Abstract
Fruiting body formation in ascomycetes is a highly complex process that is under polygenic control and is a fundamental part of the fungal sexual life cycle. However, the molecular determinants regulating this cellular process are largely unknown. Here we show that the sterile pro40 mutant is defective in a 120-kDa WW domain protein that plays a pivotal role in fruiting body maturation of the homothallic ascomycete Sordaria macrospora. Although WW domains occur in many eukaryotic proteins, homologs of PRO40 are present only in filamentous ascomycetes. Complementation analysis with different pro40 mutant strains, using full-sized or truncated versions of the wild-type pro40 gene, revealed that the C terminus of PRO40 is crucial for restoring the fertile phenotype. Using differential centrifugation and protease protection assays, we determined that a PRO40-FLAG fusion protein is located within organelles. Further microscopic investigations of fusion proteins with DsRed or green fluorescent protein polypeptides showed a colocalization of PRO40 with HEX-1, a Woronin body-specific protein. However, the integrity of Woronin bodies is not affected in mutant strains of S. macrospora and Neurospora crassa, as shown by fluorescence microscopy, sedimentation, and immunoblot analyses. We discuss the function of PRO40 in fruiting body formation.
Sexual development in filamentous ascomycetes is usually accompanied by the formation of complex fruiting bodies. During this process, a number of specialized cell types have to be generated from a more or less undifferentiated vegetative mycelium (5). This morphogenetic program is a convenient system that allows us to address the following fundamental question: how is multicellular development coordinated in eukaryotes? So far, several environmental and endogenous signals have been described to regulate fruiting body-dependent gene expression in a temporal and spatial manner (for a review, see reference 55). However, even though a decent number of genes involved in this process have been identified, the molecular determinants of fruiting body development have yet to emerge.
In this study, we have investigated the homothallic ascomycete Sordaria macrospora, which was recently developed as a model system to study fruiting body morphogenesis (27, 36, 37, 43, 52). During early sexual propagation, S. macrospora forms female gametangia, so-called ascogonia, on vegetative hyphae. In subsequent differentiation processes, ascogonia are enveloped by sterile hyphae and develop into spherical premature structures (protoperithecia), which mature into flask-like fruiting bodies (perithecia). In its final developmental stage, each perithecium contains a set of 60 to 80 asci with eight linearly ordered ascospores.
Using a forward genetic approach, we aimed to identify numerous developmental genes essential for fruiting body development in S. macrospora. Using conventional mutagenesis, several developmental mutants defective at different stages of perithecium formation were generated (37). In recent years, we have characterized several components of the pathway leading to sexual development. These include proteins involved in basic metabolism, such as fatty and amino acid biosynthesis (27, 43), fungal transcription factors, and components of putative signaling cascades (36, 37, 52). In addition, the mating type and mating type-regulated genes, such as pheromone and pheromone receptor genes, are directly involved in fruiting body development (39, 56).
Here we present the molecular characterization of the sterile pro40 mutant, which develops protoperithecia but is unable to generate any mature perithecia. A complementation analysis using an indexed S. macrospora cosmid library (54) led to the identification of the WW domain protein PRO40 as a novel factor controlling fruiting body formation. WW domains contain two conserved tryptophan residues spaced 20 to 22 amino acids apart and function as protein-protein interaction motifs (6). Our microscopic investigations using a PRO40-DsRed fusion protein revealed that PRO40 is associated with Woronin bodies in S. macrospora. These rather unexpected results are discussed in the context of the role of PRO40 in fungal sexual development.
MATERIALS AND METHODS
Strains and growth conditions.
Cloning and propagation of recombinant plasmids were performed using standard laboratory conditions (59). Escherichia coli strains XL1 Blue MRF′ (24) and DH5α (Invitrogen, Paisley, United Kingdom) were used as the hosts for plasmid amplification.
The wild-type strain (S48977) and color mutants R2 (S67813) and fus1-1 (S23442) of Sordaria macrospora were obtained from our laboratory collection. The sterile pro40 mutant (S38717) was generated as described previously (37). All fungal strains used in this study are listed in Table 1. Unless stated otherwise, standard growth conditions and DNA-mediated transformation were used for S. macrospora strains, as described previously (37, 43). In some transformation experiments, 20 mg/ml Glucanex 200G (Novozymes, Dittingen, Switzerland) was applied instead of Novozym for the generation of protoplasts. Vector pNAT4 (28), carrying the nourseothricin resistance gene nat1, was used in cotransformation experiments with hygromycin-resistant recipient strains. Transformants were selected on either nourseothricin (50 μg/ml) or hygromycin B (110 U/ml). For RNA extraction, S. macrospora strains were grown as described previously (45).
TABLE 1.
Strains used in this work
| Strain | Relevant genotype and phenotypea | Reference or sourceb |
|---|---|---|
| Sordaria macrospora strains | ||
| S48977 | Wild type | Culture collection |
| S38717 | pro40; sterile | Culture collection |
| S67813 | r2; colored mycelia and spores | Culture collection |
| S23442 | fus1-1; colored spores | Culture collection |
| S51475 | Single-spore isolate of S38717 × S23442 cross; pro40 fus1-1 | Culture collection |
| S66001 | Δku70::natrhphs | 51 |
| T76.3 | Primary Δpro40::hphrΔku70::natr transformant, homokaryotic, sterile | This work |
| S69656 | Single-spore isolate of T76.3 × S67813 cross; Δpro40::hphr; sterile | This work |
| T28.1.1G | Transformant of S38717 with pRHN3; gpd(p)::DsRed::hphr; sterile | This work |
| T58.1.2S1 | Single-spore isolate of S38717 transformed with pR-pro40; gpd(p)::pro40::DsRed::hphr; fertile | This work |
| T75.10 | T58.1.2S1 transformed with pCW15; gpd(p)::pro40::DsRed::hphrccg-1(p)::sgfp::hex-1 natr; fertile | This work |
| T89S25 | Single-spore isolate of T58.1.2S1 transformed with pNc-mtGFP (17); gpd(p)::pro40::DsRed::hphradh1(p)::atp1(presequence plus intron)::mgfp5 natr; fertile | This work |
| T99S10 | Single-spore isolate of pro40 transformed with pR-pro40 and pEGFP-KDEL; gpd(p)::pro40::DsRed::hphrgpd(p)::egfp::KDEL::natr; fertile | This work |
| T74S64 | S48977 transformed with pCW15; ccg-1(p)::sgfp::hex-1 natr; fertile | This work |
| I001 | Single-spore isolate of T74S64 × S51475 cross; pro40 ccg-1(p)::egfp::hex-1 natr; sterile | This work |
| T53.5.1B | Primary pro40(p)::pro40::mRFP1::hphr transformant, heterokaryotic, fertile | This work |
| S71343 | Single-spore isolate of T53.5.1B × S23442 cross; pro40(p)::pro40::mRFP1::hphrfus1-1; fertile | This work |
| T168 | Primary transformant of S38717 with pEH3-pro40; gpd(p)::pro40::egfp::hphr; fertile | This work |
| T187 | T58.1.2S1 transformed with the GFP-SKL plasmid (58); gpd(p)::pro40::DsRed::hphrnatrgpd(p)::egfp::SKL | This work |
| T182.1 | Single-spore isolate of S38717 transformed with pC-FLAG-pro40; gpd(p)::pro40::3xFLAG::hphr; fertile | This work |
| Neurospora crassa strains | ||
| FGSC11293 | Δso A | FGSC |
| FGSC11292 | Δso a | FGSC |
| FGSC2489 | Wild-type A (74-OR23-1VA) | FGSC |
| FGSC4200 | Wild-type a (ORS-SL6a) | FGSC |
| 74A-K93-5 | Wild-type a, St. Lawrence background | FGSC |
| FGSC8612 | hex-1 a | FGSC |
| FGSC11305 | Δpex-14 a | FGSC |
| FGSC1675 | os-1 A | FGSC |
| FGSC6103 | his-3 A | FGSC |
natr, nourseothricin resistant; hphr, hygromycin resistant; hphs, hygromycin sensitive.
Culture collection, Department for General and Molecular Botany, Ruhr-Universität, Bochum, Germany. FGSC, Fungal Genetics Stock Center.
Morphological characterization.
For determination of growth velocity, the S. macrospora wild-type strain (S48977) and the pro40 (S38717) and Δpro40 (S69656) mutants were cultivated as described previously (42). The growth front was marked every 24 h for seven consecutive days.
The densities of protoperithecia from the pro40 mutant and the wild type were determined after 7 days of cultivation on solid BMM fructification medium (12), using 12 plates per strain. Mycelia were fixed with ethanol-acetic acid (3:1) and washed three times with 70% ethanol. Protoperithecia were counted on 2.5 cm2 per plate at the edge of the plates.
For growth tests with Neurospora crassa, small pieces of mycelia (0.5 cm in diameter) were inoculated at the edges of petri dishes (150-mm diameter) containing 70 ml of medium (61). The plates were incubated at 30°C for 4 to 5 days with a 10-h light and 14-h dark rhythm. Vogel's minimal medium (VMM) (74) was supplemented with 2% sucrose or 0.1% oleic acid plus 0.05% Tergitol NP-40 (Sigma-Aldrich, St. Louis, MO) as the carbon source. Growth tests of S. macrospora were similar, with the exceptions that sucrose was replaced by glucose and incubation times were increased to up to 10 days.
Generation of a pro40 knockout strain.
For generation of a pro40 knockout strain, plasmid pDH-pro40 (Table 2) was generated as follows. Fragments of the 5′ (960 bp) and 3′ (1,032 bp) flanking regions of the pro40 open reading frame (ORF) were amplified using primers KO1 and KO2 or KO3 and KO4, with pIG1819-32 as a template (Table 2; see Table S1 in the supplemental material). The oligonucleotides contained recognition sites for SacI and ApaI or EcoRI and PstI, respectively. PCR fragments were subcloned into pDrive (QIAGEN, Hilden, Germany) and sequenced (GATC-AG, Konstanz, Germany). The SacI-ApaI 5′ fragment and the EcoRI-PstI 3′ fragment were then ligated into the corresponding sites of vector pDrive-Hyg (I. Godehardt and U. Kück, unpublished data). This vector is a modified pDrive plasmid containing a hygromycin B resistance gene under the control of the trpC promoter of Aspergillus nidulans between two multiple cloning sites. pDH-pro40 was subsequently used as a template for the amplification of the knockout cassette with primers KO1 and KO4. The 3.5-kb PCR fragment was then transformed into the S. macrospora Δku70 strain (51) to generate a pro40 knockout by homologous recombination. Single-spore isolates in which the pro40 ORF was replaced by the hph cassette and which had the wild-type genetic background were obtained as described previously by using the color mutant R2 (S67813) (51).
TABLE 2.
Plasmids used in this work
| Plasmid | Description and/or use | Reference or source |
|---|---|---|
| pDrive | Cloning vector (cloning of PCR fragments) | QIAGEN, Hilden, Germany |
| pBluescript SK(+) | Cloning vector (cloning of PCR fragments) | Stratagene, La Jolla, CA |
| pT3T7 | Cloning vector | Boehringer Mannheim, Germany |
| pAnsCos | Cosmid vector | 47 |
| D11 | 32-kb S. macrospora genomic DNA in pAnsCos1 | This work |
| pIG1819-32 | ca. 10-kb EcoRI fragment of D11 in pAnsCos1, with pro40 ORF (aa 1 to 1316) | This work |
| pIG1809-1 | ca. 3.5-kb SalI fragment of D11 in pT3T7, with pro40 ORF (aa 724 to 1316) | This work |
| pNAT4 | gpd(p)::nat1 | 28 |
| pIE46-2 | pro40 ORF (aa 1 to 1316) in pDrive | This work |
| pRHN3 | gpd(p)::DsRed | I. Godehardt and U. Kück, unpublished data |
| pR-pro40 | pro40 tagged with DsRed in NcoI site in pRHN3 | This work |
| pR-pro40-V | Derivative of pR-pro40, oligonucleotide linker (pro40-5, pro40-6) in PstI, HindIII sites | This work |
| pR-pro40-L | Derivative of pR-pro40, oligonucleotide linker (pro40-7, pro40-8) in PstI, HindIII sites | This work |
| pIG2195-1 | Derivative of pR-pro40-V, deletion (1,190 bp) of positions 2519 to 3645 of pro40 ORF | This work |
| pIG2199-8 | Derivative of pR-pro40-L, deletion (2,967 bp) of positions 1047 to 4013 of pro40 ORF | This work |
| pIG2199-9 | Derivative of pR-pro40-L, deletion (2,615 bp) of positions 1399 to 4013 of pro40 ORF | This work |
| pIG2199-10 | Derivative of pR-pro40-L, deletion (1,660 bp) of positions 2354 to 4013 of pro40 ORF | This work |
| pIG2196-11 | Derivative of pR-pro40-V, deletion (1,204 bp) of positions 2811 to 4013 of pro40 ORF | This work |
| pIE49-3 | Derivative of pIE46-2, deletion (1,042 bp) of positions 284 to 1323 of pro40 ORF | This work |
| pEH3-pro40 | pro40 tagged with egfp in NcoI site in pEH3 (44, 53) | This work |
| pC-FLAG-pro40 | pro40 tagged with three-FLAG tag | This work |
| pEGFP-KDEL | gpd(p)::ppg1 (presequence)::egfp::KDEL::nat; derivative of pSppg1-1 (38) | 42a |
| pNc-mtGFP | adh1(p)::atp1(presequence plus intron)::mgfp5 | 17 |
| GFP-SKL plasmid | gpd(p)::egfp::SKL | 58 |
| pMF272 | sgfp gene under the control of the ccg-1 promoter, with his3 selectable marker | 16 |
| pCW15 | sgfp replaced by sgfp-hex1 in pMF272 (16) | This work |
| pDrive-Hyg | trpC(p)::hph in HpaI fragment in pDrive-Nco (pDrive with NcoI linker) | I. Godehardt and U. Kück, unpublished data |
| pDH-pro40 | 1-kb 5′-untranslated region SacI-ApaI fragment and 1-kb 3′-untranslated region EcoRI-PstI fragment in pDrive-Hyg | This work |
| pMT-mRFP1 | GATEWAY vector containing the mRFP1 gene | 71 |
| pZHK2 | trpC(p)::hph | 29 |
| pZHK2-mRFP1 | mRFP1 from pMT-mRFP1 (71) in HindIII site of pZHK2 | This work |
Tagging of pro40 by homologous recombination.
To generate an S. macrospora strain that synthesizes a monomeric red fluorescent protein 1 (mRFP1)-tagged PRO40 protein under the control of its native promoter, we used a previously described strategy (77). First, two fragments of the pro40 locus, spanning 1 kb upstream (fragment 1) and downstream (fragment 2) of the stop codon, were amplified using primer pairs FPCR1/FPCR2 and FPCR3/FPCR4, respectively (see Table S1 in the supplemental material). To generate a cassette containing the mRFP1 gene followed by the hph gene as a selectable marker, plasmid pZHK2-mRFP1 was designed as follows. Using pMT-mRFP1 (71) as a template, the mRFP1 gene was amplified with primers RFP1 and RFP2, containing HindIII recognition sites. The amplicon was then cloned into HindIII-digested pZHK2, which carries the hph gene under the control of the A. nidulans trpC promoter (29). The 2,155-bp mRFP1-hph cassette (fragment 3) was amplified from plasmid pZHK2-mRFP1 with primers FPCRK1 and FPCRK2. Primer FPCRK1 primes at the 5′ end of the cassette and has a 5′ extension of 30 bp identical to the last 10 codons of the pro40 gene upstream of the stop codon. Primer FPCRK2 primes at the 3′ end of the cassette and has a 30-bp 5′ extension corresponding to the reverse complement of the sequence following the pro40 stop codon. Fragments 1 to 3 were cleaned using a PCR purification kit (Macherey-Nagel, Düren, Germany) and subjected to fusion PCR without primers, as described previously (77). To obtain a larger amount of fragment, nested primers FPCRN1 and FPCRN2 were used to generate a 4,026-bp amplicon that was precipitated and subsequently transformed into the wild type (S48977). Homologous integration was analyzed by PCR, using primer pairs FPCR5/FPCR8 and FPCR6/FPCR7 for the two flanking regions. Each pair contains one primer located in the genomic region outside the sequence used in the targeting construct and one primer located in the mRFP1-hph cassette. Purification of heterokaryotic transformants was achieved by crossing them with fus1-1 (S23442) and by generation of single-spore isolates.
Construction of plasmids.
All plasmids used in this study are listed in Table 2. For cellular localization studies, an NcoI fragment containing the entire pro40 ORF was ligated into the NcoI site of plasmid pRHN3 (Godehardt and Kück, unpublished data), leading to vector pR-pro40. Alternatively, the NcoI fragment was ligated into NcoI-digested pEH3 (44, 53), resulting in pEH3-pro40, encoding a PRO40-enhanced green fluorescent protein (EGFP) fusion protein. For biochemical analyses, oligonucleotides FLAG-1 and FLAG-2, encoding a three-FLAG tag, were introduced into the NcoI site at the end of the pro40 gene instead of the egfp gene (pC-FLAG-pro40). To localize Woronin bodies in vivo, the N. crassa protein HEX-1 was fused to the C terminus of SGFP (an S65T variant of GFP). The sgfp ORF without a stop codon was amplified from plasmid pMF272 (16), using primers RE1372 and RE1373, encoding recognition sites for XbaI and BamHI, respectively, and subcloned into pBluescript SK(+) (Stratagene, La Jolla, CA). Using primers RE1370 and RE1371, including recognition sites for BamHI and EcoRI, respectively, the N. crassa hex-1 ORF was amplified from cDNA and ligated into pBluescript SK(+). Both fragments were verified by sequencing (MWG-Biotech AG, Ebersberg, Germany), subsequently excised from pBlueskript SK(+) with the indicated enzymes, and inserted into XbaI/EcoRI-digested pMF272, resulting in plasmid pCW15 (Table 2).
Vectors for complementation analyses of the pro40 and Δpro40 mutants were generated as follows. A 10-kb EcoRI subfragment of complementing cosmid D11 was ligated into pAnsCos (47), leading to plasmid pIG1819-32. A 3.5-kb SalI fragment of this vector was cloned into pT3T7 (Boehringer Mannheim, Germany), resulting in plasmid pIG1809-1. For construction of pIE46-2, the pro40 ORF was PCR amplified from pIG1819-32, using primers Npro40 und pro40-4, and cloned into pDrive (QIAGEN, Hilden, Germany).
For complementation analyses of the Δpro40 knockout strain, several plasmids containing a shortened pro40 ORF were created from plasmid pR-pro40. For this purpose, a linker oligonucleotide derived from oligonucleotides pro40-7 and pro40-8 was digested with PstI and HindIII and then ligated into PstI/HindIII-digested pR-pro40, leading to pR-pro40-L. After digestion of this plasmid with EcoRI and KpnI, incubations with exonuclease III and S1 nuclease were performed under standard laboratory conditions (59), resulting in unidirectional deletions of pro40 from the 3′ end (Table 2). Alternatively, a linker oligonucleotide derived from oligonucleotides pro40-5 and pro40-6 was digested with PstI and HindIII and then ligated into PstI/HindIII-digested pR-pro40, leading to pR-pro40-V. This plasmid was then digested with BglII and religated, resulting in pIG2195-1, or digested with ClaI/HindIII, treated with Klenow polymerase, and religated, leading to plasmid pIG2196-11 (Table 2).
Isolation of nucleic acids, hybridization, and PCR.
DNA isolation was performed as described previously (54). RNAs were prepared according to previously described methods (22, 63). The integrity of RNA was verified by agarose gel electrophoresis and Northern blot analysis prior to cDNA synthesis. Poly(A) RNA was isolated from total RNA with the PolyATtract mRNA isolation system (Promega, Mannheim, Germany). Southern and Northern blotting was performed according to standard techniques (59), using radioactively labeled DNA probes. PCR amplification was performed using Pfu or GoTaq polymerase (Promega, Mannheim, Germany), Triple Master polymerase (Eppendorf, Hamburg, Germany), or Expand long-template polymerase (Roche Diagnostics, Mannheim, Germany) following the manufacturers’ protocols. PCR primers were synthesized by MWG-Biotech AG (Ebersberg, Germany) or Sigma-Aldrich (St. Louis, MO) (see Table S1 in the supplemental material).
Real-time PCR.
Reverse transcription of total RNA and quantitative real-time PCR were performed as described previously (45), using qPCR MasterMix Plus for SYBR green (Eurogentec, Seraing, Belgium). Each reaction was carried out in triplicate. Mean cycle threshold values were calculated from the triplicates and used for calculations of expression ratios as described previously (49). The cycle threshold values for an amplicon derived from the small-subunit rRNA were used as a reference for normalization. Real-time experiments were carried out twice with biologically independent samples. The significance of differential expression was verified with REST (the pairwise fixed-reallocation randomization test) (50).
Preparation of PNS and sucrose density centrifugation.
For subcellular fractionation, cultures were first inoculated with conidia of N. crassa (105/ml) or 20 to 25 small pieces of S. macrospora mycelia in VMM (74), with sucrose and glucose as carbon sources for N. crassa and S. macrospora, respectively. Cultures were shaken at 100 rpm at 30°C for 24 h (N. crassa) or 4 days (S. macrospora) before the hyphae were shifted to oleic acid-containing medium for an additional 12 h of growth. Mycelia were harvested by filtration, washed with water, and mixed with 1 g sea sand/g wet weight and 4 volumes of isolation buffer (150 mM Tricine, pH 7.4, 10 mM KCl, 5 mM MgCl2, 1 mM EDTA, 440 mM sucrose, 8 μM antipain, 0.3 μM aprotinin, 1 mM benzamidine, 1 μM bestatin, 10 μM chymostatin, 5 μM leupeptin, 1.5 μM pepstatin, 1 mM phenylmethylsulfonyl fluoride [PMSF]) before being ground with a pestle in a mortar. The homogenate was subjected to centrifugation three times at 500 × g for 5 min, and the supernatant was taken as the postnuclear supernatant (PNS). To obtain an organellar fraction, PNS was subjected to centrifugation at 25,000 × g for 20 min. The resulting organellar pellet was resuspended in isolation buffer, and 10 mg of protein was loaded on top of a 35-ml linear gradient of 30% to 60% (wt/wt) sucrose with a 65% (wt/wt) sucrose cushion, dissolved in 10 mM Tricine, pH 7.4, 1 mM EDTA, and subjected to centrifugation at 38,000 × g for 2 h in a Sorvall SV288 vertical rotor. The gradient was fractionated in 1-ml aliquots from the bottom to the top, and sucrose density was measured refractometrically.
Protease protection assay.
Two milligrams of a PNS prepared from strain T182.1 was subjected to centrifugation at 25,000 × g for 20 min, and the resulting organellar pellet was resuspended in 3 ml of ice-cold isolation buffer without inhibitors. The assay was started by the addition of 50 μl of a 10-mg/ml proteinase K solution in either the presence or absence of 0.2% Triton X-100. After 15 min on ice, the assay mixture was shifted to room temperature. Aliquots (500 μl) were removed after 0, 5, 10, 15, and 30 min and treated with PMSF to stop the reaction. Samples were subjected to trichloroacetic acid precipitation and analyzed by Western blotting.
Generation and usage of antisera and immunoblotting.
Antibodies against N. crassa multifunctional protein (FOX-2) (68), TIM-23 (41), HEX-1, and CAT-1 (60) were described previously. The anti-FLAG antibody was applied as described by the manufacturer (Sigma-Aldrich, St. Louis, MO). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were performed according to standard protocols (59). Immunoreactive complexes were detected with an enhanced chemiluminescence system from GE Healthcare (Freiburg, Germany).
Sequences and alignments.
DNA and protein sequence data were obtained from the public databases at NCBI (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) or by BLAST searches (3) of the almost complete genomes at the Broad Institute (http://www.broad.mit.edu/annotation/fgi/) and the Institut de Génétique et Microbiologie, Université de Paris-Sud XI/CNRS (Podospora anserina genome project [http://podospora.igmors.u-psud.fr/index.html]). Protein sequence alignments were performed using the ClustalW program (69; http://clustalw.genome.jp/). Prediction of conserved domains, posttranslational modifications, and topology was carried out with various tools of the Swiss Institute of Bioinformatics (http://www.expasy.org).
Microscopic investigations.
For microscopy, S. macrospora strains were grown on slides in glass petri dishes containing water to prevent dehydration of the samples. For this purpose, sterile slides were overlaid with a thin layer of solid BMM medium or a previously described minimal medium used for RNA extraction (45) and placed into glass petri dishes upon a spacer. After inoculation, samples were incubated at 27°C with continuous light for 2 to 7 days.
Fluorescence and light microscopic investigations were carried out with an AxioImager microscope (Zeiss, Jena, Germany) using an HBO 100 Hg or XBO 75 xenon lamp for fluorescence excitation. Fluorescence was studied using Chroma filter sets 41017 (exciter HQ470/40, emitter HQ525/50, and beamsplitter Q495Ip) and 41035 (exciter HQ546/11, emitter HQ605/75, and beamsplitter Q560Ip) (Chroma Technology Corp.) for detection of GFP and DsRed, respectively. Images were captured with a Photometrix Cool SnapHQ camera (Roper Scientific) and MetaMorph (version 6.3.1; Universal Imaging). Recorded images were edited with MetaMorph and Adobe Photoshop CS2.
Confocal laser scanning microscopy was performed using a Zeiss LSM 510 Meta microscopy system (Zeiss, Jena, Germany) based on an Axiovert microscope. GFP and DsRed were excited with the 488-nm line of an argon-ion laser and the 543-nm line of a He-Ne laser, respectively. The fluorescence emission was selected by band-pass filter BP505-530 for GFP and BP560-615 for DsRed. The multitracking mode was used in colocalization experiments.
Nucleotide sequence accession number.
Sequence data for the S. macrospora pro40 gene have been submitted to the EMBL database under accession number AJ616913.
RESULTS
Characterization of the pro40 mutant.
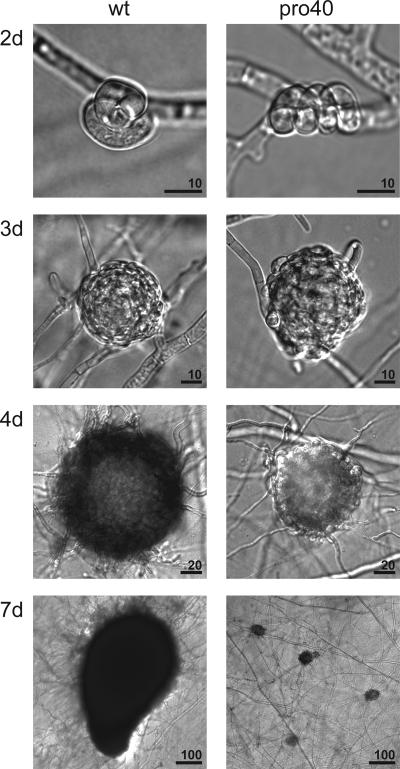
Recently, we described the generation of developmental mutants of S. macrospora with defects in fruiting body formation (37, 43, 52). During this approach, we obtained mutants of the “pro” type, forming only protoperithecia. Here we describe the pro40 mutant (S38717), which is able to form only early structures of sexual development (Fig. 1). After 3 days of growth, wild-type mycelia exhibit protoperithecia, which enlarge and develop into mature perithecia within 7 days after inoculation. During fruiting body maturation, black melanins accumulate in perithecial tissues as well as in ascospores. In contrast, development of protoperithecia stops after about 3 days in the pro40 mutant. After 7 days of growth, mycelia still display only small fruiting body precursors with light pigmentation. The density of protoperithecia in the pro40 mutant is 856 ± 165/cm2, whereas the wild type develops 592 ± 49/cm2 protoperithecia and 177 ± 17/cm2 perithecia. The lack of fruiting body development in the pro40 mutant is correlated with an increase of the vegetative growth rate on BMM fructification medium (30.4 ± 2.9 mm/day) in comparison to that of the wild type (22.8 ± 4.6 mm/day).
FIG. 1.
Morphological characterization of pro40 mutant from S. macrospora. Sexual developmental stages of the wild type (wt) and the pro40 mutant strain are shown. The images show ascogonia (wt and pro40 mutant, at 2 days [2d]), young protoperithecia (wt and pro40 mutant, at 3 days [3d]), melanized protoperithecia (wt only, at 4 days [4d]), and perithecia (wt only, at 7 days [7d]). After 7 days of growth, the pro40 mutant still exhibited only protoperithecia. Bar lengths are shown in micrometers.
Isolation and characterization of the pro40 gene.
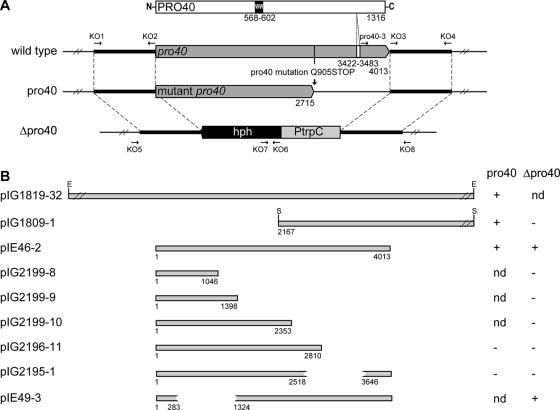
Previously, several pro mutants have been complemented by transformation with an indexed S. macrospora cosmid library (54) to identify the genes responsible for the developmental defects (37, 52). Using an identical approach, cosmid clone D11, carrying a 32-kb insert of genomic DNA, was identified due to its ability to restore the wild-type phenotype to the pro40 mutant. Several fragments of D11 were tested in subsequent complementation experiments, and we identified a 3.5-kb sequence (pIG1809-1) that is able to reestablish fruiting body formation in a sterile recipient strain (Fig. 2B).
FIG. 2.
Physical map of the pro40 gene, the predicted ORF encoding the PRO40 polypeptide, and complementation analysis. (A) Genomic organization of the pro40 gene in the wild type and the pro40 and Δpro40 mutants. The pro40 ORF is indicated by a gray arrow, and the intron is marked by a white box. Tiny black arrows represent oligonucleotides used for generation and identification of a pro40 knockout strain. A gray box and a black arrow represent the trpC promoter and the hph gene, respectively, replacing the pro40 ORF in the Δpro40 mutant. The PRO40 protein encoded by the wild-type pro40 ORF is depicted as a white box containing the WW domain as a black box. (B) Recombinant plasmids representing part of the pro40 ORF were used for complementation analysis, with the pro40 or Δpro40 mutant as the recipient. Complementation of the mutant phenotype (restoration of fertility) is indicated by plus signs, while minus signs mark noncomplementing constructs. E, EcoRI; S, SalI; ND, not determined.
For a more detailed analysis of the mutant locus, genomic DNA from the pro40 mutant and the wild type were subjected to Southern hybridization analysis, with the insert of pIG1819-32 as a probe. Comparison of the hybridization patterns of genomic DNA digested with EcoRV or XbaI revealed no differences between the pro40 mutant and the wild-type strain (data not shown). From these experiments, we concluded that no major rearrangements or deletions occurred in the pro40 mutant during the mutagenesis procedure.
To characterize the complementing 3.5-kb region, the complete sequence was determined and annotated. It contains an ORF that is homologous to the 3′ part of the N. crassa gene NCU02794.3. Quite recently, NCU02794.3 was shown to be defective in the N. crassa so mutant that is impaired in hyphal fusions (15). Our data indicated that the 3.5-kb region carries only part of a major ORF. Therefore, the flanking regions of the S. macrospora sequence were determined using DNA fragments derived from cosmid clone D11. We finally identified the pro40 gene, which is highly homologous to the so gene from N. crassa (15), showing 85.6% sequence identity on the nucleotide level for the coding region. Similar to so, pro40 comprises 4,013 bp and contains a single intron of 62 bp, as confirmed by cDNA sequencing. This intron separates the two exons, of 3,421 bp and 530 bp (Fig. 2A), which encode a predicted PRO40 polypeptide of 1,316 amino acids with a calculated mass of 141.5 kDa.
To determine the causative factor for sterility in the pro40 mutant, we constructed primers to amplify pro40 regions from the mutant strain. Sequence comparison between the wild type and the pro40 mutant revealed a C-to-T transition at nucleotide position 2713 in the pro40 mutant, changing a glutamine codon to a stop codon. Thus, only a truncated PRO40 polypeptide of 905 amino acid residues is carried by the pro40 mutant.
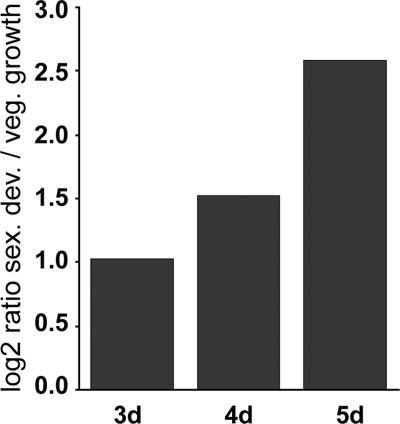
By Northern hybridization experiments, we investigated whether pro40 is transcriptionally expressed during sexual development. Most probably due to its low abundance, we were unable to detect the pro40 transcript in enriched polyadenylated mRNA from the wild type (data not shown). However, a transcript was easily detectable using quantitative real-time PCR (42). A comparative analysis using wild-type mycelia grown under conditions allowing sexual development versus vegetative growth showed that pro40 is upregulated up to fourfold during sexual development (Fig. 3).
FIG. 3.
Quantitative real-time PCR analysis of pro40 transcript levels. The wild type was grown under vegetative and sexual growth conditions (42). The data are given as base 2 logarithmic values for the mean values (calculated with REST) for two independent experiments. pro40 is differentially expressed at each time point (P, 0.001, determined with REST).
Characterization of the PRO40 polypeptide.
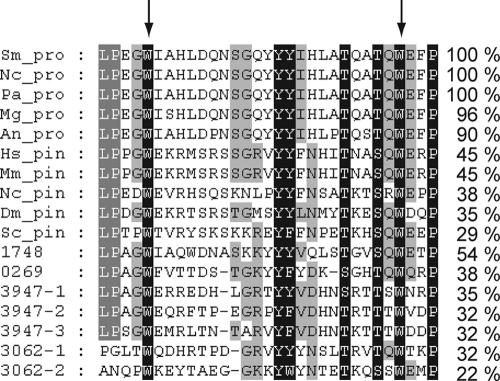
Using the deduced PRO40 protein sequence for BLAST searches, we discovered that homologs exist only in filamentous ascomycetes. The protein contains no known targeting signal for any subcellular localization. However, PRO40 has a WW domain, a protein-protein interaction motif of about 40 amino acids containing two conserved tryptophan residues spaced 20 to 22 amino acids apart (6) and found in many eukaryotic proteins. This module shows high affinity towards proline-rich ligands that allow sorting of WW domains into four groups (I to IV) (9, 66). Comparing the primary sequences of different WW domains, PRO40 might belong to group II, binding to the consensus motif PPLP. Figure 4 depicts an amino acid alignment of WW domains of fungal PRO40 homologs from A. nidulans, Magnaporthe grisea, N. crassa, P. anserina, and S. macrospora. Within these domains, the degree of identity is over 90%. WW domains themselves occur in many nonrelated eukaryotic proteins and have retained a significant level of sequence homology. For example, the WW domain of the human propyl isomerase Pin1 and its homologs in other eukaryotes is almost 50% identical to the WW domain of fungal PRO40 homologs. Moreover, the sequence identity of the PRO40 WW domain to other WW domains of N. crassa proteins lies between 22 and 54%, although there is no significant sequence identity between S. macrospora PRO40 and these WW domain-containing proteins.
FIG. 4.
Alignment of WW domains from PRO40 and other eukaryotic proteins. PRO40 homologs are characterized by the suffix “pro,” while Pin1 homologs contain the suffix “pin.” Four-digit numbers refer to the ORF numbers used in the N. crassa genome database (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html), and in cases where multiple WW domains are present in one protein, these are numbered serially. The arrows indicate the two conserved tryptophan residues (6). The degree of sequence identity within the WW domain is given on the right. Abbreviations: Sm_pro, S. macrospora PRO40 (emb CAE83713); Nc_pro, N. crassa SO (15) (gi 85111730); Nc_pin, N. crassa NCU08554.3 (gi 85109681); Pa_pro, P. anserina protien (http://podospora.igmors.u-psud.fr/index.html); Mg_pro, M. grisea MG01636.5 (gi 39951987); An_pro (A. nidulans) AN5766.3 (gi 67539212); Hs_pin, Homo sapiens Pin1 (30) (sp Q13526); Mm_pin, Mus musculus Pin1 (18) (dbj BAA87038.1); Dm_pin, Drosophila melanogaster Pin1 (7, 31) (ref NP_523428.1); Sc_pin, Saccharomyces cerevisiae ESS1 (20) (emb CAA89541.1); 0269, N. crassa NCU00269.3, SET-2 (1) (gi 85086731); 1748, N. crassa NCU01748.3 (gi 16416095); 3062, N. crassa NCU03062.3 (gi 85111298); and 3947, N. crassa NCU03947.3 (gi 11272426).
In the pro40 mutant, the mutated pro40 gene contains a base pair substitution that still allows the synthesis of a truncated protein of 905 amino acids. Complementation analyses indicated that the C-terminal part of PRO40 by itself is sufficient to restore fertility in the pro40 mutant (Fig. 2B). However, this C-terminal fragment, encoded on vector pIG1809-1, overlaps with the truncated protein putatively synthesized in the mutant. Thus, the complementation event might be due to the production of a full-length PRO40 protein assembled from the mutant-encoded N-terminal part and the vector-encoded C-terminal PRO40 polypeptide. On the other hand, the N-terminal part could be nonessential for PRO40 function.
To test both options, we constructed a strain lacking the complete pro40 ORF by homologous recombination as described previously (51; also see Materials and Methods). The knockout strain Δpro40 (S69656) is morphologically identical to the pro40 mutant. Both show the same mycelial pigmentation and the lack of any mature fruiting bodies (data not shown). Transformation of the Δpro40 mutant with plasmid pIE46-2 containing the complete pro40 ORF yielded a strain with restored fertility, confirming the developmental role of pro40 (Fig. 2B). However, transformation with plasmid pIG1809-1 did not result in fertile transformants, supporting the idea of polypeptide assembly mentioned above.
The truncated PRO40 polypeptide carried by the pro40 mutant is not sufficient to trigger complete sexual development. Therefore, parts of the PRO40 C terminus must be essential for this process. However, for mutant pro11, it has been described that fertility can partially be restored by a truncated PRO11 version shorter than the mutant-encoded PRO11 polypeptide (52). To examine whether such a phenomenon exists in the pro40 mutant, polypeptides with major truncations from the C terminus were tested for the ability to restore fertility in the pro40 or Δpro40 mutant as the recipient. In plasmids pIG2196-11, pIG2199-8, pIG2199-9, and pIG2199-10, up to 74% of the pro40 3′ end was deleted (Fig. 2B; Table 1). None of the constructs was able to complement the knockout mutation. We therefore predicted that essential domains associated with sexual development are located within the C-terminal region not present in the mutant protein. We further generated the in-frame deletion construct pIG2195-1, lacking base pairs 2519 to 3647 of the pro40 ORF but still encoding part of the PRO40 C terminus. This construct was not able to complement the sterile phenotype of the Δpro40 mutant, suggesting that the predicted domains are located within the deleted region. Interestingly, parts of the PRO40 N terminus seem not to be essential for fruiting body development, as plasmid pIE49-3 was able to restore the fertile phenotype in the Δpro40 mutant.
Subcellular distribution of PRO40.
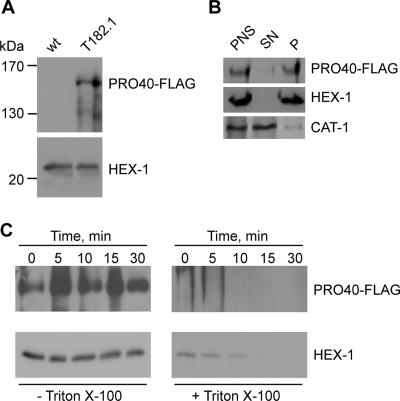
To gain a better understanding of PRO40 function in S. macrospora, we aimed to identify its subcellular localization by biochemical analysis. Since no PRO40 antibodies are available, we generated strain T182.1, expressing a C-terminally FLAG-tagged PRO40 polypeptide in the pro40 mutant background. Western analysis using an anti-FLAG antibody indicated that full-length PRO40-FLAG is synthesized in T182.1 (Fig. 5A). Furthermore, T182.1 showed restored fertility, indicating that the PRO40-FLAG fusion protein is functional in S. macrospora. By differential centrifugation, we further determined the PRO40-FLAG distribution between a 25,000 × g organellar pellet and the corresponding supernatant. As controls, the cytoplasmic catalase CAT-1 and the Woronin body protein HEX-1 were detected in different fractions with antibodies specific for the corresponding polypeptides from N. crassa (23, 60). Western analysis revealed that PRO40-FLAG is present in the organellar fraction (Fig. 5B). To assess whether PRO40 is located inside or outside organelles, we subsequently performed protease protection assays. Therefore, the 25,000 × g pellet of T182.1 was resuspended and treated with proteinase K in the presence or absence of the detergent Triton X-100. PRO40, like HEX-1, was protected from the protease in the absence of detergent, indicating its intraorganellar localization (Fig. 5C).
FIG. 5.
Subcellular distribution of PRO40-FLAG. (A) Expression of PRO40-FLAG. Correct expression of full-length FLAG-tagged PRO40 was determined by Western blotting using a monoclonal anti-FLAG antibody. The crude protein extracts analyzed were prepared from strain T182.1 harboring a construct designed to express PRO40-FLAG under the control of the gpd promoter (pro40 mutant plus PRO40-FLAG) and from the untransformed wild-type strain (wt). Expression of HEX-1 served as a loading control. (B) Differential centrifugation. A PNS of strain T182.1 was separated into a 25,000 × g organellar pellet (P) and a supernatant fraction (SN), and the resulting fractions were assayed for the presence of PRO40, HEX-1, and cytoplasmic CAT-1 by Western blot analysis. (C) Protease protection assay. The 25,000 × g organellar pellet of strain T182.1 was resuspended in homogenization buffer without inhibitors and treated with proteinase K in the absence (left) or presence (right) of Triton X-100. Aliquots were removed at the indicated time points and treated with PMSF to inactivate the proteinase. Thereafter, samples were subjected to Western blot analysis using antibodies against the FLAG epitope and HEX-1.
Localization of the PRO40 protein.
Since our biochemical assays indicated a localization of PRO40 within organelles, we further performed microscopic studies using different fluorescence markers. In a first attempt, we constructed plasmid pR-pro40, encoding a PRO40 protein with a C-terminal DsRed fusion under the control of the A. nidulans gpd promoter (57). Transformation of the pro40 mutant with the reporter plasmid resulted in transformants with restored fertility, indicating that the chimeric pro40-DsRed gene is functional in S. macrospora.
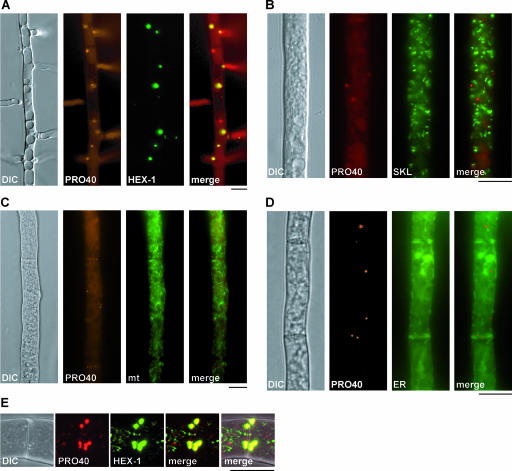
To determine whether PRO40 localizes to any particular cellular compartment, we performed colocalization studies with organellar marker proteins. The gfp gene (78), which has already been used for recombinant constructs in S. macrospora (53), was chosen as a further fluorescence marker. First, we analyzed Woronin bodies labeled with the marker protein HEX-1 (23), which was already used in the Western analysis. Since N. crassa and S. macrospora show a high level of sequence similarity, even on the nucleotide level (46), we were able to use N. crassa HEX-1 in microscopic investigations. In a similar approach, Woronin bodies were recently visualized in Aspergillus oryzae by tagging the N. crassa HEX-1 homolog AoHex1 with DsRed (35). Microscopic analysis revealed a colocalization of PRO40 and HEX-1, but this colocalization was not complete in that some PRO40 dots were not associated with HEX-1 (Fig. 6A). To rule out any cross talk between the two fluorescent signals, we conducted fluorescence microscopy with strains that express either SGFP-HEX-1 or PRO40-DsRed. Similar to Knechtle et al. (26), we used a filter set that provides an optimal signal-to-noise ratio to differentiate green and red fluorescence. In none of the obtained images did we find bleeding through of fluorescent signals (see Fig. S2 in the supplemental material).
FIG. 6.
Colocalization of PRO40 with organelles. The micrographs illustrate the fluorescence of PRO40-DsRed (PRO40), GFP-labeled Woronin bodies (HEX-1), peroxisomes (SKL), mitochondria (mt), and ER. Merged images were assembled from either the fluorescence micrographs only or fluorescence and differential interference contrast (DIC) micrographs. (A) Colocalization of PRO40-DsRed fusion protein and the SGFP-HEX-1 construct in Woronin bodies. (B) PRO40-DsRed is clearly distinguishable from GFP-labeled peroxisomes. (C) Localization of PRO40-DsRed fusion protein and mitochondrion-targeted GFP. (D) Localization of PRO40-DsRed fusion protein and ER-targeted GFP. (E) Detailed localization of SGFP-HEX-1 and PRO40-DsRed at septa. Both proteins show overlapping signals in Woronin bodies by confocal laser scanning microscopy. Bar, 10 μm.
Since N. crassa HEX-1 has been described to be transported to Woronin bodies via peroxisomes (23), we performed microscopic studies with GFP-tagged peroxisomes. For this purpose, we used a fusion of EGFP with a C-terminal SKL tripeptide, a PTS1 signal for peroxisomal matrix import (21, 65). As shown in Fig. 6B, PRO40-DsRed and EGFP-SKL labeled organelles with similar sizes but did not colocalize.
In order to determine any other organellar association, colocalization experiments with mitochondrial and endoplasmic reticulum (ER) markers were performed. First, the single-spore isolate T58.1.2S1, expressing the PRO40-DsRed fusion protein, was transformed with vector pNc-mtGFP (17), encoding mitochondrion-targeted GFP. Mitochondria are visible as tubular structures throughout the hypha that do not overlap with the red dots representing PRO40-DsRed (Fig. 6C). In a second attempt, T58.1.2S1 was transformed with pEGFP-KDEL (42a), which contains the ppg1 signal sequence (38) and encodes EGFP tagged with the C-terminal signal KDEL for retention in the ER (48). Transformants carrying pEGFP-KDEL displayed a green fluorescent network throughout the cell, as previously described for other fungi (13, 34, 75), but did not colocalize with PRO40 (Fig. 6D). The sum of our results demonstrates that PRO40 is most probably located within the Woronin bodies.
Confocal laser microscopy was further used to investigate the detailed localization of PRO40 and HEX-1 in Woronin bodies. Figure 6E shows a typical septum surrounded by these organelles. The SGFP-labeled HEX-1 and DsRed-labeled PRO40 proteins both appear uniformly distributed throughout the Woronin body. However, smaller PRO40-DsRed dots with the size of peroxisomes are visible in the hyphae that do not show overlapping signals with SGFP-HEX-1 (see Movie S3 in the supplemental material).
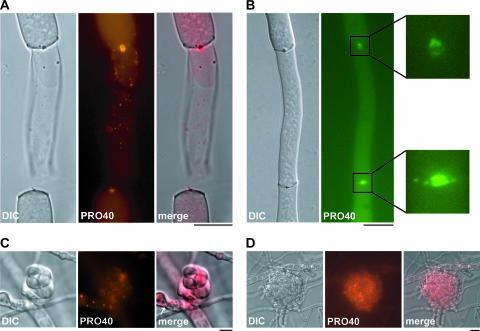
Woronin bodies are specialized organelles of filamentous ascomycetes responsible for plugging septal pores after hyphal injury (23, 67, 72). Thus, we investigated damaged hyphae and found PRO40-DsRed dots in the middle of septa, which is the site of the septal pore (Fig. 7A). The same cellular location was seen when PRO40 was fused to the EGFP autofluorescent protein. The organelle-based fluorescence in injured hyphae accumulated on both sides of the septal plugs (Fig. 7B).
FIG. 7.
Microscopic investigation of PRO40 localization in living hyphae of S. macrospora. The micrographs illustrate the fluorescence of PRO40-DsRed, PRO40-mRFP1, or PRO40-EGFP as well as the corresponding DIC and merged images. (A) Localization of PRO40-DsRed fusion protein in injured hyphae. Note the accumulation of fluorescence at the septal plug. (B) PRO40-EGFP is present at both sides of the septal plug. (Insets) Details of the plug (fourfold enlargement). (C) Localization of PRO40-DsRed fusion protein in ascogonia. The arrow indicates a PRO40-DsRed dot near a septum. (D) Localization of PRO40-mRFP1 fusion protein expressed from the pro40 promoter. Bar, 10 μm.
Since PRO40 is essential for fruiting body development, we were interested in its localization inside sexual structures. Therefore, we analyzed PRO40-DsRed fluorescence in 2- to 5-day-old hyphae of T58.1.2S1 overexpressing PRO40-DsRed. PRO40 indeed showed the same distribution in sexual structures, e.g., ascogonia, as in vegetative hyphae (Fig. 7C). To confirm that our observations were not due to overexpression effects, we performed cellular localization studies with a pro40 construct expressed from its native promoter. Using a recently described approach (77), we tagged PRO40 with mRFP1 (8) by homologous recombination at the pro40 locus, resulting in strain S71343 (see Materials and Methods). As a selectable marker, we used the hph gene expressed from the A. nidulans gpd promoter (57). mRFP1 has already been expressed successfully in filamentous fungi (71) and provides the advantage over DsRed that oligomerization is not a prerequisite for fluorescence (8). Signals obtained with S71343 were very weak and not adapted for colocalization studies. However, our analyses revealed that PRO40-mRFP1 localizes preferentially to fruiting bodies (Fig. 7D), which is consistent with our expression data.
Woronin body and peroxisome integrity in sterile mutants.
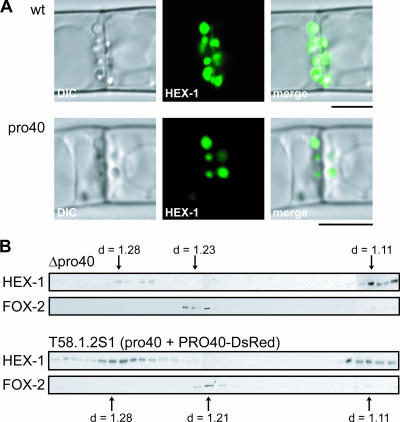
Since PRO40 appears to be associated with Woronin bodies, its absence might affect the integrity of this organelle. To address whether PRO40 is required for Woronin body biogenesis in S. macrospora, we examined the HEX-1 localization in the pro40 mutant background. For this purpose, we generated derivatives of the wild type (T74S64) and the pro40 mutant (I001) expressing the SGFP-HEX-1 fusion protein under control of the N. crassa ccg-1 promoter (16, 40). Using confocal laser microscopy, we observed identical localization of HEX-1 in the mutant and the wild type (Fig. 8A).
FIG. 8.
Peroxisome and Woronin body integrity in pro40 mutants. (A) Confocal laser scanning microscopy of SGFP-HEX-1 in S. macrospora wild type (wt) and the sterile pro40 mutant. Micrographs show DIC, SGFP-HEX-1 fluorescence, and merged images, as indicated. Bar, 5 μm. (B) Subcellular localization of organellar marker proteins in S. macrospora by immunoblot analysis of sucrose density gradient fractions. Cell lysates of oleic acid-induced mycelia of the S. macrospora Δpro40 mutant and T58.1.2S1 (pro40 mutant plus PRO40-DsRed) were layered on top of a 30 to 60% (wt/wt) sucrose density gradient and subjected to isopycnic centrifugation. Fractions were collected from the bottom (left) to the top (right) of the gradient. Equal volumes of each fraction were analyzed by SDS-PAGE and immunoblotting for the presence of Woronin bodies (HEX-1) and glyoxysomes (FOX-2). Densities were measured refractometrically. “d” refers to the sucrose density of the fractions.
Additionally, we analyzed S. macrospora Δpro40 and T58.1.2S1 (pro40 mutant expressing PRO40-DsRed) for the presence of HEX-1-containing Woronin bodies. Oleic acid-induced cells were subjected to subcellular fractionation using sucrose density gradients. Equal portions of each fraction were analyzed by SDS-PAGE and immunoblotting for the presence of Woronin bodies, using an antibody against the N. crassa HEX-1 protein. As mentioned above, S. macrospora and N. crassa show a high degree of sequence similarity (46), and thus an antibody generated against N. crassa HEX-1 also detects the S. macrospora HEX-1 homolog. The S. macrospora HEX-1 protein seems to be slightly larger (about 22 kDa) than HEX-1 of N. crassa (19 kDa) (23). We also detected peroxisomes with an antibody directed against the multifunctional protein FOX-2 (68), as peroxisomes have been discussed to be the origin of Woronin bodies (25, 76). The localization of HEX-1 and FOX-2 was not altered in the deletion strain (Δpro40) in comparison to the complemented mutant (T58.1.2S1), and only slight changes in organellar densities were observed (Fig. 8B). Similar results were obtained for an N. crassa Δso mutant compared with the wild type (data not shown). Strains FGSC11292 (Δso a) and FGSC11293 (Δso A) were obtained from the knockout project within the Neurospora genome project (10). Both strains are fertile as male partners in crosses with the wild type. However, as female partners, they produce only protoperithecia but no mature fruiting bodies and thus are sterile. Moreover, the knockout strains show a reduced formation of aerial hyphae, as already described for the so mutant (15).
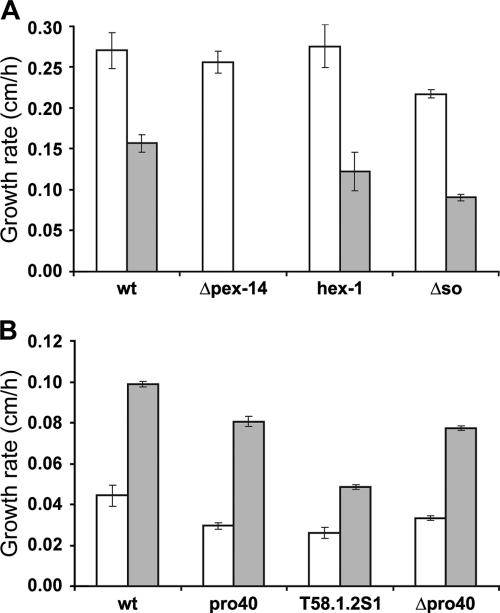
To further confirm that peroxisomal function is not impaired, N. crassa and S. macrospora strains were grown on VMM containing oleic acid solubilized in Tergitol NP-40 as the sole carbon source. The failure to grow on oleic acid has been described to be a sign of defective β-oxidation of fatty acids caused, for example, by a malfunctioning peroxisomal import machinery in yeast (2, 11). We observed only small sparse hyphae on medium containing Tergitol NP-40 alone. Thus, Tergitol cannot be used efficiently as a carbon source by the tested strains (data not shown). N. crassa Δpex-14 and hex-1 strains were tested for the ability to use oleic acid as the sole carbon source as controls for defective peroxisomes and Woronin bodies, respectively. As expected for the Δpex-14 strain, the strain was unable to grow due to a defective peroxisomal import machinery (31a), while the hex-1 strain showed growth on oleic acid as the sole carbon source (Fig. 9). Similarly, growth on oleic acid was observed for the Δso, Δpro40, and pro40 mutants, indicating that peroxisome biogenesis and function are not impaired. This was further confirmed when we compared cytoplasmic bleeding after hyphal injuries. The N. crassa hex-1 deletion strain shows extensive cytoplasmic bleeding (23, 67), while the Δso mutant displays the wild-type phenotype (14; data not shown). Taken together, our data suggest that neither SO nor PRO40 plays a role in peroxisome or Woronin body biogenesis in N. crassa and S. macrospora.
FIG. 9.
Growth of N. crassa and S. macrospora strains on various carbon sources. (A) For the N. crassa strains, 2% sucrose (white bars) or 0.1% oleic acid (gray bars) was chosen as the carbon source. (B) For S. macrospora, VMM supplemented with 2% glucose (white bars) or 0.1% oleic acid (gray bars) was used. Strains were grown in petri dishes (150-mm diameter) for 4 to 10 days, depending on the individual growth velocity. Data are means ± standard deviations for three experiments.
DISCUSSION
PRO40 is associated with Woronin bodies.
During a screening of S. macrospora mutants defective in sexual development, we identified the WW domain protein PRO40. This protein is pivotal in triggering the developmental switch from protoperithecia to perithecia. The WW domain, a short protein module of about 40 amino acids containing two conserved tryptophan residues spaced 20 to 22 amino acids apart (6), is the only detectable protein domain in PRO40.
Interestingly, cellular localization studies using biochemical assays showed a localization of PRO40 within organelles. Fluorescence microscopy further revealed that PRO40 associates with Woronin bodies in S. macrospora. Quite recently, the PRO40 homolog of N. crassa, SO, was described to contribute to the sealing efficiency of pores plugged by Woronin bodies after hyphal injury (14). In filamentous ascomycetes, these peroxisome-like organelles are responsible for an essential morphogenetic program. After damage of the fungal cell, Woronin bodies seal the septa of injured hyphae to prevent the loss of cytoplasm (33). They are typically identified as 150- to 500-nm electron-dense hexagonal structures bound by a single membrane. Core formation relies on the self-assembly of HEX-1, the major protein of the Woronin body, first identified in N. crassa (23, 79).
It remains obscure how PRO40 is targeted to the Woronin body. Early studies suggested a peroxisomal origin of Woronin bodies (25, 76), and thus it can be hypothesized that proteins are first targeted into peroxisomes by peroxisome targeting signals and then redirected to Woronin bodies. This was verified for the N. crassa HEX-1 protein, which contains a PTS1 signal for peroxisomal matrix protein import that is functional in Saccharomyces cerevisiae (21, 23, 65). We were, however, unable to detect any known peroxisomal targeting signal in the PRO40 protein sequence. Furthermore, we did not observe a colocalization with peroxisomes. Thus, PRO40 targeting to Woronin bodies might be rather different from known pathways.
PRO40 is not essential for organelle biogenesis.
In all eukaryotes, cell organelles are essential for distinct cellular differentiation processes. One example is peroxisomes, which carry out steps in lipid metabolism and free radical detoxification and are also involved in processes of development, differentiation, and morphogenesis. Impairment of organelle biogenesis is often responsible for the loss of proper cellular functions, leading to developmental defects (70).
A lack of HEX-1 has been described to lead to impairment of Woronin body biogenesis and results in diverse defects in cellular and developmental processes. For example, for A. oryzae, M. grisea, and N. crassa, hex-1 deletion strains show extensive cytoplasmic bleeding after hyphal lesions (23, 64, 67). In M. grisea, a functional Woronin body is also essential for efficient pathogenicity and survival during nitrogen starvation stress (64).
In contrast to hex-1 deletion strains, the N. crassa Δso mutant lacking the PRO40 homolog SO does not show any signs of increased cytoplasmic bleeding (our data and reference 14). This finding suggests that Woronin bodies are still present and functional. Immunoblot and microscopic analyses of pro40 mutants indeed confirmed the presence of these organelles. However, a loss or defect of one organellar component must not necessarily lead to a complete loss of the organelle but can result in disturbances of distinct morphogenetic programs. In this context, two human single-protein peroxisomal disorders, X-linked adrenoleukodystrophy and Refsum disease, have been described to lead to severe neurodegenerative defects. In both cases, the peroxisomes are of normal abundance and morphology (62, 73). This is reminiscent of the pro40 mutant, which shows wild-type-like Woronin bodies but is impaired in sexual development. Thus, PRO40 might be nonessential for Woronin body biogenesis but may contribute to the proper functioning of the organelle.
PRO40 is essential during the sexual phase.
The pro40 and so strains, from S. macrospora and N. crassa, respectively, are unable to perform sexual development beyond protoperithecium formation. Therefore, both PRO40 and SO are likely to function in the spatial and temporal context of protoperithecial development. Our finding that a PRO40-mRFP1 fusion protein localizes to fruiting bodies supports this idea. However, we cannot prove a colocalization of PRO40 and HEX-1 inside fruiting bodies, and thus, this colocalization might occur only in vegetative hyphae and PRO40 might have a quite different function during sexual development. Support for the idea that PRO40 plays a number of different roles during sexual and vegetative phases comes from results obtained for the N. crassa so mutant. Fleissner et al. (15) previously showed that although hyphal fusions are impaired in the so mutant during vegetative development, wild-type-like fusions still occur after fertilization of ascogonia with microconidia, indicating that hyphal fusion events during sexual development are not dependent on SO.
Since SO localizes not only to pores plugged by Woronin bodies but also to plugs of aging and dying hyphae (14), PRO40 likewise might localize to septal plugs in fruiting bodies in a Woronin body-independent manner. It has been described that septal structure and septal pore occlusion differ depending on developmental stage (reviewed in references 19 and 32). Furthermore, a study of Sordaria humana revealed that rather complex pore structures occur in ascogenous hyphae and croziers (4). Since we sometimes observed a dot-like localization of PRO40-mRFP1 in hyphae from which protoperithecia emerge (our unpublished results), one could speculate that PRO40 accumulates at these specialized plugs and is required to restrict cytoplasmic flow between vegetative and sexual structures.
The characterization of PRO40 as a Woronin body-associated protein required for fungal fertility raises many questions concerning its distinct functions during fungal sexual development. Although the SO WW domain is nonessential for accumulation at septal pores (14), the PRO40 WW domain as well as the C-terminal domain missing in the pro40 mutant may be required for PRO40 function during sexual development. It will therefore be our major aim to identify and characterize interaction partners of PRO40 as a prelude to a better understanding of multicellular development in eukaryotes.
Supplementary Material
Acknowledgments
We thank Ingeborg Godehardt, Regina Ricke, and Susanne Schlewinski for excellent technical assistance, Peter Strauch for his help with some experiments, Eva Szczypka and Gabriele Frenssen-Schenkel for graphical work, and P. Knechtle and P. Philippsen (Basel, Switzerland) for advice on fluorescence microscopy. H.R. thanks Ralf Erdmann (Bochum) for general support. Plasmids were kindly provided by V. Berteaux-Lecellier (Orsay, France; GFP-SKL plasmid), R. Fischer (Karlsruhe, Germany; pMT-mRFP1), M. Freitag (Eugene, OR; pMF272), and B. Westermann (Bayreuth, Germany; pNc-mtGFP).
This work was funded by the Collaborative Research Center SFB480 (projects A1 to U.K. and B10 to H.R.) of the Deutsche Forschungsgemeinschaft (Bonn, Germany). I.E. received a grant from the Studienstiftung des deutschen Volkes (Bonn-Bad Godesberg, Germany).
Footnotes
Published ahead of print on 9 March 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adhvaryu, K. K., S. A. Morris, B. D. Strahl, and E. U. Selker. 2005. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell 4:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertini, M., P. Rehling, R. Erdmann, W. Girzalsky, J. A. K. W. Kiel, M. Veenhuis, and W. H. Kunau. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89:83-92. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Beckett, A. 1981. The ultrastructure of septal pores and associated structures in the ascogenous hyphae and asci of Sordaria humana. Protoplasma 107:127-147. [Google Scholar]
- 5.Bistis, G. N., D. D. Perkins, and N. D. Read. 2003. Different cell types in Neurospora crassa. Fungal Genet. Newsl. 50:17-19. [Google Scholar]
- 6.Bork, P., and M. Sudol. 1994. The WW domain—a signaling site in dystrophin? Trends Biochem. Sci. 19:531-533. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, H. D., T. Schimansky, C. Claudianos, N. Ozsarac, A. B. Kasprzak, J. N. Cotsell, I. G. Young, H. G. Decouet, and G. L. G. Miklos. 1993. The Drosophila melanogaster flightless-I gene involved in gastrulation and muscle degeneration encodes gelsolin-like and leucine-rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc. Natl. Acad. Sci. USA 90:11386-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H. I., and M. Sudol. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 92:7819-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103:10352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdmann, R., M. Veenhuis, D. Mertens, and W. H. Kunau. 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86:5419-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esser, K. 1982. Cryptogams—cyanobacteria, algae, fungi, lichens. Cambridge University Press, London, England.
- 13.Fernandez-Abalos, J. M., H. Fox, C. Pitt, B. Wells, and J. H. Doonan. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121-130. [DOI] [PubMed] [Google Scholar]
- 14.Fleissner, A., and N. L. Glass. 2007. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 6:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleissner, A., S. Sarkar, D. J. Jacobson, M. G. Roca, N. D. Read, and N. L. Glass. 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4:920-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker, and N. D. Read. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897-910. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, F., H. Prokisch, W. Neupert, and B. Westermann. 2002. Interaction of mitochondria with microtubules in the filamentous fungus Neurospora crassa. J. Cell Sci. 115:1931-1937. [DOI] [PubMed] [Google Scholar]
- 18.Fujimori, F., K. Takahashi, C. Uchida, and T. Uchida. 1999. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem. Biophys. Res. Commun. 265:658-663. [DOI] [PubMed] [Google Scholar]
- 19.Gull, K. 1978. Form and function of septa in filamentous fungi, p. 78-93. In J. E. Smith and D. R. Berry (ed.), The filamentous fungi. III. Developmental mycology. J. Wiley and Sons, New York, NY.
- 20.Hanes, S. D., P. R. Shank, and K. A. Bostian. 1989. Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast 5:55-72. [DOI] [PubMed] [Google Scholar]
- 21.Hettema, E. H., B. Distel, and H. F. Tabak. 1999. Import of proteins into peroxisomes. Biochim. Biophys. Acta 1451:17-34. [DOI] [PubMed] [Google Scholar]
- 22.Hoge, J. H. C., J. Springer, B. Zantinge, and J. G. H. Wessels. 1982. Absence of differences in polysomal RNA from vegetative monokaryotic and dikaryotic cells of the fungus Schizophyllum commune. Exp. Mycol. 6:225-232. [Google Scholar]
- 23.Jedd, G., and N. H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226-231. [DOI] [PubMed] [Google Scholar]
- 24.Jerpseth, B., A. Greener, J. M. Short, J. Viola, and P. L. Kretz. 1992. XL1-Blue MRF′ E. coli cells: McrA−, McrCB−, McrF−, Mmr−, HsdR− derivative of XL1-Blue cells. Strateg. Mol. Biol. 5:81-83. [Google Scholar]
- 25.Keller, G. A., S. Krisans, S. J. Gould, J. M. Sommer, C. C. Wang, W. Schliebs, W. Kunau, S. Brody, and S. Subramani. 1991. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J. Cell Biol. 114:893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knechtle, P., F. Dietrich, and P. Philippsen. 2003. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14:4140-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kück, U. 2005. A Sordaria macrospora mutant lacking the leu1 gene shows a developmental arrest during fruiting body formation. Mol. Genet. Genomics 274:307-315. [DOI] [PubMed] [Google Scholar]
- 28.Kück, U., and B. Hoff. 2006. Application of the nourseothricin acetyltransferase gene (nat1) as dominant marker for the transformation of filamentous fungi. Fungal Genet. Newsl. 53:9-11. [Google Scholar]
- 29.Kück, U., and S. Pöggeler. 2004. pZHK2, a bi-functional transformation vector, suitable for two step gene targeting. Fungal Genet. Newsl. 51:4-6. [Google Scholar]
- 30.Lu, K. P., S. D. Hanes, and T. Hunter. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544-547. [DOI] [PubMed] [Google Scholar]
- 31.Maleszka, R., S. D. Hanes, R. L. Hackett, H. G. de Couet, and G. L. G. Miklos. 1996. The Drosophila melanogaster dodo (dod) gene, conserved in humans, is functionally interchangeable with the ESS1 cell division gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:447-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Managadze, D., C. Würtz, M. Sichting, G. Niehaus, M. Veenhws, and H. Rottensteiner. 2007. The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glycosomes and Woronin bodies. Traffic doi: 10.1111/j.1600-0854.2006.00560.x. [DOI] [PubMed]
- 32.Markham, P. 1994. Occlusions of septal pores in filamentous fungi. Mycol. Res. 98:1089-1106. [Google Scholar]
- 33.Markham, P., and A. J. Collinge. 1987. Woronin bodies of filamentous fungi. FEMS Microbiol. Rev. 46:1-11. [Google Scholar]
- 34.Maruyama, J., S. Kikuchi, and K. Kitamoto. 2006. Differential distribution of the endoplasmic reticulum network as visualized by the BipA-EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet. Biol. 43:642-654. [DOI] [PubMed] [Google Scholar]
- 35.Maruyama, J. I., P. R. Juvvadi, K. Ishi, and K. Kitamoto. 2005. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 331:1081-1088. [DOI] [PubMed] [Google Scholar]
- 36.Masloff, S., S. Jacobsen, S. Pöggeler, and U. Kück. 2002. Functional analysis of the C-6 zinc finger gene pro1 involved in fungal sexual development. Fungal Genet. Biol. 36:107-116. [DOI] [PubMed] [Google Scholar]
- 37.Masloff, S., S. Pöggeler, and U. Kück. 1999. The pro1(+) gene from Sordaria macrospora encodes a C-6 zinc finger transcription factor required for fruiting body development. Genetics 152:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayrhofer, S., and S. Pöggeler. 2005. Functional characterization of an alpha-factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with its cognate receptor in Saccharomyces cerevisiae. Eukaryot. Cell 4:661-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayrhofer, S., J. M. Weber, and S. Pöggeler. 2006. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics 172:1521-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNally, M. T., and S. J. Free. 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 14:545-551. [DOI] [PubMed] [Google Scholar]
- 41.Mokranjac, D., S. A. Paschen, C. Kozany, H. Prokisch, S. C. Hoppins, F. E. Nargang, W. Neupert, and K. Hell. 2003. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22:816-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowrousian, M., and P. Cebula. 2005. The gene for a lectin-like protein is transcriptionally activated during sexual development, but is not essential for fruiting body formation in the filamentous fungus Sordaria macrospora. BMC Microbiol. 5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Nowrousian, M., S. Frank, S. Koers, P. Strauch, T. Weitner, C. Ringelberg, J. C. Dunlap, J. J. Loros, and V. Kück. The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 43.Nowrousian, M., S. Masloff, S. Pöggeler, and U. Kück. 1999. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol. Cell. Biol. 19:450-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowrousian, M., M. Piotrowski, and U. Kück. 6 November 2006, posting date. Multiple layers of temporal and spatial control regulate accumulation of the fruiting body-specific protein APP in Sordaria macrospora and Neurospora crassa. Fungal Genet. Biol. doi: 10.1016/j.fgb.2006.09.009. [DOI] [PubMed]
- 45.Nowrousian, M., C. Ringelberg, J. C. Dunlap, J. L. Loros, and U. Kück. 2005. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genomics 273:137-149. [DOI] [PubMed] [Google Scholar]
- 46.Nowrousian, M., C. Würtz, S. Pöggeler, and U. Kück. 2004. Comparative sequence analysis of Sordaria macrospora and Neurospora crassa as a means to improve genome annotation. Fungal Genet. Biol. 41:285-292. [DOI] [PubMed] [Google Scholar]
- 47.Osiewacz, H. D. 1994. A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr. Genet. 26:87-90. [DOI] [PubMed] [Google Scholar]
- 48.Pelham, H. R. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15:483-486. [DOI] [PubMed] [Google Scholar]
- 49.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pöggeler, S., and U. Kück. 2006. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378:1-10. [DOI] [PubMed] [Google Scholar]
- 52.Pöggeler, S., and U. Kück. 2004. A WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryot. Cell 3:232-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pöggeler, S., S. Masloff, B. Hoff, S. Mayrhofer, and U. Kück. 2003. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43:54-61. [DOI] [PubMed] [Google Scholar]
- 54.Pöggeler, S., M. Nowrousian, S. Jacobsen, and U. Kück. 1997. An efficient procedure to isolate fungal genes from an indexed cosmid library. J. Microbiol. Methods 29:49-61. [Google Scholar]
- 55.Pöggeler, S., M. Nowrousian, and U. Kück. 2006. Fruiting-body development in ascomycetes, p. 325-355. In U. Kües and R. Fischer (ed.), The Mycota I. Springer-Verlag, Berlin, Germany.
- 56.Pöggeler, S., M. Nowrousian, C. Ringelberg, J. J. Loros, J. C. Dunlap, and U. Kück. 2006. Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genomics 275:492-503. [DOI] [PubMed] [Google Scholar]
- 57.Punt, P. J., M. A. Dingemanse, A. Kuyvenhoven, R. D. Soede, P. H. Pouwels, and C. A. van den Hondel. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101-109. [DOI] [PubMed] [Google Scholar]
- 58.Ruprich-Robert, G., V. Berteaux-Lecellier, D. Zickler, A. Panvier-Adoutte, and M. Picard. 2002. Identification of six loci in which mutations partially restore peroxisome biogenesis and/or alleviate the metabolic defect of pex2 mutants in Podospora. Genetics 161:1089-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 60.Schliebs, W., C. Würtz, W. H. Kunau, M. Veenhuis, and H. Rottensteiner. 2006. A eukaryote without catalase-containing microbodies: Neurospora crassa exhibits a unique cellular distribution of its four catalases. Eukaryot. Cell 5:1490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott, W. A. 1977. Unsaturated fatty acid mutants of Neurospora crassa. J. Bacteriol. 130:1144-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, K. D., S. Kemp, L. T. Braiterman, J. F. Lu, H. M. Wei, M. Geraghty, G. Stetten, J. S. Bergin, J. Pevsner, and P. A. Watkins. 1999. X-linked adrenoleukodystrophy: genes, mutations, and phenotypes. Neurochem. Res. 24:521-535. [DOI] [PubMed] [Google Scholar]
- 63.Sokolovsky, V., R. Kaldenhoff, M. Ricci, and V. E. A. Russo. 1990. Fast and reliable mini-prep RNA extraction from Neurospora crassa. Fungal Genet. Newsl. 37:41. [Google Scholar]
- 64.Soundararajan, S., G. Jedd, X. L. Li, M. Ramos-Pamplona, N. H. Chua, and N. I. Naqvi. 2004. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 16:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramani, S. 1993. Protein import into peroxisomes and biogenesis of the organelle. Annu. Rev. Cell Biol. 9:445-478. [DOI] [PubMed] [Google Scholar]
- 66.Sudol, M., and T. Hunter. 2000. NeW wrinkles for an old domain. Cell 103:1001-1004. [DOI] [PubMed] [Google Scholar]
- 67.Tenney, K., I. Hunt, J. Sweigard, J. I. Pounder, C. McClain, E. J. Bowman, and B. J. Bowman. 2000. Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31:205-217. [DOI] [PubMed] [Google Scholar]
- 68.Thieringer, R., and W. H. Kunau. 1991. The beta-oxidation system in catalase-free microbodies of the filamentous fungus Neurospora crassa. Purification of a multifunctional protein possessing 2-enoyl-CoA hydratase, l-3-hydroxyacyl-CoA dehydrogenase, and 3-hydroxyacyl-CoA epimerase activities. J. Biol. Chem. 266:13110-13117. [PubMed] [Google Scholar]
- 69.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Titorenko, V. I., and R. A. Rachubinski. 2004. The peroxisome: orchestrating important developmental decisions from inside the cell. J. Cell Biol. 164:641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toews, M. W., J. Warmbold, S. Konzack, P. Rischitor, D. Veith, K. Vienken, C. Vinuesa, H. Wei, and R. Fischer. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383-389. [DOI] [PubMed] [Google Scholar]
- 72.Trinci, A. P., and A. J. Collinge. 1974. Occlusion of the septal pores of damaged hyphae of Neurospora crassa by hexagonal crystals. Protoplasma 80:57-67. [DOI] [PubMed] [Google Scholar]
- 73.van den Brink, D. M., and R. J. Wanders. 2006. Phytanic acid: production from phytol, its breakdown and role in human disease. Cell Mol. Life Sci. 63:1752-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogel, H. J. 1956. A convenient growth medium for Neurospora (medium N). Microb. Genet. Bull. 13:42-43. [Google Scholar]
- 75.Wedlich-Söldner, R., I. Schulz, A. Straube, and G. Steinberg. 2002. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 13:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wergin, W. P. 1973. Development of Woronin bodies from microbodies in Fusarium oxysporum f.sp. lycopersici. Protoplasma 76:249-260. [DOI] [PubMed] [Google Scholar]
- 77.Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies, C. P. C. De Souza, X. W. Dou, A. Perez-Balaguer, and S. A. Osmani. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, T. T., L. Cheng, and S. R. Kain. 1996. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 24:4592-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan, P., G. Jedd, D. Kumaran, S. Swaminathan, H. Shio, D. Hewitt, N. H. Chua, and K. Swaminathan. 2003. A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 10:264-270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.