Abstract
This study describes the distribution of Hepatitis E virus (HEV) in a naturally infected swine population and the genetic relatedness of HEV strains on swine farms in Spain. Of fecal and serum samples collected from 131 pigs and manure-ditch samples collected from 17 farms, HEV was detected in 16%, 14%, and 59%, respectively, for an overall prevalence rate of 23%. The maximum prevalence rates for feces and serum were in pigs 5 to 12 wk old. A high prevalence of the virus in feces (18%) was observed in sows. Gene sequencing was performed on 6 strains from feces, serum, and manure ditch: the nucleotide identities varied from 81.5% to 99% when compared with those of other strains of genotype 3 isolated from swine. This is the first study in Europe to show the variation in virus distribution by age in feces and serum in a naturally infected swine population.
Résumé
Cette étude décrit la distribution du virus de l’hépatite E (HEV) dans une population de porcs naturellement infectés et la parenté génétique de souches de HEV provenant de fermes porcines en Espagne. À partir d’échantillons de fèces et de sérum prélevés de 131 porcs, et de fosses à purin sur 17 fermes, le HEV a été détecté, respectivement, dans 16 %, 14 % et 59 % des échantillons, pour une prévalence globale de 23 %. Les taux maximums de prévalence pour les fèces et le sérum ont été retrouvés chez les porcs de 5 à 12 semaines d’âge. Une prévalence élevée de virus dans les fèces (18 %) a été observée chez les truies. Le séquençage génétique a été effectué sur 6 isolats provenant des fèces, du sérum et de fosse à purin : l’identité nucléotidique a varié de 81,5 % à 99 % en comparaison avec d’autres souches du génotype 3 isolées de porcs. Il s’agit de la première étude européenne à démontrer la variation de la distribution du virus selon l’âge dans les fèces et le sérum d’une population de porcs infectés naturellement.
(Traduit par Docteur Serge Messier)
Hepatitis E is the main cause of enterically transmitted non-A, non-B hepatitis in developing countries. The mortality rate for this infection is generally low (1%) but is up to 25% in pregnant women. A nonenveloped, single-stranded RNA virus of approximately 7.2 kb, the Hepatitis E virus (HEV) is encoded by 3 open reading frames (ORFs) in the genome (1). At least 4 major genotypes of HEV have been identified. Genotype 1 includes human isolates from Asia and North Africa, and genotype 2 comprises human isolates from Mexico and some African countries. Genotypes 3 and 4 include human and swine strains isolated in industrialized countries as well as developing areas (2).
Hepatitis E has been considered endemic in Asia, South America, and Africa. Despite the low number of sporadic acute cases in developed regions among people with no history of travel to endemic areas, a surprisingly high level of HEV antibodies has been reported, suggesting that the virus is more widespread than previously recognized (3,4). This finding has raised the suspicion of an animal reservoir for the virus and that hepatitis E is a zoonosis. The first animal strain was isolated from a pig in the United States in 1997 (5). More studies in other developed countries have reported high seroprevalence rates and new isolates of swine origin (6,7). Sequence analysis has shown that indigenously acquired human and swine isolates in industrialized countries are clustered in the same genotype (3 or 4) (7), supporting the strong suspicion of hepatitis E as a zoonosis. Furthermore, swine isolates are related more to human strains from the same geographic region than to swine strains from different areas (8). Since pigs experimentally infected with human strains have shed the virus for several weeks in feces (9), swine production may represent a significant environmental reservoir of infection. Routes of transmission remain unclear, although in 2004 Kasorndorkbua et al (10) reported experimental evidence of fecal–oral transmission between pigs in the same pen. However, the way in which HEV passes from pigs to humans remains unknown. It has been reported that swine veterinarians in the United States are 1.51 times more likely to have positive results of HEV antibody tests than are normal blood donors in the same geographic area (11).
The aim of this study was to detect HEV and to investigate the dynamics of virus distribution in feces and serum at various ages in a naturally infected swine population in Spain. In addition, the strains isolated were to be genetically characterized and compared with other swine and human HEV strains from different areas of the world.
Sample size was established taking into account the national census of the Ministry of Agriculture of Spain in 2002, which reported 1 129 055 pigs for the Valencia community. The expected prevalence was 13.7% (12), with a confidence of 95% and an accepted error of 6%. Included in the study, conducted from January 2002 to August 2004, were 131 pigs from 21 farms.
Fecal and blood samples were collected at the same time from 131 animals at different production stages: 20 piglets 0 to 4 wk old, 22 weaners 5 to 12 wk old, 20 feeders 13 to 20 wk old, 27 finishers 21 to 24 wk old, 4 boars, and 38 sows with suckling piglets. The number of animals sampled at each farm was determined on the basis of the farm’s size. Additionally, samples of manure ditch were taken from 17 farms. The fecal samples were obtained directly from the rectum and kept in sterile containers. They were diluted at 10% (w/v) in phosphate-buffered saline, pH 7.2, and centrifuged to collect supernatant for RNA extraction. The remaining supernatant was stored at −80°C. The blood samples were obtained by puncture of the jugular vein and collected in tubes without anticoagulant. After separation, the serum was used for RNA extraction. The remaining sample was stored at −80°C. Samples of manure ditch were centrifuged directly without dilution and the supernatants used for RNA extraction.
We performed RNA extraction, reverse transcription (RT), and nested polymerase chain reaction according to the method described by Fernández-Barredo et al (13), using primers designed by Huang et al (8). Sequence alignments were generated by ClustalW, and sequence analysis was performed with the use of Bioedit Sequence Alignment Editor. The swine sequences reported in this work were compared with 36 sequences selected from the 4 genotypes available in the GenBank database, which included strains of human origin (AY204877 Chad, AF051352 Egypt, AF141652 China, X98292 India, AF170450 Vietnam, X99441 Madras [India], AF065061 Morocco, M80581 Sar55 [Pakistan], M73218 Burma [India], AF076239 India, AH006999 HEVBCN [Spain], AF058684 HEVBCN6 [Spain], M74506 Mexico, AF060668 US1 [USA], AF060669 US2 [USA], AF195062 VH2 [Spain], AF195061 VH1 [Spain], AJ272108 China T1, AF151962 China T11, AF296160 TW261E [Taiwan], AB097812 JA1 [Japan]), swine origin (AF082843 US-SW, AF336292 NLSW28 [The Netherlands], AF503511 UK P354, AF503512 UK P143, AF195063 E11 [Spain], AY858936 G3-8f [Mexico], AY858920 S2-29f [Mexico], AY858917 S1-29f [Mexico], AY858901 T4-7s [Taiwan], AF516179 KOR2 [Korea], AY115488 Canada, AB073910 swJ681 [Japan], AF117280 TW32SW [Taiwan], AB097811 swJ13-1 [Japan]), and avian origin (AY043166); the last sequence had only recently been identified. A phylogenetic tree was constructed by the neighbor-joining method with a substitution model of the Kimura 2-parameter, based on the partial nucleotide sequence of the ORF2 region (222 nucleotides). Bootstrap values were determined on 1000 resamplings of the data sets. Phylogenetic and molecular evolutionary genetics analyses were conducted by means of MEGA, version 3.0 (www.megasoftware.net).
We detected HEV at 16 of the 21 farms (76%) and in 21 (16%) of the 131 fecal samples, 18 (14%) of the 131 serum samples, and 10 (59%) of the 17 manure-ditch samples. As Table I shows, the frequency of detection in feces was highest among the pigs 5 to 12 wk old (41%) and lowest among the pigs 13 to 20 wk old (5%). Whereas HEV was not detected in feces from the boars, it was detected in 18% of the fecal samples from the sows. A similar pattern was observed for the serum samples, with the highest rate among the pigs 5 to 12 wk old (32%) and much lower rates in the next 2 age groups; however, apart from the boars, whose serum samples were free of HEV, the sows had the lowest rate in serum, at 5%. The overall rate of HEV detection among the pigs, including positive results for either type of sample or both types, was 23% (30/131), and the highest overall rate by age was 41%, in the animals 5 to 12 wk old. In 9 pigs HEV was detected in both feces and serum; 7 were aged 5 to 12 wk, and the other 2 were sows. No clinical signs specific for HEV infection were recorded for any of the pigs studied.
Table I.
Detection of Hepatitis E virus (HEV) RNA in feces and serum of pigs of various ages
| % of HEV-positive samples
|
||
|---|---|---|
| Age, wk (and no.of pigs) | Feces | Serum |
| 0–4 (20) | 2 (10) | 4 (20) |
| 5–12 (22) | 9 (41) | 7 (32) |
| 13–20 (20) | 1 (5) | 2 (10) |
| 21–24 (27) | 2 (7) | 3 (11) |
| > 25 (boars) (4) | 0 (0) | 0 (0) |
| > 25 (sows) (38) | 7 (18) | 2 (5) |
| Total (131) | 21 (16) | 18 (14) |
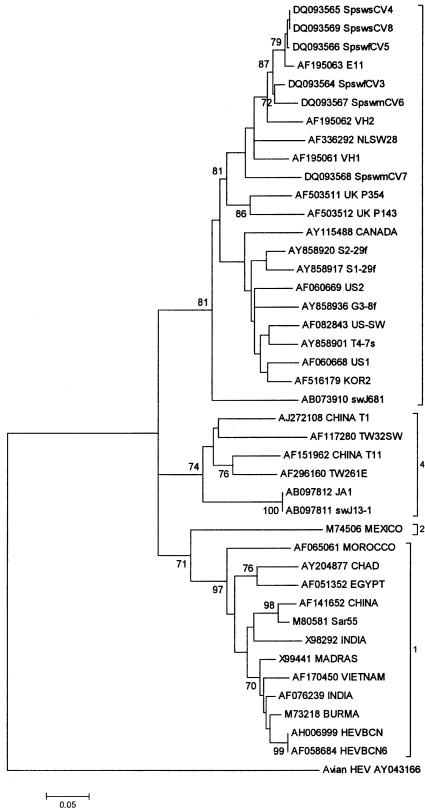
Partial sequences of 222 base pairs of the HEV capsid protein gene were obtained from 6 isolates and deposited in GenBank: 2 sequences from serum samples (SpswsCV4 [accession no. DQ093565] and SpswsCV8 [DQ093569]), 2 from fecal samples (SpswfCV3 [DQ093564] and SpswfCV5 [DQ093566]), and 2 from manure-ditch samples (SpswmCV6 [DQ093567] and SpswmCV7 [DQ093568]). Each isolate was from a different farm. The sequences were compared with others from the 4 known genotypes and were found to cluster in genotype 3 (Figure 1), together with porcine strains from Europe and North America as well as a Japanese swine strain. In this genotype, the Spanish sequences clustered in a minor branch with European porcine strains and with human strains from acute autochthonous hepatitis E in Spain. The 6 new sequences had nucleotide similarity (Table II) with other swine sequences in genotype 3 ranging from 81.5% between SpswmCV7 and a Japanese swine strain to 98.6% between SpswsCV4 and SpswsCV8 and E11 (from sewage from a porcine slaughterhouse in Spain). The closest relationship (94.1% nucleotide similarity) between the 6 new sequences and human strains in genotype 3 was between SpswsCV4 and VH2, from a patient with acute hepatitis in Barcelona, Spain. Nucleotide translation to amino acids and multiple alignments demonstrated no variation among all the partial sequences in genotype 3 studied, human or swine, except for US2, which differed in only 1 amino acid. The similarity of swine Spanish strains and other strains in the other genotypes varied from 91% with the Mexican strain to 96% with human and porcine strains in genotype 4.
Figure 1.
Phylogenetic relationships among swine and human strains of Hepatitis E virus (HEV) representing the 4 major genotypes, based on a 222-nucleotide fragment of open reading frame 2 of the genome. The scale bar represents an evolutionary distance of 0.05 nucleotides per site.
Table II.
Nucleotide similarity of a 222-base-pair fragment of open reading frame 2 of the HEV genome of swine and human strains of genotype 3, as compared with 6 strains isolated in this study
| Study strains (GenBank accession no. of the sequence); % similarity of fragment
|
||||||
|---|---|---|---|---|---|---|
| Selected strainsa | SpswfCV3 (DQ093564) | SpswsCV4 (DQ093565) | SpswfCV5 (DQ093566) | SpswmCV6 (DQ093567) | SpswmCV7 (DQ093568) | SpswsCV8 (DQ093569) |
| AF082843 US-SW | 85.5 | 87.3 | 86.9 | 84.6 | 83.7 | 87.3 |
| AF060668 US1 | 84.6 | 85.5 | 86.0 | 83.7 | 83.3 | 85.5 |
| AF060669 US2b | 85.5 | 85.5 | 86.0 | 83.7 | 85.1 | 85.5 |
| AF336292 NLSW28 | 90.5 | 90.9 | 90.5 | 88.7 | 86.4 | 90.9 |
| AF503511 UK P354 | 86.9 | 85.1 | 85.5 | 86.4 | 85.5 | 85.1 |
| AF503512 UK P143 | 85.5 | 83.7 | 84.2 | 84.6 | 86.9 | 83.7 |
| DQ093564 SpswfCV3 | — | 96.8 | 97.2 | 96.3 | 90.0 | 96.8 |
| DQ093565 SpswsCV4 | 96.8 | — | 99.5 | 95.0 | 89.6 | 100 |
| DQ093566 SpswfCV5 | 97.2 | 99.5 | — | 95.4 | 90.0 | 99.5 |
| DQ093567 SpswmCV6 | 96.3 | 95.0 | 95.4 | — | 89.1 | 95.0 |
| DQ093568 SpswmCV7 | 90.0 | 89.6 | 90.0 | 89.1 | — | 89.6 |
| DQ093569 SpswsCV8 | 96.8 | 100 | 99.5 | 95.0 | 89.6 | — |
| AF195063 E11 | 97.2 | 98.6 | 98.1 | 94.5 | 89.1 | 98.6 |
| AF195062 VH2 | 93.6 | 94.1 | 93.6 | 91.8 | 91.4 | 94.1 |
| AF195061 VH1 | 92.7 | 92.3 | 91.8 | 90.0 | 89.1 | 92.3 |
| AY858936 G3-8f | 83.3 | 85.1 | 85.5 | 83.3 | 84.2 | 85.1 |
| AY858920 S2-29f | 86.9 | 87.8 | 87.3 | 85.1 | 84.6 | 87.8 |
| AY858917 S1-29f | 86.0 | 86.9 | 86.4 | 86.0 | 84.2 | 86.9 |
| AY858901 T4-7s | 83.7 | 83.7 | 83.3 | 81.9 | 82.4 | 83.7 |
| AF516179 KOR2 | 86.0 | 86.9 | 87.3 | 86.0 | 85.5 | 86.9 |
| AY115488 CANADA | 85.1 | 85.1 | 85.5 | 86.0 | 83.7 | 85.1 |
| AB073910 swJ681 | 81.9 | 82.8 | 83.3 | 82.8 | 81.5 | 82.8 |
Those in italics are of swine origin and those underlined are of human origin.
For this strain, the deduced amino acid sequence of the fragment was 99% identical to that of all 6 of this study’s strains, whereas for all of the other selected strains the similarity was 100%.
It has been reported that HEV in pigs is widespread in the world, involving not only countries in which human HEV infection is common, such as India (14), but also countries in which hepatitis E is rare, such as Canada, Britain, the United States, the Netherlands, New Zealand, and Spain (6–8,13,15,16). In several of these industrialized countries, there have been seroprevalence studies involving a large number of animals, but the sample size for RNA detection was much smaller than ours. Moreover, the special situation of Spain as the European doorway to Africa, in which only genotype 1 and 2 strains have been identified in humans, makes it interesting to determine the relationship between these swine strains and other human and swine strains from different areas of the world.
In Spain, a 25% HEV seroprevalence in pigs was reported, but no HEV RNA was detected in any of the serum or fecal samples (17). In another study, a seroprevalence of 13.7% was reported, but HEV RNA was again not isolated from serum (12); however, the genome was identified in pools of feces from a single farm in all the age groups studied (pigs 3, 5, and 8 wk old, fattening pigs, and primiparous sows). Ours is the first report from Europe about HEV detection in a naturally infected population through study of fecal and serum samples collected at the same time from each animal, which permitted us to evaluate at what age pigs should be more infectious for humans and other pigs because of virus shedding in the feces.
Data from the study of individual fecal samples are scarce and vary greatly depending on the study. Banks et al (18) in Britain reported 11 pigs (26%) to be positive out of 42, all tested between 12 and 15 wk of age. Van der Poel et al (6) in the Netherlands studied 9 pigs but did not detect HEV RNA. Garkavenko et al (16) in New Zealand, studying pigs between 7 and 17 wk old, found that 17 (38%) of 45 shed HEV, all 17 being 10 to 12 wk old. The frequency of positive fecal samples in our study was only 16%. A possible explanation for the higher rates in the other studies is a bias in age distribution, as the pigs aged 5 to 12 weeks had the highest rate of HEV detection in feces (41%) in our study.
In our study, 18% of sows had HEV in their feces and thus could be the source of new infections on the same farm and also of spread to other farms, since many farms sell piglets for feeding. The mechanism by which HEV almost disappears from the feces of 5-mo-old pigs (whose rate was only 7%) and then emerges in sows is uncertain. Stress during farrowing and suckling might increase susceptibility to HEV infection. Studies involving a larger number of samples from sows would be desirable to clarify this point.
The overall rate of HEV detection in serum in our study (14%) is the highest described to date among studies encompassing all ages but lower than the overall rate of detection in feces in our study. Since it has been reported that HEV viremia is transient (5), one would expect the real prevalence of HEV infection to be even higher that that obtained when testing pig serum for the presence of HEV RNA instead of feces. The high detection rates in this study show that the effectiveness of spreading is high with this virus.
The nucleotide sequences reported in this study are the first obtained in Spain from serum of pigs naturally infected by HEV as well as from manure ditches at farms. The latest reports of acute autochthonous hepatitis E in Spain (3,19) made it necessary to study a larger sample than in previous investigations to evaluate the level of endemicity of HEV in Spain. In addition, the sequences are useful for establishing, by phylogenetic analysis, the relationship to Spanish human strains and to human and swine strains from different geographic areas. The high nucleotide identity (94.1%) between human and swine sequences in genotype 3 observed in this study supports the evidence that HEV is a zoonotic agent. Furthermore, we found 100% homology in amino acid identity of the studied fragment when comparing human and porcine strains from genotype 3, which shows a close relationship among these strains, since 90.54% of the mutations are silent in the fragment studied. The recent finding of HEV of only genotype 3 in pigs from areas where HEV in the human population belongs to genotype 1 raises the hypothesis that only strains of genotypes 3 and 4 have zoonotic potential (20). Since strains from pigs do not produce any symptoms, it would be reasonable to think that HEV could be more widespread among healthy people, as seroprevalence studies in normal blood donors have reported (21), especially in people working in contact with pigs (farmers, veterinarians).
In summary, this study has contributed to the knowledge of HEV distribution by age in serum and feces of a naturally infected swine population. It has also shown that HEV is detected at a high rate not only in feces of young pigs but also in feces of breeding sows. Moreover, HEV strains circulating in Spanish swine farms are highly homologous with Spanish human strains, which raises the high possibility of HEV transmission from swine to humans.
Acknowledgments
This work was supported by grants from Generalitat Valenciana (GV05/132), Escuela Valenciana para Estudios de la Salud (Consellería de Sanidad, 053/2005), and CEU Cardenal Herrera University (PRUCH 04/8, 06/21). Dr. Fernández-Barredo holds a grant from CEU Cardenal Herrera University. We are thankful to Dr. Malcolm Banks, Veterinary Laboratories Agency, Weybridge, England, for his kind help in the critical review of the manuscript.
References
- 1.Tam AW, Smith MM, Guerra ME, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Gracia MT, Garcia-Valdivia MS, Galan F, Rodriguez-Iglesias MA. Detection of hepatitis E virus in patients sera in southern Spain. Acta Virol. 2004;48:197–200. [PubMed] [Google Scholar]
- 4.Perez-Gracia MT, Rodriguez-Iglesias M. Hepatitis E virus: current status. Med Clin (Barc) 2003;121:787–792. [PubMed] [Google Scholar]
- 5.Meng XJ, Purcell RH, Halbur PG, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Poel WH, Verschoor F, van der Heide R, et al. Hepatitis E virus sequences in swine related to sequences in humans, the Netherlands. Emerg Infect Dis. 2001;7:970–976. doi: 10.3201/eid0706.010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banks M, Bendall R, Grierson S, Heath G, Mitchell J, Dalton H. Human and porcine hepatitis E virus strains, United Kingdom. Emerg Infect Dis. 2004;10:953–955. doi: 10.3201/eid1005.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang FF, Haqshenas G, Guenette DK, et al. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol. 2002;40:1326–1332. doi: 10.1128/JCM.40.4.1326-1332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halbur PG, Kasorndorkbua C, Gilbert C, et al. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol. 2001;39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasorndorkbua C, Guenette DK, Huang FF, Thomas PJ, Meng XJ, Halbur PG. Routes of transmission of swine hepatitis E virus in pigs. J Clin Microbiol. 2004;42:5047–5052. doi: 10.1128/JCM.42.11.5047-5052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng XJ, Wiseman B, Elvinger F, et al. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J Clin Microbiol. 2002;40:117–122. doi: 10.1128/JCM.40.1.117-122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemente-Casares P, Pina S, Buti M, et al. Hepatitis E virus epidemiology in industrialized countries. Emerg Infect Dis. 2003;9:448–454. doi: 10.3201/eid0904.020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Barredo S, Galiana C, Garcia A, Vega S, Gómez MT, Perez-Gracia MT. Detection of hepatitis E virus shedding in feces of pigs at different stages of production using reverse transcription-polymerase chain reaction. J Vet Diagn Invest. 2006;18:462–465. doi: 10.1177/104063870601800506. [DOI] [PubMed] [Google Scholar]
- 14.Arankalle VA, Chobe LP, Walimbe AM, Yergolkar PN, Jacob GP. Swine HEV infection in south India and phylogenetic analysis (1985–1999) J Med Virol. 2003;69:391–396. doi: 10.1002/jmv.10301. [DOI] [PubMed] [Google Scholar]
- 15.Yoo D, Willson P, Pei Y, et al. Prevalence of hepatitis E virus antibodies in Canadian swine herds and identification of a novel variant of swine hepatitis E virus. Clin Diagn Lab Immunol. 2001;8:1213–1219. doi: 10.1128/CDLI.8.6.1213-1219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garkavenko O, Obriadina A, Meng J, et al. Detection and characterisation of swine hepatitis E virus in New Zealand. J Med Virol. 2001;65:525–529. [PubMed] [Google Scholar]
- 17.Pina S, Buti M, Cotrina M, Piella J, Girones R. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J Hepatol. 2000;33:826–833. doi: 10.1016/s0168-8278(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 18.Banks M, Heath GS, Grierson SS, et al. Evidence for the presence of hepatitis E virus in pigs in the United Kingdom. Vet Rec. 2004;154:223–227. doi: 10.1136/vr.154.8.223. [DOI] [PubMed] [Google Scholar]
- 19.Buti M, Clemente-Casares P, Jardi R, et al. Sporadic cases of acute autochthonous hepatitis E in Spain. J Hepatol. 2004;41:126–131. doi: 10.1016/j.jhep.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Cooper K, Huang FF, Batista L, et al. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol. 2005;43:1684–1688. doi: 10.1128/JCM.43.4.1684-1688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukuda S, Sunaga J, Saito N, et al. Prevalence of antibodies to hepatitis E virus among Japanese blood donors: identification of three blood donors infected with a genotype 3 hepatitis E virus. J Med Virol. 2004;73:554–561. doi: 10.1002/jmv.20125. [DOI] [PubMed] [Google Scholar]