF540563 | 100% 167/167 | ||
| DQ347593 | 100% 170/170 | |||
| DQ347594 | 95% 356/372 | |||
| DQ347595 | B. taurus Histidine triad nucleotide binding protein 1 | Hydrolase | NM_175812 | 95% 350/366 |
| DQ347596 | 95% 340/356 | |||
| DQ347597 | B. taurus coatomer protein complex, subunit ε | retrograde Golgi-to-ER transport | X76980 | 99% 364/366 |
| DQ347598 | B. taurus neural precursor cell expressed, developmental down-regulated 8 | linkage of ubiquitin | AF227256 | 99% 173/174 |
| DQ347599 | H. sapiens tyrosine 3-monooxygenase | signal transduction | BC010352 | 91% 77/84 |
| DQ347600 | H. sapiens ATPase Na/K transporting, subunit b3 | Osmoregulation | NM_001035393 | 99% 212/214 |
| DQ347601 | 98% 175/178 | |||
| DQ347634 | B. taurus cyclin G-associated kinase | Cell cycle | NM_001046084 | 95% 226/236 |
| DQ347637 | 96% 130/135 | |||
| DQ347603 | B. taurus elongation factor 1 α | Translation | AJ238405 | 89% 305/341 |
| DQ347604 | B. taurus ribosomal prot. L3 | Translation | NM_174715 | 100% 167/167 |
| DQ347605 | B. taurus ribosomal prot. L6 | NM_001031756 | 99% 133/134 | |
| DQ347606 | H. sapiens ribosomal prot. L7 | NM_001014928 | 99% 423/426 | |
| DQ347607 | B. taurus ribosomal prot. L10/QM | NM_174760 | 99% 166/167 | |
| DQ347608 | B. taurus ribosomal prot. L17 | AB099021 | 96% 256/265 | |
| DQ347609 | B. taurus ribosomal prot. L23 | NM_001035014 | 99% 330/331 | |
| DQ347610 | B. taurus ribosomal prot. S5 | NM_001015531 | 98% 465/470 | |
| DQ347611 | H. sapiens ribosomal prot. S11 | NM_001024568 | 99% 289/290 | |
| DQ347613 | B. taurus ribosomal prot S8 | NM_001025317 | 99% 333/334 | |
| DQ347612 | B. taurus ribosomal prot. S12 | BC102500 | 98% 386/390 | |
| DQ347614 | B. taurus ribosomal prot. 40S S26-2-like | NM_001015561 | 98% 331/336 | |
| DQ347615 | B. taurus ribosomal prot. S27 | XM_880953 | 99% 287/289 | |
| DQ347616 | B. taurus ribosomal prot. large P2 | NM_174788 | 98% 443/452 | |
| DQ347617 | B. taurus U1 snRNP-specific prot. | XM_872762 | 99% 331/333 | |
| DQ347602 | B. taurus ATPase H+ transporter V0 subunit b | Energy metabolism | NM_001038038 | 100% 634/634 |
| DQ347636 | B. taurus cytochrome C oxidase 7A2 | Energy metabolism | NM_175807 | 94%199/210 |
| DQ347618 | B. taurus mitoch. genome ATP6 | Energy metabolism | AY526085 | 100% 251/251 |
| DQ347619 | B. taurus mitoch. genome ND4 | 99% 250/251 | ||
| DQ347620 | B. taurus mitoch. genome COX3 | 98% 374/381 | ||
| DQ347621 | B. taurus mitoch. genome COX2 | 99% 225/226 | ||
| DQ347622 | B. taurus mitoch. genome COX1 | 97% 260/268 | ||
| DQ347623 | B. taurus mitoch. genome COX3 | 98% 259/264 | ||
| DQ347624 | B. taurus mitoch. genome COX2 | 99% 219/220 | ||
| DQ347625 | B. taurus mitoch. genome COX2 | 99% 226/227 | ||
| DQ347626 | B. taurus mitoch. genome COX1 | 100% 240/240 | ||
| DQ347627 | B. taurus mitoch. genome COX2 | 99% 429/430 | ||
| DQ347629 | B. taurus BAC library clone rp42-147e22 | Unknown | AC092858 | 98% 400/406 |
| DQ347631 | 95% 141/147 | |||
| DQ347628 | B. taurus BAC library clone rp42-518P7 | Unknown | AC129959 | 94% 218/230 |
| DQ347630 | 98% 193/195 | |||
| DQ347632 | B. taurus BAC library clone CH240-472P12 | Unknown | AC150855 | 90% 142/157 |
| DQ347633 | Unknown EST | Unknown | No similarity found | |
| DQ347635 |
Quantification of specific transcripts by real-time PCR
Real-time RT-PCR analysis was performed for 12 genes in order to make a relative quantification of the RNA levels in the 3 studied bovine developmental stages and to check for differences in RNA levels between in vivo and in vitro produced embryos.
Primers were designed for a selection of 12 identified genes, based on their gene function. The primer information (gene symbol, gene name, amplicon accession number, GenBank accession number and % identities with Genbank sequences) is listed in Table 2. Gene-specific amplification was confirmed for the 12 primer pairs by sequencing, by a single peak in melt-curve analysis and a single band with the expected size in agarose gel-electrophoresis. No primer-dimer formation was detected and the standard curves derived from 10-fold serial dilutions of pooled cDNA gave correlation coefficients greater than 0.99 and efficiencies between 82 and 105%. The RNA expression levels of the selected genes were measured in duplicate in 3 to 6 single in vitro and in vivo produced embryos from the 3 studied stages of preimplantation development (2-cell, 8-cell and blastocyst).
Table 2.
Details of the primers used for PCR analysis
| Gene Symbol | Gene Name | Amplicon Acc. Number | Genbank Acc. Number | % Identity |
| ACTN1 | Actinin, alpha 1 | DQ347561 | XM_603102 | 100% |
| ATP1B3 | Na/K ATPase, beta 3 subunit | DQ347562 | EST SSH | 100% |
| ATP6V0B | H+ ATPase, V0 subunit B | DQ347563 | XM_582011 | 100% |
| COPE | coatomer protein complex, subunit epsilon | DQ347564 | NM_176673 | 100% |
| EEF1A1 | Elongation factor 1 alpha | DQ347565 | AJ238405 | 100% |
| FN1 | Fibronectin 1 | DQ347566 | K00800 | 100% |
| FTH1 | Ferritin heavy chain polypeptide 1 | DQ347567 | NM_174062 | 99% |
| HINT1 | Histidine triad nucleotide binding protein 1 | DQ347568 | NM_175812 | 98% |
| KRT18 | Cytokeratin 18 | DQ347569 | XM_582930 | 100% |
| MYL6 | Myosin light chain protein 6 | DQ347572 | NM_175780 | 100% |
| RPL10 | Ribosomal protein L10/QM | DQ347575 | NM_174760 | 100% |
| SLC25A5 | Adenin nucleotide translocator 2 | DQ347576 | NM_174659 | 100% |
To correct for any variation in both mRNA content and differences in enzymatic efficiencies, the quantitative results were normalised to the geometric mean of the 3 best reference genes as described in our previous study [25]. The normalised values for each target transcript were analysed using the 95% confidence intervals.
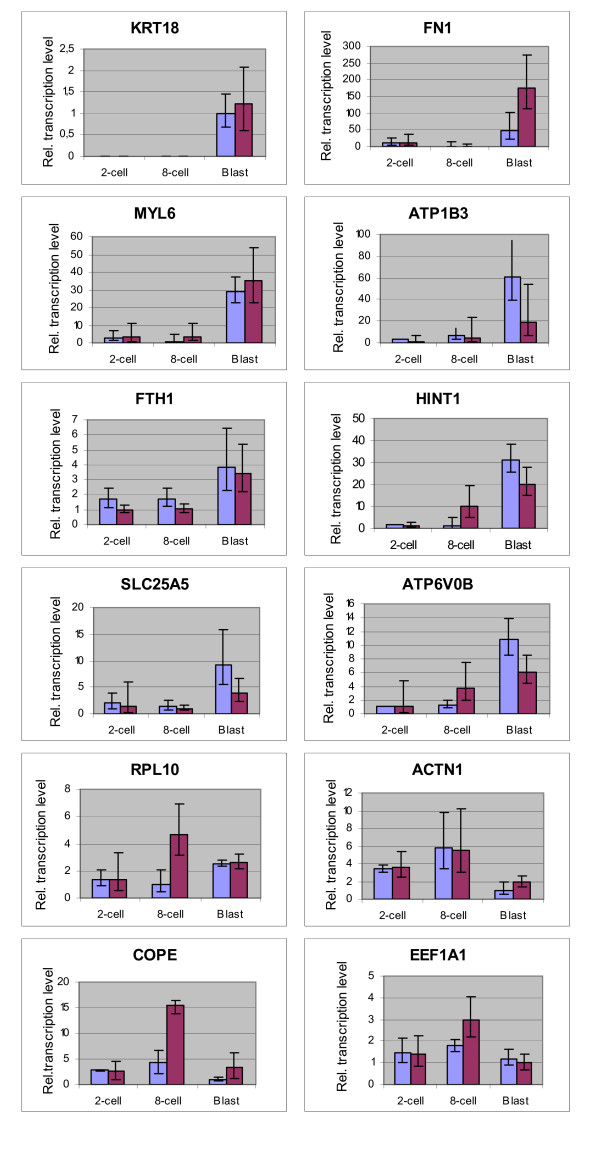
The mean relative abundances of the gene transcripts studied are shown in Figure 1.
Figure 1.
Comparison of the relative expression levels of 12 genes at 3 stages of bovine embryo development. Mean relative abundance of 12 transcripts at 3 different developmental stages (2-cell, 8-cell and blastocyst) of in vitro (lilac-coloured bars)and in vivo (claret-coloured bars) produced bovine embryos, determined by real-time qPCR.
KRT18 was not detected up to the blastocyst stage, and no differences were observed between in vitro and in vivo produced embryos. For MYL6 and FN1 the RNA levels were significantly higher at the blastocyst stage compared to the earlier stages. Remarkable is the 3.5-fold higher FN1 RNA expression in in vivo produced blastocysts compared with their in vitro produced counterparts.
The differences between the blastocysts and the 2/8-cell stage embryos for FTH1 were smaller but significant in both in vitro and in vivo produced embryos.
ATP1B3, HINT1, SLC25A5, ATP6V0B and RPL10 had significantly lower RNA levels in in vitro produced 2/8-cell embryos compared to the in vitro produced blastocysts, but surprisingly no significant differences exist between the in vivo produced 8-cell and in vivo produced blastocyst embryos. This is due to a lower RNA level in the in vivo produced blastocysts (ATP1B3 and SLC25A5) on the one side and a higher RNA level at the 8-cell stage (RPL10) on the other side or a combination of both (HINT1 and ATP6V0B).
In contrast, 3 genes (ACTN1, COPE and EEF1A1) showed a relative expression pattern that was not in agreement with the results obtained by SSH. The RNA levels for those 3 genes were highest at the 8-cell stage and decreased significantly at the blastocyst stage. Especially for COPE but also for EEF1A1 the in vivo produced 8-cell embryos had a significantly higher RNA level than their in vitro produced counterparts.
Immunofluorescent labelling
Immunofluorescent labelling was performed for KRT18, FN1 and MYL6 on in vitro produced embryos of different developmental stages (2–4 cell, 5–8 cell, morula day 5 p.i., morula day 7 p.i., blastocyst day 7 p.i. and hatched blastocyst day 8 p.i) to verify whether the protein expression showed the same pattern as the RNA expression. The results of the immunofluorescent labelling experiments are shown in Figure 2.
Figure 2.
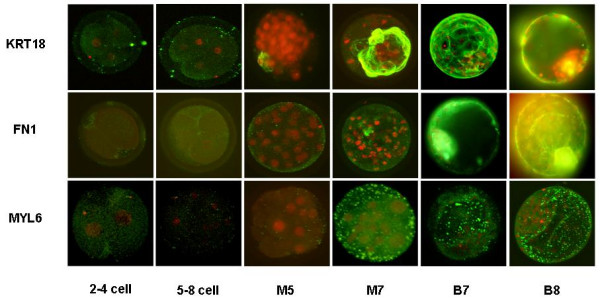
Results of the immunofluorescent experiments for KRT18, FN1 and MYL6. Confocal laser scanning images of in vitro produced bovine embryos labelled with primary mouse antibodies for KRT18, FN1 and MYL6 respectively in combination with FITC-labelled secondary goat-anti-mouse antibodies. The nuclei are stained with propidium iodide. Different stages of preimplantation embryo development were analysed (2–4 cell, 5–8 cell, M5: morula day 5 p.i., M7: morula day 7 p.i., B7: blastocyst day 7 p.i., B8: blastocyst day 8 p.i.).
No KRT18 expression was measured in the 2–4 cell and 5–8 cell embryos except for a few small positive spots on the surface of the embryos, which may be caused by remainders of cumulus cells. The first onset of the KRT18 protein expression was detected at the morula day 5 p.i. and at the morula day 7 p.i. a ring of KRT18 positive cells was observed. At the blastocyst stage, KRT18 was predominantly expressed in the TE cells, whereas little or no expression was measured in the ICM. As shown in Figure 3A, KRT18 was detected at the cell-cell contact sites of the TE cells.
Figure 3.
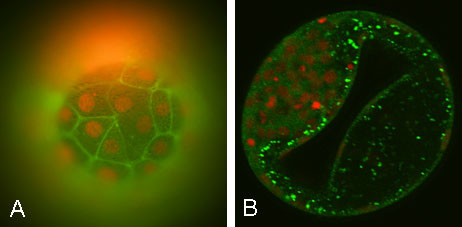
KRT18 and MYL6 expression in in vitro produced bovine blastocysts. A) Image of the surface of a bovine blastocyst day 8 p.i.labelled for KRT18. The nuclei were stained with propidium iodide. KRT18 was expressed at the cell-cell contact sites of the trophectoderm cells. B) Image of a midplane section through a bovine blastocyst day 8 p.i. labelled with antibodies for MYL6. The nuclei were stained with propidium iodide. MYL6 proteins were located in the trophectoderm cells surrounding the blastocoel cavity.
FN1 was not detected in the first developmental stages. The first onset of FN1 expression was seen in 7-day old morulae and it was clearly present at the blastocyst stages. But in contrast to KRT18, FN1 was very distinct in the ICM, forming filamentous structures between the TE and the ICM.
No MYL6 protein expression was seen before 7 days p.i MYL6 was strongly expressed at the late morula, the blastocyst and hatched blastocyst stage. In Figure 3B one can appreciate that MYL6 is located in the TE cells surrounding the blastocoel cavity.
The images of the positive and negative control experiments are supplemented in additional file 1.
The results of the immunofluorescent experiments were consistent with the RNA expression patterns during preimplantation embryo development.
Discussion
In the present study a subtractive cDNA library was constructed between blastocyst embryos as the tester population and a pool of 2-cell and 8-cell embryos as the driver population, in order to enrich for genes differentially expressed at the blastocyst stage.
Sequence information was obtained for 65 randomly picked clones. Out of these 65 ESTs representing the blastocyst embryos, 10 were homologous to mitochondrial genes and 14 to ribosomal genes. Mitochondria play an essential role in many events during early development. In cattle, the mtDNA copy number increases during blastocyst expansion and hatching, consistent with an increase in mitochondrial RNAs. This indicates that the transcriptional activation of the mitochondrial genome coincides with the pronounced structural and functional differentiation of the mitochondria during preimplantation development [26]. The largest group of ESTs was identified as genes involved in protein synthesis and included several ribosomal proteins. The high degree of ribosomal protein mRNAs corresponds with previous data showing that between the 2-cell stage and the blastocyst stage, the level of ribosomal protein mRNA is said to increase about 20-fold [27]. The protein content of in vivo derived preattachment cattle embryos increases 2-fold from the 2-cell through to the blastocyst stage [28]. This rise in protein content is correlated with rapid cell divisions at the late morula and early blastocyst stage [29,30].
Real-time qPCR was used to verify the SSH results and at the same time to compare the gene expression between in vitro produced embryos and in vivo golden standard embryos. The geometric mean of GAPD, YWHAZ and SDHA was used for normalisation. Vandesompele and colleagues [31] demonstrated that the conventional use of a single gene for normalisation leads to relative large errors and validated the geometric mean of multiple carefully selected housekeeping genes as an accurate normalisation factor. In Goossens and colleagues [25] it was demonstrated that GAPD, YWHAZ and SDHA were stably expressed reference genes, suitable for normalisation of quantitative data within different preimplantation embryo stages. The problem of measuring RNA expression levels throughout preimplantation development is confounded by the fact that cell numbers and cell sizes are constantly changing during this developmental interval. To allow ontogenic analysis, the embryos were compared as a single unit and the reference genes will correct for the differences between the embryos. Other normalisation strategies, like normalisation against total cell number, normalisation against RNA mass quantity or the use of exogenous control RNA (spike) [32,33] are not assumption free. Spikes correct for differences in RNA extraction and differences in enzymatic efficiencies but do not account for the quality and quantity of input sample.
In most studies about gene expression in embryos, only the RNA expression was considered, but the differences in RNA expression do not always imply differences on protein level, as there may be regulation at the translation level as well. By immunofluorescent labelling using mouse antibodies, the protein expression of KRT18, FN1 and MYL6 was evaluated for the first time during bovine preimplantation embryo development. Furthermore, the location of the protein expression in the embryo can tell us more about the specific function of the protein during the process of blastocyst formation.
Both KRT18 mRNA and protein expression were found to be absent at the 2-cell and 8-cell stage and abundant at relatively high levels at the blastocyst stage, demonstrating the embryonic origin of this transcript. KRT18 was only expressed in the cell-cell contact sites of the TE, but not in those of the ICM. This makes that KRT18 can be seen as a marker for differentiation, adding to previous described markers for trophoblast differentiation in mice as there are Cdx2 and Eomes [34-36]. As TE is a kind of specialized epithelium, genes involved in de novo differentiation of an epithelium, including gene families encoding for cell polarity, cell junctions, cytoskelet formation and ion transport [37,38] are involved in TE differentiation. KRT18 is a cytoskeletal protein and is found, together with KRT8, in simple epithelia where they heterodimerise to form the intermediate filaments. They influence the 3-D formation of cell-cell or cell-substrate contacts in embryonic visceral endoderm [39-41]. Stanton and Green [42] reported in mouse embryos a progressive rise for both KRTs from the 2-cell to the blastocyst stage, paralleling the development of the TE. Targeted deletion of KRT18 in the mouse leads to trophoblast fragility and early embryonic lethality [43]. Other cytoskeletal proteins such as α-catenin, β-catenin, occludin, zonula occludens protein-1 and connexin43 are also expressed at the cell-cell contact sides of the TE cells [44], suggesting a co-localisation with KRT18.
FN1 was found to be significantly higher expressed in blastocysts compared to the earlier stages, and this difference was even 3.5× higher in the in vivo produced embryos compared to the in vitro produced ones. FN1 protein was predominantly expressed in the ICM and formed filamentous structures between the TE and the ICM. The differential expression of FN1 in blastocyst stage embryos of several mammalian species, including bovine, has previously been reported by other authors [4,22,45-47] and this gene has been associated with blastocyst formation. It is an adhesive extracellular matrix component and performs a vital role during cell proliferation, cell adhesion and cell mobility. It exists as a homodimer and is composed of several domains such as a heparin binding domain, a fibrin binding domain, a collagen binding domain and a cell recognition domain. This provides FN1 with the opportunity to interact with and bind to several ligands such as cells, heparin, fibrin, collagen, immunoglobulins and DNA [48]. Because events such as cell proliferation, cell adhesion and cell mobility are essential during early embryogenesis, FN1 does qualify as an important participant during this phase of embryo development. Besides, Aplin and colleagues [46] reported that FN1 acts as a bridging ligand between the collagen matrix and integrins at the cytotrophoblast surface, mediating anchorage and/or migratory activity in the process of implantation. The vital importance of FN1 for normal embryo development has been demonstrated convincingly since mouse embryos lacking this protein die shortly after gastrulation from mesodermal defects [49]. The significant difference in FN1 mRNA expression levels which we noticed between in vivo and in vitro produced bovine embryos has also been reported by Mohan and colleagues [10]. Lower RNA levels exhibited by in vitro produced embryos may be responsible for the poor quality of these embryos compared to their in vivo counterparts.
Myosin light chains associate with the motor protein myosin and are believed to play a role in the regulation of its actin-based ATPase activity. RNA levels of MYL6, the smooth muscle isoform of myosin light chain, were significantly higher at the blastocyst stage compared to the earlier stages. No significant differences were seen between in vivo and in vitro produced embryos. MYL6 is involved in the cytoskeletal organisation [50] which is in agreement with the observed protein expression in the TE cells surrounding the blastocoel cavity. The expression of MYL6 proteins around the blastocoel may also contribute to the statement of Niimura [51] who inferred that contractions in mouse blastocysts occur by activation of myosin light chain kinase resulting in the phosphorylation of myosin light chains and causing the efflux of blastocoelic fluid.
ATP1B3 shows a significant difference in RNA levels between the 2/8-cell stage and the blastocyst stage of in vitro produced embryos, but remarkably this difference is negligible for in vivo produced embryos. The Na/K ATPase consists of a catalytic α subunit and a noncatalytic, glycosylated β subunit, each encoded by multigene families [52]. The β subunit is required for structural and functional maturation of the α subunit [53]. Blastocyst formation involves the establishment of a transtrophectoderm ion gradient mediated by the Na/K ATPase which pumps water through water channels called aquaporins [28]. Accumulation of fluid in the blastocoel is essential for differentiation of the ICM and TE cell types [54]. The gene expression patterns of several Na/K ATPase subunits have been reported previously in mouse [55] and bovine [56], and Adjaye and colleagues [57] identified ATP1B3 as a marker specific for the TE in human blastocysts using a cDNA microarray but as far as we know, this is the first study in which the RNA expression levels of ATP1B3 in bovine embryos are described. The higher ATP1B3 RNA levels in in vitro produced blastocysts might be an explanation for the faster blastulation of in vitro produced embryos compared to in vivo produced ones. Van Soom and colleagues [58] have demonstrated that in vivo morulae display a more firm and prolonged compaction and that they start blastulation at a later embryonic age and cell number, moreover the addition of serum to the culture medium seemed to enhance blastocyst development [59]. In vivo morulae develop more gradually to the blastocyst stage and this might go together with a slower rise in ATP1B3 expression. Another explanation for the higher ATP1B3 expression in vitro might be a compensation for the lower expression of another NA/K ATPase subunit or another functional analogue, but this was not further investigated.
The differences between the blastocysts and the 2- and 8-cell stage embryos for FTH1 were smaller but significant and no differences were observed between in vivo and in vitro produced embryos. FTH1 has an important role in the control of intracellular iron distribution and the constitution of long term iron stores. The ferroxidase activity associated with the H subunit is necessary for iron uptake by the ferritin molecule. FTH1-/- mice embryos die between 3.5 and 9.5 days of development [60] demonstrating that the ferritin H subunit is indispensable for embryonic development. FTH1 was also present in the human blastocyst SSH library as reported by Morozov et al. [61].
The mRNA levels for ATP6V0B, HINT1, RPL10 and SLC25A5 were higher at the blastocyst stage compared to the earlier stages for in vitro produced embryos, but no significant differences existed between the in vivo produced 8-cell and in vivo produced blastocyst embryos. The qPCR results of those genes for in vitro produced embryos confirmed the results of the subtractive cDNA library.
The RNA expression patterns of the remaining genes (ACTN1, COPE and EEF1A1) were not in agreement with the subtractive cDNA cloning results. Those genes were abundant at an equal amount or higher at the earlier stages. Those false positives prove the requirement for the verification of the subtractive cDNA outcome and are in agreement with the reported 30% false positives obtained by subtractive cDNA cloning in previous studies [10]. The significantly higher RNA levels in vivo produced 8-cell embryos for COPE but also for EEF1A1 is an important observation. COPE is a subunit of the coatomer protein complex and is involved in the retrograde Golgi-to-ER transport of dilysine-tagged proteins, whereas EEF1A1 is responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome. Both genes are involved in protein synthesis. Around the 8–16 cell stage the embryonic genome becomes active, resulting in the synthesis of embryonic proteins. The lower expression of genes involved in protein synthesis (COPE, EEF1A1 and RPL10) in in vitro 8-cell embryos might indicate that the embryonic protein synthesis is affected or delayed in in vitro embryos.
Differences in gene expression between in vivo and in vitro produced embryos reflect the effects of the in vitro culture system on the transcriptional activity [11]. Preimplantation embryos are capable of developing in a wide range of culture conditions. However, this adaptation of the embryo to suboptimal conditions may result in a lower embryo quality. It will be a challenge for the future to determine the importance of the affected genes for the process of embryo development and to change the culture conditions in vitro in order to induce the expression of those genes to levels identical to those under in vivo conditions, and in this way improve the quality of the embryo.
Conclusion
The results of this study add to the knowledge of the process of bovine blastocyst formation. Several genes were identified as candidate markers involved in blastocyst formation. The fact that some of those candidate genes were differentially expressed between in vivo and in vitro produced embryos confirms the belief that culture conditions influence embryonic gene expression. Further functional analyses of these candidate genes may help to optimise in vitro embryo culture systems in order to improve the quality of in vitro produced bovine embryo.
Methods
In vitro embryo production
Bovine embryos were produced in vitro as described by Yuan and colleagues [62]. Briefly, bovine oocytes were aspirated from ovaries collected at a local slaughterhouse. Immature cumulus-oocyte complexes were selected from follicular fluid, washed three times in HEPES-TALP and matured for 22 to 26 hr in groups of 100 in 500 μl maturation medium at 39°C in a humidified 5% CO2 incubator. After maturation, the oocytes were inseminated with frozen-thawed sperm from one bull (1 × 106 spermatozoa/ml). After 20 hr the cumulus cells and spermatozoa were mechanically removed from the presumptive zygotes, which were placed in groups of 25 in 50 μl droplets of synthetic oviduct fluid supplemented with 5% fetal calf serum and cultured up to the desired stages at 39°C in 5% CO2, 5% O2 and 90% N2. The embryos were collected at the indicative time period after fertilisation: 2-cell (24–28 hr p.i.), 8-cell (48–52 hr p.i.), blastocyst (day 8). All embryos were washed three times in PBS, pooled and frozen at -80°C until RNA extraction.
In vivo embryo production
The in vivo embryo production protocol has been approved by the local ethical committee and was done after superovulation and artificial insemination of 4 Holstein cows for the collection of 2-cell and 8-cell embryos and 2 Holstein heifers for the collection of blastocysts. Oestrus cycles of the donor cows and heifers were synchronized by two intramuscular injections (on day 1 and on day 12) with 526 mg of cloprostenol (Estrumate® – Shering-Plough, Belgium), followed by oestrus detection 2 days later. Donor cows were intramuscularly injected with 8 equal doses of Stimufol® (Université de Liège, Belgium) (500 μg porcine FSH-100 μg porcine LH) over 4 days, where as heifers received a lower total dosis of Stimufol® (Université de Liège, Belgium) (250 μg porcine FSH-50 μg porcine LH) in 8 decreasing doses over 4 days. Stimufol administration was started at day 10 in the cows and at day 11 and 12 in the heifers in order to group the embryo collections. To induce luteolysis, 789 mg cloprostenol (Estrumate® – Shering-Plough, Belgium) was injected along with the 5th injection of pFSH/pLH. All donors were checked for oestrus 48 h after the cloprostenol injection and artificially (AI) inseminated twice with two straws of semen of a bull of established fertility on 12 and 24 hr after the onset of the oestrus. The same bull was used for in vivo and in vitro insemination. The 2-cell and 8-cell embryos were recovered 36 hours respectively 84 hours after AI. After collection of the reproductive tract at the slaughterhouse, the fallopian tube and the uterus were both flushed with approximately 250 ml PBS supplemented with 2% FCS. Blastocysts were recovered by non-surgical uterine flushing on day 7 and 8 after AI. After epidural anaesthesia with 5 ml Procaine HCL 4%® (procainehydrochloride, VMD, Belgium), each horn was flushed with 0.5 l of PBS.
All in vivo embryos were washed four times in Hepes-Talp and subsequently three times in PBS before freezing at -80°C.
Construction of the SSH library
mRNA extraction, cDNA synthesis and amplification
Poly(A)+ RNA was isolated from a pool of 40 IVP blastocysts (day 8) as a tester population and a pool of 40 IVP embryos at the 2-cell and 40 at the 8-cell stage as a driver population, using the Oligotex Direct Mini Kit (Qiagen, the Netherlands) according to the manufacturer's instructions.
Due to the small amounts of mRNA extracted from bovine embryos, both tester and driver mRNA were reverse transcribed and amplified using the SMART PCR cDNA synthesis Kit (Takara Bio Inc., France). This kit is optimised for the amplification of cDNA without altering the original expression ratios [63,64] and is widely used in combination with the subtractive hybridisation procedure to study gene expression during embryo development [4,10,30,65,66].
Suppression Subtractive Hybridisation
The SSH was performed with the PCR-Select cDNA Subtraction Kit (Takara Bio Inc., France) following the manufacturer's instructions. Amplified double-stranded cDNA from the tester and driver was RsaI digested, and 2 different adaptors were ligated to 2 fractions of the tester population. A first hybridisation was performed with an excess of driver cDNA, followed by a second hybridisation between the mixed tester fractions in the presence of an excess driver. After the second hybridisation, tester cDNA was subjected to two rounds of PCR to enrich the tester specific cDNA fragments. The amplified products were cloned into the pCR 2.1 vector (Invitrogen, Belgium) and transformed into competent DH5α E. coli cells (Invitrogen, Belgium).
Clones were randomly picked, the DNA-inserts were sequenced (Thermo Sequenase Primer Cycle Sequencing Kit, Amersham Bioscience, the Netherlands) with the ALF Express sequencer (Amersham Bioscience, the Netherlands) and identified using the BLAST algorithm [67].
Quantification of specific transcripts by real-time PCR
RNA extraction and cDNA amplification
For the quantification of the mRNA expression levels of 12 genes obtained by SSH, total RNA was isolated from 3 to 6 in vivo and in vitro produced single embryos per developmental stage (2-cell, 8-cell and blastocyst) using the PicoPure RNA Isolation Kit (Arcturus, USA) according to the manufacturer's instructions. An in solution DNase digestion followed by an RT minus control was performed as described by Goossens and colleagues [25]. The first-strand cDNA synthesis and linear amplification were done using the WT-Ovation RNA Amplification system (NuGEN, The Netherlands) as described in the manufacturer's instructions. After the RT reaction and the linear amplification step, the cDNA was 50 times diluted in 10 mM Tris HCl pH 8.0.
Primer design
Sequence-specific primers were designed for 12 identified genes by the Primer 3 software [68] (Table 2). The specificity of the primers was tested using a BLAST analysis against the genomic NCBI database. PCR amplicons were characterised using Mfold [69] in order to account for any secondary structures which might influence the PCR efficiency. The PCR products were cloned (pCR 2.1 vector, Invitrogen, Belgium) and sequenced for verification (Thermo Sequenase Primer Cycle Sequencing Kit, Amersham Bioscience, the Netherlands) with a ALF Express sequencer (Amersham Bioscience, the Netherlands).
Real-time PCR reactions
All PCR reactions were performed in a 15 μl reaction volume on the iCycler iQ Real-Time PCR Detection System (Bio-Rad, Belgium) using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Belgium) and 200 nM of each specific primer. The PCR program consisted of an initial UDG incubation step at 50°C for 2 minutes and an initial denaturation step at 95°C for 3 minutes to activate the Taq DNA polymerase, followed by 45 cycles of denaturation at 95°C for 20 seconds and a combined primer annealing/extension at the specific annealing temperature for 40 seconds during which fluorescence was measured. A melt curve was produced to confirm a single gene-specific peak and to detect primer-dimer formation by heating the samples from 70 to 95°C in 0.5°C increments with a dwell time at each temperature of 10 seconds while continuously monitoring the fluorescence. PCR efficiencies were calculated using a relative standard curve derived from a pooled cDNA mixture (a 10-fold dilution series with five measuring points). Each reaction was run in duplicate in the same run, whereby a no-template control was included.
The geometric mean of three reference genes, YWHAZ, GAPD and SDHA, was used to calculate an accurate normalisation factor as described by Goossens and colleagues [25]. The mean quantity of each transcript (raw data) was divided by the respective normalisation factor to obtain a normalised value for each transcript. The sample with the lowest value was assigned the value 1. The normalised target values were divided by the calibrator normalised target values to generate the relative expression levels.
The normalised gene expression levels were analysed by calculating 95% confidence intervals after logarithmic transformations, whereby non-overlapping intervals denote significant differences at the 0.05 level.
Immunofluorescent labelling
Pools of 10 embryos per developmental stage (2–4 cell (24–28 hr p.i.), 5–8 cell (48–52 hr p.i.), morula day 5 p.i., morula day 7 p.i., blastocyst day 7 p.i. and hatched blastocyst day 8 p.i) were selected from the culture media, washed 3 times in PBS and fixed with 4% paraformaldehyde (Sigma, Belgium) in PBS for 1 hour at 4°C.
After washing in polyvinyl pyrrolidone (PVP; 1 mg PVP/ml PBS), the embryos were permeabilized with 0.5% Triton X-100 (Sigma, Belgium) in PBS for 30 min at room temperature. Non-specific binding sites were blocked with 10% goat serum in PVP for 30 min. After 3 washes in PVP the embryos were incubated with the mouse primary antibodies against FN1, KRT18 and MYL6 respectively (Table 3) for 2 hours at 37°C, washed again 3 times in PVP and then incubated with FITC-labeled goat-anti-mouse secondary antibodies (Molecular Probes, Invitrogen, Belgium) for 1 hour at 37°C in the dark.
Table 3.
Primary antibodies used for immunofluorescent experiments
| Antigen | Primary antibody | Isotype | Supplier | Dilution | Secondary antibody | Supplier | Dilution |
| KRT18 | Anti KRT18 C-04 | Mouse IgG1 | Abcam | 1/100 | Goat-anti- mouse FITC | Molecular probes | 1/100 |
| FN1 | Anti FN1 CSI 005–17 | Mouse IgG1 | Abcam | 1/100 | Goat-anti- mouse FITC | Molecular probes | 1/100 |
| MYL6 | Anti MYL6 A01 | Mouse polyclonal | Abnova | 1/50 | Goat-anti- mouse FITC | Molecular probes | 1/100 |
The nuclei were stained with 0.5% propidium iodide (PI; Molecular Probes, Invitrogen, Belgium) for 30 min at room temperature. After 2 washes in PVP the embryos were mounted in a drop of glycerol with 1,4-diazabicyclo (2.2.2) octane (25 mg/ml) on slides with vaseline bridges. Negative (by replacing the primary antibody with goat serum) and double negative controls (only PI staining) were performed simultaneously to check for non-specific binding of the secondary antibody and for auto-fluorescence. A monolayer of cultured cumulus cells was used as a positive control. The results of the control experiments were supplemented in additional file 1.
Samples were examined by fluorescence microscopy (Leica DMR, Van Hopplynus, Belgium) and by confocal laser scanning microscopy (Leica TCS SP2 laser scanning spectral confocal system linked to a Leica DM IRB inverted microscope, Leica Microsystems GmbH, Germany). A krypton-argon ion laser was used for the simultaneous excitation of fluorescence for proteins and PI for DNA. All the labelling experiments were performed in duplicate.
Authors' contributions
KG performed all the experimental procedures and was the primary author of the manuscript. AVS contributed to the design of the IVF experiments. MVP participated in the study design and provided real-time support. LV helped with the production of the in vivo embryos and the evaluation of the immunofluorescent experiments. JV provided expert input in data analysis and statistics. AVZ and LJP participated in the design of the project, helped to draft the manuscript and supervised the study. All authors read and approved the final manuscript.
Supplementary Material
Negative and positive controls for immunofluorescent labelling experiments. Negative (by replacing the primary antibody with goat serum) and double negative controls (only PI staining) were performed to check for non-specific binding of the secondary antibody and for auto-fluorescence. A monolayer of cultured cumulus cells was used as a positive control. (A: negative control for KRT18, B: double negative control for KRT18, C: positive control for KRT18; A': negative control for FN1, B': double negative control for FN1, C': positive control for FN1; A": negative control for MYL6, B": double negative control for MYL6, C": positive control for MYL6).
Acknowledgments
Acknowledgements
The authors like to thank Machteld Baetens, Johanna Mestach and Griet Spaepen for their excellent technical assistance, Ruben Van Gansbeke for helping with sample collection in the slaughterhouses and Bart Mateusen for his help with the confocal microscope system.
Contributor Information
Karen Goossens, Email: Karen.Goossens@UGent.be.
Ann Van Soom, Email: Ann.VanSoom@UGent.be.
Mario Van Poucke, Email: Mario.VanPoucke@UGent.be.
Leen Vandaele, Email: Leen.Vandaele@UGent.be.
Jo Vandesompele, Email: Joke.Vandesompele@UGent.be.
Alex Van Zeveren, Email: Alex.VanZeveren@UGent.be.
Luc J Peelman, Email: Luc.Peelman@UGent.be.
References
- Wrenzycki C, Herrmann D, Lucas-Hahn A, Gebert C, Korsawe K, Lemme E, Carnwath JW, Niemann H. Epigenetic reprogramming throughout preimplantation development and consequences for assisted reproductive technologies. Birth Defects Res C Embryo Today. 2005;75:1–9. doi: 10.1002/bdrc.20035. [DOI] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Niemann H, Wrenzycki C. Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology. 2000;53:21–34. doi: 10.1016/S0093-691X(99)00237-X. [DOI] [PubMed] [Google Scholar]
- Ponsuksili S, Tesfaye D, El-Halawany N, Schellander K, Wimmers K. Stage-specific expressed sequence tags obtained during bovine development by differential display RT-PCR and suppression subtractive hybridization. Prenat Diagn. 2002;22:1135–1142. doi: 10.1002/pd.501. [DOI] [PubMed] [Google Scholar]
- Van Soom A, de Kruif A. A comparative study of in vivo and in vitro derived bovine embryos. P Int C An Repr. 1992;3:1365–1367. [Google Scholar]
- Thompson JG. Comparison between in vivo -derived and in vitro-produced pre-elongation embryos from domestic ruminants. Reprod Fertil Dev. 1997;9:341–354. doi: 10.1071/R96079. [DOI] [PubMed] [Google Scholar]
- Abe H, Otoi T, Tachikawa S, Yamashita S, Satoh T, Hoshi H. Fine structure of bovine morulae and blastocysts in vivo and in vitro. Anat Embryol. 1999;199:519–527. doi: 10.1007/s004290050249. [DOI] [PubMed] [Google Scholar]
- Holm P, Callesen H. In vivo versus in vitro produced bovine ova: similarities and differences relevant for practical application. Reprod Nutr Dev. 1998;28:579–594. doi: 10.1051/rnd:19980601. [DOI] [PubMed] [Google Scholar]
- Lonergan P, Gutierrez-Adan A, Rizos D, Pintado B, de la Fuente J, Boland MP. Relative messenger RNA abundance in bovine oocytes collected in vitro or in vivo before and 20 hr after the preovulatory luteinizing hormone surge. Mol Reprod Dev. 2003;66:297–305. doi: 10.1002/mrd.10357. [DOI] [PubMed] [Google Scholar]
- Mohan M, Hurst AG, Malayer JR. Global gene expression analysis comparing bovine blastocysts flushed on day 7 or produced in vitro. Mol Reprod Dev. 2004;68:288–298. doi: 10.1002/mrd.20086. [DOI] [PubMed] [Google Scholar]
- Tesfaye D, Ponsuksili S, Wimmers K, Gilles M, Schellander K. A comparative expression analysis of gene transcripts in post-fertilization developmental stages of bovine embryos produced in vitro or in vivo. Reprod Domest Anim. 2004;39:396–404. doi: 10.1111/j.1439-0531.2004.00531.x. [DOI] [PubMed] [Google Scholar]
- Davis W, Jr, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol. 1996;174:190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- Ma J, Svoboda P, Schultz RM, Stein P. Regulation of zygotic gene activation in the preimplantation mouse embryo: Global activation and repression of gene expression. Biol Reprod. 2001;64:1713–1721. doi: 10.1095/biolreprod64.6.1713. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Bolton V, Monk M. Developmental expression of specific genes detected in high-quality cDNA libraries from single human preimplantation embryos. Gene. 1999;237:373–383. doi: 10.1016/S0378-1119(99)00329-7. [DOI] [PubMed] [Google Scholar]
- Ko MS, Kitchen JR, Wang X, Threat TA, Wang X, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, Cui YS, Sano Y, Tanaka T, Liang YL, Mason S, Paonessa PD, Sauls AD, DePalma GE, Sharara R, Rowe LB, Eppig J, Morrell C, Doi H. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Ponsuksili S, Wimmers K, Adjaye J, Schellander K. A source for expression profiling in single preimplantation bovine embryos. Theriogenology. 2002;57:1611–1624. doi: 10.1016/S0093-691X(02)00661-1. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Wabnitz P, Halter R, Klocke R, Carnwath J, Kues W, Wrenzycki C, Paul D, Niemann H. Application of cDNA arrays to monitor mRNA profiles in single preimplantation mouse embryos. Biotechniques. 2002;33:376–385. doi: 10.2144/02332rr04. [DOI] [PubMed] [Google Scholar]
- Carter MG, Hamatani T, Sharov AA, Carmack CE, Qian Y, Aiba K, Ko NT, Dudekula DB, Brzoska PM, Hwang SS, Ko MS. In situ-synthesized novel microarray optimized for mouse stem cell and early developmental expression profiling. Genome Res. 2003;13:1011–1021. doi: 10.1101/gr.878903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Daikoku T, Wang H, Matsumoto H, Carter MG, Ko MS, Dey SK. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc Natl Acad Sci USA. 2004;101:10326–10331. doi: 10.1073/pnas.0402597101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Barnes FL, Hue I, Sirard MA. Subtractive hybridization used to identify mRNA associated with the maturation of bovine oocytes. Mol Reprod Dev. 2000;57:167–175. doi: 10.1002/1098-2795(200010)57:2<167::AID-MRD8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biol Reprod. 2003;68:31–39. doi: 10.1095/biolreprod.102.007674. [DOI] [PubMed] [Google Scholar]
- El-Halawany N, Ponsuksili S, Wimmers K, Gilles M, Tesfaye D, Schellander K. Quantitative expression analysis of blastocyst-derived gene transcripts in preimplantation developmental stages of in vitro -produced bovine embryos using real-time polymerase chain reaction technology. Reprod Fertil Dev. 2004;16:753–762. doi: 10.1071/RD04041. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST--database for "expressed sequence tags". Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. http://www.ncbi.nlm.nih.gov/projects/dbEST/ [DOI] [PubMed] [Google Scholar]
- Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol. 2005;5:27. doi: 10.1186/1471-213X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LC, Thundathil J, Filion F. Role of the mitochondrial genome in preimplantation development and assisted reproductive technologies. Reprod Fertil Dev. 2005;17:15–22. doi: 10.1071/RD04084. [DOI] [PubMed] [Google Scholar]
- Jakobsen AS, Avery B, Dieleman SJ, Knijn HM, Vos PL, Thomsen PD. Transcription of ribosomal RNA genes is initiated in the third cell cycle of bovine embryos. Mol Reprod Dev. 2006;73:196–205. doi: 10.1002/mrd.20389. [DOI] [PubMed] [Google Scholar]
- Mohan M, Ryder S, Claypool PL, Geisert RD, Malayer JR. Analysis of gene expression in the bovine blastocyst produced in vitro using suppression-subtractive hybridization. Biol Reprod. 2002;67:447–453. doi: 10.1095/biolreprod67.2.447. [DOI] [PubMed] [Google Scholar]
- Morgan PM, Kane MT. Protein content of rabbit embryos: one cell to peri-implantation blastocyst. J Reprod Fertil. 1993;97:101–106. doi: 10.1530/jrf.0.0970101. [DOI] [PubMed] [Google Scholar]
- Pacheco-Trigon S, Hennequet-Antier C, Oudin JF, Piumi F, Renard JP, Duranthon V. Molecular characterization of genomic activities at the onset of zygotic transcription in mammals. Biol Reprod. 2002;67:1907–1918. doi: 10.1095/biolreprod67.6.1907. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:7. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnison M, Pfeffer PL. Isolation of genes associated with developmentally competent bovine oocytes and quantitation of their levels during development. Biol Reprod. 2004;71:1813–1821. doi: 10.1095/biolreprod.104.032367. [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Patel OV, Ireland JJ, Smith GW. Quantitative analysis of messenger RNA abundance for ribosomal protein L-15, cyclophilin-A, phosphoglycerokinase, beta-glucuronidase, glyceraldehyde 3-phosphate dehydrogenase, beta-actin, and histone H2A during bovine oocyte maturation and early embryogenesis in vitro. Mol Reprod Dev. 2006;73:267–278. doi: 10.1002/mrd.20333. [DOI] [PubMed] [Google Scholar]
- Kunath T, Strumpf D, Rossant J. Early trophoblast determination and stem cell maintenance in the mouse – A review. Placenta. 2004;25:32–38. doi: 10.1016/j.placenta.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka T, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Barcroft LC. Regulation of blastocyst formation. Front Biosci. 2001;6:708–730. doi: 10.2741/Watson. [DOI] [PubMed] [Google Scholar]
- Stanton JAL, Macgregor AB, Green DPL. Gene expression in the mouse preimplantation embryo. Reproduction. 2003;125:457–468. doi: 10.1530/rep.0.1250457. [DOI] [PubMed] [Google Scholar]
- Jackson BW, Grund C, Schmid E, Burki K, Franke WW, Illmensee K. Formation of cytoskeletal elements during mouse embryogenesis. Intermediate filaments of the cytokeratin type and desmosomes in preimplantation embryos. Differentiation. 1980;17:161–179. doi: 10.1111/j.1432-0436.1980.tb01093.x. [DOI] [PubMed] [Google Scholar]
- Duprey P, Morello D, Vasseur M, Babinet C, Condamine H, Brulet P, Jacob F. Expression of the cytokeratin endo A gene during early mouse embryogenesis. Proc Natl Acad Sci USA. 1985;82:8535–8539. doi: 10.1073/pnas.82.24.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E. Intermediate filaments as mechanical integrators of cellular space. Nature. 1980;283:249–256. doi: 10.1038/283249a0. [DOI] [PubMed] [Google Scholar]
- Stanton JL, Green DP. A set of 1542 mouse blastocyst and pre-blastocyst genes with well-matched human homologues. Mol Hum Reprod. 2002;8:149–166. doi: 10.1093/molehr/8.2.149. [DOI] [PubMed] [Google Scholar]
- Hesse M, Franz T, Tamai Y, Taketo MM, Magin TM. Targeted deletion of keratins 18 and 19 leads to trophoblast fragility and early embryonic lethality. EMBO J. 2000;19:5060–5070. doi: 10.1093/emboj/19.19.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M, Larue L, Schwarz H, Kemler R. Cell-junctional and cytoskeletal organization in mouse blastocysts lacking E-cadherin. Dev Biol. 1997;185:261–271. doi: 10.1006/dbio.1997.8560. [DOI] [PubMed] [Google Scholar]
- Shigemoto T. Fibronectin induces pseudopod formation and cell migration by mobilizing internal Ca(2+) in blastoderm cells from medaka embryos. Jpn J Physiol. 1999;49:527–539. doi: 10.2170/jjphysiol.49.527. [DOI] [PubMed] [Google Scholar]
- Aplin JD, Haigh T, Jones CJ, Church HJ, Vicovac L. Development of cytotrophoblast columns from explanted first-trimester human placental villi: role of fibronectin and integrin alpha5beta1. Biol Reprod. 1999;60:828–838. doi: 10.1095/biolreprod60.4.828. [DOI] [PubMed] [Google Scholar]
- Darribere T, Schwarzbauer JE. Fibronectin matrix composition and organization can regulate cell migration during amphibian development. Mech Dev. 92:239–250. doi: 10.1016/S0925-4773(00)00245-8. [DOI] [PubMed] [Google Scholar]
- Yamada KM. Adhesive recognition sequences. J Biol Chem. 1991;266:12809–12812. [PubMed] [Google Scholar]
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:54–58. doi: 10.1093/nar/gki031. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=full_report&list_uids=4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura S. Time-lapse videomicrographic analyses of contractions in mouse blastocysts. J Reprod Dev. 2003;49:413–423. doi: 10.1262/jrd.49.413. [DOI] [PubMed] [Google Scholar]
- Blanco G, Mercer RW. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. APS Ren Physiol. 1998;275:633–650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- Geering K, Beggah A, Good P, Girardet S, Roy S, Schaer D, Jaunin P. Oligomerization and maturation of Na-K-ATPase: Functional interaction of the cytoplasmic NH2 terminus of the beta subunit with the alpha subunit. J Cell Biol. 1996;133:1193–1204. doi: 10.1083/jcb.133.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AJ. The cell biology of blastocyst development. Mol Reprod Dev. 1992;33:492–504. doi: 10.1002/mrd.1080330417. [DOI] [PubMed] [Google Scholar]
- MacPhee DJ, Jones DH, Barr KJ, Betts DH, Watson AJ, Kidder GM. Differential involvement of Na+,K+-ATPase isozymes in preimplantation development of the mouse. Dev Biol. 2000;222:486–498. doi: 10.1006/dbio.2000.9708. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Herrmann D, Niemann H. Timing of blastocyst expansion affects spatial messenger RNA expression patterns of genes in bovine blastocysts produced in vitro. Biol Reprod. 2003;68:2073–2080. doi: 10.1095/biolreprod.102.012104. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, Gosden RG, Lehrach H. Primary differentiation in the human blastocyst: Comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23:1514–1525. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- Van Soom A, Boerjan ML, Bols PEJ, Vanroose G, Lein A, Coryn M, de Kruif A. Timing of compaction and inner cell allocation in bovine embryos produced in vivo after superovulation. Biol Reprod. 1997;57:1041–1049. doi: 10.1095/biolreprod57.5.1041. [DOI] [PubMed] [Google Scholar]
- Pinyopummintr T, Bavister BD. Development of bovine embryos in a cell-free culture medium: effects of type of serum, timing of its inclusion and heat inactivation. Theriogenology. 1994;41:1241–1249. doi: 10.1016/0093-691X(94)90481-W. [DOI] [PubMed] [Google Scholar]
- Ferreira C, Bucchini D, Martin ME, Levi S, Arosio P, Grandchamp B, Beaumont C. Early embryonic lethality of H ferritin gene deletion in mice. J Biol Chem. 2000;275:3021–3024. doi: 10.1074/jbc.275.5.3021. [DOI] [PubMed] [Google Scholar]
- Morozov G, Verlinsky O, Rechitsky S, Ivakhnenko V, Goltsman E, Gindilis V, Strom C, Kuliev A, Verlinsky Y. Construction and sequence analysis of subtraction complementary DNA libraries from human preimplantation embryos. J Assist Reprod Genet. 1999;16:212–215. doi: 10.1023/A:1020368908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YQ, Van Soom A, Leroy JL, Dewulf J, Van Zeveren A, de Kruif A, Peelman LJ. Apoptosis in cumulus cells, but not in oocytes, may influence bovine embryonic developmental competence. Theriogenology. 2005;63:2147–2163. doi: 10.1016/j.theriogenology.2004.09.054. [DOI] [PubMed] [Google Scholar]
- Endege WO, Steinmann KE, Boardman LA, Thibodeau SN, Schlegel R. Representative cDNA libraries and their utility in gene expression profiling. Biotechniques. 1999;26:542–548. doi: 10.2144/99263cr04. [DOI] [PubMed] [Google Scholar]
- Bui LC, Leandri RD, Renard JP, Duranthon V. SSH adequacy to preimplantation mammalian development: scarce specific transcripts cloning despite irregular normalisation. BMC Genomics. 2005;6:155. doi: 10.1186/1471-2164-6-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard MA, Dufort I, Vallee M, Massicotte L, Gravel C, Reghenas H, Watson AJ, King WA, Robert C. Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: preliminary analysis using a bovine embryonic array. Reprod Fertil Dev. 2005;7:47–57. doi: 10.1071/RD04113. [DOI] [PubMed] [Google Scholar]
- Wang S, Cowan CA, Chipperfield H, Powers RD. Gene expression in the preimplantation embryo: in-vitro developmental changes. Reprod Biomed Online. 2005;10:607–616. doi: 10.1016/s1472-6483(10)61668-9. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. http://www.ncbi.nlm.nih.gov/blast/ [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. http://www.bioinfo.rpi.edu/applications/mfold/old/dna/form1.cgi [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Negative and positive controls for immunofluorescent labelling experiments. Negative (by replacing the primary antibody with goat serum) and double negative controls (only PI staining) were performed to check for non-specific binding of the secondary antibody and for auto-fluorescence. A monolayer of cultured cumulus cells was used as a positive control. (A: negative control for KRT18, B: double negative control for KRT18, C: positive control for KRT18; A': negative control for FN1, B': double negative control for FN1, C': positive control for FN1; A": negative control for MYL6, B": double negative control for MYL6, C": positive control for MYL6).