Abstract
Heterotrimeric G proteins consist of three subunits (α, β, and γ). α- and β- subunits have been previously cloned in plants, but the γ-subunit has remained elusive. To isolate the γ-subunit of a plant heterotrimeric G protein an Arabidopsis thaliana yeast two-hybrid library was screened by using a tobacco G-β-subunit as the bait protein. One positive clone (AGG1) was isolated several times; it displays significant homology to the conserved domains of mammalian γ-subunits. The predicted AGG1 protein sequence contains all of the typical characteristics of mammalian γ-subunits such as small size (98 amino acids, 10.8 kDa), presence of a C-terminal CAAX box to direct isoprenyl modification, and an N-terminal α-helix region capable of forming a coiled-coil interaction with the β-subunit. Northern and Southern analyses showed that AGG1 is a single-copy gene in Arabidopsis with a similar expression pattern to the Arabidopsis β-subunit, AGB1 [Weiss, C. A., Garnaat, C. W., Mukai, K., Hu, Y. & Ma, H. (1994) Proc. Natl. Acad. Sci. USA 91, 9554–9558]. By using the yeast two-hybrid system, we show that AGG1 strongly interacts with tobacco and Arabidopsis β-subunits. The in vivo results have been confirmed by using in vitro methods to prove the interaction between AGG1 and the Arabidopsis β-subunit. As previously observed in mammalian systems, both the coiled-coil domain and the WD repeat regions of the β-subunit are essential for AGG1 interaction. Also in agreement with previous observations, the removal of the N-terminal α-helix of the AGG1 greatly reduces but does not completely block the interaction.
Heterotrimeric guanine nucleotide-binding (G) proteins are involved in a myriad of different signal-transduction pathways, from sexual mating to perception of low light. G proteins consist of three subunits (α, β, and γ) and associate with receptors containing seven-transmembrane-spanning domains (also known as G protein-coupled receptors, or GPCRs). On binding a ligand, the GPCR changes conformation, activating the G protein by promoting the α-subunit's exchange of GDP for GTP. The nucleotide exchange causes the heterotrimer to dissociate into a GTP-bound α-subunit and a βγ-dimer. In this active state, the two components interact with a number of downstream effectors that will ultimately elicit the cellular response. Finally, the intrinsic GTPase activity of the α-subunit will hydrolyze the molecule of GTP, thereby allowing the reconstitution of the inactive heterotrimer.
In plants, heterotrimeric G proteins have been associated with several physiological responses (1–4). The cloning of an α-subunit homologue in Arabidopsis thaliana was first reported by Ma et al. in 1990 (5), and four years later the first plant β-subunit homologue was reported by Weiss et al. (6). There have been 10 α-subunits and 7 β-subunits cloned from various plant species including Arabidopsis, tomato, rice, maize, and tobacco. However, the third subunit (γ) of the plant heterotrimeric G proteins has remained elusive.
It is reasonable to expect that plant γ-subunits will display the same characteristics as their mammalian counterparts, including the small size and the C-terminal CAAX motif (A, aliphatic amino acid; X, any amino acid) that directs prenylation of the invariant cysteine residue (7). Additionally, γ-proteins form a strong interaction with the β-subunit, requiring denaturing conditions to separate the dimer (8). A coiled-coil domain on the N terminus of both β- and γ-subunits provides stability to the dimer (9).
The main problem hindering the cloning of a plant G protein γ-subunit has been the lack of amino acid similarity among the known G protein γ-subunits, making it impossible to use homology-based cloning procedures. The strong association of γ- and β-subunits (8) is one of the few characteristics that can be exploited to isolate γ-subunits or their cDNAs. The first mammalian γ-subunits were isolated based on their interaction with G-β and the complete heterotrimer (10, 11). We have made use of this strong interaction between the two subunits to clone the first plant γ-subunit, by screening an A. thaliana yeast two-hybrid library that used a β-subunit as bait.
Materials and Methods
Yeast Two-Hybrid Screen.
Tobacco G protein β-subunit TGB1 was cloned into pAS-CYH, and the construct was introduced into Saccharomyces cerevisiae strain Y190 as described by Gietz and Schiestl (12). Y190 containing the TGB1-pAS-CYH plasmid was then retransformed with the Arabidopsis yeast two-hybrid library CD4–22 obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The library was constructed from poly(A)+RNA extracted from 3-day-old etiolated Arabidopsis seedlings (13). Transformants were plated on to synthetic complete medium without uracil, tryptophan, leucine, or histidine, but containing 25 mM 3-amino triazole (14). After a 5-day incubation, colony lift-filter assays to determine β-galactosidase activity were performed on each plate as described in the yeast protocols handbook (CLONTECH). For every positive colony, the library plasmid was isolated. Confirmation of positive interactions (secondary screening) was performed by transforming Y190 with (i) the library plasmid plus TGB1-pAS-CYH plasmid and (ii) the library plasmid plus a negative control plasmid containing human lamin C protein fused to the Gal4 DNA-binding domain. Library clones that tested positive for i but not ii were sequenced.
Southern and Northern Blot Analysis.
Total RNA and genomic DNA were isolated from Arabidopsis (ecotype Columbia) as described in Etheridge et al. (15). From each tissue, 20 μg of total RNA was separated electrophoretically on a 1% agarose gel containing 2.2 M formaldehyde before being transferred to a nylon membrane (Hybond-N, Amersham Pharmacia) by capillary action in 20× SSC. Genomic DNA (5 μg) was digested with BamHI, EcoRI, HindIII, XbaI, or SacII, separated on a 0.8% agarose gel, and transferred to nylon membrane.
Membranes were prehybridized for 3 h at 42°C in a solution containing 5× SSC, 50% (vol/vol) formamide, 5× Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 0.5% SDS, and 100 μg/ml denatured fragmented salmon sperm DNA. Full-length AGG1 cDNA was used as a template in a labeling reaction using [α-32P]dCTP and the Gigaprime DNA Random Priming Kit (Bresatec, Adelaide, Australia). Labeled AGG1 probe was added to the hybridization solution to a final concentration of 1 × 106 cpm/ml. Hybridization was performed overnight at 42°C. Membranes were washed in 2× SSC/0.1% SDS for 15 min at room temperature, in 2× SSC/0.1% SDS at 50°C for 15 min, and then in 0.2× SSC/0.1% SDS at 50°C for 10 min. Alternatively, low-stringency hybridizations were performed by reducing the percentage of formamide to 30% during hybridization and omitting the last wash step. To ensure that equal quantities of RNA were present in each lane of the Northern blot, the membrane was stripped and reprobed with a wheat 25S ribosomal cDNA (16).
Genomic Library Screening.
A Lambda Zap II Arabidopsis genomic library (Stratagene) was screened by standard methods (17) by using a 32P-labeled full-length AGG1 cDNA as a probe. Positive phages were induced to undergo in vivo excision. The resultant plasmid inserts were tested for the presence of AGG1 sequence by PCR. Positive clones were then sequenced.
In Vitro Binding Assay.
The in vitro binding procedure was performed according to Rozenblatt-Rosen et al. (18) with minor modifications. Briefly, glutathione S-transferase (GST) and an AGG1:GST fusion protein were expressed in Escherichia coli strain BL21. Recombinant GST and AGG1:GST fusion protein were purified by immobilization on glutathionine sepharose 4B affinity matrix. The matrix was washed once in binding/washing buffer (20 mM Tris, pH 8/0.2% Triton X-100/2 mM EDTA/150 mM NaCl/1 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin).
Arabidopsis G protein β-subunit RNA was synthesized by using a T7 polymerase transcription kit (RiboScribe, Epicentre Technologies, Madison, WI). RNA transcript (1 μg) was translated in the presence of [35S]methionine by using a rabbit-reticulocyte translation system (Promega). Labeled product (8 μl) was added to glutathionine sepharose 4B matrix containing 4 μg of either GST or AGG1:GST protein. After a 2-h incubation at 4°C, each matrix was washed twice with 1 ml of washing buffer. Bound proteins were eluted by boiling in 2× SDS loading buffer and separated on a SDS/12% PAGE gel. The resultant gel was fixed, dried, and exposed to Kodak β-max x-ray film.
β-Galactosidase Quantitative Assay.
Yeast cells were grown until early- to mid-log phase in synthetic complete drop-out liquid medium (14), and the OD600 of each culture was recorded. Two 1-ml aliquots of each culture were centrifuged at 14,000 × g for 2 min, and the pellets were resuspended in 200 μl of Z buffer without β-mercaptoethanol (19). The yeast cells were lysed by three freeze/thaw cycles. The lysed cells were mixed with 500 μl of Z buffer and 150 μl of 4 mg/ml O-nitrophenyl β-d-galactopyranoside. When the solution began to turn yellow, the incubation time was recorded, and the reaction was stopped by adding 400 μl of 1 M Na2CO3. The solution was then cleared by short centrifugation, and the OD420 of the supernatant was measured. Relative β-galactosidase activity was calculated by using the formula: relative β-galactosidase activity = 1,000 × OD420/(time × OD600). Note that time is measured in minutes.
Bioinformatics.
DNA-sequence analysis including blast searches, multiple-sequence alignments, and predicted translations were performed at the Australian National Genomic Information Service (ANGIS) web site (http://bris1.angis.org.au). Coiled-coil structure prediction of AGG1, AGB1 and TGB1 was performed by using the ANGIS pepcoil program.
Results and Discussion
Isolation and Sequence Analysis of AGG1.
Known G protein γ-subunits show little amino acid sequence conservation, but they have several conserved structural features and bind strongly to corresponding Gβ-subunits. We therefore used a plant Gβ as bait in a yeast two-hybrid screen to identify interacting proteins that were later analyzed for structural features common to Gγ-subunits.
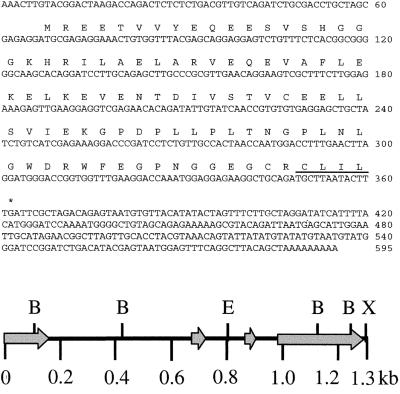
The tobacco β-subunit TGB1 was used as bait to screen 4.2 million transformants from an Arabidopsis seedling yeast two-hybrid library (CD4–22; ref. 13). After both primary and secondary screenings, 14 positive clones were obtained. These clones were sequenced and analyzed for the following structural features conserved in all known G protein γ-subunits (20): (i) coding for a small 6- to 13-kDa peptide (21); (ii) coiled-coil domain at the N terminus (9); and (iii) a C terminus CAAX motif (A, aliphatic amino acid; X, any amino acid; ref. 7). Four clones contained an identical coding region meeting these criteria. The longest (595 bp) of these four clones was designated AGG1 (Fig. 1). Northern blot analysis of AGG1 identified a single transcript of approximately the same length, indicating that we had isolated a near-full-length clone. The remaining 10 positive clones included the 26S ribosomal RNA gene, sedoheptulose-1,7-bisphosphatase, and several genes of unknown function that were not investigated any further.
Figure 1.
AGG1 cDNA sequence and genomic map. Nucleotide and deduced amino acid sequence of AGG1 (Upper). The prenyl group-binding site (CAAX box) is underlined. Restriction map of the AGG1 genomic clone (Lower) displays the BamHI (B), EcoRI (E) and XbaI (X) restriction endonuclease sites. Arrows indicate exons including the 5′ and 3′ untranslated regions.
The deduced AGG1 product is a 98-aa (10.8-kDa) peptide. A region close to the N terminus (amino acids 17–53), constituting over a third of the protein, was identified by secondary structure-prediction computer algorithms as strongly favoring coiled-coil formation (predicted probability of 1). A prenyl-group-binding site (CAAX box) was located on the protein C terminus (Fig. 1). In addition, the predicted amino acid sequence of AGG1 displays significant similarity to three short, highly conserved regions in mammalian γ-subunits (Fig. 2).
Figure 2.
Multiple alignment of the protein sequences of AGG1 and all of the mammalian γ-subunits cloned to date. Shaded areas represent highly conserved regions in all mammalian γ-subunits (22). The site of isoprenyl modification (CAAX box) is boxed. The total number of amino acids is listed at the end of each protein sequence. Swiss-Prot accession numbers for the mammalian proteins are P02698 (1), P16874 (2), P29798 (3), P50150 (4), P30670 (5), P30671 (7), P50154 (8), P43426 (9), P50151 (10), P50152 (11), and Q28024 (12).
The yeast two-hybrid system is a valuable tool for the detection of many protein–protein interactions and the cloning of their respective cDNAs (23). This technique has been used successfully to identify proteins that interact with heterotrimeric G proteins. For example, the G protein regulator GAIP was cloned by using a human Gα-subunit as bait (24).
AGG1 Is Differentially Expressed.
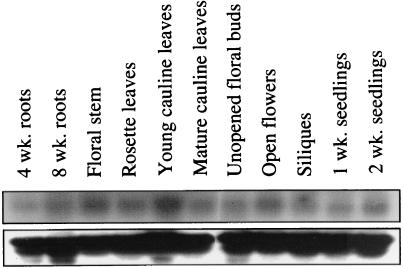
RNA gel blots hybridized to an AGG1 probe show that the gene is expressed in all tissues examined, although at different levels (Fig. 3). The highest levels of expression were found in young cauline leaves, open flowers, and floral stem, whereas roots, rosette leaves, siliques, and unopened floral buds contained lower levels of transcript. The expression pattern observed for AGG1 is similar to that reported for the Arabidopsis β-subunit (AGB1; ref. 5), showing comparable expression levels in roots, leaves, and flowers.
Figure 3.
Expression analysis of AGG1 by using total RNA extracted from A. thaliana (ecotype Columbia). RNA (20 μg) from each tissue was loaded onto a 1% formaldehyde gel and transferred to a nylon membrane. The blot was hybridized with 32P-labeled AGG1 cDNA and washed at high stringency (Upper). The blot was then stripped and rehybridized to a 25S wheat ribosomal probe (Lower).
In general, there is an increase in AGG1 mRNA levels in young tissues during development (compare 1-week-old vs. 2-week-old seedlings and 4-week-old vs. 8-week-old roots), but transcript levels in cauline leaves halved after leaf maturation. The α-subunit (GPα-1) displays a similar expression pattern, because it is highly expressed in developing tissues with reduced mRNA levels after maturation (25, 26). Interestingly, GPα1 displays very weak expression in the floral buds, which contrasts with the relatively high transcript levels of AGG1 and AGB1 in the same tissue (6). On the basis of expression analysis, it seems that AGG1, AGB1, and GPα-1 are involved in the same or similar signal-transduction pathways, although empirical proof that the three subunits can combine to form a functional heterotrimeric G protein has not yet been provided.
AGG1 Is a Single Copy Gene with No Close Relatives.
At the time this work was performed, the AGG1 genomic sequence was not publicly available as part of the Arabidopsis Genome Initiative; therefore, we used the full-length AGG1 clone to probe an Arabidopsis genomic library (Stratagene). A genomic clone was isolated and sequenced. The AGG1 gene spans 1.31 kb and contains three introns of 512, 139, and 75 bp that border four small exons of 157, 53, 45, and 333 bp (Fig. 1). Recently, the Arabidopsis sequencing project revealed the presence of the AGG1 gene on chromosome 3. Inspection of the sequence uncovered an in-frame ATG codon 120 bp upstream of the proposed AGG1 start codon. To establish whether the upstream ATG is part of the AGG1 transcript, we performed reverse transcription–PCR experiments with primers adjacent to the two putative start-of-transcription sites and a downstream primer. These experiments failed to amplify a product including the upstream ATG, whereas a product was consistently obtained including the ATG at the proposed start of transcription (data not shown).
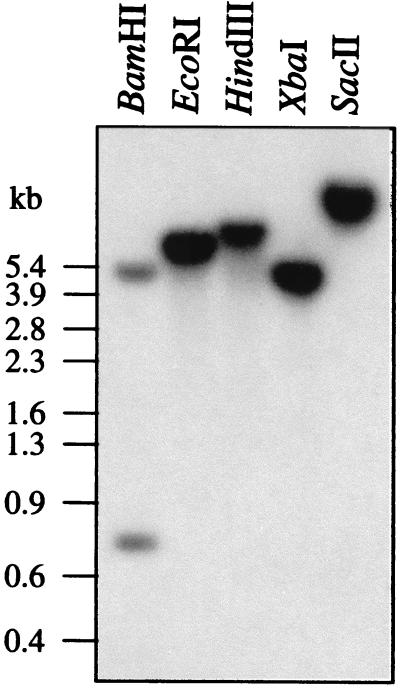
The full-length AGG1 cDNA clone was used, under high-stringency conditions, to probe a Southern blot containing Arabidopsis genomic DNA digested with BamHI, EcoRI, HindIII, XbaI, and SacII (Fig. 4). A single signal was detected in every lane except for the BamHI lane, in which two bands were observed. The discrepancy between the number of bands expected from the genomic restriction map in the BamHI line and the actual banding pattern observed can be explained by the short-sequence overlap of the genomic fragments and the cDNA probe used in the hybridization. No new bands were detected when the Southern blot was reprobed under low-stringency conditions (data not shown). These results indicate that in Arabidopsis, AGG1 is a single-copy gene with no closely related family members.
Figure 4.
Southern Analysis of AGG1. A. thaliana (ecotype Columbia) genomic DNA was digested with BamHI, EcoRI, HindIII, XbaI, or SacII. The blot was hybridized with 32P-labeled AGG1 cDNA overnight and washed at high stringency.
It is calculated that in humans there are approximately 1,000 different GPCRs that mediate responses to a large collection of stimuli, from the detection of odorants in the olfactory system to the sensing of dopamines in the brain. G proteins are able to provide specific signal-transduction pathways to each receptor by forming functionally different α, β and γ combinations. To date, at least 23 different Gα-subunits, 6 Gβ-subunits, and 11 Gγ-subunits have been identified in humans, providing a total of 1,518 possible heterotrimer combinations, although there are data indicating that not all combinations are compatible (8, 21, 27, 28). GPCRs do not seem to be so ubiquitous in plant systems; over 20 putative GPCRs have been reported to date (29–32). Two Gα-subunits have been reported in soybean (33, 34), and two β-subunits have been reported in wild oats (35). Furthermore, analysis of the Arabidopsis genome sequence reveals several distinct regions that display homology to the G protein α-subunit. It seems that plants use G proteins to regulate several signal-transduction pathways, although it is unlikely they are used to the same extent as their animal counterparts.
AGG1 Exhibits a Strong Interaction with AGB1 and TGB1.
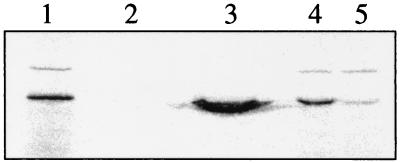
The interaction between Arabidopsis AGG1 and AGB1 was tested in vitro. Recombinant GST and AGG1:GST fusion proteins were produced in E. coli and immobilized on a sepharose-affinity matrix through the GST protein. Our results show that, when AGG1 is present in the matrix, it shows a strong ability to bind 35S-labeled AGB1, whereas GST alone is not able to bind any detectable AGB1 (Fig. 5).
Figure 5.
In vitro binding of AGB1 to the AGG1:GST fusion protein. [35S]Met-labeled AGB1 (lane 1) was incubated with a glutathionine sepharose 4B matrix bound to 4 μg of either GST alone or AGG1:GST fusion protein. After a 2-h incubation, each matrix was washed twice with 1 ml of washing buffer. Bound proteins were eluted by boiling the matrix in 2× SDS loading buffer and separated on a SDS/12% PAGE gel. The resultant gel was fixed, dried, and exposed to Kodak β-max X-ray film. Lanes 2 and 3 show the elution products from the GST matrix and the AGG1:GST matrix incubations. Lanes 4 and 5 contain 5 μl (out of 20 μl) of the unbound products from the in vitro binding reactions in lanes 2 and 3, respectively.
In addition to the qualitative detection of protein–protein interaction, the yeast two-hybrid system can also be used to determine the relative strength of the interaction. This quantitative measure was exploited to determine the relative ability of various regions of AGG1, AGB1, and TGB1 to interact with each other.
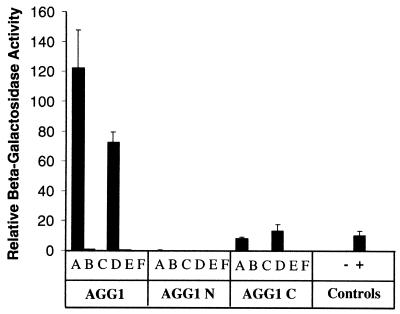
The complete coding region of AGG1 was cloned into pACT2 yeast-expression vector, behind the GAL4 activation domain. This construct was cotransformed into yeast with either AGB1 or TGB1 bait constructs, and the β-galactosidase activity was measured. Both AGB1 and TGB1 interacted very strongly with AGG1 (Fig. 6), resulting in β-galactosidase activity 11.2 and 6.6 times higher, respectively, than in the positive controls simian virus 40 and p53 (36). These results agree with data from animal studies that have demonstrated that although the Gβγ-complex is made up of two polypeptides, it can be considered a functional monomer, because the two subunits can only be dissociated with strong denaturants (8). As expected, AGG1 interacts with the Arabidopsis β-subunit more efficiently than with the tobacco subunit, as observed in the β-galactosidase activity levels.
Figure 6.
Protein–protein interaction studies between AGG1 and the Arabidopsis and tobacco Gβ-subunits. Yeast two-hybrid constructs containing AGG1 (full length), AGG1 N (1–52), or AGG1 C (53–98) were cotransformed into yeast with each of the following: AGB1 full length (A), AGB1 N (1–41) (B), AGB1 C () (C), TGB1 full length (D), TGB1 N (1–59) (E), and TGB1 C () (F). In all cases, “N” denotes the protein's coiled-coil domain, whereas “C” denotes the remaining C terminus. Three independently transformed yeast cultures were assayed in duplicate for β-galactosidase activity. The average activity values and standard error bars are included. The negative and positive controls were AGG1 plus the human lamin C protein and simian virus 40 plus p53, respectively.
To understand the nature of the interaction between the β- and γ-subunits fully, it is necessary to consider the complexity of the three-dimensional structure of the β-subunit and the topology of the interaction between β and γ. The β-subunit contains two highly differentiated regions, an N-terminal domain that forms an α-helix structure and a collection of seven WD repeats. The crystal structure of the G protein heterotrimer has revealed that the WD core region of β forms a propeller-like structure with seven blades arranged in a ring, leaving the α-helix domain extending outside the propeller. The N terminus of γ forms a coiled-coil structure with the N-terminal helix of β, whereas the rest of the protein extends outside the propeller but makes contacts with residues in blades 5, 6, and 7 (37, 38).
We wanted to determine the importance of the different domains within AGG1, AGB1, and TGB1 in the interaction. For that purpose, the domains of each protein theoretically involved in the formation of the coiled-coil structure, determined by a computer algorithm, were separated from the remaining C termini, and each fragment was cloned into yeast two-hybrid vectors. The different deletion constructs were then cotransformed into yeast, and their ability to interact with each other was measured. Our results show that removal of the relatively short N terminus (coiled-coil domain) from the β-subunit eliminates the interaction with AGG1 (Fig. 6, AGG1 C and F). This result is in agreement with studies of mammalian G proteins that found the β-subunits' coiled-coil domain to be essential for γ association (9, 39). Similarly, the β-subunit's coiled-coil domain alone is clearly not sufficient for the strong interaction with close-to-background levels of activity detected (Fig. 6, AGG1 B and E). Given the highly compact structure of Gβ, it is not surprising that any truncation of the protein prevents the correct assembly of the βγ-dimer (37). Recently, Wall et al. (40) have shown that the strength of the βγ association seems to come from multiple interactions between individual or groups of amino acids from both subunits.
In contrast with the results observed for the β-subunits, the removal of the N-terminal domain from AGG1 reduces the strength of the interaction with both β-subunits but does not completely block it (Fig. 6, AGG1 C A and D). These results are similar to the observation by Mende et al. (41) that the removal of 15 amino acids of the N terminus of a mammalian Gγ-subunit strongly impairs but does not entirely prevent the formation of βγ-dimers.
Conclusions
We have isolated the AGG1 cDNA from A. thaliana by using the yeast two-hybrid system and a tobacco Gβ-subunit as bait. The predicted AGG1 protein sequence displays significant homology to the conserved domains of mammalian γ subunits. More importantly, the AGG1 protein contains all of the defining features of a γ-subunit including size, presence of an N-terminal coiled-coil domain, C-terminal CAAX box, and strong interaction with the β-subunit both in vivo and in vitro. On the basis of these results, we conclude that AGG1 is an Arabidopsis heterotrimeric G protein γ-subunit.
These results provide additional support to the existence of heterotrimeric G proteins in plants. Furthermore, we have shown that the AGG1 and AGB1 subunits display identical characteristics to their mammalian counterparts, exhibiting a strong interaction in which the coiled-coil domain plays a vital role. The cloning of all three subunits of the heterotrimer provides a strong foundation to advance our knowledge of plant heterotrimeric G proteins and their involvement in plant signal transduction.
Acknowledgments
We are grateful to Ms. Naomi Etheridge for help provided with RNA extractions and β-galactosidase quantification and to Dr. R. Birch for critical reading of the manuscript.
Abbreviations
- GST
glutathione S-transferase
- GPCR
G protein-coupled receptor
Footnotes
References
- 1.Armstrong F, Blatt M R. Plant J. 1995;8:187–198. [Google Scholar]
- 2.Fairley-Grenot K, Assmann S M. Plant Cell. 1991;3:1037–1044. doi: 10.1105/tpc.3.9.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legendre L, Heinstein P F, Low P S. J Biol Chem. 1992;267:20140–20147. [PubMed] [Google Scholar]
- 4.Warpeha K M F, Hamm H E, Rasenick M M, Kaufman L S. Proc Natl Acad Sci USA. 1991;88:8925–8929. doi: 10.1073/pnas.88.20.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H, Yanofsky M F, Meyerowitz E M. Proc Natl Acad Sci USA. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss C A, Garnaat C W, Mukai K, Hu Y, Ma H. Proc Natl Acad Sci USA. 1994;91:9554–9558. doi: 10.1073/pnas.91.20.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey P J. Curr Opin Biotechnol. 1994;6:219–225. doi: 10.1016/0955-0674(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt C J, Thomas T C, Levine M A, Neer E J. J Biol Chem. 1992;267:13807–13810. [PubMed] [Google Scholar]
- 9.Garritsen A, van Galen P J M, Simonds W F. Proc Natl Acad Sci USA. 1993;90:7706–7710. doi: 10.1073/pnas.90.16.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurley J B, Fong H K W, Teplow D B, Dreyer W J, Simon M I. Proc Natl Acad Sci USA. 1984;81:6948–6952. doi: 10.1073/pnas.81.22.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinshaw J D, Kalman V K, Moomaw C R, Slaughter C A. J Biol Chem. 1989;264:15758–15761. [PubMed] [Google Scholar]
- 12.Gietz R D, Schiestl R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 13.Kim J, Harter K, Theologis A. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston J R, editor. Molecular Genetics of Yeast. A Practical Approach. Oxford Univ. Press, New York: IRL; 1994. [Google Scholar]
- 15.Etheridge N, Trusov Y, Verbelen J P, Botella J R. Plant Mol Biol. 1999;39:1113–1126. doi: 10.1023/a:1006137221259. [DOI] [PubMed] [Google Scholar]
- 16.Gerlach W L, Bedbrook J R. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 2.60–2.81. [Google Scholar]
- 18.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 157–159. [Google Scholar]
- 20.Bohm A, Gaudet R, Sigler P B. Curr Opin Biotechnol. 1997;8:480–487. doi: 10.1016/s0958-1669(97)80072-9. [DOI] [PubMed] [Google Scholar]
- 21.Morishita R, Nakayama H, Isobe T, Matsuda T, Hashimoto Y, Okano T, Fukada Y, Mizuno K, Ohno S, Kozawa O, et al. J Biol Chem. 1995;270:29469–29475. doi: 10.1074/jbc.270.49.29469. [DOI] [PubMed] [Google Scholar]
- 22.Ray K, Kunsch C, Bonner L M, Robishaw J D. J Biol Chem. 1995;270:21765–21771. doi: 10.1074/jbc.270.37.21765. [DOI] [PubMed] [Google Scholar]
- 23.Fields S, Sternglanz R. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 24.De Vries L, Mousli M, Wurmser A, Farquhar M G. Proc Natl Acad Sci USA. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss C A, Huang H, Ma H. Plant Cell. 1993;5:1513–1528. doi: 10.1105/tpc.5.11.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Weiss C A, Ma H. Int J Plant Sci. 1994;155:3–14. [Google Scholar]
- 27.Gudermann T, Kalkbrenner F, Dippel E, Laugwitz K, Schultz G. In: Signal Transduction in Health and Disease. Corbin J, Francis S, editors. Philadelphia: Lippincott–Raven; 1997. , Ser. 31, pp. 253–262. [Google Scholar]
- 28.Pronin A N, Gautam N. Proc Natl Acad Sci USA. 1992;89:6220–6224. doi: 10.1073/pnas.89.13.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 30.Plakidou-Dymock S, Dymock D, Hooley R. Curr Biol. 1998;8:315–324. doi: 10.1016/s0960-9822(98)70131-9. [DOI] [PubMed] [Google Scholar]
- 31.Josefsson L G, Rask L. Eur J Biochem. 1997;249:415–420. doi: 10.1111/j.1432-1033.1997.t01-1-00415.x. [DOI] [PubMed] [Google Scholar]
- 32.Wise A, Thomas P G, White I R, Millner P A. FEBS Lett. 1994;356:233–237. doi: 10.1016/s0014-5793(94)80076-6. [DOI] [PubMed] [Google Scholar]
- 33.Kim W Y, Cheong N E, Lee D C, Je D Y, Bahk J D, Cho M J, Lee S Y. Plant Physiol. 1995;108:1315–1316. doi: 10.1104/pp.108.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gotor C, Lam E, Cejudo F J, Romero L C. Plant Mol Biol. 1996;32:1227–1234. doi: 10.1007/BF00041411. [DOI] [PubMed] [Google Scholar]
- 35.Jones H D, Smith S J, Desikan R, Plakidou-Dymock S, Lovegrove A, Hooley R. Plant Cell. 1998;10:245–253. doi: 10.1105/tpc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Fields S. FASEB J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- 37.Clapham D E, Neer E J. Annu Rev Pharmacol Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- 38.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko M P, Reiner O, Smith T F, Neer E J. Biochemistry. 1996;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- 40.Wall M A, Posner B A, Sprang S R. Structure (London) 1998;6:1169–1183. doi: 10.1016/s0969-2126(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 41.Mende U, Schmidt C J, Yi F, Spring D J, Neer E J. J Biol Chem. 1995;270:15892–15898. doi: 10.1074/jbc.270.26.15892. [DOI] [PubMed] [Google Scholar]