Abstract
Summary: The genome of the human food-borne pathogen Listeria monocytogenes is predicted to encode a high number of surface proteins. This abundance likely reflects the ability of this bacterium to survive in diverse environments, including soil, food, and the human host. This review focuses on the various mechanisms by which listerial proteins are attached at the bacterial surface and their many functions, including peptidoglycan metabolism, protein processing, adhesion to host cells, and invasion of host tissues. Extensive in silico analysis of the domains or motifs present in these mosaic proteins reveals that diverse structural features allow the surface proteome to interact with diverse bacterial or host components. This diversity offers new clues about the molecular bases of Listeria pathogenesis.
INTRODUCTION
Bacterial surface proteins constitute a diverse group of molecules involved in various important processes, such as bacterial growth, sensing of and protection from environmental stresses, adhesion, invasion of host cells, signaling, and interaction with the immune system. Because of these various functions, an in-depth characterization of the surface protein repertoire is required to better understand the factors that contribute to the success of a bacterial pathogen in colonizing different environmental niches and its mammalian host. This is especially the case for the food-borne pathogen Listeria monocytogenes, whose ability to survive in diverse environments, including food and the cytosol of eukaryotic cells, relies in part on its complex surface proteome. Annotation of the first L. monocytogenes genome (strain EGDe) (67) predicted a total of 2,853 proteins, among which were 133 surface proteins that were precisely classified according to their different anchoring systems and potential structural domains (30). Accumulation of genome sequence information, new predictive bioinformatics approaches, comparative genomics, proteomics, X-ray three-dimensional structure determination of specific domains, and the recent characterization of mutants in animal models has since then provided new insight into the L. monocytogenes “surfaceome.” We present here an updated characterization of listerial surface proteins present in the L. monocytogenes model strain EGDe (67), focusing on their highly modular nature. We have performed an in-depth annotation of surface proteins and combined these in silico analyses with recently published experimental data. This review emphasizes the diversity of modules dedicated to cell envelope association and that of other modules involved in more specific functions, such as cell wall metabolism or host-pathogen interactions. Many of these modules are structurally related to domains found in eukaryotic proteins and have evolved to bind a variety of bacterial or host receptors.
To annotate protein domains, we have combined sequence information from the database dedicated to the analysis of the genomes of L. monocytogenes (strain EGDe) and of its nonpathogenic relative Listeria innocua (strain CLIP 11262) (http://genolist.pasteur.fr/ListiList [67]) with that from the Pfam database (http://www.sanger.ac.uk/Software/Pfam [11]). We also used information from the NCBI Conserved Domain database (194) and the Interpro database (98). Sequence comparisons were performed using a PSI-BLAST analysis.
THE L. MONOCYTOGENES CELLULAR ENVELOPE
A prerequisite to understand how proteins remain attached to the bacterial envelope is a good knowledge of the biochemical characteristics of this compartment. Most of the descriptions of the composition and suspected structure of the L. monocytogenes membrane, peptidoglycan, and associated polymers were made about 20 to 40 years ago (57, 65, 171, 180, 181). However, this topic now is receiving further attention, which should help to refine the overall structure (for a review, see reference 145).
Membrane
The listerial membrane is ∼90 Å thick and is composed of 55 to 60% protein, 30 to 35% lipid, and 1.3 to 2.3% carbohydrate (glucose, galactose, ribose, and arabinose) (65). Lipids include 80 to 85% phospholipids, such as phosphatidylglycerol, diphosphatidylglycerol, and phosphoglycolipid (185). The L. monocytogenes membrane fatty acid composition is dominated to an unusual extent by branched-chain fatty acids (>90% of the total fatty acid content) (6) and is modulated upon temperature variation. For instance, upon cold shock the anteiso-C15:0 content rises to maintain optimal membrane fluidity (6, 51, 132). However, nothing is yet known about the variation of the plasma membrane composition during Listeria cellular infection and how it affects anchoring of membrane proteins.
Peptidoglycan
A striking characteristic of the Listeria peptidoglycan is its similarity with that of gram-negative bacteria, such as Escherichia coli (82, 162). It is a polymer of alternating units of the disaccharide N-acetylmuramic acid (MurNAc)-(β-1,4)-N-acetylglucosamine (GlcNAc), cross-linked by peptidic bridges. Muropeptides, which in L. monocytogenes are l-alanyl-γ-d-glutamyl-meso-diaminopimelyl-d-alanine-d-alanine, are bound to the MurNAc residue and are connected by a direct link between the d-Ala residue of one lateral peptide and the meso-diaminopimelyl residue of the other stem peptide (44, 57, 89, 162). Partial deacetylation of GlcNAc residues by PgdA enzyme is another characteristic of Listeria peptidoglycan (21, 89) (see below).
Cell Wall Secondary Polymers: Teichoic Acids and Lipoteichoic Acids
L. monocytogenes contains two different polyanionic polymers decorating the cell wall: the teichoic acids (TAs), which are covalently bound to the peptidoglycan, and the lipoteichoic acids (LTAs), which are embedded into the plasma membrane by a diacylglycerolipid (128, 130). These polymers play important functions in metal cation homeostasis, anchoring of surface proteins, and transport of ions, nutrients, and proteins and are main determinants of surface immunogenicity, conferring most of the basis of the serotype diversity known in L. monocytogenes. Of note, d-Ala esterification of TAs and LTAs is important for L. monocytogenes pathogenicity (1, 118).
TAs are electronegative polymers of ribitol-phosphates substituted with d-Ala residues and diverse sugars, which vary depending on the serotype (57-59, 145, 155). For instance, in serovar 1/2 (e.g., serotypes 1/2a and 1/2b), GlcNAc and rhamnose are present as substituents on the ribitol, whereas in serovar 4, GlcNAc is integral to the TA chains. Interestingly, serotype 4b strains are unique in bearing both galactose and glucose substituents on the GlcNAc of TA. This particularity is related to the presence in these strains of specific genes, such as gtcA, gltA, and gltB (106, 144).
The distinct glycosylations of TAs are probably of importance for their specific recognition by listerial proteins or phage endolysins (113) (see below). LTAs are polymers of glycerophosphate substituted with Ala, galactose, and lipid residues. The glycerophosphate chain of LTAs is uncovalently anchored to the outer leaflet of the plasma membrane through a lipid anchor, composed of galactose bound to glycerol and substituted with fatty acids (69, 79, 145). The LTAs are both membrane associated and secreted in the growth medium (58). Their X-ray structure suggests that they adopt a compact, organized micellar form with a length of 10 to 20 nm (101).
CLASSIFICATION OF LISTERIAL SURFACE PROTEINS ACCORDING TO THEIR ANCHORING MECHANISMS
To be specifically localized at the cell surface, proteins from gram-positive bacteria need to translocate across the membrane and then associate with a cell surface component. Surface localization of proteins relies on the presence of specific domains or motifs known to mediate secretion and attachment to the cell envelope. The first step of transport across the membrane occurs by two main pathways, both of which require a specific N-terminal secretion signal: the general secretory Sec pathway and, to a lesser extent, the Tat pathway (176). However several alternative pathways have been discovered, such as the SecA2 pathway (27, 108) or specialized pathways for secretion of specific proteins, such as the pseudopilin export pathway, various ABC transporter pathways, the holin systems (176), and the ESAT-6/WXG100 secretion system (135, 136). The presence of these secretion systems in L. monocytogenes has been extensively reviewed recently by Desvaux and Hebraud (43). In this review, we focus only on proteins exported through the Sec and SecA2 pathways, as well as flagellin, which is secreted by the specialized flagellar export machinery. The numerous nonsecreted integral membrane proteins that presumably also expose specific domains on the outside of the membrane bilayer are not discussed in this review.
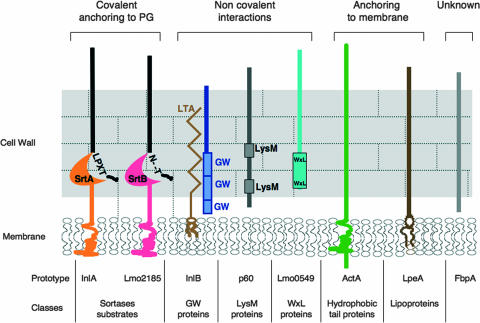
Through extensive in silico analysis of the genome of L. monocytogenes EGDe, Trost et al. (179) were able to predict a total of 525 sequences coding for proteins carrying a signal peptide, which corresponds to 18.4% of the total number of protein-coding genes. Among them, the “surface proteins” discussed in this review are those carrying motifs enabling strong or weak interactions with at least one component of the bacterial envelope (Fig. 1).
FIG. 1.
The different types of surface proteins found in L. monocytogenes. A prototype of each family is given.
Proteins Covalently Linked to the Peptidoglycan: Sortase Substrates
LPXTG sorting signal.
The best known class of L. monocytogenes surface proteins is that of the LPXTG proteins, which in strain EGDe comprises 41 members. They are characterized by a short C-terminal sorting signal predicted to direct covalent attachment to the peptidoglycan (Fig. 2) (Pfam PF00746, http://www.sanger.ac.uk/Software/Pfam). Originally characterized in Streptococcus pyogenes protein M and Staphylococcus aureus protein A, this signal consists of an LPXTG motif, followed by an hydrophobic domain of about 20 amino acids and a tail of positively charged residues (60, 128). The sorting signal is the substrate of sortase, a membrane-bound transpeptidase, which cleaves the LPXTG motif between the threonine and glycine residues and catalyzes the formation of an amide link between the carboxyl group of the threonine and cell wall precursors (123, 177). For a thorough analysis of the sorting mechanism, we refer the reader to an excellent recent review on sortases (121). The most studied LPXTG protein in L. monocytogenes is internalin (InlA), which promotes bacterial entry into epithelial cells (61, 73). InlA harbors an LPTTG motif and is anchored covalently by the sortase SrtA to meso-diaminopimelic acid residues of the peptidoglycan (17, 44, 62). The recently discovered virulence factor LPXTG protein Vip is also localized on the bacterial surface by an SrtA-dependent pathway (32). Using a novel nongel proteomic method, Pucciarelli et al. (146) were able to identify 13 SrtA substrate proteins (Fig. 2), through comparison of the peptidoglycan contents from wild-type EGDe and sortase-defective mutants grown in rich medium. The remaining LPXTG proteins predicted by the genome may not be produced under the growth conditions used in that study.
FIG. 2.
Sortase substrates. A schematic representation of L. monocytogenes LPXTG and NXXTX proteins is shown. The numbers within domains indicate the number of repeats. Nineteen proteins are absent in L. innocua and are indicated in green letters. Proteins detected in the L. monocytogenes cell wall fraction (146) are indicated by an arrow. The two sortase B substrates, Lmo2185 and Lmo2186, are drawn on the right of LPXTG proteins. AA, amino acids. (Adapted from reference 30 with permission from Elsevier.)
The L. monocytogenes genome encodes the highest number of LPXTG proteins among all gram-positive bacteria whose genome sequences are known. InlA and 18 other family members remarkably display an N-terminal leucine-rich repeat (LRR) domain and belong to the so-called “internalin family” (Fig. 2) (30, 73), which also includes the GW protein InlB, the WxL protein Lmo0549 (see below), and four proteins without any obvious surface anchoring motif, among which is InlC (73). The LRR domain consists of tandemly arranged repeats of 22 amino acids each. In InlA, InlB, and 10 other surface internalins, the LRR domain is flanked at its C terminus by a conserved LRR-adjacent domain (Fig. 2; also see below). InlA, InlB, InlG, InlH, InlE, and InlF also possess a region of B repeats. The crystal structures of the LRR domains of InlA, InlB, InlH, and InlE have been solved, revealing a curved structure ideally shaped for protein-protein interaction (120, 133, 164). However, while the LRR domains of InlA and InlB are structurally related, they bind to very different mammalian receptors, namely, E-cadherin (Ecad) and Met (125, 167). Therefore, one cannot assume that members of the internalin family share similar functions solely on the basis of their structural classification. Of the internalin-like proteins, only InlA and InlB have so far been identified as invasins (see below).
NXXTX sorting signal.
A second sortase type, sortase B (SrtB), was originally identified in S. aureus and specifically recognizes and cleaves a sorting signal different from LPXTG (121, 124). Staphylococcal SrtB anchors its only known substrate, IsdC, by cleaving a C-terminal NPQTN motif, between threonine and asparagine, and linking threonine to the pentaglycine cross bridges. The genes encoding SrtB and IsdC are both part of the isd locus, which is involved in bacterial heme iron uptake (121, 124). L. monocytogenes has also an alternative sortase B, whose coding sequence maps to an operon containing two genes encoding putative substrates, Lmo2185 (formerly SvpA) and Lmo2186 (16). Both proteins share homology to S. aureus IsdC and bear as putative sorting motifs NAKTN (Lmo2185) or NKVTN or NPKSS (Lmo2186), respectively. Inactivation of srtB impairs sorting of Lmo2185 (16). Lmo2185 and Lmo2186 are the only two SrtB substrates present in the L. monocytogenes cell wall proteome, consistent with the fact that there is no other protein with an NXXTX sorting signal encoded in the L. monocytogenes genome (146).
Work with S. aureus and analysis of other complete gram-positive genomes suggest that SrtB enzymes may have evolved to specifically target some proteins involved in iron uptake (49, 121). IsdC and Lmo2186 each contain one NEAT (near transporter) domain, and Lmo2185 contains three NEAT domains that are expected to play a role in iron transport (Fig. 2) (5). Interestingly, the L. monocytogenes operon containing the lmo2185, lmo2186, and srtB genes is regulated by the iron-responsive transcriptional repressor Fur and is induced under iron-deficient conditions (131). However, neither Lmo2185 nor Lmo2186 allows hemin, hemoglobin, or ferrichrome utilization (131). The exact functions of these two surface proteins, which are conserved in all Listeria species, remain to be characterized more precisely.
Proteins with Noncovalent Association to the Cell Wall
A second group of cell surface proteins in gram-positive bacteria comprises those that are thought to bind to the cell wall by noncovalent interactions, most often via repeated domains (Table 1) (for extensive recent reviews, see references 42, 89, and 165).
TABLE 1.
L. monocytogenes EGDe proteins predicted to carry motifs promoting noncovalent association to the cell wall
| Proteina | Name | Cell wall association domain(s) | Predicted function and/or domain |
|---|---|---|---|
| Lmo0434 | InlB | GW modules | Bacterial invasion into host cells, LRR |
| Lmo1076 | Auto | GW modules | Autolysin, N-acetylglucosaminidaseb |
| Lmo1215 | GW modules | Autolysin, N-acetylglucosaminidaseb | |
| Lmo1216 | GW modules | Autolysin, N-acetylglucosaminidaseb | |
| Lmo1521 | GW modules | Autolysin, N-acetylmuramoyl-l-alanine amidaseb | |
| Lmo2203 | GW modules | Autolysin, N-acetylglucosaminidaseb | |
| Lmo2713 | GW modules | Unknown | |
| Lmo2558 | Ami | GW modules | Autolysin, N-acetylmuramoyl-l-alanine amidaseb |
| Lmo2591 | GW modules | Autolysin, N-acetylglucosaminidaseb | |
| Lmo0582 | P60 (Iap) | LysM | γ-d-Glutamyl-l-m-Dpm peptidaseb |
| Lmo0880 | LysM and LPXTG | Unknown, collagen binding domainc | |
| Lmo1303 | LysM | Unknown | |
| Lmo1941 | LysM | Unknown | |
| Lmo2522 | LysM | Unknown, C-terminal domain with similarities to lysozymeb | |
| Lmo2691 | MurA | LysM | Autolysin, N-acetylglucosaminidaseb |
| Lmo549 | WxL | Unknown, CscC like, LRRd | |
| Lmo551 | WxL | Unknown, CscB liked | |
| Lmo585 | WxL | Unknown, CscB liked | |
| Lmo587 | WxL | Unknown, CscC liked | |
| Lmo1851 | Peptidoglycan binding domain | Unknown, peptidase domaine |
GW modules.
The first characterized protein of this type in L. monocytogenes was InlB, a protein of the internalin family required for L. monocytogenes entry into many eukaryotic cell types (15, 47, 73). The InlB C-terminal domain comprises three highly conserved tandem repeats of ∼80 amino acids, which are termed GW modules as they begin with the dipeptide Gly-Trp (25). GW modules are necessary and sufficient to anchor InlB to the bacterial surface by binding LTAs (86). These modules also interact with eukaryotic molecules, namely, glycosaminoglycans (GAGs) and gC1q-R (87, 119).
Eight additional proteins carrying a variable number of GW modules are encoded in the L. monocytogenes EGDe genome; seven are putative autolysins containing cell wall hydrolase domains, while Lmo2713 is of unknown function (Table 1; Fig. 3A). Among the autolysins are Ami and Auto, which also are implicated in interactions with eukaryotic cells (32, 127). Auto is the only autolysin of this subfamily that is absent in L. innocua. Interestingly, the aut gene, encoding Auto, is in a locus comprising several genes presumably involved in TA synthesis, some of which are absent in L. innocua. Whether these genes products are required for association of Auto with the cell wall is at present unknown.
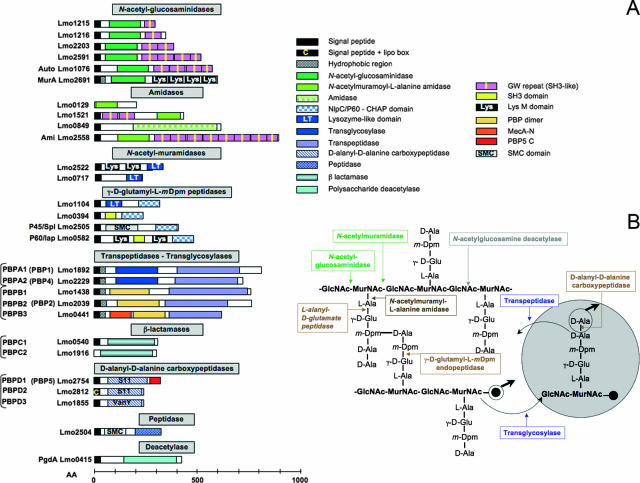
FIG. 3.
Surface proteins predicted to be involved in cell wall metabolism. (A) Schematic representation of L. monocytogenes surface proteins potentially involved in cell wall synthesis, hydrolysis, or modification. AA, amino acids. Most of the enzymatic activities are deduced from domain sequence similarities and require biochemical experimental validation. (B) Cell wall synthetase and hydrolase activities on Listeria peptidoglycan. Glycan chains of GlcNAc and MurNAc are interlinked by direct peptide linkage between the meso-diaminopimelic acid (m-Dpm) residue of one wall peptide and the d-alanyl residue at position 4 of the adjacent wall peptide. A peptidoglycan precursor linked to the undecaprenyl pyrophosphate group (black circle) is represented by a spotted circle. The precursor is incorporated to the preexisting peptidoglycan by the formation of two bonds: (i) a transglycosylase splits the pyrophosphate bond between the undecaprenyl group and the MurNAc of a nascent glycan strand and forms a glycosidic bond between MurNAc and the hydroxyl group of the GlcNAc of the precursor molecule, and (ii) a transpeptidase forms a dd-peptide bond between the carboxyl group of the d-Ala of the precursor and the amino group of an m-Dpm residue present in a peptide moiety of the cell wall. An example of each type of bond attacked by different specific murein hydrolases is also shown. (Adapted from reference 82 with permission.)
Association of the GW domain with LTA occurs whether the protein is produced within the bacterium or added externally (25, 86). It displays specificities, as GW modules of InlB do not bind to the surface of L. innocua or to that of Streptococcus pneumoniae (86). In addition, the strength of the association seems to increase with the number of GW modules. Thus, an InlB variant bearing the eight GW modules of the autolysin Ami binds more efficiently to the cell surface (25). The InlB GW modules are structurally related to Src homology 3 (SH3) domains found in many eukaryotic adaptor proteins, but the binding site for proline-rich ligands of bona fide SH3 domains is obstructed (119, 133). Interestingly, two non-GW surface proteins, the autolysin P60 and Lmo0394 (Fig. 3), also contain an SH3-related domain (Pfam PF08239). Whether this domain interacts with cell wall polymers is unknown. A related bacterial SH3 domain (Pfam PF08460) mediates binding to the cell wall of lysostaphin, a bacteriocin secreted by Staphylococcus simulans (9).
GW modules like those in Listeria promote surface localization of several staphylococcal surface autolysins, such as AtlC from Staphylococcus caprae (2), AtlE from Staphylococcus epidermidis (77), and Aas from Staphylococcus saprophyticus (78). Domains containing 20-amino-acid repeat units beginning with GW but weakly related to Listeria GW modules are present in S. pneumoniae surface proteins, such as LytA (55, 56, 183) and PspA (197), or in clostridial proteins, such as CspA of Clostridium acetobutylicum (158) or the toxin ToxB of Clostridium difficile (56). These modules are responsible for binding of the protein to choline residues that decorate the TAs and LTAs in these species (29, 63).
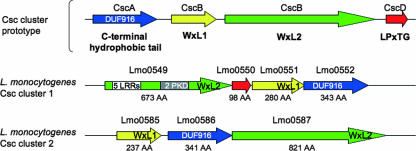
WxL domain.
A novel type of cell wall association domain of 160 to 190 amino acids, termed the WxL domain since it contains two conserved sequence motifs with the Trp-x-Leu signature, was recently identified in surface proteins of Lactobacillus plantarum and Enterococcus faecalis (28, 168). WxL proteins are present in many low-GC gram-positive bacteria and belong to the so-called Csc family of surface proteins. Csc gene clusters typically encode CscA, a protein with a conserved DUF916 domain of unknown function and a C-terminal transmembrane anchor; CscB and CscC, which display a C-terminal WxL domain; and CscD, a small LPXTG protein (Fig. 4). Csc proteins are proposed to form a multicomponent complex at the bacterial surface, although there is still no evidence for such a hypothesis (168). The EGDe L. monocytogenes genome contains two Csc-like gene clusters, among which four genes encode WxL domain-containing proteins (Lmo0549, Lmo0551, Lmo0585, and lmo0587). Lmo0549 contains an N-terminal LRR region and hence is a member of the internalin family. Two E. faecalis WxL proteins, EF2686 and EF2250, are also internalin-like proteins. The WxL domain of EF2686 promotes the association of the protein at the bacterial surface by interacting with peptidoglycan (28). It is tempting to speculate that listerial WxL proteins are attached at the bacterial surface by a similar mechanism, although this hypothesis remains to be experimentally addressed.
FIG. 4.
The two Csc Clusters in L. monocytogenes. The first line shows a characteristic four-component Csc cluster, with the predicted surface-anchoring domain of Csc proteins indicated below. The two L. monocytogenes Csc clusters are schematically represented, with genes corresponding to Csc components indicated in blue (CscA like), yellow (CscB like), green (CscC like), and red (CscD like), respectively. The number of amino acids (AA) in each gene product is indicated. Cluster 2 does not contain any cscD-like gene. PKD, PKD repeats; DUF 919 (Pfam PF06030), domain of unknown function. (Adapted from reference 168 with permission.)
LysM domain.
Six listerial proteins, including P60 and MurA, carry one to four copies of a domain of ∼40 amino acids called the lysine motif (LysM, Pfam PF01476) domain (Table 1). The LysM domain is found in a variety of enzymes involved in bacterial cell wall degradation and is also present in many other bacterial proteins with various enzymatic or binding activities (10). Several LysM-containing proteins, such as staphylococcal immunoglobulin G (IgG) binding proteins and Escherichia coli intimin, are involved in bacterial pathogenesis. LysM domains are also present in some eukaryotic proteins, possibly as a result of horizontal gene transfer from bacteria. The LysM domain is thought to be a general peptidoglycan binding module. Indeed, the C-terminal domain of the Enterococcus faecalis N-acetylglucosaminidase AtlA, which contains six LysM modules, bind to highly purified peptidoglycan (50). The LysM domain can adopt a beta-alpha-alpha-beta conformation, with the beta strands forming an antiparallel beta sheet and the two alpha helices packing on one side of this sheet (10). These structures show no similarity to other bacterial cell surface domains. The peptidoglycan binding activity of LysM domains has not been demonstrated for any listerial proteins. However, proteomic analyses of L. monocytogenes extracts suggest that LysM domain-containing proteins are surface exposed. The autolysins P60 and MurA, which contain two and four LysM domains, respectively, are present in purified cell wall fractions (34). Lmo1303 and Lmo2522, which contain one and two LysM domains, respectively, are detected in supernatant fractions, indicating that these proteins can translocate across the plasma membrane (179). One LysM domain is also found in Lmo1941, a protein with a transmembrane hydrophobic domain that is detected in the L. monocytogenes membrane fraction (193). Finally, a LysM domain is present in Lmo0880, a cell wall LPXTG protein (Fig. 2) (146). In this case, the LysM domain could play a role in the topological distribution within the cell wall of a protein attached by a sortase A-dependent mechanism.
Peptidase peptidoglycan binding domain.
One protein, Lmo1851, is predicted to possess an N-terminal signal peptide (179), a transmembrane domain, and a C-terminal domain that is also found at the N or C termini of a variety of peptidases (Table 1) (Fig. 5). This domain of ∼70 amino acids is composed of three alpha helices and may have a general peptidoglycan binding function (see INTERPRO entry IPR002477 and Pfam PF01471). Many of the proteins having this domain are uncharacterized. Lmo1851 is detected in the L. monocytogenes membrane fraction (193), suggesting that it may be associated with the membrane via its N-terminal hydrophobic region and stabilized inside the cell wall via its C-terminal peptidoglycan binding domain. However, this hypothesis requires experimental validation.
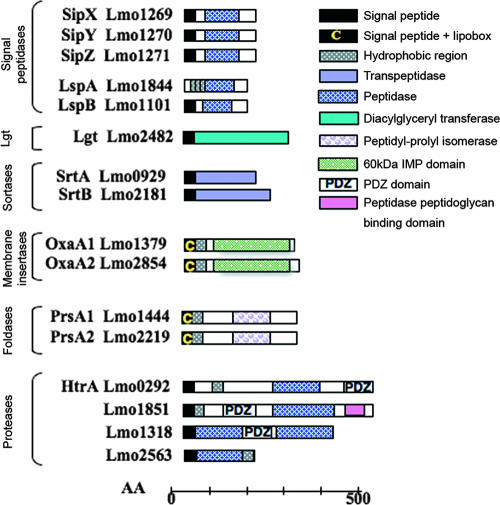
FIG. 5.
Surface proteins predicted to be involved in protein processing, folding, and anchoring at the cell surface. A schematic representation of L. monocytogenes surface proteins potentially involved in modifications or degradation of proteins at the surface is shown. AA, amino acids.
Membrane-Bound Proteins
Proteins with hydrophobic tails.
Proteins carrying a signal peptide can be retained in the membrane bilayer by hydrophobic segments generally present at the N- or C-terminal part of the protein. Proteins associated with the membrane by their C terminus contain a carboxyl-terminal stretch of hydrophobic residues followed by few charged residues thought to serve as a stop-transfer signal. This class includes the surface protein ActA, which promotes Listeria intracellular motility (94); two CscA-like proteins as mentioned above (Fig. 4); and four proteins of unknown function (Table 2). It also includes Lm0058, an ortholog of EssA, a component of the Wss(WGX100) specific secretion pathway (43). Finally, it includes two proteins, Lmo0528 and Lmo0529, which are similar to a UDP-glucose dehydrogenase and UDP-glucose glycosyltransferase, respectively. Whether these proteins play a role in exopolysaccharide production at the bacterial surface is an interesting possibility that has to be tested.
TABLE 2.
L. monocytogenes EGDe proteins with a carboxyl-terminal hydrophobic tail
| Proteina | Name | Predicted function |
|---|---|---|
| Lmo0058 | Similar to EssA | |
| Lmo0082 | Unknown | |
| Lmo0204 | ActA | Actin polymerization, intracellular motility |
| Lmo0528 | Similar to UDP-glucose 6-dehydrogenase | |
| Lmo0529 | Similar to UDP-glycosyltransferase | |
| Lmo0552 | Unknown, CscA likeb | |
| Lmo0586 | Unknown, CscA likeb | |
| Lmo0701 | Unknown | |
| Lmo0821 | Unknown | |
| Lmo2061 | Unknown |
Proteins absent from L. innocua are indicated in boldface.
See Fig. 4.
Proteins may also be tethered to the cell membrane by an amino-terminal hydrophobic stretch, which may be the signal peptide itself if it remains uncleaved (176). Sortases SrtA and SrtB, as well as several proteins involved in protein folding or in cell wall synthesis, such as penicillin binding proteins (PBPs), belong to this class of proteins (Fig. 3 and 5). Whether they remain attached to the bacterial surface needs to be experimentally verified. Nevertheless, some of them have been detected in the L. monocytogenes membrane fraction by proteomics (193).
Lipoproteins.
Lipoproteins are anchored to the membrane by covalent N-terminal lipidation. This process is directed by a specific signal peptide sequence characterized by a lipobox with a conserved cysteine residue (174). Two steps are necessary and sufficient for maturation of lipoproteins. Following signal peptide-directed export of the prolipoprotein, the prolipoprotein diacylglycerol transferase (Lgt) catalyzes the transfer of a diacylglycerol moiety from phosphatidylglycerol present in the membrane to the thiol of the conserved cysteine in the prolipoprotein. Subsequently, the signal peptide is removed by a specific lipoprotein signal peptidase (SPase) II (Lsp) enzyme, which cleaves within the lipobox to release the mature lipoprotein. Based on identification of a lipobox, it was possible to predict 68 lipoproteins in the L. monocytogenes EGDe genome sequence, the largest family of surface proteins (67). Recently, Baumgartner and colleagues were able to experimentally verify the genome predictions for 26 lipoproteins, which were specifically released in supernatants of a Δlgt strain (13). Listerial lipoproteins include 28 putative substrate binding proteins (SBPs) of ABC transporter systems, 19 proteins predicted to be involved in different enzymatic activities or other function, and 21 proteins of unknown function, the genes for six of which (Lmo0255, Lmo0460, Lmo0617, Lmo1340, Lmo2594, and Lmo2595) are absent from the L. innocua genome (Table 3).
TABLE 3.
Lipoproteins of L. monocytogenes strain EGDea
| Functional group | Proteinb | Name | Predicted function |
|---|---|---|---|
| Substrate binding proteins of ABC TSc | Lmo0135 | Similar to oligopeptide binding lipoproteins, ABC TS | |
| Lmo0152 | Similar to oligopeptide binding lipoproteins, ABC TS | ||
| Lmo0153 | Similar to Zn(II) binding lipoproteins, ABC TS | ||
| Lmo0181 | Similar to sugar binding proteins, ABC TS | ||
| Lmo0285 | Similar to substrate binding lipoproteins, ABC TS | ||
| Lmo0541 | Similar to iron compound binding lipoproteins, ABC TS | ||
| Lmo0768 | Similar to sugar binding proteins, ABC TS | ||
| Lmo0859 | Similar to sugar binding proteins, ABC TS | ||
| Lmo1016 | GbuC | Similar to glycine betaine binding protein, ABC TS | |
| Lmo1041 | Similar to molybdate binding proteins, ABC TS | ||
| Lmo1073 | Similar to metal ion binding proteins, ABC TS | ||
| Lmo1388 | TcsA | Similar to substrate binding lipoproteins, ABC TS; CD4+ T-cell-stimulating antigen | |
| Lmo1426 | OpuC | Glycine betaine/carnitine/choline binding proteins, ABC TS | |
| Lmo1671 | Similar to adhesion proteins and Mn/Zn binding proteins, ABC TS | ||
| Lmo1730 | Similar to sugar binding proteins, ABC TS | ||
| Lmo1738 | Similar to amino acid binding proteins, ABC TS | ||
| Lmo1847 | LpeA | Similar to manganese binding lipoprotein, ABC TS | |
| Lmo1959 | Similar to ferrichrome binding proteins, ABC TS | ||
| Lmo2007 | Similar to sugar binding proteins, ABC TS | ||
| Lmo2125 | Similar to maltose/maltodextrin binding lipoproteins, ABC TS | ||
| Lmo2184 | Similar to ferrichrome binding lipoproteins, ABC TS | ||
| Lmo2196 | OppA | Similar to oligopeptide binding lipoprotein, ABC TS | |
| Lmo2349 | Similar to amino acid binding lipoproteins, ABC TS | ||
| Lmo2417 | Similar to substrate binding lipoproteins, ABC TS, and to pheromone cOB1 | ||
| Lmo2431 | Similar to ferrichrome binding lipoproteins, ABC TS | ||
| Lmo2499 | Similar to phosphate binding lipoproteins, ABC TS | ||
| Lmo2569 | Similar to dipeptide binding proteins, ABC TS | ||
| Lmo2839 | Similar to sugar binding proteins, ABC TS | ||
| Enzymatic activities | Lmo0013 | QoxA | Similar to AA3-600 quinol oxidase subunit II |
| Lmo0355 | Similar to flavocytochrome c fumarate reductase chain A | ||
| Lmo0517 | Similar to phosphoglycerate mutase | ||
| Lmo0945 | Similar to metallo-beta-lactamase, DNA binding and competence protein (ComEC and ComEA of B. subtilis) | ||
| Lmo1379 | OxaA | Similar to membrane insertase OxaAd | |
| Lmo1444 | PrsA | Similar to foldase PrsAd | |
| Lmo1800 | Similar to protein tyrosine-phosphatase | ||
| Lmo1903 | Similar to thioredoxin | ||
| Lmo2219 | PrsB | Similar to foldase PrsAd | |
| Lmo2446 | Similar to glycosidase | ||
| Lmo2578 | Similar to hydrolase | ||
| Lmo2636 | Similar to thiamine biosynthesis lipoprotein ApbE | ||
| Lmo2642 | Similar to serine/threonine protein phosphatase | ||
| Lmo2812 | Similar to d-alanyl-d-alanine carboxypeptidasee | ||
| Lmo2854 | OxaB | Similar to membrane insertase OxaAd | |
| Other | Lmo0366 | Similar to lipoprotein involved in iron transport | |
| Lmo1757 | Similar to sex pheromone staph-cAM373 | ||
| Lmo2331 | Similar to Gp32 bacteriophage A118 protein | ||
| Lmo2637 | Similar to sex pheromone cAD1 | ||
| Lmo0047 | Unknown | ||
| Lmo0207 | Unknown | ||
| Lmo0255 | Unknown | ||
| Lmo0303 | Unknown | ||
| Lmo0324 | Unknown | ||
| Lmo0460 | Unknown | ||
| Lmo0510 | Unknown | ||
| Lmo0617 | Unknown | ||
| Lmo0791 | Unknown | ||
| Lmo0821 | Unknown | ||
| Lmo0953 | Unknown | ||
| Lmo1068 | Unknown | ||
| Lmo1265 | Unknown | ||
| Lmo1340 | Unknown | ||
| Lmo1649 | Unknown | ||
| Lmo1653 | Unknown | ||
| Lmo2079 | Unknown | ||
| Lmo2080 | Unknown | ||
| Lmo2416 | Unknown | ||
| Lmo2594 | Unknown | ||
| Lmo2595 | Unknown |
Adapted from reference 13 with permission. Of note, Lmo1340 was identified as a new lipoprotein, and Lmo0810 was removed from the first lipoprotein annotation (67), as it was incorrectly annotated as a lipoprotein.
Proteins absent from L. innocua are indicated in boldface.
TS, transporter systems.
See Fig. 5.
See Fig. 3.
Nonconventional Secreted Surface Proteins
Proteomic studies identified proteins that have no predicted signal sequence, and are assumed to have cytoplasmic functions, in the L. monocytogenes cell wall or supernatant fractions (160, 179). Among them are glycolytic enzymes, chaperones, and heat shock proteins, as well as proteins involved in detoxification and adaptation to atypical conditions, nucleic acid metabolism, transcription, and translation. Although these cytosolic proteins may be recovered in these fractions due to cell lysis, increasing evidence suggests that some of them may be specifically transported to the surface by alternative pathways. In L. monocytogenes the auxiliary secretory protein SecA2 is involved in export of a subset of listerial proteins, some of which lack recognizable Sec sequences (107). Recent reports identified superoxide dismutase as a protein dependent on SecA2 for secretion and surface association in L. monocytogenes (7). Superoxide dismutase is also secreted via the SecA2 pathway in Mycobacterium tuberculosis (27). Another study reports the unambiguous presence at the bacterial surface of FbpA (Lmo1829), a protein that does not possess any characteristic of a surface-exposed protein. FbpA is also exported via the SecA2-dependent secretion pathway (48). Orthologs of FbpA, including PavA of S. pneumoniae and Fbp54 of S. pyogenes, are also cell surface proteins (40, 81). Together these data suggest that SecA2-dependent export is a new type of secretion pathway that is partly responsible for L. monocytogenes virulence (7, 48, 107).
The autolysin-like protein Lmo0129 (Fig. 3) does not contain any obvious signal peptide, although it is detected in Listeria supernatant fractions (179). Interestingly, the gene encoding Lmo0129 is adjacent to a gene encoding a putative holin of the TcdE family, Lmo0128 (43), raising the possibility that Lmo0129 could be exported by a holin-like pathway (reviewed in references 70 and 191).
Another possible mechanism is deduced from electron microscopy studies performed 40 years ago that revealed the presence of small vesicles attached to the listerial membrane. These vesicles could be derived from small membrane extrusions (65). This observation is reminiscent of a well-described phenomenon in gram-negative bacteria. Many of them release vesicles from their outer membranes as secretory vehicles for proteins, lipids, and immunomodulatory compounds (189; for a review, see reference 99). These vesicles mediate bacterial binding and invasion, cause cytotoxicity, and modulate the host immune response. It is tempting to speculate that such phenomenon occurs in gram-positive bacteria. L. monocytogenes would be a nice model with which to test this hypothesis.
Finally, some proteins may also reach the cell surface through cosecretion and interaction with other surface proteins. Together, these results highlight the possibility of Sec-independent mechanisms of secretion and surface association that clearly deserve future attention.
FUNCTIONS OF SURFACE PROTEINS AND THEIR RELEVANCE IN PATHOGENESIS
Interfering with the surface-anchoring pathways represents a powerful tool for analyzing the diverse classes of surface proteins and their role during the Listeria infectious process. This approach has proven to be successful for LPXTG proteins and lipoproteins. An srtA mutant, in which LPXTG proteins are missorted, displays a severe attenuation phenotype following oral infection, with a more pronounced effect than an internalin mutant, pointing to the role of several LPXTG proteins in pathogenesis (16, 17). Similarly, interfering with the processing of lipoproteins affects intracellular survival of L. monocytogenes (13, 148). However, from these global approaches it is not known whether the alteration of a predominant single protein or the cumulative effect of alterations of several proteins is responsible for the attenuated phenotype. In order to answer this question, it is necessary to examine each protein individually or in groups of functional categories. An in-depth examination of domains present in predicted surface proteins may help in this classification prior to the essential experimental validation.
Cell Wall Metabolism
The cell wall is a highly dynamic structure that expands during cell growth and must be cleaved during cell division and lysis. Cell wall remodeling is also critical in numerous cellular processes, including formation of flagella, sporulation, biofilm formation, chemotaxis, genetic competence, and protein secretion. Growth and breakage of the peptidoglycan involve several enzymes, synthetases, and hydrolases, whose activities must be coordinated in time and space (82). Autolysins break the glycosidic bonds between the peptidoglycan monomers and also break the peptide cross-bridges that link the rows of sugars together (169). Transglycosylases (also referred to as glycosyltransferases) insert and link new peptidoglycan monomers into the breaks in the peptidoglycan. Finally, transpeptidases reform the peptide cross-links between the rows and layers of peptidoglycan (Fig. 3B).
Knowledge of the enzymatic activities involved in cell wall assembly and turnover is important, as peptidoglycan synthetases are targets for many antibiotics and cell wall components are potent determinants of inflammation. L. monocytogenes cell wall derivatives are recognized by membrane-bound immune sensors, such as the macrophage scavenger receptor class A and Toll-like receptors (TLRs), or by intracytosolic sensors, such as Nod proteins, thereby stimulating host immunity. Macrophage scavenger receptor class A- and TLR2-deficient mice are highly susceptible to L. monocytogenes infection (85, 178). Nod2-deficient mice are susceptible to oral bacterial infections (92). Evidence for sensing of L. monocytogenes by Nod1 in vitro has also been recently reported (134). Together, these observations suggest that enzymes promoting the release of bacterial components are likely to be important modulators of the host innate (20) as well as adaptative (122) immune responses.
Autolysins.
By digesting the cell wall peptidoglycan, autolysins are involved in numerous cellular processes, including cell growth and division, cell wall turnover, motility, chemotaxis, protein secretion, differentiation, and pathogenicity (140, 169). These hydrolytic enzymes are also responsible for the active release of immunogenic cell wall components. There are several types of autolysins, categorized by the nature of the bonds hydrolyzed in the peptidoglycan (Fig. 3B). Based on domain homology search data and a few studies using zymogram assays, we can expect the existence of several autolysins in L. monocytogenes. However, a direct demonstration of cleavage activities of these enzymes at specific bonds will require further biochemical studies.
(i) N-Acetylglucosaminidases and N-acetylmuramidases.
Two types of enzymes hydrolyze the sugar backbone of peptidoglycan between the alternating MurNAc and GlcNAc residues of the glycan chains. N-Acetylmuramidases, which are related to lysozyme, cleave the β-1,4 glycosidic bond between MurNAc and GlcNAc (Fig. 3B), whereas N-acetylglucosaminidases hydrolyze the bond between GlcNAc and MurNAc by liberating free reducing groups of GlcNAc. Three listerial proteins, Lmo0717, Lmo1104, and Lmo2522, possess a putative N-terminal N-acetylmuramidase domain. Six proteins, among which are the GW protein Auto and the LysM protein MurA (Fig. 3A), possess a domain with similarities to the catalytic domain of N-acetylglucosaminidases. Both Auto and MurA display cell wall hydrolase properties (31, 37). However, an aut deletion mutant has a morphology similar to that of the wild-type strain and has no defect in septation and cell division, while a murA mutant grows as long chains during exponential growth. Interestingly, Auto is required for entry of L. monocytogenes into different nonphagocytic eukaryotic cell lines, and an aut mutant displays a reduced virulence (31). How Auto specifically contributes to pathogenicity is unknown, though it may be linked to the maintenance of the cell surface architecture and/or to the release of immunologically active cell wall components.
(ii) Amidases.
The Listeria genome encodes four proteins with a putative amidase domain known to hydrolyze the bond between the glycan chain and the peptide side chain of peptidoglycan. Two are GW proteins, Ami and Lmo1521, and two are proteins without any obvious modules for targeting at the bacterial surface, Lmo0129 and Lmo0849. Ami is the only amidase for which autolysin activity was reported (25). Like in InlB, the GW modules appear to promote Ami adhesion to host cells (127). Since the InlB GW modules bind cellular matrix proteoglycans (87), it is possible that Ami GW modules exert a similar function. Thus, Ami may act as a complementary adhesin during infection.
(iii) γ-d-Glutamyl-(l)-meso-diaminopimelate peptidases.
Another important group of L. monocytogenes autolysins is the P60 subfamily. P60, encoded by the iap (invasion-associated protein) gene, is a major 60-kDa autolysin first described as being involved in the L. monocytogenes invasion process (100). P60-defective mutants display an abnormal morphology, characterized by the presence of filamented cells containing fully formed septa (72, 196). The four members of the P60 family, Spl (P45), Lmo0394, Lmo1104, and P60 itself, share in their C-terminal region an NlpC/P60 domain (Pfam PF00877) related to the CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) domain, which is present in many proteins predicted to function in peptidoglycan hydrolysis (12, 151). For instance, this domain is present in Bacillus subtilis LytE and LytF and in S. aureus LytA and LytN autolysins (4). All CHAP superfamily enzymes characterized to date hydrolyze diverse substrates that contain a γ-d-glutamyl moiety. Although not yet biochemically validated, it is proposed that the P60 family fulfils the same function as Bacillus sphaericus endopeptidase II, which hydrolyzes the γ-d-glutamyl-meso-diaminopimelate linkage in the cell wall peptides (Fig. 3B) (169).
Besides P60, P45 (Spl) is the only protein of this family in L. monocytogenes known to exhibit peptidoglycan-lytic activity (163). Interestingly, P45 combines the C-terminal CHAP domain with an N-terminal domain related to the SMC (structural maintenance of chromosomes) family of proteins. The SMC proteins are essential for successful chromosome transmission during replication and segregation of the genome in all organisms (80). They form heterodimers and are core components of large multiprotein complexes. Lmo2504 is another putative surface protein with a C-terminal peptidase domain and an SMC-related N-terminal domain (Fig. 3). It is possible that P45 and Lmo2504 somehow couple the peptidoglycan remodeling to the segregation of the nucleoid, although further studies are required to test this hypothesis. Lmo1104 also combines the C-terminal CHAP domain with another N-terminal module. In this case, the N-terminal region shows similarity to a lytic transglycolsylase domain (Pfam PF01464). From this, Lmo1104 may thus have a dual function in hydrolyzing the cell wall. Lmo1104 is the only putative autolysin of the group that is absent in L. innocua.
A link between this class of surface proteins and Listeria pathogenicity has been established in the case of P60. Rough mutants expressing lower levels of P60 enter less efficiently into certain eukaryotic cells, suggesting a role for P60 in invasion (100). A P60-deficient mutant is attenuated in virulence after intravenous infection of mice and is impaired in its intracellular motility due to mislocalization of the actin-polymerizing factor ActA (107, 138). In addition, P60 plays an important role in the immune response against L. monocytogenes. P60-specific antibodies act as opsonins and might play a role in preventing systemic infections in immunocompetent individuals (95). P60 is also a major protective antigen that induces both T-CD8 and Th1 protective immune responses (64, 75). Finally, it was recently reported that P60 contributes to subversion of NK cell activation and production of gamma interferon (83).
(iv) Phage endolysins.
The genome of L. monocytogenes EGDe contains one prophage inserted in the comK locus (67, 111). This cryptic phage is highly homologous to phage A118 (111, 112, 114), a temperate bacteriophage attacking L. monocytogenes serovar 1/2. Like A118, the Φ EGDe element possesses a gene encoding a putative endolysin (Lmo2278, homologous to Ply118 [115]) followed by a gene encoding a holin (homologous to Hol118 [115]) thought to trigger secretion of the endolysin (188). The Φ EGDe endolysin is not characterized but could function like Ply118 endolysin, an l-alanyl-d-glutamate peptidase that cuts the amide bonds between l-Ala and d-Gln within the peptidoglycan peptide bridges (Fig. 3A) (115). Interestingly, endolysin Ply500 from phage A500 has the same enzymatic activity as Ply118, although it targets the cell wall of Listeria serovars 4, 5, and 6. The specificities of the endolysins are conferred by their different cell wall binding domains, which are thought to recognize Listeria-specific surface carbohydrates (113). Supporting this idea, phage PSA endolysin has a cell wall binding domain closely related to that of Ply500 and targets the same serovars, while having different enzymatic activity than Ply500 (96).
PBPs: transglycosylases, transpeptidases, β-lactamases, and d-alanyl-d-alanine carboxypeptidases.
Peptidoglycan assembly is a multistep process carried out by different enzymatic activities. Transglycosylase enzymes catalyze the insertion of peptidoglycan precursors into the growing end of the bacterial cell wall. Subsequently, transpeptidase enzymes join the peptide of a precursor with that of the preexisting peptidoglycan in order to cross-link the sugar chains (Fig. 3B). Penicillins and cephalosporins block the formation of the peptide cross-links and induce osmotic lysis of the bacterium by binding to the transpeptidase enzyme. Bacteria may resist the action of these β-lactam antibiotics by mutating transpeptidases or by expressing β-lactamase, an enzyme hydrolyzing β-lactam rings. All these enzymes belong to the family of PBPs.
A first study reported that L. monocytogenes had five PBPs, as detected by their ability to bind benzylpenicillin (186). By searching the genome, Guinane and colleagues recently found seven surface proteins with similarities to PBPs (71). Our own in silico research identified 10 PBP-like proteins, which we classified into four families as represented in Fig. 3A. The first family, including PBPA1 and PBPA2 (formerly PBP1 and PBP4 [186]), corresponds to class A PBPs, which are bifunctional proteins with an N-terminal transglycosylase domain and a C-terminal transpeptidase domain. The second family, including PBPB1, PBPB2, (formerly PBP2 [186]), and PBPB3, corresponds to class B PBPs, which have a C-terminal transpeptidase domain and an N-terminal PBP dimerization domain (Pfam PF03717) involved in interaction with other proteins during cell morphogenesis. The third family encompasses two putative PBPs, PBPC1 and PBPC2, which contain a β-lactamase class C domain (Pfam PF00144). PBPC1 is expected to be at the bacterial surface, while PBPC2 lacks any known cell surface association domain. Finally, the fourth family includes three proteins, PBPD1, PBPD2, and PBPD3, that contain a d-alanyl-d-alanine carboxypeptidase domain. This domain may catalyze the removal of the C-terminal d-alanine residue from peptidoglycan pentapeptides (Fig. 3B). PBPD1 (formerly PBP5 [186]) and PBPD2 are related to the peptidase S11 family (Pfam PF00768), represented by E. coli PBP5, while PBPD3 is related to the VanY dd-carboxypeptidase family (Pfam PF02557).
L. monocytogenes clinical isolates are resistant to cephalosporins but are, fortunately, susceptible to penicillin and ampicillin, which constitute the major treatment for listeriosis in combination with the aminoglycoside gentamicin. However, several recent findings provide evidence of the emergence of multidrug-resistant L. monocytogenes strains, including strains with resistance to penicillin and ampicillin (38, 143, 157, 170). Dissemination of these resistant traits in clinical strains of L. monocytogenes could have important medical consequences. It is therefore import to characterize listerial PBPs.
Recent studies have confirmed the enzymatic activity of PBPD1 (Lmo2754) (97) and of PBPA2 (Lmo2229) (198). Moreover, analysis of mutant strains suggests that PBPB3 (Lmo0441) and, to a lesser extent, PBPA2 (Lmo2229) are central to β-lactam resistance in Listeria (71). Of interest is that PBPB3 shares similarities with MecA, the key determinant of β-lactam resistance in methicillin-resistant S. aureus strains (109). Furthermore, disruption of some pbp genes attenuates L. monocytogenes virulence, probably due to alteration of the cell wall metabolism (71).
Deacetylase.
A last important peptidoglycan-modifying enzyme is PgdA (Lmo0415), an enzyme recently characterized in L. monocytogenes as a GlcNAc deacetylase (21). pgdA mutants of L. monocytogenes, like those of S. pneumoniae (187), are hypersensitive to lysozyme, one of the first-line defense mechanisms in the human host against invading bacteria. Inactivation of pgdA also dramatically attenuates virulence and induces a massive beta interferon response in a TLR2- and Nod1-dependent manner. Peptidoglycan deacetylation is therefore essential for resistance against the innate immune response (21).
Enzymes involved in assembly and modification of TAs and LTAs.
In addition to the assembly of cross-linked peptidoglycan, several enzymes are required for the synthesis of polyanionic TAs and LTAs, which make up an important polyanionic network with ion-exchange properties at the bacterial surface. However, polymerization of TAs and LTAs occurs intracellularly (14). Therefore, enzymes involved in cell wall polymer synthesis are expected to be cytosolic and not surface exposed, although a signal peptide is proposed to be present in two of them in L. monocytogenes, TagO (Lmo2519) and TagB (Lmo1088) (179). Enzymes involved in d-alanine esterification, a common modification found in TAs and LTAs of many gram-positive bacteria, are cytosolic or membrane-bound proteins (130). The synthesis of d-alanyl-LTA requires four proteins that are encoded by the dlt operon (1): dltA (lmo0974) encodes a cytoplasmic d-alanine-d-alanyl carrier protein ligase (designated DltA or Dcl); dltB (lmo0972) encodes the d-alanyl carrier protein Dcp, which is secreted; and dltC (lmo0973) and dltD (lmo0971) encode membrane-associated proteins, each having a signal peptide according to the Pfam annotation. However, the current model is that DltD would function intracellularly, providing binding sites for DltA and Dcp on the cytoplasmic leaflet, while DltB would form a channel for the secretion of d-alanyl-Dcp towards the cell wall, where d-alanylation occurs (130). This pathway still needs clarification.
Protein Processing, Folding, and Anchoring
Following translocation across the Sec channel, most proteins have their signal peptide cleaved by an SPase. The L. monocytogenes genome encodes five membrane-bound SPases (Fig. 5). Three of them, SipX, SipY, and SipZ, are type I SPases, whose predicted function is to remove the signal peptides of preproteins exported by the general secretory pathway. SipZ appears to be the major SPase I for listerial secreted virulence factors (22). Two other SPases, LspA and the as yet undescribed LspB, belongs to class II SPases involved in maturation of lipoproteins. LspA processes at least lipoprotein LpeA (148), while LspB remains to be characterized.
As described above, mature proteins exhibiting a specific C-terminal sorting signal are processed by sortases, while lipoproteins are processed by Lgt (Fig. 5) (13, 16, 17, 62). Proteins that will remain at the surface inserted in the membrane will be processed by components that catalyze efficient lateral transport into or across the plasma membranes. These include the conserved YidC/Oxa1/Alb3 proteins family that are believed to function as membrane insertases, possibly functioning also as membrane chaperones (reviewed in references 139 and 182). Two L. monocytogenes lipoproteins, Lmo1379 (OxaA) and Lmo2854 (OxaB), share homology with YidC/Oxa1 and may thus target membrane-bound surface proteins (43). Future work is needed to validate this hypothesis.
The majority of proteins that are translocated across the cytoplasmic membrane are delivered to the membrane-cell wall interface essentially unfolded. They must then be folded into their native configuration without blocking the translocation machinery in the membrane, by forming inadequate interactions with the cell wall or insoluble aggregates. Misfolded or aberrant proteins have to be cleared off the translocase and the cell wall. Bacteria encode membrane- and cell wall-associated foldases and proteases that act as folding catalysts and quality control machines (159). Several genes in the L. monocytogenes genome encode proteins likely to be involved in folding and degradation of polypeptides at the bacterial surface (Fig. 5). PrsA (Lmo1444) and PrsB (Lmo2219) are lipoproteins related to the PrsA protein of B. subtilis. PrsA is a parvulin-type peptidyl-prolyl cis/trans isomerase, which assists posttranslocational folding of exported proteins and their stabilization at the bacterial surface. Interestingly, lmo2219 is preceded by a PrfA box, in contrast to lmo1444 (67), and is regulated by PrfA, the major regulator of Listeria virulence gene expression (13, 126). In addition, lmo2219 is upregulated during the intracytosolic phase of the Listeria cellular infectious process (39). We therefore suggest that PrsB could play a specific role in bacterial adaptation to the host.
lmo0292 (degP) encodes an HtrA-like serine protease related to B. subtilis YycK (172). HtrA is a protease that can also function as a chaperone. The listerial HtrA-like protease is essential for optimal growth under stress conditions and is involved in listerial pathogenesis (172, 195). Interestingly, like the gene encoding PrsB, the htrA gene is upregulated intracellularly (39). In addition to its N-terminal protease domain (Pfam PF00089), HtrA possesses a C-terminal PDZ domain. PDZ domains consist of ∼80 amino acids compactly arranged in a globular structure thought to function in protein or peptide binding (Pfam PF00595). They are found in diverse signaling proteins, in both bacteria and eukaryotes. We found two other proteins with protease and PDZ domains in the L. monocytogenes genome, Lmo1851 and Lmo1318 (Fig. 5). Lmo1851 carries a peptidase domain of the S41 family (Pfam PF03572), to which the ClpP protease belongs. Lmo1318 is a hypothetical zinc metalloprotease of the S2P/M50 peptidase family (Pfam PF02163), such as the membrane-bound E. coli protease EcfE (90). Related to Lmo1318 is Lmo2563, which is also predicted to be a S2P/M50 peptidase albeit without a PDZ domain and sharing similarities with the B. subtilis sporulation factor SpoIVFB (35). Interestingly, S2P/M50 proteases, by catalyzing cleavage of membrane-bound proteins, seem to be involved in a variety signal transduction pathways, some off which play roles in host-pathogen interaction (117). Whether Lmo1851, Lmo1318, and Lmo2563 function as cell surface proteases will require further investigation.
Adhesion to Host Cells
As their name implies, adhesins are specialized surface proteins that mediate bacterial adhesion. They specifically recognize receptors on the surface of target host cells, determining tissue tropism of the pathogen. Among these host receptors are components of the extracellular matrix, a complex structural entity surrounding and supporting cells within mammalian tissues (24). The extracellular matrix is composed of three major classes of biomolecules: (i) structural proteins, including collagen and elastin; (ii) specialized proteins, such as fibrillin, fibronectin, and laminin; and (iii) proteoglycans, which are high-molecular-weight components composed of a protein core to which long chains of repeating disaccharide units termed GAGs are attached. Besides proteoglycans, other high-molecular-weight glycosylated proteins, such as mucins, are likely to play an important role in the bacterial adherence to epithelia. Mucins, either secreted or membrane bound, form a major part of a protective biofilm on the surface of epithelial cells (142, 154).
Sequence comparisons with previously characterized proteins suggest that some domains found in listerial proteins may play a role in adhesion to host cells.
Collagen binding domain.
Five LPXTG proteins (Lmo0159, Lmo0160, Lmo0880, Lmo2158, and Lmo2576) are predicted to have a collagen binding domain (Pfam PF05737) (Fig. 2), which is found in the collagen binding adhesins of several other gram-positive pathogens (137, 199). The prototype member of this family is the collagen binding adhesin Cna of S. aureus. Cna participates in the infectious process of pathogenic S. aureus (52), suggesting that the ability to interact with collagen provides a general advantage to the bacteria in pathogenesis. Cna is composed of an N-terminal A region that carries the collagen binding activity and a C-terminal region of B repeats (CnaB-like repeats) (see below) (137). The Cna A region is composed of three subdomains, N1, N2, and N3 (199). Crystal structure determination of the N1 and N2 domains suggest a model in which both domains, each taking an Ig-like fold, cooperate to wrap around and hug the collagen triple helix (199). The collagen binding domain present in listerial proteins shows similarity to N2, which is the minimal collagen binding domain of Cna. Further work is required to explore the function of these listerial proteins, especially their possible binding to collagen.
Fibronectin binding domain.
FbpA (Lmo1829) is a Listeria surface fibronectin binding protein required for intestinal and liver colonization of L. monocytogenes following oral infection (48). FbpA has strong homology to atypical fibronectin binding proteins, such as PavA of S. pneumoniae (81) and Fbp54 of S. pyogenes (40). FbpA binds to immobilized human fibronectin in a dose-dependent and saturable manner and increases adherence of wild-type L. monocytogenes to HEp-2 cells in the presence of exogenous fibronectin. Like FbpA, Lmo0721 is another reported listerial fibronectin binding protein that lacks conventional secretion/anchoring signals (66). Whether it is exposed at the bacterial surface has not been formally demonstrated.
Proteoglycan binding domain.
As mentioned above, InlB GW repeats bind not only to LTAs present at the bacterial surface but also to GAGs. GW repeats binding to GAGs releases InlB from the bacterial surface (87). Furthermore, the presence of GAGs on the cell surface significantly increases InlB-dependent invasion, suggesting that binding of GW repeats to cellular GAGs could enhance interaction of InlB with its cellular receptor, Met. It is not known whether GW modules present in autolysins also bind GAGs. However, the purified GW domain of Ami binds eukaryotic cells, suggesting that a similar interaction may explain the role of Ami in listerial adhesion to host cells (127).
RGD motif.
Three listerial surface proteins contain an arginine-glycine-aspartate (RGD) motif: ActA, the LPXTG protein Lmo1666, and the lipoprotein Lmo0460. The RGD motif is found in various proteins of the extracellular matrix and is known to interact with integrins. Although the RGD motif in ActA does not seem to contribute to Listeria adhesion to host cells (3), it may be a potential integrin binding site in Lmo1666 and Lmo0460.
Mucin binding domains.
Twelve LPXTG proteins (Fig. 2) and one protein with a membrane anchor domain, Lmo0576, contain a MucBP (mucin binding protein) domain (Pfam PF06458). Identified in MUB, an extracellular mucus binding protein of Lactobacillus reuteri 1063, this domain is arranged as tandem repeats that were shown to trigger adhesion to mucus material (152). MucBP repeats are present in 1 to 14 copies in listerial proteins. According to the Pfam database, MucBP repeats are present only in Lactobacillales and Listeria bacterial species. Sequence analysis indicates that the MucBP repeats vary in size, for instance, ranging from ∼60 amino acids in listerial proteins to almost 200 amino acids in MUB proteins of L. reuteri (19, 152). The difference in size between Listeria MucBP and Lactobacillus MUB repeats can be explained by the presence of a distinct N-terminal region in the MUB domain. MUB repeats and MucBP repeats may therefore be different functional units (19). At present, there is no experimental evidence for a role of listerial MucBP binding to mucins.
Ig-like fold domains.
The Ig fold is an important structure, playing essential roles in the vertebrate immune response, cell adhesion, and many other processes. Novel three-dimensional structures of domains from distantly related proteins have revealed that unrelated sequences may take a similar Ig fold, which is characterized by the presence of β sheets. Thus, nearly all Listeria LPXTG proteins possess Ig-like domains that fall into one of five distinct sequence families: the Cna collagen binding domain described above, the Cna protein B-type domain, the LRR-adjacent domain, the PKD repeat domain, and the bacterial Ig-like domain (Big 3). Two main functions may be attributed to these domains. They may be directly involved in adhesion by interacting with carbohydrates on the bacterial or host cell surface. Alternatively, they may serve as a stalk that projects a ligand binding region from the bacterial surface, thus facilitating bacterial interaction with host cells. For instance, the receptor binding domain of intimin in enteropathogenic Escherichia coli, invasin in Yersinia, or internalin in Listeria is presented at the tip of an elongated structure, in which the repeat region is used as a spacer domain or a stabilizer for the whole structure (133).
(i) CnaB-like repeat domain.
As mentioned above, staphylococcal Cna has a nonrepetitive collagen binding A region followed by a repetitive B region. The B region has one to four 23-kDa repeat units [B(1) to B(4)], depending on the strain of origin. Each B repeat has two domains, D1 and D2, each exhibiting an inverse Ig-like fold. Modeling studies suggest an accordion structure in which the B repeat units could project the A region from the cell surface and aid in binding to collagen (41). Six listerial LPXTG proteins contain repeats related to the D domain of CnaB repeats (Pfam PF05738) (Fig. 2), among which four possess a collagen binding domain (Lmo0159, Lmo0160, Lmo2178, and Lmo2576).
(ii) LRR-adjacent Ig-like domain.
Eleven LPXTG proteins of the internalin family contain the LRR-adjacent Ig-like domain (Pfam PF08191) (Fig. 2). It is a small beta-strand domain fused to the C-terminal end of the LRR region (133, 164). This domain belongs to the family of Ig-like domains in that it consists of two sandwiched beta sheets. The beta strands in one of the sheets is, however, much smaller than those in most standard Ig-like domains. The function of this domain appears to be mainly structural, forming a common rigid entity with the LRR. It is thought to facilitate the presentation of the adjacent LRR domain for protein-protein-interactions.
(iii) PKD repeat domain.
Eleven LPXTG proteins contain the PKD repeat domain (Pfam PF00801) (Fig. 2). The PKD repeat domain was first identified in the human polycystic kidney disease 1 (PKD1) protein as 16 Ig-like repeats, each of ∼80 amino acid in length. PKD1 is involved in adhesive protein-protein and protein-carbohydrate interactions. PKD1 Ig-like repeats II to XVI are involved in strong calcium-independent homophilic interactions in vitro (84), suggesting that PKD1 may be involved in cell-cell adhesion through its cluster of Ig-like repeats. However, the function of PKD1 repeats in listerial proteins remains totally unknown. For instance, InlI, the longest internalin-like protein (Fig. 2), possesses eight PKD repeats, but no detectable role could be attributed to InlI in vitro or in virulence assays (156).
(iv) Big 3 domain.
Four LPXTG proteins contain a domain with an Ig-like fold called the Big 3 domain (Pfam PF07523) (Fig. 2). This domain is present in one copy in the internalin-like Lmo2026, Lmo0732, and Lmo0171 proteins and is repeated seven times in the Lmo0842 protein. Interestingly, Lmo0842 repeats are related to those found in the Alp family in streptococci, which comprises alpha, Rib, R28, and Alp2 proteins, which are important surface proteins known to confer protective immunity (110). Alp repeats are themselves related to repeats of ∼80 amino acids found in biofilm-associated proteins (Bap) in staphylococci and in Esp in enterococci (102), as well as to HYR (hyalin repeat) domains identified in several eukaryotic proteins (33). By BLAST analysis, we found that PKD repeats present in Listeria LPXTG proteins are also weakly similar to the Bap family repeats. Together, these data suggest an evolutionary relationship between all these repeated domains.
Invasion
Two surface proteins, the LPXTG protein internalin A (InlA) and the GW protein InlB, are major L. monocytogenes invasins, promoting bacterial entry into mammalian cells (47, 61; for a review, see reference 73). InlA is sufficient for bacterial uptake into epithelial cells and is required for Listeria to cross the intestinal and placental barriers (104). InlA-dependent entry is mediated by human Ecad, a cell adhesion molecule that is required for the integrity of adherens junction of polarized epithelial cells and is connected to cytoskeletal proteins. InlA-Ecad interaction is species specific and occurs between InlA and human or guinea pig Ecad but not mouse or rat Ecad (73, 104). The Pro residue at position 16 in human Ecad is essential for InlA recognition. InlB mediates entry into a variety of cell types by interaction with the hepatocyte growth factor receptor Met (167). InlB also interacts via its GW modules with gC1qR, a ubiquitous molecule first identified as the receptor for C1q, the first component of the complement cascade, and with GAGs (26, 73, 119). In vivo, InlB is required for efficient mouse liver colonization (91) and plays a role in invading human placenta in conjunction with InlA (105). Recent studies reveal species specificity for InlB and show that InlB does not recognize or stimulate guinea pig or rabbit Met (91). The LPXTG protein Vip (Lmo0320) is another surface virulence factor required for L. monocytogenes entry into some mammalian cells (32). Vip interacts with the mammalian heat shock protein Gp96, a member of the Hsp protein family that plays significant roles in protein folding and in the modulation of both the innate and adaptive immune responses. Vip-Gp96 interaction is critical for bacterial entry into cells. Vip could thus appear as a new type of virulence factor, exploiting Gp96 as a receptor triggering a signaling pathway required for cell invasion and/or to subvert the host immune response during infection (32).
Motility
Flagellar movement.
Bacterial movement is produced through the action of the flagella. The flagellum comprises a long filament that acts as a propeller, which is anchored to the cell envelope by a flexible hook and basal body (116). The growth of the bacterial flagellar filament occurs at its distal end by self-assembly of flagellin transported from the cytoplasm through the channel of the flagellum (116). Listerial flagellin is produced and assembled at the cell surface when L. monocytogenes is grown at between 20 and 25°C, whereas its production is markedly reduced at 37°C. However, it is important to note that flagellin expression is still high at 37°C in some laboratory-adapted strains and in approximately 20% of clinical isolates (192). Flagellin is encoded by the flaA gene (lmo0690), which is located in a 38-kb locus that comprises 26 other genes encoding putative flagellar machinery proteins (18, 43). Listerial flagellin is posttranslationally modified, being glycosylated with β-O-linked GlcNAc (161). Whether glycosylation is essential for flagellar assembly or in some way facilitates interactions of Listeria with either other cells or environmental substrates is currently unknown.
Flagellin from L. monocytogenes stimulates TLR5 (76), which is expressed by intestinal epithelial cells, monocytes, and dendritic cells. TLR5 activation induces the production of tumor necrosis factor (TNF) alpha, an important cytokine in host resistance to enteric listeriosis. In addition, flagella seem to facilitate the adhesion to and invasion of eukaryotic host cells by L. monocytogenes (45). However, the relationship between flagellin and virulence is not clear. On one hand, no differences in virulence, bacterial clearance, or induction of L. monocytogenes-specific T-lymphocyte responses between L. monocytogenes strain 10403S and an isogenic flaA deletion strain were observed in the mouse infection model, suggesting that flagellin is not essential for pathogenesis (192). On the other hand, inactivation of the regulator of flaA expression, MogR, induces overexpression of FlaA and causes defects in bacterial division, intracellular spread, and virulence in mice (166). Finally, virulence of flaA mutants is diminished relative to that of the wild-type strain in tumor necrosis factor alpha receptor-p55−/− mice, suggesting that flagella participate in the triggering of tumor necrosis factor secretion, leading to protection against infection (45).
Intracellular movement.
The well-known listerial surface protein ActA mediates polymerization of host actin to propel the bacterium through the cytoplasm of its host (for recent reviews, see references 68 and 73). ActA is the first bacterial protein identified to promote actin nucleation, by mimicking its eukaryotic counterparts, the Wiskott-Aldrich syndrome (WASP) family of proteins. The N-terminal region of ActA has homology to the C-terminal region of WASP, and both ActA and the WASP proteins act as nucleation-promoting factors for the Arp2/3 complex. The other major ligand of ActA is the Ena/vasodilator-stimulated phosphoprotein, which binds the central proline-rich domain of ActA and accelerates L. monocytogenes actin-based motility. A growing number of intracellular pathogens, such as Rickettsia conorii, Shigella flexneri, Mycobacterium marinum, Burkholderia pseudomallei, and vaccinia virus, are being recognized as displaying surface proteins that trigger actin polymerization for intra- and intercellular movement (68, 173).
Other Functions
Noninvasin internalin-like proteins.
Data resulting from a Listeria DNA macroarray analysis identified six genes encoding surface proteins of the internalin family always found in all L. monocytogenes strains associated with listeriosis: inlA, inlB, inlH, inlE, inlI, and inlJ (46). Besides the invasion factors InlA and InlB, InlH (Lmo0263) and InlJ (Lmo2821) were associated with Listeria pathogenesis (156, 164). Screening of a bank of transposon mutants by signature-tagged mutagenesis in the mouse model revealed another internalin-like protein, Lmo2026 (formerly ORF626), as a factor possibly involved in listerial multiplication in the brain (8). The functions of InlH, InlJ, and Lmo2026 are unknown and do not seem to be related to the internalization process. LRR-containing proteins are widely represented in eukaryotes and participate in many biologically important processes, such as hormone-receptor interactions, enzyme inhibition, cell adhesion, cellular trafficking, regulation of gene expression, apoptosis signaling, and pathogen recognition. LRR domains appear to function primarily in providing a versatile structural framework for the formation of protein-protein interactions (93). Many bacteria, either pathogenic or commensal, also possess LRR domains (see Pfam PF00560 and PF07723). Some representatives play a role in host-pathogen interactions, such as YopM in Yersinia pestis, the IpaH family in Shigella flexneri, SspH in Salmonella enterica serovar Typhimurium, Slr in group A streptococci, and IlsA in Bacillus cereus (53, 54, 74, 150, 184).
Listerial internalins are likely to play a wide range of functions, not only those dedicated to host-pathogen interactions. For instance, Lmo0327 is an internalin-like protein associated with an autolytic activity. However, this conclusion is based on phenotypes of an lmo0327 mutant (141), and further work is required to confirm whether Lmo0327, which has no domain related to peptidoglycan hydrolysis, is a bona fide autolysin.
Transporters.
Among the 28 lipoproteins that are putative SBPs of ABC transporter systems (Table 3), two have been associated with Listeria pathogenesis, OppA (Lmo2196) and LpeA (Lmo1847). OppA, which mediates the transport of oligopeptides, is required for bacterial growth at low temperature and intracellular survival in cultured macrophages as well as in organs of mice during the early phase of infection in vivo (23). Recent data suggest that OppA is subject to a PrfA-dependent posttranslational modification (13). Four other listerial proteins, Lmo0152, Lmo0135, Lmo2044, and Lmo2569, are structurally related to OppA, having an SBP bac-5 domain (Pfam PF00496) shared by proteins involved in active transport of solutes across the cytoplasmic membrane.
LpeA contains an SBP domain of the bac-9 family (Pfam PF01297). It is related to several streptococcal adhesins, including PsaA of S. pneumoniae, FimA of Streptococcus parasanguis, ScaA of Streptococcus gordonii, SsaB of Streptococcus sanguis, and EfmA of Enterococcus faecium (149). These proteins belongs to a family of surface-associated proteins designated lipoprotein receptor-associated antigen I (LraI). The LraI family constitutes a new family of extracellular SBPs specific for metal (Zn or Mn) ions. LpeA displays a metal binding site formed by His-67, His-139, Glu-205, and Asp-280, as described for PsaA. Interestingly, LpeA-defective bacteria efficiently adhere to eukaryotic cells but fail to penetrate in vitro into epithelial and hepatocyte cells, while they survive better in macrophages than do wild-type bacteria and display a weak exacerbation of virulence (149). LpeA might act directly through a cell receptor or indirectly through its hypothetical function as an ABC transporter, potentially influencing the cell sensing or an intracellular cascade associated with cell entry through the uptake of polypeptides, oligopeptides, and multiple sugars. We found two other listerial proteins related to LpeA, Lmo0153 and Lmo1671, but nothing is yet known about their function.
CONCLUSIONS AND FUTURE DIRECTIONS
Listeria surface proteins appear mosaic, often combining unique and repeated segments. Each domain could fold independently, but all domains likely function in concert with each other. Several questions remain to be addressed, in three major directions.
Pathogenic Potential
Several surface proteins contribute to L. monocytogenes virulence (Table 4). However, relatively few have received extensive biochemical characterization. In addition, functional analysis is still required for many uncharacterized proteins that may bind eukaryotic molecules. It will be especially interesting to address whether these proteins play redundant functions, work together, and/or act in macromolecular complexes. It will be also of major importance to address aspects of host specificities. InlA and InlB entry pathways are not functional in all animal models due to species specificity of interaction with their cellular receptors (91, 104). Such host specificities could occur for other surface proteins.
TABLE 4.
Surface proteins involved in L. monocytogenes virulence
| Protein | Name | Bacterial surface association type | Function(s) | Reference(s) for experimental validation |
|---|---|---|---|---|
| Lmo0433 | InlA | LPXTG | Adhesion and invasion | Reviewed in references 73 and 104 |
| Lmo0263 | InlH | LPXTG | Unknown | 164 |
| Lmo2821 | InlJ | LPXTG | Unknown | 156 |
| Lmo2026 | Lmo2026 | LPXTG | Unknown | 8 |
| Lmo0320 | Vip | LPXTG | Invasion | 32 |
| Lmo0434 | InlB | GW | Invasion and signaling | Reviewed in references 15 and 73 |
| Lmo1076 | Auto | GW | Cell wall turnover and invasion | 31 |
| Lmo2558 | Ami | GW | Cell wall turnover and adhesion | 127 |
| Lmo0582 | P60 | LysM | Cell wall turnover and invasion | 100 |
| Lmo1847 | LpeA | Lipoprotein | Metal transport and invasion | 149 |
| Lmo2196 | OppA | Lipoprotein | Oligopeptide transport and intracellular survival | 23 |
| Lmo0204 | ActA | Hydrophobic tail | Actin-based intracellular motility | Reviewed in reference 68 |
| Lmo1829 | FbpA | Unknown | Adhesion | 48 |
Spatio-Temporal Expression
Relatively few genes encoding surface proteins are known to be modulated during the Listeria infectious process in cultured cells in vitro (39, 88), and only a subset of surface proteins have been detected by proteomic approaches (13, 34, 146). It is likely that surface proteins are produced at different time points of the growth phase, with their expression depending upon various environmental changes, such as temperature, nutrients, or host factors. Identification of growth conditions during which they are expressed is a prerequisite for recognizing their mechanisms of function. Moving from cellular to tissue microbiology should help us to discover when these bacterial effectors are expressed and subsequently play a role in L. monocytogenes pathogenesis.
Localization
The spatial and temporal localization of a given protein in the bacterial cell wall is probably critical for its optimal function. For instance, Auto and MurA autolysins both have a glucosaminidase domain, but mutant bacteria elongate in one case and not in the other, suggesting that their glucosaminidase activities operate on distinct sets of muropeptides or regions of the peptidoglycan via different cell wall binding domains. Another example is provided by the intracellular motility factor ActA, whose asymmetric localization at one pole of the bacterium triggers the unidirectional movement of L. monocytogenes (94). It was recently suggested that the polarization of ActA might be a direct consequence of the differential cell wall growth rates along the bacterium (147). InlA has a polarized distribution similar to that of ActA and may thus follow the same rule (103, 147).
The distribution of cell wall-associated proteins is likely to also depend on the localization of the secretion apparatus. For S. pyogenes, controversial studies indicate that secretion could occur at the single Exportal site (153) or at multiple sites (36, 175), with a key role of signal peptide sequences in addressing secretion to a subcellular localization (36). It will be very interesting to test whether such information is present on Listeria signal sequences.
Finally, some proteins may be buried in the cell wall, such as InlB, which is not accessible to antibodies at the surface unless the peptidoglycan is digested with a muramidase (86). However, InlB acts as an invasion protein that needs to be accessible to interact with its host receptor, suggesting that external factors regulate its accessibility, possibly by acting on GW repeats. This hypothesis is supported by the demonstration that GW repeats bind to cellular proteoglycans and that these interactions are required for efficient entry (87). Other internalin-like proteins, such as E. faecalis EF2686 (ElrA), S. pyogenes Slr, and S. agalactiae BrlD are also buried in the cell wall (28, 190). The accessibility of these proteins at the bacterial surface may be modulated, affecting their function during the infectious process in vivo. It is thus of interest to understand what contributes to the topological distribution of proteins at the surface.
Strain and Species Variability
Although a number of surface proteins similar to those of strain EGDe has been reported to exist in the three other L. monocytogenes strains whose genomes are known (129), it is clear that each strain displays specific repertoires of surface proteins. It will be of interest to address to what extent surface proteins present in “pathovariants” are playing a role in successful infection. A large project of sequencing various different strains, from different environments and with different host tropisms, will soon provide important data to measure gene content variability (Listeria monocytogenes Sequencing Project. Broad Institute of Harvard and MIT, http://www.broad.mit.edu). It should reveal the core set of Listeria virulence factors, especially among surface proteins.
Acknowledgments
We address our sincere apologies to colleagues whose work has not been cited in this paper due to space constraints. We are very grateful to Francisco Garcia-del Portillo, Lothar Jänsch, and Uwe Kärst for sharing unpublished results and for helpful discussions. We thank Francisco Garcia-del Portillo, Serge Mostowy, Olivier Dussurget, and Samantha Sixin Lu for critical reading of the manuscript.
Work in P. Cossart's laboratory received financial support from the Pasteur Institute (GPH no. 9), INRA (AIP291), INSERM, ANR, and the Ministère de l'Education Nationale, de la Recherche et de la Technologie. P. Cossart is an international research scholar of the Howard Hughes Medical Institute. H. Bierne belongs to the Institut National de la Recherche Agronomique staff.
REFERENCES
- 1.Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Allignet, J., S. Aubert, K. G. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez-Dominguez, C., J. A. Vazquez-Boland, E. Carrasco-Marin, P. Lopez-Mato, and F. Leyva-Cobian. 1997. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect. Immun. 65:78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anantharaman, V., and L. Aravind. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade, M. A., F. D. Ciccarelli, C. Perez-Iratxeta, and P. Bork. 2002. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 3:RESEARCH0047. [DOI] [PMC free article] [PubMed]
- 6.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archambaud, C., M. A. Nahori, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281:31812-31822. [DOI] [PubMed] [Google Scholar]
- 8.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 11.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 13.Baumgartner, M., U. Karst, B. Gerstel, M. Loessner, J. Wehland, and L. Jansch. 2007. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhavsar, A. P., and E. D. Brown. 2006. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 60:1077-1090. [DOI] [PubMed] [Google Scholar]
- 15.Bierne, H., and P. Cossart. 2002. InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. J. Cell Sci. 115:3357-3367. [DOI] [PubMed] [Google Scholar]
- 16.Bierne, H., C. Garandeau, M. G. Pucciarelli, C. Sabet, S. Newton, F. Garcia-del Portillo, P. Cossart, and A. Charbit. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. Garcia-del Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 18.Bigot, A., H. Pagniez, E. Botton, C. Frehel, I. Dubail, C. Jacquet, A. Charbit, and C. Raynaud. 2005. Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity. Infect. Immun. 73:5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boekhorst, J., Q. Helmer, M. Kleerebezem, and R. J. Siezen. 2006. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152:273-280. [DOI] [PubMed] [Google Scholar]
- 20.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 8:46-53. [DOI] [PubMed] [Google Scholar]
- 21.Boneca, I. G., O. Dussurget, D. Cabanes, M. A. Nahori, S. Sousa, M. Lecuit, E. Psylinakis, V. Bouriotis, J. P. Hugot, M. Giovannini, A. Coyle, J. Bertin, A. Namane, J. C. Rousselle, N. Cayet, M. C. Prevost, V. Balloy, M. Chignard, D. J. Philpott, P. Cossart, and S. E. Girardin. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. USA 104:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnemain, C., C. Raynaud, H. Reglier-Poupet, I. Dubail, C. Frehel, M. A. Lety, P. Berche, and A. Charbit. 2004. Differential roles of multiple signal peptidases in the virulence of Listeria monocytogenes. Mol. Microbiol. 51:1251-1266. [DOI] [PubMed] [Google Scholar]
- 23.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosman, F. T., and I. Stamenkovic. 2003. Functional structure and composition of the extracellular matrix. J. Pathol. 200:423-428. [DOI] [PubMed] [Google Scholar]
- 25.Braun, L., S. Dramsi, P. Dehoux, H. Bierne, G. Lindahl, and P. Cossart. 1997. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25:285-294. [DOI] [PubMed] [Google Scholar]
- 26.Braun, L., B. Ghebrehiwet, and P. Cossart. 2000. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 19:1458-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453-464. [DOI] [PubMed] [Google Scholar]
- 28.Brinster, S., S. Furlan, and P. Serror. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 189:1244-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buey, R. M., B. Monterroso, M. Menendez, G. Diakun, P. Chacon, J. A. Hermoso, and J. F. Diaz. 2007. Insights into molecular plasticity of choline binding proteins (pneumococcal surface proteins) by SAXS. J. Mol. Biol. 365:411-424. [DOI] [PubMed] [Google Scholar]
- 30.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 31.Cabanes, D., O. Dussurget, P. Dehoux, and P. Cossart. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601-1614. [DOI] [PubMed] [Google Scholar]
- 32.Cabanes, D., S. Sousa, A. Cebria, M. Lecuit, F. Garcia-del Portillo, and P. Cossart. 2005. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J 24:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callebaut, I., D. Gilges, I. Vigon, and J. P. Mornon. 2000. HYR, an extracellular module involved in cellular adhesion and related to the immunoglobulin-like fold. Protein Sci. 9:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calvo, E., M. G. Pucciarelli, H. Bierne, P. Cossart, J. P. Albar, and F. Garcia-Del Portillo. 2005. Analysis of the Listeria cell wall proteome by two-dimensional nanoliquid chromatography coupled to mass spectrometry. Proteomics 5:433-443. [DOI] [PubMed] [Google Scholar]
- 35.Campo, N., and D. Z. Rudner. 2006. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol. Cell 23:25-35. [DOI] [PubMed] [Google Scholar]
- 36.Carlsson, F., M. Stalhammar-Carlemalm, K. Flardh, C. Sandin, E. Carlemalm, and G. Lindahl. 2006. Signal sequence directs localized secretion of bacterial surface proteins. Nature 442:943-946. [DOI] [PubMed] [Google Scholar]
- 37.Carroll, S. A., T. Hain, U. Technow, A. Darji, P. Pashalidis, S. W. Joseph, and T. Chakraborty. 2003. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J. Bacteriol. 185:6801-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charpentier, E., and P. Courvalin. 1999. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 43:2103-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courtney, H. S., Y. Li, J. B. Dale, and D. L. Hasty. 1994. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deivanayagam, C. C., R. L. Rich, M. Carson, R. T. Owens, S. Danthuluri, T. Bice, M. Hook, and S. V. Narayana. 2000. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 8:67-78. [DOI] [PubMed] [Google Scholar]
- 42.Desvaux, M., E. Dumas, I. Chafsey, and M. Hebraud. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256:1-15. [DOI] [PubMed] [Google Scholar]
- 43.Desvaux, M., and M. Hebraud. 2006. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 30:774-805. [DOI] [PubMed] [Google Scholar]
- 44.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 45.Dons, L., E. Eriksson, Y. Jin, M. E. Rottenberg, K. Kristensson, C. N. Larsen, J. Bresciani, and J. E. Olsen. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 48.Dramsi, S., F. Bourdichon, D. Cabanes, M. Lecuit, H. Fsihi, and P. Cossart. 2004. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 53:639-649. [DOI] [PubMed] [Google Scholar]
- 49.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156:289-297. [DOI] [PubMed] [Google Scholar]
- 50.Eckert, C., M. Lecerf, L. Dubost, M. Arthur, and S. Mesnage. 2006. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J. Bacteriol. 188:8513-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgcomb, M. R., S. Sirimanne, B. J. Wilkinson, P. Drouin, and R. D. Morse. 2000. Electron paramagnetic resonance studies of the membrane fluidity of the foodborne pathogenic psychrotroph Listeria monocytogenes. Biochim. Biophys. Acta 1463:31-42. [DOI] [PubMed] [Google Scholar]
- 52.Elasri, M. O., J. R. Thomas, R. A. Skinner, J. S. Blevins, K. E. Beenken, C. L. Nelson, and M. S. Smeltzer. 2002. Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30:275-280. [DOI] [PubMed] [Google Scholar]
- 53.Fallman, M., and A. Gustavsson. 2005. Cellular mechanisms of bacterial internalization counteracted by Yersinia. Int. Rev. Cytol. 246:135-188. [DOI] [PubMed] [Google Scholar]
- 54.Fedhila, S., N. Daou, D. Lereclus, and C. Nielsen-LeRoux. 2006. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol. Microbiol. 62:339-355. [DOI] [PubMed] [Google Scholar]
- 55.Fernandez-Tornero, C., E. Garcia, R. Lopez, G. Gimenez-Gallego, and A. Romero. 2002. Two new crystal forms of the choline-binding domain of the major pneumococcal autolysin: insights into the dynamics of the active homodimer. J. Mol. Biol. 321:163-173. [DOI] [PubMed] [Google Scholar]
- 56.Fernandez-Tornero, C., R. Lopez, E. Garcia, G. Gimenez-Gallego, and A. Romero. 2001. A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 8:1020-1024. [DOI] [PubMed] [Google Scholar]
- 57.Fiedler, F. 1988. Biochemistry of the cell surface of Listeria strains: a locating general view. Infection 16(Suppl. 2):S92-S97. [DOI] [PubMed] [Google Scholar]
- 58.Fischer, W. 1988. Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 29:233-302. [DOI] [PubMed] [Google Scholar]
- 59.Fischer, W., T. Mannsfeld, and G. Hagen. 1990. On the basic structure of poly(glycerophosphate) lipoteichoic acids. Biochem. Cell Biol. 68:33-43. [DOI] [PubMed] [Google Scholar]
- 60.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 61.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 62.Garandeau, C., H. Reglier-Poupet, I. Dubail, J. L. Beretti, P. Berche, and A. Charbit. 2002. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 70:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia, J. L., A. R. Sanchez-Beato, F. J. Medrano, and R. Lopez. 1998. Versatility of choline-binding domain. Microb. Drug Resist. 4:25-36. [DOI] [PubMed] [Google Scholar]
- 64.Geginat, G., T. Nichterlein, M. Kretschmar, S. Schenk, H. Hof, M. Lalic-Multhaler, W. Goebel, and A. Bubert. 1999. Enhancement of the Listeria monocytogenes p60-specific CD4 and CD8 T cell memory by nonpathogenic Listeria innocua. J. Immunol. 162:4781-4789. [PubMed] [Google Scholar]
- 65.Ghosh, B. K., and K. K. Carroll. 1968. Isolation, composition, and structure of membrane of Listeria monocytogenes. J. Bacteriol. 95:688-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilot, P., Y. Jossin, and J. Content. 2000. Cloning, sequencing and characterisation of a Listeria monocytogenes gene encoding a fibronectin-binding protein. J. Med. Microbiol. 49:887-896. [DOI] [PubMed] [Google Scholar]
- 67.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 68.Gouin, E., M. D. Welch, and P. Cossart. 2005. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8:35-45. [DOI] [PubMed] [Google Scholar]
- 69.Greenberg, J. W., W. Fischer, and K. A. Joiner. 1996. Influence of lipoteichoic acid structure on recognition by the macrophage scavenger receptor. Infect. Immun. 64:3318-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grundling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 98:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guinane, C. M., P. D. Cotter, R. P. Ross, and C. Hill. 2006. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob. Agents Chemother. 50:2824-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutekunst, K. A., L. Pine, E. White, S. Kathariou, and G. M. Carlone. 1992. A filamentous-like mutant of Listeria monocytogenes with reduced expression of a 60-kilodalton extracellular protein invades and grows in 3T6 and Caco-2 cells. Can. J. Microbiol. 38:843-851. [DOI] [PubMed] [Google Scholar]
- 73.Hamon, M., H. Bierne, and P. Cossart. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4:423-434. [DOI] [PubMed] [Google Scholar]
- 74.Haraga, A., and S. I. Miller. 2006. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell. Microbiol. 8:837-846. [DOI] [PubMed] [Google Scholar]
- 75.Harty, J. T., and E. G. Pamer. 1995. CD8 T lymphocytes specific for the secreted p60 antigen protect against Listeria monocytogenes infection. J. Immunol. 154:4642-4650. [PubMed] [Google Scholar]
- 76.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 77.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 78.Hell, W., H. G. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 79.Hether, N. W., P. A. Campbell, L. A. Baker, and L. L. Jackson. 1983. Chemical composition and biological functions of Listeria monocytogenes cell wall preparations. Infect. Immun. 39:1114-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hirano, T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell. Biol. 7:311-322. [DOI] [PubMed] [Google Scholar]
- 81.Holmes, A. R., R. McNab, K. W. Millsap, M. Rohde, S. Hammerschmidt, J. L. Mawdsley, and H. F. Jenkinson. 2001. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41:1395-1408. [DOI] [PubMed] [Google Scholar]
- 82.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Humann, J., R. Bjordahl, K. Andreasen, and L. L. Lenz. 2007. Expression of the p60 autolysin enhances NK cell activation and is required for Listeria monocytogenes expansion in IFN-gamma-responsive mice. J. Immunol. 178:2407-2414. [DOI] [PubMed] [Google Scholar]
- 84.Ibraghimov-Beskrovnaya, O., N. O. Bukanov, L. C. Donohue, W. R. Dackowski, K. W. Klinger, and G. M. Landes. 2000. Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. Hum. Mol. Genet. 9:1641-1649. [DOI] [PubMed] [Google Scholar]
- 85.Ishiguro, T., M. Naito, T. Yamamoto, G. Hasegawa, F. Gejyo, M. Mitsuyama, H. Suzuki, and T. Kodama. 2001. Role of macrophage scavenger receptors in response to Listeria monocytogenes infection in mice. Am. J. Pathol. 158:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jonquieres, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34:902-914. [DOI] [PubMed] [Google Scholar]
- 87.Jonquieres, R., J. Pizarro-Cerda, and P. Cossart. 2001. Synergy between the N- and C-terminal domains of InlB for efficient invasion of non-phagocytic cells by Listeria monocytogenes. Mol. Microbiol. 42:955-965. [DOI] [PubMed] [Google Scholar]
- 88.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamisango, K., I. Saiki, Y. Tanio, H. Okumura, Y. Araki, I. Sekikawa, I. Azuma, and Y. Yamamura. 1982. Structures and biological activities of peptidoglycans of Listeria monocytogenes and Propionibacterium acnes. J. Biochem. (Tokyo) 92:23-33. [DOI] [PubMed] [Google Scholar]
- 90.Kanehara, K., K. Ito, and Y. Akiyama. 2002. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 16:2147-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khelef, N., M. Lecuit, H. Bierne, and P. Cossart. 2006. Species specificity of the Listeria monocytogenes InlB protein. Cell. Microbiol. 8:457-470. [DOI] [PubMed] [Google Scholar]
- 92.Kobayashi, K. S., M. Chamaillard, Y. Ogura, O. Henegariu, N. Inohara, G. Nunez, and R. A. Flavell. 2005. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307:731-734. [DOI] [PubMed] [Google Scholar]
- 93.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 94.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 95.Kolb-Maurer, A., S. Pilgrim, E. Kampgen, A. D. McLellan, E. B. Brocker, W. Goebel, and I. Gentschev. 2001. Antibodies against listerial protein 60 act as an opsonin for phagocytosis of Listeria monocytogenes by human dendritic cells. Infect. Immun. 69:3100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Korndorfer, I. P., J. Danzer, M. Schmelcher, M. Zimmer, A. Skerra, and M. J. Loessner. 2006. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J. Mol. Biol. 364:678-689. [DOI] [PubMed] [Google Scholar]
- 97.Korsak, D., W. Vollmer, and Z. Markiewicz. 2005. Listeria monocytogenes EGD lacking penicillin-binding protein 5 (PBP5) produces a thicker cell wall. FEMS Microbiol. Lett. 251:281-288. [DOI] [PubMed] [Google Scholar]
- 98.Kriventseva, E. V., M. Biswas, and R. Apweiler. 2001. Clustering and analysis of protein families. Curr. Opin. Struct. Biol. 11:334-339. [DOI] [PubMed] [Google Scholar]
- 99.Kuehn, M. J., and N. C. Kesty. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645-2655. [DOI] [PubMed] [Google Scholar]
- 100.Kuhn, M., and W. Goebel. 1989. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect. Immun. 57:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Labischinski, H., D. Naumann, and W. Fischer. 1991. Small and medium-angle X-ray analysis of bacterial lipoteichoic acid phase structure. Eur. J. Biochem. 202:1269-1274. [DOI] [PubMed] [Google Scholar]
- 102.Lasa, I., and J. R. Penades. 2006. Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 157:99-107. [DOI] [PubMed] [Google Scholar]
- 103.Lebrun, M., J. Mengaud, H. Ohayon, F. Nato, and P. Cossart. 1996. Internalin must be on the bacterial surface to mediate entry of Listeria monocytogenes into epithelial cells. Mol. Microbiol. 21:579-592. [DOI] [PubMed] [Google Scholar]
- 104.Lecuit, M. 2005. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin. Microbiol. Infect. 11:430-436. [DOI] [PubMed] [Google Scholar]
- 105.Lecuit, M., D. M. Nelson, S. D. Smith, H. Khun, M. Huerre, M. C. Vacher-Lavenu, J. I. Gordon, and P. Cossart. 2004. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc. Natl. Acad. Sci. USA 101:6152-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lei, X. H., F. Fiedler, Z. Lan, and S. Kathariou. 2001. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J. Bacteriol. 183:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. USA 100:12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45:1043-1056. [DOI] [PubMed] [Google Scholar]
- 109.Lim, D., and N. C. Strynadka. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870-876. [DOI] [PubMed] [Google Scholar]
- 110.Lindahl, G., M. Stalhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Loessner, M. J., and R. Calendar. 2006. The bacteriophages, p. 593-601. Oxford University Press, New York, NY.
- 112.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324-340. [DOI] [PubMed] [Google Scholar]
- 113.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 114.Loessner, M. J., and S. Scherer. 1995. Organization and transcriptional analysis of the Listeria phage A511 late gene region comprising the major capsid and tail sheath protein genes cps and tsh. J. Bacteriol. 177:6601-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 116.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 117.Makinoshima, H., and M. S. Glickman. 2006. Site-2 proteases in prokaryotes: regulated intramembrane proteolysis expands to microbial pathogenesis. Microbes Infect. 8:1882-1888. [DOI] [PubMed] [Google Scholar]
- 118.Mandin, P., H. Fsihi, O. Dussurget, M. Vergassola, E. Milohanic, A. Toledo-Arana, I. Lasa, J. Johansson, and P. Cossart. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57:1367-1380. [DOI] [PubMed] [Google Scholar]
- 119.Marino, M., M. Banerjee, R. Jonquieres, P. Cossart, and P. Ghosh. 2002. GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands. EMBO J. 21:5623-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 1999. Structure of the lnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes. Mol. Cell 4:1063-1072. [DOI] [PubMed] [Google Scholar]
- 121.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mazmanian, S. K., and D. L. Kasper. 2006. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 6:849-858. [DOI] [PubMed] [Google Scholar]
- 123.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 124.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 126.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 127.Milohanic, E., R. Jonquieres, P. Cossart, P. Berche, and J. L. Gaillard. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 39:1212-1224. [DOI] [PubMed] [Google Scholar]
- 128.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Newton, S. M., P. E. Klebba, C. Raynaud, Y. Shao, X. Jiang, I. Dubail, C. Archer, C. Frehel, and A. Charbit. 2005. The svpA-srtB locus of Listeria monocytogenes: fur-mediated iron regulation and effect on virulence. Mol. Microbiol. 55:927-940. [DOI] [PubMed] [Google Scholar]
- 132.Nichols, D. S., K. A. Presser, J. Olley, T. Ross, and T. A. McMeekin. 2002. Variation of branched-chain fatty acids marks the normal physiological range for growth in Listeria monocytogenes. Appl. Environ. Microbiol. 68:2809-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Niemann, H. H., W. D. Schubert, and D. W. Heinz. 2004. Adhesins and invasins of pathogenic bacteria: a structural view. Microbes Infect. 6:101-112. [DOI] [PubMed] [Google Scholar]
- 134.Opitz, B., A. Puschel, W. Beermann, A. C. Hocke, S. Forster, B. Schmeck, V. van Laak, T. Chakraborty, N. Suttorp, and S. Hippenstiel. 2006. Listeria monocytogenes activated p38 MAPK and induced IL-8 secretion in a nucleotide-binding oligomerization domain 1-dependent manner in endothelial cells. J. Immunol. 176:484-490. [DOI] [PubMed] [Google Scholar]
- 135.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 136.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 137.Patti, J. M., J. O. Boles, and M. Hook. 1993. Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry 32:11428-11435. [DOI] [PubMed] [Google Scholar]
- 138.Pilgrim, S., A. Kolb-Maurer, I. Gentschev, W. Goebel, and M. Kuhn. 2003. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect. Immun. 71:3473-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pohlschroder, M., E. Hartmann, N. J. Hand, K. Dilks, and A. Haddad. 2005. Diversity and evolution of protein translocation. Annu. Rev. Microbiol. 59:91-111. [DOI] [PubMed] [Google Scholar]
- 140.Popowska, M. 2004. Analysis of the peptidoglycan hydrolases of Listeria monocytogenes: multiple enzymes with multiple functions. Pol. J. Microbiol. 53(Suppl.):29-34. [PubMed] [Google Scholar]
- 141.Popowska, M., and Z. Markiewicz. 2006. Characterization of Listeria monocytogenes protein Lmo0327 with murein hydrolase activity. Arch. Microbiol. 186:69-86. [DOI] [PubMed] [Google Scholar]
- 142.Porchet, N., and J. P. Aubert. 2004. MUC genes: mucin or not mucin? That is the question. Med. Sci. (Paris) 20:569-574. [DOI] [PubMed] [Google Scholar]
- 143.Prazak, M. A., E. A. Murano, I. Mercado, and G. R. Acuff. 2002. Antimicrobial resistance of Listeria monocytogenes isolated from various cabbage farms and packing sheds in Texas. J. Food Prot. 65:1796-1799. [DOI] [PubMed] [Google Scholar]
- 144.Promadej, N., F. Fiedler, P. Cossart, S. Dramsi, and S. Kathariou. 1999. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J. Bacteriol. 181:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pucciarelli, M. G., H. Bierne, and F. García-del Portillo. 2007. Listeria monocytogenes: pathogenesis and host response, p. 81-110. Springer, Berlin, Germany.
- 146.Pucciarelli, M. G., E. Calvo, C. Sabet, H. Bierne, P. Cossart, and F. Garcia-del Portillo. 2005. Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomic analysis. Proteomics 5:4808-4817. [DOI] [PubMed] [Google Scholar]
- 147.Rafelski, S. M., and J. A. Theriot. 2006. Mechanism of polarization of Listeria monocytogenes surface protein ActA. Mol. Microbiol. 59:1262-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reglier-Poupet, H., C. Frehel, I. Dubail, J. L. Beretti, P. Berche, A. Charbit, and C. Raynaud. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 278:49469-49477. [DOI] [PubMed] [Google Scholar]
- 149.Reglier-Poupet, H., E. Pellegrini, A. Charbit, and P. Berche. 2003. Identification of LpeA, a PsaA-like membrane protein that promotes cell entry by Listeria monocytogenes. Infect. Immun. 71:474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Reid, S. D., A. G. Montgomery, J. M. Voyich, F. R. DeLeo, B. Lei, R. M. Ireland, N. M. Green, M. Liu, S. Lukomski, and J. M. Musser. 2003. Characterization of an extracellular virulence factor made by group A Streptococcus with homology to the Listeria monocytogenes internalin family of proteins. Infect. Immun. 71:7043-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 152.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 153.Rosch, J., and M. Caparon. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513-1515. [DOI] [PubMed] [Google Scholar]
- 154.Rose, M. C. 1992. Mucins: structure, function, and role in pulmonary diseases. Am. J. Physiol. 263:L413-L429. [DOI] [PubMed] [Google Scholar]
- 155.Ruhland, G. J., M. Hellwig, G. Wanner, and F. Fiedler. 1993. Cell-surface location of Listeria-specific protein p60—detection of Listeria cells by indirect immunofluorescence. J. Gen. Microbiol. 139:609-616. [DOI] [PubMed] [Google Scholar]
- 156.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73:6912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Safdar, A., and D. Armstrong. 2003. Antimicrobial activities against 84 Listeria monocytogenes isolates from patients with systemic listeriosis at a comprehensive cancer center (1955 to 1997). J. Clin. Microbiol. 41:483-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sanchez-Beato, A. R., C. Ronda, and J. L. Garcia. 1995. Tracking the evolution of the bacterial choline-binding domain: molecular characterization of the Clostridium acetobutylicum NCIB 8052 cspA gene. J. Bacteriol. 177:1098-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sarvas, M., C. R. Harwood, S. Bron, and J. M. van Dijl. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694:311-327. [DOI] [PubMed] [Google Scholar]
- 160.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 161.Schirm, M., M. Kalmokoff, A. Aubry, P. Thibault, M. Sandoz, and S. M. Logan. 2004. Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J. Bacteriol. 186:6721-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 164.Schubert, W. D., G. Gobel, M. Diepholz, A. Darji, D. Kloer, T. Hain, T. Chakraborty, J. Wehland, E. Domann, and D. W. Heinz. 2001. Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain. J. Mol. Biol. 312:783-794. [DOI] [PubMed] [Google Scholar]
- 165.Scott, J. R., and T. C. Barnett. 2006. Surface proteins of gram-positive bacteria and how they get there. Annu. Rev. Microbiol. 60:397-423. [DOI] [PubMed] [Google Scholar]
- 166.Shen, A., and D. E. Higgins. 2006. The MogR transcriptional repressor regulates nonhierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shen, Y., M. Naujokas, M. Park, and K. Ireton. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501-510. [DOI] [PubMed] [Google Scholar]
- 168.Siezen, R., J. Boekhorst, L. Muscariello, D. Molenaar, B. Renckens, and M. Kleerebezem. 2006. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 170.Srinivasan, V., H. M. Nam, L. T. Nguyen, B. Tamilselvam, S. E. Murinda, and S. P. Oliver. 2005. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog. Dis. 2:201-211. [DOI] [PubMed] [Google Scholar]
- 171.Srivastava, K. K., and I. H. Siddique. 1973. Quantitative chemical composition of peptidoglycan of Listeria monocytogenes. Infect. Immun. 7:700-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stack, H. M., R. D. Sleator, M. Bowers, C. Hill, and C. G. Gahan. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 71:4241-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Stevens, J. M., E. E. Galyov, and M. P. Stevens. 2006. Actin-dependent movement of bacterial pathogens. Nat. Rev. Microbiol. 4:91-101. [DOI] [PubMed] [Google Scholar]
- 174.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 148:2065-2077. [DOI] [PubMed] [Google Scholar]
- 175.Swanson, J., K. C. Hsu, and E. C. Gotschlich. 1969. Electron microscopic studies on streptococci. I. M antigen. J. Exp. Med. 130:1063-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ton-That, H., S. K. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J. Biol. Chem. 275:9876-9881. [DOI] [PubMed] [Google Scholar]
- 178.Torres, D., M. Barrier, F. Bihl, V. J. Quesniaux, I. Maillet, S. Akira, B. Ryffel, and F. Erard. 2004. Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect. Immun. 72:2131-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Trost, M., D. Wehmhoner, U. Karst, G. Dieterich, J. Wehland, and L. Jansch. 2005. Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5:1544-1557. [DOI] [PubMed] [Google Scholar]
- 180.Uchikawa, K., I. Sekikawa, and I. Azuma. 1986. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J. Biochem. (Tokyo) 99:315-327. [DOI] [PubMed] [Google Scholar]
- 181.Ullmann, W. W., and J. A. Cameron. 1969. Immunochemistry of the cell walls of Listeria monocytogenes. J. Bacteriol. 98:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.van der Laan, M., N. P. Nouwen, and A. J. Driessen. 2005. YidC—an evolutionary conserved device for the assembly of energy-transducing membrane protein complexes. Curr. Opin. Microbiol. 8:182-187. [DOI] [PubMed] [Google Scholar]
- 183.Varea, J., J. L. Saiz, C. Lopez-Zumel, B. Monterroso, F. J. Medrano, J. L. Arrondo, I. Iloro, J. Laynez, J. L. Garcia, and M. Menendez. 2000. Do sequence repeats play an equivalent role in the choline-binding module of pneumococcal LytA amidase? J. Biol. Chem. 275:26842-26855. [DOI] [PubMed] [Google Scholar]
- 184.Venkatesan, M. M., J. M. Buysse, and A. B. Hartman. 1991. Sequence variation in two ipaH genes of Shigella flexneri 5 and homology to the LRG-like family of proteins. Mol. Microbiol. 5:2435-2445. [DOI] [PubMed] [Google Scholar]
- 185.Verheul, A., N. J. Russell, T. H. R. Van, F. M. Rombouts, and T. Abee. 1997. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 63:3451-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Vicente, M. F., J. C. Perez-Daz, F. Baquero, M. Angel de Pedro, and J. Berenguer. 1990. Penicillin-binding protein 3 of Listeria monocytogenes as the primary lethal target for beta-lactams. Antimicrob. Agents Chemother. 34:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Vollmer, W., and A. Tomasz. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 70:7176-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vukov, N., I. Moll, U. Blasi, S. Scherer, and M. J. Loessner. 2003. Functional regulation of the Listeria monocytogenes bacteriophage A118 holin by an intragenic inhibitor lacking the first transmembrane domain. Mol. Microbiol. 48:173-186. [DOI] [PubMed] [Google Scholar]
- 189.Wai, S. N., B. Lindmark, T. Soderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe, and B. E. Uhlin. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 190.Waldemarsson, J., T. Areschoug, G. Lindahl, and E. Johnsson. 2006. The streptococcal Blr and Slr proteins define a family of surface proteins with leucine-rich repeats: camouflaging by other surface structures. J. Bacteriol. 188:378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 192.Way, S. S., L. J. Thompson, J. E. Lopes, A. M. Hajjar, T. R. Kollmann, N. E. Freitag, and C. B. Wilson. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell. Microbiol. 6:235-242. [DOI] [PubMed] [Google Scholar]
- 193.Wehmhoner, D., G. Dieterich, E. Fischer, M. Baumgartner, J. Wehland, and L. Jansch. 2005. “LaneSpector,” a tool for membrane proteome profiling based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis/liquid chromatography-tandem mass spectrometry analysis: application to Listeria monocytogenes membrane proteins. Electrophoresis 26:2450-2460. [DOI] [PubMed] [Google Scholar]
- 194.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, L. Y. Geer, W. Helmberg, Y. Kapustin, D. L. Kenton, O. Khovayko, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, K. D. Pruitt, G. D. Schuler, L. M. Schriml, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, T. O. Suzek, R. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2006. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 34:D173-D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wonderling, L. D., B. J. Wilkinson, and D. O. Bayles. 2004. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wuenscher, M. D., S. Kohler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zawadzka-Skomial, J., Z. Markiewicz, M. Nguyen-Disteche, B. Devreese, J. M. Frere, and M. Terrak. 2006. Characterization of the bifunctional glycosyltransferase/acyltransferase penicillin-binding protein 4 of Listeria monocytogenes. J. Bacteriol. 188:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zong, Y., Y. Xu, X. Liang, D. R. Keene, A. Hook, S. Gurusiddappa, M. Hook, and S. V. Narayana. 2005. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 24:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]