Abstract
Aldehyde dehydrogenases (ALDHs) are critical enzymes in the metabolism of endogenous and exogenous aldehydes. The human genome contains nineteen putatively functional ALDH genes; ALDH3B1 belongs to the ALDH3 family. While recent studies have linked the ALDH3B1 locus to schizophrenia, nothing was known, until now, about the properties and significance of the ALDH3B1 protein. The aim of this study was to characterize the ALDH3B1 protein. Human ALDH3B1 was baculovirus-expressed and found to be catalytically active towards medium- and long-chain aliphatic aldehydes and the aromatic aldehyde benzaldehyde. Western blot analyses indicate that ALDH3B1 is highly expressed in kidney and liver and moderately expressed in various brain regions. ALDH3B1-transfected HEK293 cells were significantly protected against cytotoxicity induced by the lipid-peroxidation product octanal when compared to vector-transfected cells. This study shows for the first time the functionality, expression and protective role of ALDH3B1 and indicates a potential physiological role of ALDH3B1 against oxidative stress.
Keywords: ALDH3B1; aldehyde metabolism; schizophrenia; 3,4-dihydroxyphenylacetaldehyde; 4-hydroxy-2-nonenal; oxidative stress; lipid peroxidation
Introduction
Aldehyde dehydrogenases (ALDHs) comprise a superfamily of NAD(P)+-dependent enzymes that catalyze the oxidation of numerous endogenous and exogenous aldehydes to their respective carboxylic acids [1]. Aldehydes are generated from such precursors as alcohols, lipids, neurotransmitters, drugs and environmental agents. They are highly electrophilic, relatively long-lived species that react with biomolecules such as proteins and nucleic acids, leading to various pathological effects. For example, aldehydes can be cytotoxic, mutagenic and even carcinogenic [2]. ALDHs efficiently oxidize and, in most circumstances, detoxify a significant number of chemically-diverse aldehydes which would otherwise be harmful. Not unexpectedly, mutations in ALDH genes are the molecular basis of many disease states, well illustrated by those resulting from inborn errors of aldehyde metabolism, such as Sjögren-Larsson syndrome (SLS), type II hyperprolinemia, γ-hydroxybutyric aciduria and pyridoxine-dependent seizures [1; 3]. In addition, mutations in ALDH genes contribute to other clinically-relevant diseases, such as cancer and Alzheimer’s disease [2; 4].
To date, 19 distinct ALDH genes have been identified in the human genome [5]. ALDH3B1 is located on chromosome 11q13.2 and encodes a protein of approximately 52 kDa. Recent studies have linked a single nucleotide polymorphism (SNP) at the ALDH3B1 locus to the development of paranoid schizophrenia [6; 7]. While this gene has been cloned [8; 9], nothing was known about the properties and significance of the ALDH3B1 protein. Here we report the initial characterization of ALDH3B1. To assess the putative functionality of ALDH3B1, the conservation of critical ALDH residues essential for catalysis and cofactor binding were evaluated. ALDH3B1 catalytic activity with various aldehyde substrates was determined utilizing a baculovirus expression system. Further, the tissue distribution of ALDH3B1 was studied in mouse using Western blot analyses. Finally, a potential physiological role of ALDH3B1 in the protection against oxidative stress was revealed by utilizing stably-transfected HEK293 cells.
Materials and Methods
Protein Sequence Alignment
ALDH protein sequences were obtained from NCBI and aligned using the Clustal W program [10]. Sequence comparisons were performed utilizing the BLAST program [11].
Baculovirus expression
Human ALDH3B1 cDNA cloned into the pT7T3D-pac vector (EST clone ID 1951332; Incyte Genomics, Palo Alto, CA) was expanded and sequenced (GenBank accession no. EF411198). The full-length ALDH3B1 cDNA, digested from pT7T3D-pac, was ligated into the pBlueBac 4.5 baculovirus expression vector at Xho I and Hind III (Invitrogen, Carlsbad, CA). Restriction and sequence analyses verified correct insertion. Viruses were plaque-purified and amplified in Sf9 insect cells (Spodoptera frugiperda), as described [12]. Plaques were tested for ALDH3B1 protein expression by Western blot. ALDH3B1 protein was not detected in non-infected cells. Sf9 cells (at a density of 1 × 106) were infected with the human ALDH3B1 baculovirus at a multiplicity of 1 for 48 h. Infected cells were harvested by centrifugation at 1000 g for 5 min, washed with PBS and lysed by sonication for subsequent analysis.
Enzymatic activity assays
Cell lysates from ALDH3B1 baculovirus-infected Sf9 cells and from non-infected Sf9 cells were assayed for enzymatic activity. Cells were resuspended,sonicated and centrifuged at 100,000 g for 1 h, as described previously [13]. Protein concentrations were estimated by BCA kit (Pierce, Rockford, IL). The enzymatic activity of cell lysates was measured spectrophotometrically by monitoring the production of NAD(P)H at 340 nm during the oxidation of aldehyde substrates, as described previously [13]. Assays were conducted at 25°C in a 1 ml reaction volume of 75 mM sodium pyrophosphate (pH 8), cofactor (2.5 mM NADP+ or 1 mM NAD+), 1 mM pyrazole and 50 μl cell lysate (approximately 300 – 600 μg total protein). The addition of 100 μl of various concentrations of aldehyde substrates (unless otherwise specified, purchased from Sigma, St. Louis, MO) initiated the reaction. 4-Hydroxy-2-nonenal (4-HNE) was purchased from Cayman Chemical Company (Ann Arbor, MI). Malondialdehyde (MDA) was synthesized as described previously [14]. 3,4-Dihydroxyphenylacetaldehyde (DOPAL) was synthesized as described previously [15], with minor modification. Briefly, DOPAL-bisulfite was produced by incubating dopamine with monoamine oxidase (Sigma) and sodium bisulfite. Free DOPAL was then dissociated at pH 8.8 and extracted with ether. DOPAL concentration was determined spectrophotometrically using alcohol dehydrogenase (Sigma) with NADH. For each substrate, three or more concentrations were assayed. All assays were performed in triplicate and enzyme activity rates were normalized to protein concentration to determine specific activity (nmol of NAD(P)H/min per mg protein).
Production of ALDH3B1-specific antibodies
To produce an antibody against human ALDH3B1, a strictly conserved amino acid sequence was identified by BLAST (MDPLGDTLRRLREAFHAG, amino acids 1-18; Figure 1) and a synthetic peptide produced. Following keyhole limpet hemocyanin (KLH) conjugation, a two-rabbit antibody production was initiated (Alpha Diagnostics International, San Antonio, TX). Sera were collected, characterized for reactivity and serum that was optimal in specificity was affinity purified. To generate a specific antibody against both mouse and human ALDH3B1, an amino acid sequence strictly conserved in both proteins was identified by BLAST analysis (ALAQDLHKSAFE, amino acids 47-58; Figure 1). A synthetic peptide was produced, conjugated to KLH, and a two-rabbit antibody production protocol initiated (Cocalico Biologicals, Reamstown, PA). Sera were collected, characterized for reactivity and affinity purified. While the sera were reactive against mouse ALDH3B1, there was no reactivity against human ALDH3B1. The specificity of this antibody against mouse ALDH3B1 was confirmed by incubating the antibody with and without the conjugated peptide before exposure to Western blot membranes of mouse tissue lysates. Incubation with the peptide resulted in blockade of the Western blot band at 52 kDa, the molecular weight of mouse ALDH3B1 (data not shown).
Figure 1.

Alignment of mammalian class 3 ALDH protein amino acid sequences. Shading indicates highly conserved amino acids that are discussed in the text. Underlined sequences represent peptides used to generate ALDH3B1-specific antibodies. Numbering is based on human ALDH3B1.
Western blot analysis
Organs were harvested from male C57BL/129sv mice (60-90 days old), processed and subjected to Western blot analysis, as described previously [16]. Anti-mouse ALDH3B1 (1:500) and anti-human ALDH3B1 (1:200) antibody binding was detected using peroxidase-conjugated goat anti-rabbit IgG (1:5000) (Calbiochem, San Diego, CA) and proteins were visualized using chemiluminescence (NEN Life Science Products, Boston, MA) and hyperfilm (Amersham Biosciences, Piscataway, NJ). Equivalent loading of protein samples was confirmed by re-probing membranes with mouse monoclonal anti-β-actin IgG (1:10,000; Sigma) followed by peroxidase-conjugated rabbit anti-mouse IgG (1:5000; Sigma).
Generation of HEK293 cell lines stably transfected with ALDH3B1
The human embryonic kidney cell line (HEK293) was grown in Dulbecco’s Modified Eagle Medium with high glucose (Invitrogen, Carlsbad, CA) and 7% FBS (Gemini, West Sacramento, CA) at 37 °C in a 5% CO2 incubator. For transfection, the human ALDH3B1 cDNA was digested from the pBlueBac/hALDH3B1 plasmid and ligated into the pcDNA3.1(+) plasmid at EcoR I and Not I sites. HEK293 cells were transfected with either pcDNA3.1 or pcDNA3.1/ALDH3B1 by a standard calcium phosphate precipitation method, as described previously [17]. Stable cell populations were selected using geneticin and allowed to form colonies that were further expanded. ALDH3B1 expression was screened using Western blot analysis. ALDH3B1/HEK293 clone 8 was used for all subsequent experiments in this study.
Sulforhodamine B Assay
The sulforhodamine (SRB) assay is a sensitive measure of cytotoxicity that detects the binding of SRB to basic amino acids of cellular proteins and colorimetric analysis provides an estimate of cell number. Cells (1 × 104) were plated in 96-well culture plates, allowed to attach overnight, and then treated in triplicate with octanal (0-1mM) in serum-free medium for 24 h. Cell viability was determined by the SRB assay, as described previously [18].
Statistical Analysis
All values are expressed as mean ± S.E. Groups were compared by Student’s unpaired t-test (SigmaPlot, version 9.0, 2004). P<0.05 was considered to be significant.
Results
Sequence homology and conservation of critical ALDH residues
Human ALDH3B1 is 85% identical to the mouse ALDH3B1 protein, 81 to 95% identical to chimp, cow, dog, rat, and rhesus monkey ALDH3B1 proteins, and 48% and 66% identical to frog and chicken ALDH3B1 proteins, respectively. Human ALDH3B1 is 83% identical to the human ALDH3B2 protein and 53% and 55% identical to human ALDH3A1 and ALDH3A2 proteins, respectively.
Sequence alignment of human and mouse ALDH3B1 with other ALDH3 proteins revealed conservation of ALDH residues critical for cofactor binding and catalysis (Figure 1). These include the catalytically-essential amino acids, Cys-244 and Glu-334 (numbering based on human ALDH3B1), as well as residues involved in cofactor binding such as Gly-188 and Gly-193 [19; 20].
Enzymatic activity
The catalytic activity of human ALDH3B1 was assessed using protein expressed in Sf9 cells. Initially, lysates from infected and non-infected Sf9 cells were examined by Western blot analysis using the anti-human ALDH3B1 antibody. No bands were detected in extracts from non-infected Sf9 cells whereas a single band with an apparent molecular weight of 52 kDa, consistent with the size of human ALDH3B1, was detected in cell lysates from Sf9 cells infected with human ALDH3B1 (Figure 2).
Figure 2.
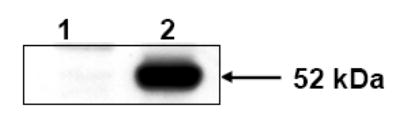
Expression of human ALDH3B1 protein in Sf9 cells. Non-infected Sf9 control cells (lane 1) and Sf9 cells infected with ALDH3B1 recombinant baculovirus (lane 2) were assayed for human ALDH3B1 antigen by Western blotting following SDS/PAGE (n=3).
Cell lysates from human ALDH3B1-infected Sf9 cells were used for enzyme activity assays and compared to cell lysates from non-infected Sf9 cells (control) (Table 1). In these studies, ALDH3B1 was shown to be capable of oxidizing medium and long chain (six carbons and higher) saturated and unsaturated aldehydes, including the lipid peroxidation-derived aldehydes hexanal, 4-HNE, nonanal, octanal, trans-2-hexenal, trans-2-nonenal, and trans-2-octenal. In addition, ALDH3B1 also metabolized the aromatic aldehyde, benzaldehyde. By contrast, short-chain aldehydes such as betaine aldehyde, MDA and propionaldehyde did not appear to be metabolized by ALDH3B1. Additionally, limited ALDH3B1 activity towards acetaldehyde and the dopamine-derived DOPAL was demonstrated, indicating these aldehydes are poor substrates. With most substrates ALDH3B1 appeared to be capable of utilizing either NAD+ or NADP+ as cofactor, however, some preferences were seen. For instance, greater fold increases in ALDH3B1 specific activity (over control) were seen with the substrates benzaldehyde and trans-2-hexenal when NAD+ was used as cofactor, as opposed to NADP+. However, with the substrates hexanal, 4-HNE, nonanal, octanal, trans-2-nonenal and trans-2-octenal, NADP+ appeared to be preferred. In addition, an apparent preference was demonstrated for the saturated aldehydes hexanal, nonanal and octanal over the corresponding unsaturated aldehydes of the same carbon length.
Table 1.
Enzymatic specific activity (nmol NAD(P)H/min per mg protein) of ALDH3B1-infected (ALDH3B1) and uninfected (control) Sf9 cell lysates with various aldehyde substrates.
| Cofactor-NAD+ | Cofactor-NADP+ | ||||
|---|---|---|---|---|---|
| Substrate | ALDH3B1 | control | ALDH3B1 | control | |
| Acetaldehyde | 50 μM | 1.06 ± 0.01 | 1.30 ± 0.06 | 0.17 ± 0.03 | 0.50 ± 0.11 |
| 250 μM | 1.23 ± 0.02 | 1.76 ± 0.05 | 0.20 ± 0.08 | 0.10 ± 0.04 | |
| 1 mM | 1.80 ± 0.09 | 2.23 ± 0.01 | *0.23 ± 0.03 | 0.08 ± 0.03 | |
| Benzaldehyde | 50 μM | *1.72 ± 0.05 | 1.23 ± 0.08 | 0.13 ± 0.04 | 0.09 ± 0.04 |
| 250 μM | *3.78 ± 0.38 | 1.68 ± 0.09 | *0.66 ± 0.06 | 0.18 ± 0.12 | |
| 1 mM | *6.43 ± 0.15 | 3.14 ± 0.18 | *1.87 ± 0.18 | 0.32 ± 0.08 | |
| 5 mM | *7.56 ± 0.13 | 6.04 ± 0.46 | *6.33 ± 0.17 | 0.27 ± 0.08 | |
| Betaine Aldehyde | 50 μM | 0.42 ± 0.10 | 0.92 ± 0.07 | 1.99 ± 0.63 | 1.24 ± 0.63 |
| 250 μM | 0.44 ± 0.11 | 0.93 ± 0.07 | 0.61 ± 0.34 | 1.76 ± 0.32 | |
| 1 mM | 0.46 ± 0.05 | 1.27 ± 0.04 | 0.97 ± 0.06 | 0.61 ± 0.20 | |
| DOPAL | 0.66 μM | 1.02 ± 0.13 | 2.36 ± 0.19 | 0.48 ± 0.23 | 0.22 ± 0.17 |
| 6.6 μM | 1.61 ± 0.52 | 2.32 ± 0.01 | *0.65 ± 0.05 | 0.37 ± 0.05 | |
| 19.8 μM | 2.77 ± 0.31 | 3.02 ± 0.17 | 0.17 ± 0.04 | 0.56 ± 0.15 | |
| Hexanal | 50 μM | 5.58 ± 0.22 | 4.88 ± 0.27 | *1.65 ± 0.17 | 0.68 ± 0.13 |
| 250 μM | *14.19 ± 0.37 | 12.43 ± 0.01 | *4.20 ± 0.08 | 0.72 ± 0.20 | |
| 1 mM | 14.40 ± 0.47 | 15.10 ± 0.12 | 2.61 ± 0.19 | 2.0 ± 0.37 | |
| 4-HNE | 50 μM | 4.01 ± 0.87 | 2.32 ± 0.18 | *1.38 ± 0.16 | 0.44 ± 0.05 |
| 100 μM | 4.22 ± 0.14 | 3.21 ± 0.67 | *3.52 ± 0.30 | 1.88 ± 0.14 | |
| 250 μM | *4.60 ± 0.18 | 3.18 ± 0.24 | *2.96 ± 0.15 | 0.56 ± 0.12 | |
| 1 mM | *5.41 ± 0.09 | 2.35 ± 0.54 | *5.14 ± 0.16 | 0.60 ± 0.07 | |
| MDA | 50 μM | 0.71 ± 0.03 | 1.34 ± 0.13 | 0.52 ± 0.24 | 3.0 ± 0.18 |
| 250 μM | 0.92 ± 0.03 | 1.49 ± 0.06 | 0.52 ± 0.36 | 1.46 ± 1.00 | |
| 1 mM | 0.87 ± 0.21 | 1.75 ± 0.07 | 0.72 ± 0.38 | 1.56 ± 0.98 | |
| Nonanal | 10 μM | 16.56 ± 0.45 | 23.0 ± 0.06 | *5.46 ± 0.50 | 2.66 ± 0.10 |
| 50 μM | *17.11 ± 0.28 | 10.29 ± 0.42 | *11.55 ± 0.31 | 2.0 ± 0.23 | |
| 100 μM | *13.56 ± 0.76 | 9.36 ± 0.32 | *8.85 ± 0.66 | 0.76 ± 0.20 | |
| Octanal | 10 μM | *10.77 ± 0.39 | 7.48 ± 0.28 | *3.77 ± 0.17 | 0.91 ± 0.29 |
| 50 μM | *20.22 ± 0.14 | 12.45 ± 0.06 | *12.40 ± 1.62 | 1.03 ± 0.09 | |
| 100 μM | *20.94 ± 0.20 | 14.69 ± 0.13 | *12.90 ± 0.19 | 1.32 ± 0.51 | |
| Propionaldehyde | 50 μM | 1.51 ± 0.04 | 2.04 ± 0.05 | 0.07 ± 0.04 | 0.13 ± 0.07 |
| 250 μM | 1.80 ± 0.07 | 1.98 ± 0.07 | 0.40 ± 0.06 | 0.28 ± 0.16 | |
| 1 mM | 2.73 ± 0.08 | 2.63 ± 0.23 | 0.65 ± 0.17 | 0.28 ± 0.08 | |
| 10 mM | 5.28 ± 0.10 | 4.97 ± 0.89 | 2.69 ± 0.37 | 1.75 ± 0.04 | |
| Trans-2-Hexenal | 50 μM | 1.04 ± 0.13 | 1.60 ± 0.15 | 0.14 ± 0.05 | 0.17 ± 0.12 |
| 250 μM | *2.36 ± 0.10 | 1.42 ± 0.07 | *0.80 ± 0.06 | 0.13 ± 0.05 | |
| 1 mM | *4.53 ± 0.29 | 1.13 ± 0.14 | *1.98 ± 0.17 | 0.16 ± 0.08 | |
| Trans-2-Nonenal | 10 μM | *3.58 ± 0.01 | 1.88 ± 0.10 | *3.0 ± 0.27 | 0.28 ± 0.09 |
| 50 μM | *3.59 ± 0.39 | 1.78 ± 0.01 | *6.90 ± 0.05 | 0.62 ± 0.08 | |
| 250 μM | *3.85 ± 0.19 | 1.82 ± 0.04 | *2.31 ± 0.65 | 0.34 ± 0.29 | |
| Trans-2-Octenal | 50 μM | *4.85 ± 0.09 | 2.04 ± 0.94 | *1.18 ± 0.16 | 0.15 ± 0.05 |
| 250 μM | *5.87 ± 0.09 | 1.80 ± 0.09 | *1.55 ± 0.07 | 0.18 ± 0.06 | |
| 1 mM | *4.24 ± 0.68 | 0.85 ± 0.09 | *0.79 ± 0.13 | 0.61 ± 0.12 | |
Specific activity significantly greater than that in control. Data represent the mean ± S.E. from triplicate measurements.
Tissue Expression
Western blot analysis of organs harvested from male C57BL/129sv mice indicate that ALDH3B1 is highly expressed in both mouse kidney and liver (Figure 3A). Only low expression of ALDH3B1 was observed in mouse lung. Because ALDH3B1 has been hypothesized to play a role in schizophrenia, its expression in various brain regions was investigated. ALDH3B1 is expressed in moderate amounts in mouse cortex, striatum and hippocampus (Figure 3B). Mouse brainstem and cerebellum also express ALDH3B1, albeit to a lesser extent.
Figure 3.
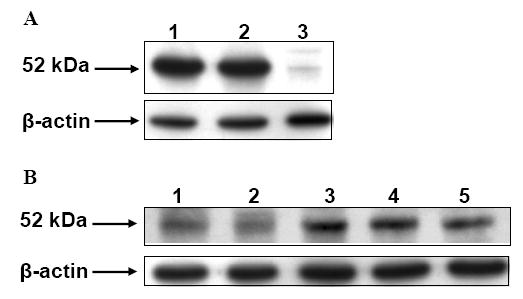
ALDH3B1 expression in mouse tissues. Organs and tissues from male C57BL/129sv mice were harvested and processed for Western blot analysis. (A) ALDH3B1 expression in kidney (lane 1), liver (lane 2) and lung (lane 3). (B) ALDH3B1 expression in regions of the brain. Brainstem (lane 1), cerebellum (lane 2), cortex (lane 3), striatum (lane 4) and hippocampus (lane 5). n=3
Protection against aldehyde-induced cytotoxicity
To investigate the potential role of ALDH3B1 in vivo, stably-transfected HEK293 cell lines were created. Figure 4 shows the expression of human ALDH3B1 in transfected clones. A low level of endogenous expression of ALDH3B1 is also evident in the vector-transfected clone (Figure 4). ALDH3B1/HEK293 clone 8 was used in subsequent experiments. To determine if ALDH3B1 may play a role in oxidative stress, ALDH3B1- and vector-transfected HEK293 cells were exposed to the lipid peroxidation product octanal and analyzed for cell survival. Figure 5 indicates a protective role for ALDH3B1 against octanal-induced cytotoxicity. Cell survival of ALDH3B1-transfected cells was significantly higher than vector-transfected cells at octanal concentrations at and exceeding 10 μM (Figure 5). For instance, at 1 mM octanal, 60% of the vector-transfected cells died whereas only 17% of the ALDH3B1-transfected cells died. These findings correlate with the enzymatic activity data presented in Table 1 in which ALDH3B1 was active with octanal.
Figure 4.
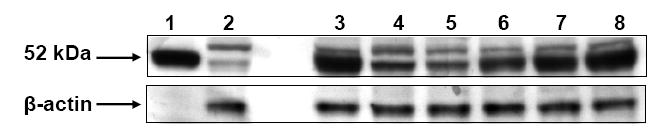
Human ALDH3B1 expression in HEK293 cells. Cell extracts from HEK293 clones stably transfected with human ALDH3B1 (lanes 3-8) or transfected with vector alone (lane 2) were subjected to SDS/PAGE and analyzed for ALDH3B1 expression by Western blotting. Lane 1 (positive control) represents cell extracts from ALDH3B1-infected Sf9 cells. n=3
Figure 5.
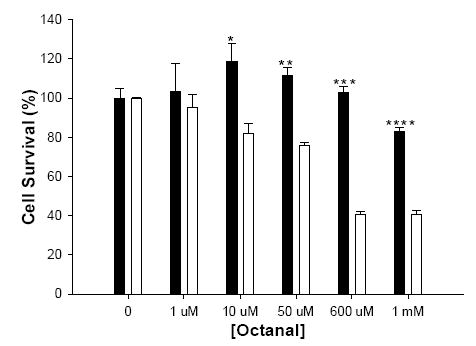
Survival of HEK293 cells treated with octanal. ALDH3B1(clone 8)- (■) and vector- (□) stably transfected HEK293 cells were treated with octanal (0-1 mM) for 24 h and assayed for cell survival by the SRB assay. (*P<.05; **P<.01; ***P<.001; ****P<.0001) Data represent the mean ± S.E. from triplicate assays.
Discussion
Aldehydes are produced during numerous processes including those associated with oxidative stress. Indeed, reactive oxygen species (ROS) cause lipid peroxidation, a process that results in the degradation of cellular membranes and the generation of endogenous toxins, such as free radicals and over 200 species of electrophilic aldehydes [14]. These aldehydes mediate various toxic effects, such as GSH depletion, protein/DNA adduction and impaired cellular homeostasis, and they have been implicated in many pathological states, including neurodegenerative diseases [21] and alcoholic liver disease [22]. ALDH-mediated detoxification of aldehyde products of lipid peroxidation, therefore, represents a critical defense.
Increasing evidence suggests that the ALDH3 family, composed of ALDH3A1, ALDH3A2, ALDH3B2, and ALDH3B1, is important in the defense against aldehydes and related oxidative processes. In the cornea, ALDH3A1 protects ocular structures against UV-induced damage through the metabolism of lipid peroxidation-derived aldehydes [23], the absorption of UV-light [24] and the scavenging of ROS [18; 25]. Indeed, Aldh3a1-null mice develop cataracts [24]. ALDH3A2 catalyzes the oxidation of fatty aldehydes derived from such precursors as fatty alcohols and ether glycerolipids [1]. Loss of ALDH3A2 catalytic function, from genetic mutations in the ALDH3A2 gene, results in SLS, a disorder characterized by congenital ichthyosis, mental retardation, and ocular abnormalities [26]. The significance of ALDH3B2 is unknown; the presence of an in-frame stop codon downstream from the first methionine, suggests it may be a pseudogene [9]. Similarly, until now, nothing was known about the functionality or significance of ALDH3B1. The results described in the present study support the notion that ALDH3B1 may be involved in the defense against oxidative stress and, specifically, lipid peroxidation-derived aldehydes.
ALDH3B1 shares a high sequence homology with ALDH3 isozymes and contains the residues essential for catalysis and cofactor binding, including the invariant catalytic nucleophile, Cys-244. Glu-334, also highly conserved, is important in activating the thiol of Cys-244 to a thiolate, which forms a tetrahedral intermediate with the substrate aldehyde [19]. Gly-241 appears necessary for correct positioning of Cys-244 [27]. Asp-248, conserved in only ALDH3 isozymes, is thought to maintain active-site geometry [19]. Gly-212 marks the boundary between the catalytic and coenzyme-binding domains. Gly-188 and Gly-193 are essential residues of the NAD(P)+-binding site of the ALDH Rossmann fold (GxxxxG) [20]. Lys-138, Glu-334 and Phe-336 also play important roles in cofactor binding by positioning the nicotinamide ring and hydrogen bonding to the adenine ribose of NAD(P)+ [27; 28]. Ile-335 and Lys-214 maintain the geometry of the cofactor-binding domain [28]. Asn-115, essential for dehydrogenase activity, transiently stabilizes the carbonyl oxygen of the tetrahedral intermediate during catalysis [28]. Hydride transfer to the nicotinamide ring of the cofactor leads to the collapse of the tetrahedral intermediate to a thiolester, which is hydrolyzed by a water molecule activated by Glu-210 with concomitant release of the carboxylic acid product [19]. Arg-26, Gly-106, Pro-117, Gly-132, Pro-338, Gly-384, Asn-389 and Gly-404 are highly conserved residues believed to lie in critical turn areas in ALDH3 isozymes [28; 29].
The presence of the above-mentioned critical residues in the primary sequence of ALDH3B1 indicated that this protein may be a catalytically active enzyme. Accordingly, our findings indicate that the human ALDH3B1 gene encodes a metabolically-active enzyme that has distinct specificity for various aldehyde substrates, including many derived from lipid peroxidation such as hexanal, 4-HNE, nonanal, octanal, trans-2-hexenal, trans-2-nonenal, and trans-2-octenal. As shown for octanal, ALDH3B1 is catalytically-active and may play an important physiological role in detoxifying these aldehydes. While future studies will also examine the activity of purified ALDH3B1, the use of cell lysates in this study has the advantage of reflecting the contribution of the enzyme to aldehyde oxidation in a more realistic cellular environment. Our observed high expression of ALDH3B1 in liver and kidney provides further support for the hypothesis that ALDH3B1 may play a role in the detoxification of aldehydes produced during oxidative stress.
A SNP in intron 2 (rs581105; T/G) of the human ALDH3B1 gene has recently been associated with schizophrenia [6; 7]. Introns are involved in alternative splicing of genes and intronic SNPs have been implicated in diseases, including schizophrenia, due to altered pre-mRNA splicing and subsequent exon deletion [30]. An alternative splice variant of human ALDH3B1 has been found that lacks both exons 3 and 4, resulting in a shorter protein isoform (b). While isoform b has the same amino and carboxyl termini as isoform a, amino acids 55-91 are missing. It is, therefore, likely that isoform b has altered enzymatic activity, which potentially could have pathophysiological implications. Further investigation of the ALDH3B1 intronic SNP, its potential connection to isoform b and significance in schizophrenia is necessary in order to draw any conclusions. The ALDH3B1 SNP genotype has been proposed to be associated with schizophrenia through an alteration in dopamine metabolism [6]. While our findings indicate that ALDH3B1 is present in the brain, the dopamine-derived aldehyde, DOPAL, seems to be a poor substrate. On the other hand, ALDH3B1 may protect the brain by detoxifying other aldehydes, such as those produced during lipid peroxidation. Indeed, this protective role of ALDH3B1 is evident from our results in ALDH3B1-transfected HEK293 cells treated with the lipid peroxidation product, octanal. Despite a low level of endogenous expression of ALDH3B1 in vector-transfected cells (not surprising given the kidney origin of HEK293 cells), high levels of ALDH3B1 over-expression were achieved in transfected clones and significant protection against octanal-induced cytotoxicity was demonstrated. Other ALDHs, including ALDH3A1, have been shown to protect cells against cytotoxicity caused by lipid peroxidation-derived aldehydes [23].
Although the major function of ALDHs is the NAD(P)+-dependent oxidation of aldehydes, these enzymes appear to possess multiple catalytic and non-catalytic properties [5]. For example, ALDH1A1, ALDH2, ALDH3A1 and ALDH4A1 can catalyze ester hydrolysis and ALDH2 may have nitrate reductase activity [5; 31]. In addition, ALDH proteins are capable of non-catalytic interactions with various compounds. In this regard, ALDH2 has been shown to bind acetaminophen while ALDH1A1 binds both exogenous (daunorubicin, quinoline drugs) and endogenous compounds (androgen, cholesterol, thyroid hormone) [1; 32]. ALDHs may also play a critical role in cellular homeostasis by maintaining redox balance. For example, ALDHs may scavenge hydroxyl radicals via the thiol groups of their Cys and Met residues [25]. Additionally, ALDH isozymes may contribute to cellular antioxidant capacity by generating NAD(P)H, which is critical for the regeneration of GSH and may also function as a direct antioxidant by reducing glutathiyl (GS•) and tyrosyl radicals [33]. Finally, structural functions have been postulated for some ALDHs, including ALDH1A1 and ALDH3A1, both believed to contribute to and maintain corneal transparency [25]. Given the diversity of the ALDH family, it is possible that ALDH3B1 has multiple catalytic and non-catalytic functions aside from aldehyde metabolism. Future studies will elucidate such functions of ALDH3B1 and provide further insight into the physiological role and significance of this protein.
Acknowledgments
The work described in this report was supported by NIH Grant EY11490.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36:279–299. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl R. Aldehyde dehydrogenases and their role in carcinogenesis. Crit Rev Biochem Mol Biol. 1992;27:283–335. doi: 10.3109/10409239209082565. [DOI] [PubMed] [Google Scholar]
- 3.Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, Willemsen MA, Omran H, Tacke U, Uhlenberg B, Weschke B, Clayton PT. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med. 2006;12:307–309. doi: 10.1038/nm1366. [DOI] [PubMed] [Google Scholar]
- 4.Kamino K, Nagasaka K, Imagawa M, Yamamoto H, Yoneda H, Ueki A, Kitamura S, Namekata K, Miki T, Ohta S. Deficiency in mitochondrial aldehyde dehydrogenase increases the risk for late-onset Alzheimer’s disease in the Japanese population. Biochem Biophys Res Commun. 2000;273:192–196. doi: 10.1006/bbrc.2000.2923. [DOI] [PubMed] [Google Scholar]
- 5.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Q, Jia YB, Zhang BY, Zou K, Tao YB, Wang YP, Qiang BQ, Wu GY, Shen Y, Ji HK, Huang Y, Sun XQ, Ji L, Li YD, Yuan YB, Shu L, Yu X, Shen YC, Yu YQ, Ju GZ. Association study of an SNP combination pattern in the dopaminergic pathway in paranoid schizophrenia: a novel strategy for complex disorders. Mol Psychiatry. 2004;9:510–521. doi: 10.1038/sj.mp.4001472. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Jia Y, Zhang X, Xu Q, Shen Y, Li Y. Multi-locus association study of schizophrenia susceptibility genes with a posterior probability method. Sci China C Life Sci. 2005;48:263–269. doi: 10.1007/BF03183620. [DOI] [PubMed] [Google Scholar]
- 8.Hsu LC, Chang WC, Yoshida A. Cloning of a cDNA encoding human ALDH7, a new member of the aldehyde dehydrogenase family. Gene. 1994;151:285–289. doi: 10.1016/0378-1119(94)90672-6. [DOI] [PubMed] [Google Scholar]
- 9.Hsu LC, Chang WC, Yoshida A. Human aldehyde dehydrogenase genes, ALDH7 and ALDH8: genomic organization and gene structure comparison. Gene. 1997;189:89–94. doi: 10.1016/s0378-1119(96)00839-6. [DOI] [PubMed] [Google Scholar]
- 10.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 12.Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA DNA. Cell Biol. 2003;22:329–338. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- 13.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376:615–623. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson GE, Tottmar O. Biogenic aldehydes in brain: on their preparation and reactions with rat brain tissue. J Neurochem. 1987;48:1566–1572. doi: 10.1111/j.1471-4159.1987.tb05702.x. [DOI] [PubMed] [Google Scholar]
- 16.Vasiliou V, Lee J, Pappa A, Petersen DR. Involvement of p65 in the regulation of NF-kappaB in rat hepatic stellate cells during cirrhosis. Biochem Biophys Res Commun. 2000;273:546–550. doi: 10.1006/bbrc.2000.2993. [DOI] [PubMed] [Google Scholar]
- 17.Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lassen N, Pappa A, Black WJ, Jester JV, Day BJ, Min E, Vasiliou V. Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med. 2006;41:1459–1469. doi: 10.1016/j.freeradbiomed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Hempel J, Kuo I, Perozich J, Wang BC, Lindahl R, Nicholas H. Aldehyde dehydrogenase. Maintaining critical active site geometry at motif 8 in the class 3 enzyme. Eur J Biochem. 2001;268:722–726. doi: 10.1046/j.1432-1327.2001.01926.x. [DOI] [PubMed] [Google Scholar]
- 20.Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics. 1999;9:421–434. [PubMed] [Google Scholar]
- 21.Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci USA. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch OR, Pani G, Borrello S, Colavitti R, Cravero A, Farre S, Galeotti T. Oxidative stress and antioxidant defenses in ethanol-induced cell injury. Mol Aspects Med. 2004;25:191–198. doi: 10.1016/j.mam.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–1189. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 24.Lassen N, Bateman JB, Estey T, Kuszak JR, Piatigorsky J, Duester G, Vasiliou V. Cataract phenotype in Aldh1a1-/-/Aldh3a1-/- double knockout mice. Invest Ophthalmol Vis Sci; The Association for Research in Vision and Ophthalmology (ARVO) 2006 Annual Meeting; 2006. p. 4105. E-Abstract. [Google Scholar]
- 25.Estey T, Piatigorsky J, Lassen N, Vasiliou V. ALDH3A1: a corneal crystallin with diverse functions. Exp Eye Res. 2006 doi: 10.1016/j.exer.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Rizzo WB, Carney G. Sjogren-Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2) Hum Mutat. 2005;26:1–10. doi: 10.1002/humu.20181. [DOI] [PubMed] [Google Scholar]
- 27.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 28.Perozich J, Nicholas H, Wang BC, Lindahl R, Hempel J. Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 1999;8:137–146. doi: 10.1110/ps.8.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hempel J, Liu ZJ, Perozich J, Rose J, Lindahl R, Wang BC. Conserved residues in the aldehyde dehydrogenase family. Locations in the class 3 tertiary structure. Adv Exp Med Biol. 1997;414:9–13. doi: 10.1007/978-1-4615-5871-2_2. [DOI] [PubMed] [Google Scholar]
- 30.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 31.Sladek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17:7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 32.Graves PR, Kwiek JJ, Fadden P, Ray R, Hardeman K, Coley AM, Foley M, Haystead TA. Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol Pharmacol. 2002;62:1364–1372. doi: 10.1124/mol.62.6.1364. [DOI] [PubMed] [Google Scholar]
- 33.Kirsch M, De GH. NAD(P)H, a directly operating antioxidant? FASEB J. 2001;15:1569–1574. doi: 10.1096/fj.00-0823hyp. [DOI] [PubMed] [Google Scholar]


