Abstract
The study objectives were to determine the prevalence and serotypes of non-O157 Shiga toxin-producing Escherichia coli (STEC) in pens of feedlot cattle and on corresponding beef carcasses. We collected 25 fecal samples from 84 pens in 21 Alberta feedlots and 40 carcass swabs from each preslaughter pen for analysis by culture and polymerase chain reaction (PCR). Non-O157 STEC were recovered from feces from 12 (14%) of the 84 pens and 12 (57%) of the 21 feedlots by examination of 1 E. coli isolate positive for 4-methylumbelliferyl-β-d-glucuronide per sample. Twelve non-O157 serotypes were detected, but 7 of the 15 STEC isolates lacked the accessory virulence genes eae and hlyA. Although 115 (7%) of the carcass broths were PCR-positive, no STEC isolates were recovered from the 1650 carcasses sampled. Our data indicate that multiple non-O157 STEC serotypes may be present in cattle feces, yet are unlikely to be recovered from the corresponding beef carcasses when 20 colonies per sample from PCR-positive broth cultures are analyzed.
Résumé
Cette étude avait comme objectifs de déterminer la prévalence et les sérotypes des isolats d’Escherichia coli non-O157 producteurs de toxine Shiga (STEC) retrouvés dans les enclos de bovins d’embouche et sur les carcasses correspondantes. Vingt-cinq échantillons de fèces ont été ramassés dans 84 enclos de 21 parcs d’engraissement en Alberta et 40 écouvillonnages de carcasse effectués pour chaque enclos d’attente pré-abattage afin de procéder à une analyse par culture et réaction d’amplification en chaîne par la polymérase (PCR). Des STEC non-O157 ont été isolés à partir des fèces provenant de 12 (14 %) des 84 enclos et 12 (57 %) des 21 parcs d’engraissement par examen de 1 isolat par échantillon d’E. coli positif pour le test du 4-méthylumbelliferyl-β-d-glucuronide. Douze sérotypes non-O157 ont été détectés, mais chez 7 des 15 isolats de STEC les gènes de virulence accessoires eae et hlyA étaient manquants. Bien que 115 (7 %) des bouillons provenant des carcasses étaient positifs par PCR, aucun isolat de STEC ne fut récupéré des 1650 carcasses échantillonnées. Nos résultats indiquent que de nombreux isolats de STEC non-O157 peuvent être présents dans les fèces de bovins, mais qu’ils sont rarement isolés des carcasses correspondantes lorsque 20 colonies par échantillon provenant de bouillons positifs par PCR sont analysées.
(Traduit par Docteur Serge Messier)
Shiga toxin-producing Escherichia coli (STEC), also referred to as shigatoxigenic or verotoxigenic E. coli (VTEC), are important agents of human foodborne disease worldwide (1). Human infections are often asymptomatic or result in uncomplicated diarrhea, but they may progress to hemorrhagic colitis, hemolytic–uremic syndrome, and death. Although all strains of STEC by definition produce Shiga toxins (Stx), this diverse group of E. coli has other important virulence factors, and more than 100 serotypes are associated with human disease (1). In North America, O157:H7 is the most important STEC serotype, but other STEC serotypes cause disease to a limited extent in North America and to a much greater extent in other parts of the world. Since national surveillance programs tend to focus on O157:H7, the incidence of cases associated with non-O157 STEC is often impossible to estimate (1). However, an Alberta study showed that testing for free toxin rather than just culturing for serotype O157: H7 would increase the diagnosis of STEC 3-fold in clinical specimens from humans (2).
The feces of animals, particularly healthy cattle, are considered the main source of STEC. The consumption of food and water contaminated with feces and direct contact with animal feces are primary routes of human infection (1). Although E. coli O157:H7 has been studied extensively in cattle, there have been few studies of non-O157 STEC in North American cattle. A recent study in the United States indicated that approximately 30% of cattle were shedding STEC in their feces (3). Another US study indicated a similar prevalence in cattle and showed that the prevalence of non-O157 STEC on postintervention carcasses approached 9% (4). A previous study in Alberta indicated evidence of serotypes O157, O111, and O26 in feedlots but was limited to few feedlots and did not investigate carcasses (5). The beef industry has made significant changes in the processing environment in order to reduce beef contamination and improve beef safety. Feces and the hides of cattle are important sources of contamination during processing, and there is a correlation between preharvest fecal and hide prevalence of E. coli O157:H7 and carcass contamination (6). Although preharvest interventions could potentially reduce carcass contamination, no preharvest interventions for STEC are proven to improve beef safety. The objectives of this study were to estimate the pen prevalence and serotypes of non-O157 STEC in Alberta feedlots and the prevalence of STEC on corresponding beef carcasses in federally inspected abattoirs.
This study used a subset of the samples collected for a larger study investigating antimicrobial resistance and potential foodborne pathogens in Alberta feedlots and beef carcasses. Of relevance to this study were feedlot fecal samples and corresponding carcass swabs collected in coolers of federally inspected Alberta abattoirs. CanFax, a service providing market information on the Canadian beef industry (www.canfax.ca/default.htm), was used to identify feedlots. To ensure that appropriate pens would be available at the time of sampling, only feedlots with a capacity of more than 5000 head were considered. All Alberta feedlots of this size in the CanFax database were eligible for sampling and were stratified by capacity: 5001 to 10 000, 10 001 to 15 000, 15 001 to 20 000, or more than 20 000 head. The number of feedlots randomly selected per stratum was weighted by the number of feedlots in each stratum. CanFax maintained the list of eligible feedlots to ensure anonymity and contacted the selected feedlots to identify 21 that were willing to participate. Fecal samples were collected over 2 sampling periods: period 1, March through June 2004; and period 2, September through December 2004. At each feedlot visit, the pen shortest on feed (SOF) and the pen closest to slaughter (preslaughter [PS]) were sampled. When the animals in PS pens were slaughtered, samples were later collected from carcasses in the abattoir coolers.
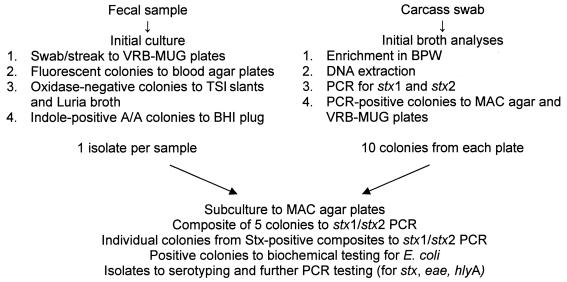
From each pen, 25 samples (about 50 g each) of freshly voided feces were collected from throughout the pen floor and placed in sterile bags. Samples were sent on ice in coolers by overnight courier to Washington State University for culture of generic E. coli. Sterile swabs were used to inoculate feces (~ 0.1 g) onto 90-mm plates containing violet red bile agar containing 50 mg/L of 4-methylumbelliferyl-β-d-glucuronide (VRB-MUG; Hardy Diagnostics, Santa Maria, California, USA), which were then streaked for isolation and incubated overnight at 37°C. Up to 3 purple/red colonies that were fluorescent under ultraviolet illumination (312 nm) were picked as presumptive E. coli, streaked for purity on a Columbia blood agar plate, and incubated overnight at 37°C. Oxidase-negative colonies were inoculated into triple sugar iron (TSI) slants and Luria broth (Hardy Diagnostics) and incubated overnight at 37°C. Colonies that were indole-positive and had an acid/acid reaction in TSI were inoculated into brain–heart infusion (Becton, Dickinson, Sparks, Maryland, USA) agar plugs, incubated overnight at 37°C, and stored. One E. coli isolate per sample was sent by courier to the Agri-Food Laboratories Branch in Edmonton, Alberta, for further testing (Figure 1).
Figure 1.
Outline of procedures for identifying Shiga toxin-producing Escherichia coli in fecal samples and carcass swabs. VRB-MUG — violet red bile agar containing 50 mg/L of 4-methylumbelliferyl-β-d-glucuronide; TSI — triple sugar iron; A/A — acid/acid-reacting; BHI — brain–heart infusion; BPW — buffered peptone water; PCR — polymerase chain reaction; stx1 — Shiga toxin 1; stx2 — Shiga toxin 2; MAC — MacConkey.
Arrangements were made with processors to collect carcass samples in the cooler according to their hazard analysis and critical control point plans. Cattle from the PS pens in participating feedlots were those tested in the plant. Plant personnel pulled carcasses of animals from each of the PS pens during each period onto a cooler line for sampling. When available, samples from up to 40 carcasses from each study pen were tested for STEC. Carcasses were sampled by following US Department of Agriculture/Food Safety Inspection Service procedures (7). In brief, a single sterile premoistened sponge was used to sample 3 sites (flank, brisket, and rump) on each carcass side (approximately 300 cm2 in total). Commercial carcass-sampling kits (Qualicum Scientific, Ottawa, Ontario) were used according to the manufacturer’s instructions. Once the sites were swabbed, the sponge was placed in a sterile bag, and 15 mL of sterile buffered peptone water (BPW) was added. The bags were immediately placed in a cooler containing ice packs and sent by courier to the Agri-Food Laboratories Branch in Edmonton for analysis (Figure 1).
At the laboratory, an additional 40 mL of BPW was added to each carcass sample by means of a Dilumat Dispenser (AES Laboratories, Combourg, France) to bring the total volume to 60 mL. The samples were mixed by vigorous shaking and incubated at 35°C for 24 h, with the sponges completely immersed in broth. Positive and negative controls were included with each batch of samples. Positive control samples consisted of a sponge immersed in 60 mL of BPW inoculated with 1 to 24 colony-forming units of E. coli O157:H7 (American Type Culture Collection 35150). After incubation, 300 μL of the BPW was transferred to a 1.5-mL tube containing 700 μL of sterile purified water. The positive control also was processed to monitor the DNA extraction procedure. After vortexing, the tube was centrifuged at 18 000 × g for 5 min and the supernatant discarded. The pellet was resuspended in 100 μL of sterile purified water.
To extract DNA, we used a Magnesil KF Genomic System kit (Promega, Madison, Wisconsin, USA) and a KingFisher (KF) ML semiautomated magnetic particle processor (Thermo Electron Corporation, Vantaa, Finland). Briefly, 700 μL of lysis buffer and 200 μL of magnetic beads were added to well 1 of the KF strip tube, 1 mL of salt wash was added to well 2, 1 mL of ethanol wash was added to wells 3 and 4 (reagents supplied in kits), and 200 μL of sterile purified water was added to well 5 for elution. The resuspended pellet was added to well 1, and the strip tube was placed in the KF processor. The processor was run using the ML program supplied in the manufacturer’s software. After extraction was complete, either the DNA was transferred to a 1.5-mL tube and a polymerase chain reaction (PCR) run immediately, or else the DNA was stored at 2°C to 8°C for no longer than 5 d before analysis.
Extracted DNA was tested for the presence of the genes for Shiga toxin 1 (stx1) and Shiga toxin 2 (stx2) with the use of primers in a multiplex assay (8). Master mix (reagents from Invitrogen, Burlington, Ontario) was prepared to provide the following concentrations in a 50-μL reaction volume: 1X PCR buffer, 200 μM of each deoxyribonucleotide triphosphate, 2.5 mM of MgCl2, 1 μM of stx1 primers, 0.5 μM of stx2 primers, 0.02 U/μL of platinum Taq polymerase, and 0.10 mg/mL of bovine serum albumin. An internal control, designed in-house to amplify with the stx1 primers and titrated so as not to interfere with amplification of the target DNA, also was added, to a final concentration of 8 pg/mL, to monitor for inhibitors. The master mix was dispensed in 40-μL aliquots and stored at −20°C. A positive control of genomic DNA from an E. coli O157:H7 field isolate and a negative water control were included in each assay. Template DNA, 10 μL, was added to 40 μL of master mix, and the reaction tubes were placed in an MJ Research DNA Engine thermal cycler (BioRad, Mississauga, Ontario). An initial heating step at 94°C for 5 min was followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min. A final extension step of 72°C for 5 min was followed by a hold step at 10°C.
Amplified product was detected by electrophoresis on a 1.2% agarose gel with use of a 100-base pair (bp) standard (Invitrogen) as a size marker and visualized by ethidium-bromide staining. Images were captured with a DC290 digital camera and Kodak 1D image analysis software (Kodak, Rochester, New York, USA). Samples with a band (or bands) of 614 or 779 bp were considered positive. With 10-μL disposable inoculation loops, PCR-positive broths were streaked onto MacConkey agar (MAC) and VRB-MUG plates, which were then incubated at 42°C for 24 h. After incubation, 10 isolated colonies from each of the MAC and VRB-MUG plates were transferred to MAC plates, which were divided so that there were 5 colonies per row and 4 rows per plate. The plates were incubated at 35°C for 24 h. Each row of 5 colonies was grouped into a composite for PCR testing.
Isolates of MUG-positive E. coli (presumptively non-O157) from the sampled feces were received from Washington State University on agar slants, plated similarly onto MAC plates, and incubated at 35°C for 24 h. Each row of 5 colonies was grouped into a composite for PCR testing. For DNA extraction, these composites were picked and emulsified in 200 μL of Prepman Ultra reagent (Applied Biosystems, Foster City, California, USA). After the samples were boiled for 10 min and then allowed to cool to room temperature, the tubes were spun at 18 000 × g for 5 min. Supernatant, 50 μL, was added to 450 μL of 12 mM Tris buffer, pH 8.0. The diluted samples were tested by PCR for stx1 and stx2 genes as previously described. Individual colonies from Stx-positive composites were picked to 500 μL of 12 mM Tris buffer, boiled for 10 min, and then immediately chilled in a chiller block. Testing for stx1 and stx2 genes was again performed. Colonies positive for either gene (or both) were streaked onto blood agar plates and incubated at 35°C for 24 h to confirm purity. Pure cultures were verified as E. coli by means of biochemical tests (API 32E; bioMerieux Canada, St. Laurent, Quebec). Isolates also were tested for E. coli attaching and effacing genes (eaeO111 eaeO26) by PCR (9) and were inoculated into sterile sheep blood and and stored at −70°C. Later, isolates were sent to the Public Health Agency of Canada for serotyping and PCR testing for the stx, eae, and enterohemorrhagic E. coli (EHEC) hemolysin (hlyA) genes at the Laboratory for Foodborne Zoonoses, Guelph, Ontario.
Prevalence proportions and 95% exact binomial confidence intervals (CIs) for proportions were calculated in SAS (version 9.1, Cary, North Carolina, USA). Fisher’s exact tests were used to investigate homogeneity of proportions, and a P-value of 0.05 or less was used for hypothesis testing (SAS).
Of the 21 commercial feedlots, located throughout Alberta, that were studied, 11 had a capacity of 5001 to 10 000 head, 4 a capacity of 10 001 to 15 000 head, 3 a capacity of 15 001 to 20 000 head, and 3 a capacity of more than 20 000 head. Approximate pen size averaged 250 head and ranged from 150 to 500 head. Feedlots included both calf and yearling lots, and all feedlots fed cattle until ready for slaughter. We studied 84 pens: 42 in each period and 21 of each pen type (SOF and PS) in each period. Of the 2099 fecal samples analyzed, 25 were from 83 pens and 24 from 1 pen.
Of the 2099 fecal samples, E. coli was cultured from 2068, and 1925 viable cultures were available for Stx testing. There were 207 composites of isolates for each of the 2 sampling periods: 9 (4%) of the period-1 composites and 6 (3%) of the period-2 composites, for a total of 15 (4%), were positive for Stx genes, 3 for stx1 only, 9 for stx2 only, and 3 for both genes. The 15 positive composites consisted of E. coli isolates from 63 individual samples. Shiga toxin genes were detected in at least 1 individual isolate from all 15 positive composites and in 15 (24%) of the 63 individual isolates. Of the 1925 E. coli isolates available for testing, 15 (0.8%) were STEC; 3 (20%) of the STEC isolates were positive for stx1 only, 11 (73%) were positive for stx2 only, and 1 (7%) was positive for both genes. One of the STEC isolates was positive for eaeO111; none were positive for eaeO26. One STEC isolate (stx2+, eaeO111+) could not be recovered after storage; PCR results for the other 14 isolates were confirmed by the Public Health Agency of Canada, and serotyping was done.
Non-O157 STEC isolates were recovered from 12 (14%) of the 84 pens and 12 (57%) of the 21 feedlots studied — from 6 (14%) of the 42 pens in each period (CI 5% to 28%) and from 8 (19%) of the 42 SOF pens (CI 9% to 34%) versus 4 (10%) of the 42 PS pens (CI 3% to 23%). The pen prevalence was not significantly different between periods or between SOF pens and PS pens. Multiple serotypes were recovered from 2 pens in 2 different feedlots: 2 O2:H27 isolates and 1 O111 isolate from an SOF pen in March, and 1 isolate each of O113:H21 and O?:H29 from an SOF pen in April (Table I). In no feedlot were non-O157 STEC recovered from more than 1 pen.
Table I.
Serotype, virulence genes, and sources of non-O157 Shiga toxin-producing Escherichia coli (STEC) recovered from feces of cattle sampled from 84 pens in 21 Alberta feedlots in 2004
| Virulence genes
|
|||||||
|---|---|---|---|---|---|---|---|
| Serotype | stx1 | stx2 | hlyA | eae | No. of pensa | No. of feedlotsa | Month (no. of isolates, pen typeb) |
| O2:H27 | − | + | + | − | 2 | 2 | March (2, SOF)
November (1, PS) |
| O113:H21 | − | + | − | − | 2 | 2 | April (1, SOF)
December (1, SOF) |
| O165:H25 | − | + | − | + | 1 | 1 | April (1, SOF) |
| O?:NM | + | − | + | − | 1 | 1 | April (1, SOF) |
| O?:H5 | + | − | − | − | 1 | 1 | March (1, PS) |
| O?:H29 | − | + | − | − | 1 | 1 | April (1, SOF) |
| O145:NM | − | + | + | + | 1 | 1 | June (1, PS) |
| O15:H16 | − | + | − | − | 1 | 1 | September (1, SOF) |
| O51:NM | + | − | − | − | 1 | 1 | November (1, SOF) |
| O?:H16 | − | + | − | − | 1 | 1 | October (1, PS) |
| O139:H19 | + | + | + | − | 1 | 1 | December (1, SOF) |
| (O111) | − | + | NA | + | 1 | 1 | March (1, SOF) |
NA — not available: the isolate could not be recovered for serotyping and hlyA testing after storage but had been positive for the stx2 and eaeO111 genes.
Multiple isolates were recovered from 2 pens in 2 different feedlots.
At each feedlot visit, the pen shortest on feed (SOF) and the pen closest to slaughter (within 2 wk preslaughter [PS]) were sampled.
Of the 1650 carcass swab samples analyzed for STEC, 832 were collected during period 1 (March through June 2004) and 818 during period 2 (September 2004 through January 2005). Initial PCR screening of enrichment broths showed that 115 (7.0%) were positive for Stx genes, 49 (43%) for stx1 only, 63 (55%) for stx2 only, and 3 (3%) for both genes. Of the 832 broths in period 1, 82 (10%) were positive (CI 8% to 12%); of the 818 broths in period 2, 33 (4%) were positive (CI 3% to 6%). For 15 of the 42 pens, all of the broths were negative; in the other 27 pens the proportion of positive broths ranged from 2% to 50% (Table II). The proportion of positive broths did not differ significantly on the basis of whether non-O157 STEC were recovered from PS fecal samples. Viable bacteria that were presumptively considered E. coli were cultured from only 65 of the 115 positive broths, and only 10 of the 65 isolate composites were positive for Stx genes. Although isolates of E. coli, Serratia fonticola, and Enterobacter cloacaea were recovered, none were positive for stx or eae or for E. coli on the basis of biochemical analysis. Therefore, STEC were not recovered from any carcass sample.
Table II.
Results of initial STEC testing of carcasses from the 42 PS pens
| Feedlot | Perioda | Total no. of samples | No. (and %) STEC-positiveb |
|---|---|---|---|
| A | 1 | 40 | 6 (15) |
| A | 2 | 40 | 0 (0) |
| B | 1 | 40 | 0 (0) |
| B | 2 | 40 | 0 (0) |
| Cc | 1 | 35 | 1 (3) |
| C | 2 | 40 | 0 (0) |
| D | 1 | 40 | 9 (22) |
| D | 2 | 40 | 2 (5) |
| E | 1 | 40 | 0 (0) |
| E | 2 | 40 | 0 (0) |
| F | 1 | 40 | 6 (15) |
| F | 2 | 40 | 4 (10) |
| G | 1 | 40 | 1 (2) |
| G | 2 | 40 | 1 (2) |
| Hc | 1 | 39 | 0 (0) |
| H | 2 | 40 | 3 (7) |
| I | 1 | 40 | 1 (2) |
| I | 2 | 35 | 1 (3) |
| J | 1 | 40 | 1 (2) |
| J | 2 | 40 | 10 (25) |
| K | 1 | 39 | 0 (0) |
| K | 2 | 40 | 2 (5) |
| L | 1 | 40 | 18 (45) |
| L | 2 | 40 | 0 (0) |
| M | 1 | 40 | 4 (10) |
| Mc | 2 | 40 | 3 (7) |
| N | 1 | 40 | 2 (5) |
| N | 2 | 40 | 0 (0) |
| O | 1 | 40 | 0 (0) |
| O | 2 | 40 | 1 (2) |
| P | 1 | 39 | 0 (0) |
| P | 2 | 40 | 0 (0) |
| Q | 1 | 40 | 0 (0) |
| Q | 2 | 40 | 3 (8) |
| R | 1 | 40 | 20 (50) |
| R | 2 | 23 | 1 (4) |
| S | 1 | 40 | 1 (3) |
| Sc | 2 | 40 | 0 (0) |
| T | 1 | 40 | 8 (20) |
| T | 2 | 40 | 1 (3) |
| U | 1 | 40 | 4 (10) |
| U | 2 | 40 | 1 (3) |
| Total | 1650 | 115 (7) |
For carcass sampling, period 1 was March through June 2004, and period 2 was September 2004 through January 2005.
Presumptively positive according to the results of testing with polymerase chain reaction; no STEC isolates were later recovered from any sample.
Non-O157 STEC were recovered from fecal samples collected from the PS pen, as shown in Table I.
We found that multiple non-O157 STEC strains could be detected in feces from feedlot cattle, but none were recovered from the corresponding carcasses by the methods we used. Although the number of E. coli fecal isolates tested was limited to 1 per sample and 25 per pen, we found STEC in 14% of the pens and 57% of the feedlots. The diversity in serotypes and virulence genes among the non-O157 STEC recovered from the feces is not unusual and may represent potential variability in the risk to human health (3). Despite PCR evidence of STEC in approximately 7% of the carcass samples, no isolates of STEC were recovered from the samples through analysis of 20 colonies from the PCR-positive broth cultures. This is perhaps not surprising given the relatively low preharvest prevalence of non-O157 STEC, the reported effectiveness of postharvest interventions, and the extremely low prevalence and numbers of STEC previously reported for carcasses (1,4,6,10).
This study was limited to 84 pens in 21 Alberta feedlots, and the selection of this study population was not completely random. However, the participants were targeted so as to have a representative study population reflecting the majority of the Alberta beef industry. The sampling and diagnostic strategies were developed in-house for recovering STEC from cattle populations or carcass samples, but validated estimates of diagnostic (field) sensitivity and specificity are not available. Specificity should be extremely high, given the number of confirmation and characterization procedures. However, sensitivity, particularly at the level of the individual sample, would be compromised by the fact that only 1 E. coli isolate was tested from each fecal sample, and STEC often constitute a very small proportion of the coliforms in feces, even in clinical specimens from humans (11). Procedures such as selective enrichment and immunomagnetic separation have been shown to improve the sensitivity for recovering specific STEC strains (11,12). However, the methods used in this study enable the detection of multiple STEC serotypes. Although we are surely underestimating the sample-level prevalence of non-O157 STEC, the primary objective of our study was to determine the pen-level prevalence and to characterize non-O157 STEC present in feedlot pens.
Although the low sample-level prevalence of non-O157 STEC in feces (0.8%) likely reflects the number of E. coli tested, the pen and feedlot prevalence estimates are consistent with previous estimates (1). Estimates of STEC prevalence in the literature vary widely, in part owing to differences in sampling and diagnostic testing methods (1,3). The estimate of pen prevalence of non-O157 STEC in feces in this study (14%) is less than that reported previously for STEC in Alberta feedlot pens (92%) with the use of composite fecal samples (5) and in slaughter lots of feedlot cattle (80%) sampled at Alberta abattoirs (13). However, the previous studies identified molecular markers in feces and did not attempt to recover individual STEC isolates. In addition, these previous studies, unlike ours, used methods that would have detected E. coli O157:H7, which is considered the most common STEC in North America (1). On Prince Edward Island, STEC isolates were recovered from fecal swabs from 4% of slaughtered beef cattle (14). In a recent study of Alberta and Saskatchewan feedlots, the prevalence of serotype O157:H7 within PS pens ranged from 0% to 90% (15). In a longitudinal US study, STEC isolates were recovered from all cattle herds; the within-herd prevalence ranged from 5% to 33%, and there was no apparent seasonal difference in prevalence (3). Although the prevalence of E. coli O157:H7 is higher in the summer (16), and reports of a similar trend for STEC exist (4,17), in this study, with a limited sample size, there was a relatively similar prevalence of non-O157 STEC in the 2 sampling periods.
A variety of serotypes and variation in the presence of virulence genes among the non-O157 STEC isolates from feces were found in this study. Our results are consistent with those of previous studies of STEC isolates from cattle (3) and beef carcasses (10), which also showed a tremendous diversity among isolates. Most of the isolates were positive for stx2, which is more commonly associated with severe human disease than is stx1 (1). However, only about half of the isolates contained genes for EHEC hemolysin or E. coli attaching and effacing genes, which also are associated with human disease (1). Serogroup or serotype is the characteristic most commonly used to assess the potential for STEC to cause human disease, and only the O113, O145, and O111 isolates that were detected are among the STEC serogroups most frequently associated with human disease (1). Although the non-O157 STEC recovered may not be equally virulent for humans given the diversity among strains, it has been suggested that virtually all STEC strains might be considered potential human pathogens to some extent (18).
Although evidence of STEC was found in approximately 7% of carcass samples on initial testing, perhaps it is not surprising that STEC isolates were not recovered from the samples. Isolation rates for E. coli O157:H7 and other STEC from beef carcasses vary widely but are often extremely low (1,6). A recent US study showed that the prevalence of non-O157 STEC on postintervention carcasses approached 9% (4), and another study indicated that the prevalence of STEC (O157 and non-O157) on postprocessing carcasses was approximately 10% (10). It is interesting that the prevalence from PCR screening of carcass broths in this study was similar to the estimates of prevalence from those studies, even though the study populations, sampling procedures, and laboratory methods were quite different. For example, much larger areas of the carcasses were sampled in those studies than in this study: 2500 cm2 (4) and 750 cm2 (10) versus 300 cm2. The inability to isolate STEC from carcasses in this study could be due to the smaller sampling area or the fact that a majority of Stx-positive postintervention carcasses contain extremely low numbers of STEC cells (4), or both factors. Furthermore, a relatively low prevalence of STEC was found before harvest in this study, and recent research indicates that in-plant (i.e., postharvest) processing procedures and interventions dramatically reduce the overall levels of bacteria found preharvest (4,6,10). Although the low prevalence of STEC in this study limited the effective sample size for comparisons, there was no evidence that carcass prevalence was higher when STEC was recovered from preharvest fecal samples.
Acknowledgments
This work was supported by the Food Safety Division of Alberta Agriculture, Food and Rural Development. We thank Barbara Dakin, Pat Layton, Denise Patterson, Suzanne Gibson, and Cathy Sheppard for their technical assistance. We are grateful to the Laboratory for Foodborne Zoonoses for serotyping and PCR testing. This study would not have been possible without the participating Alberta cattle producers and abattoirs. This is contribution number 06-274-J from the Kansas Agricultural Experiment Station.
References
- 1.Mainil JG, Daube G. Verotoxigenic Escherichia coli from animals, humans and foods: Who’s who? J Appl Microbiol. 2005;98:1332–1344. doi: 10.1111/j.1365-2672.2005.02653.x. [DOI] [PubMed] [Google Scholar]
- 2.Ramotar K, Henderson E, Szumski R, Louie TJ. Impact of free verotoxin testing on epidemiology of diarrhea caused by verotoxin-producing Escherichia coli. J Clin Microbiol. 1995;33:1114–1120. doi: 10.1128/jcm.33.5.1114-1120.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renter DG, Morris JG, Sargeant JM, et al. Prevalence, risk factors, O-serogroups, and virulence profiles of Shiga toxin- producing bacteria from cattle production environments. J Food Prot. 2005;68:1156–1565. doi: 10.4315/0362-028x-68.8.1556. [DOI] [PubMed] [Google Scholar]
- 4.Barkocy-Gallagher GA, Arthur TM, Rivera-Betancourt M, et al. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J Food Prot. 2003;66:1978–1986. doi: 10.4315/0362-028x-66.11.1978. [DOI] [PubMed] [Google Scholar]
- 5.Renter DG, Checkley SL, Campbell J, King R. Shiga toxin-producing Escherichia coli in the feces of Alberta feedlot cattle. Can J Vet Res. 2004;68:150–153. [PMC free article] [PubMed] [Google Scholar]
- 6.Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW. Correlation of enterohemor-rhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci U S A. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.US Department of Agriculture. Appendix F — Guidelines for E. coli testing for process control verification in cattle and swine slaughter establishments. Final rule. Federal Register. July 25. 1996;61:38929–38938. [Google Scholar]
- 8.Gannon VP, King RK, Kim JY, Thomas EJ. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louie M, Read S, Simor AE, et al. Application of multiplex PCR for detection of non-O157 verocytotoxin-producing Escherichia coli in bloody stools: identification of serogroups O26 and O111. J Clin Microbiol. 1998;36:3375–3377. doi: 10.1128/jcm.36.11.3375-3377.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthur TM, Barkocy-Gallagher GA, Rivera-Betancourt M, Koohmaraie M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli on carcasses in commercial beef cattle processing plants. Appl Environ Microbiol. 2002;68:4847–4852. doi: 10.1128/AEM.68.10.4847-4852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettelheim KA, Beutin L. Rapid laboratory identification and characterization of verocytotoxigenic (Shiga toxin producing) Escherichia coli (VTEC/STEC) J Appl Microbiol. 2003;95:205–217. doi: 10.1046/j.1365-2672.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins C, Pearce MC, Smith AW, et al. Detection of Escherichia coli serogroups O26, O103, O111 and O145 from bovine faeces using immunomagnetic separation and PCR/DNA probe techniques. Lett Appl Microbiol. 2003;37:207–212. doi: 10.1046/j.1472-765x.2003.01379.x. [DOI] [PubMed] [Google Scholar]
- 13.Van Donkersgoed J, Graham T, Gannon V. The prevalence of verotoxins, Escherichia coli O157:H7, and Salmonella in the feces and rumen of cattle at processing. Can Vet J. 1999;40:332–338. [PMC free article] [PubMed] [Google Scholar]
- 14.Schurman RD, Hariharan H, Heaney SB, Rahn K. Prevalence and characteristics of Shiga toxin-producing Escherichia coli in beef cattle slaughtered on Prince Edward Island. J Food Prot. 2000;63:1583–1586. doi: 10.4315/0362-028x-63.11.1583. [DOI] [PubMed] [Google Scholar]
- 15.Van Donkersgoed J, Hancock D, Rogan D, Potter AA. Escherichia coli O157:H7 vaccine field trial in 9 feedlots in Alberta and Saskatchewan. Can Vet J. 2005;46:724–728. [PMC free article] [PubMed] [Google Scholar]
- 16.Renter DG, Sargeant JM. Enterohemorrhagic Escherichia coli O157: epidemiology and ecology in bovine production environments. Anim Health Res Rev. 2002;3:83–94. [PubMed] [Google Scholar]
- 17.Cobbold RN, Rice DH, Szymanski M, Call DR, Hancock DD. Comparison of Shiga-toxigenic Escherichia coli prevalences among dairy, feedlot, and cow-calf herds in Washington state. Appl Environ Microbiol. 2004;70:4375–4378. doi: 10.1128/AEM.70.7.4375-4378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gyles C, Johnson R, Gao A, et al. Association of enterohemorrhagic Escherichia coli hemolysin with serotypes of Shiga-like-toxin-producing Escherichia coli of human and bovine origins. Appl Environ Microbiol. 1998;64:4134–4141. doi: 10.1128/aem.64.11.4134-4141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]