Abstract
Summary: Viruses have long been studied not only for their pathology and associated disease but also as model systems for molecular processes and as tools for identifying important cellular regulatory proteins and pathways. Recent advances in mass spectrometry methods coupled with the development of proteomic approaches have greatly facilitated the detection of virion components, protein interactions in infected cells, and virally induced changes in the cellular proteome, resulting in a more comprehensive understanding of viral infection. In addition, a rapidly increasing number of high-resolution structures for viral proteins have provided valuable information on the mechanism of action of these proteins as well as aided in the design and understanding of specific inhibitors that could be used in antiviral therapies. In this paper, we discuss proteomic studies conducted on all eukaryotic viruses and bacteriophages, covering virion composition, viral protein structures, virus-virus and virus-host protein interactions, and changes in the cellular proteome upon viral infection.
INTRODUCTION
Viruses have long been studied not only for their pathology and associated disease but also as model systems for molecular processes and as tools for identifying important cellular regulatory proteins and pathways. Over the last 50 years, viral studies have provided many important insights into our understanding of positive and negative gene regulation, repressor-operator interactions, DNA replication, transcriptional elongation and termination, chaperone activity, immune signaling, RNA splicing, and oncogenic transformation. Although numerous biologically important findings have come from studying the structure, function, and protein interactions of individual viral proteins, only a small number of viral proteins have been studied in a small percentage of viruses. There is a large, untapped body of information on viruses and virus-host interactions that will continue to reveal new insights into the understanding of basic biological processes as well as contribute to useful technological innovations.
The development of proteomic methods has revolutionized our ability to assess protein interactions and cellular changes on a global scale, allowing the discovery of previously unknown connections. For example, proteomic methods used to generate genome-wide protein interaction maps for budding yeast uncovered a number of novel protein complexes (38, 58). Viruses are suitable targets for genome-wide analyses since their relatively small size makes them a readily tractable system and because there is a large number of fully sequenced genomes available (including many adenovirus, ichnovirus, influenza virus, herpesvirus, papillomavirus, rotavirus, and reovirus isolates). In addition, many are amenable to genetic manipulation. Indeed, the first completely sequenced genome from any organism was that of bacteriophage φX174, reported in 1977 (88). Sequences of larger viral genomes began to enter the public databases several years later, with the 172-kb Epstein-Barr virus (EBV) genome in 1984 and the 230-kb human cytomegalovirus genome in 1990 (4, 18). There has since been an explosion in the number of sequenced viral genomes, totaling 2,323 eukaryotic viruses and 411 phages in the NCBI genome database (http://www.ncbi.nlm.nih.gov/genomes/static/vis.html) at the time of writing of this review. Although most viral genomes are considerably smaller than cellular genomes, there is a growing list of remarkable “giant” viruses whose genomes are similar in size and coding potential to some cellular organisms (23). The giant viruses are from a diverse mix of viral families including bacteriophage, phycodnavirus, nimavirus, and poxvirus, which infect bacteria, algae, invertebrates, and vertebrates, respectively. The recently discovered mimivirus challenges our views of the boundaries separating viruses and cellular organisms, since its 1.2-megabase genome surpasses the size of several bacterial genomes, and its 1,262 potential proteins include a translation system not previously found in viruses (81). Our understanding of the large viruses and their host cell interactions would benefit enormously from the use of proteomic methods to analyze the contributions of the numerous encoded viral proteins in an unbiased way.
In this review, we discuss the work that has been done to date using proteomic approaches to study viruses from any viridae infecting eukaryotic or prokaryotic cells. This includes determining the protein composition of virions, the structure and protein interactions of viral proteins, and the effects of viral infection and individual viral proteins on the cellular proteome.
VIRION PROTEOMICS
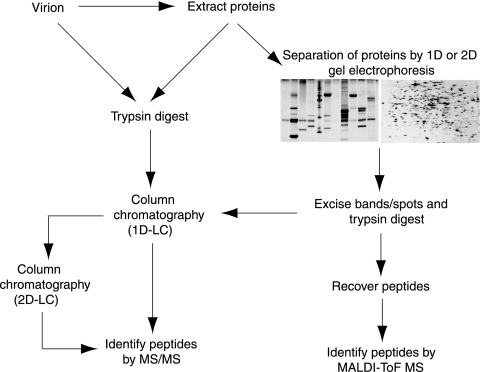
Knowledge of the protein composition of the infectious viral particle or virion is an important prerequisite for functional studies, as it focuses the analysis on specific proteins and their roles during infection. Determining the virion composition may be straightforward for small viruses that lack envelopes, since they are comprised of a small number of viral proteins with limited or no capacity to package host proteins in the viral capsid. However, determining the makeup of virions with more complex structures can be challenging. Enveloped viruses have considerable potential to incorporate both viral and host proteins into their membrane(s) as well as inside the envelope, and these can be present at low levels, making their detection difficult. In recent years, two mass spectrometry approaches, matrix-assisted laser desorption ionization (MALDI)-time of flight (TOF) mass spectrometry and liquid chromatography (LC)-linked tandem mass spectrometry (LC-MS/MS), have been widely used to analyze the composition of purified virions, leading to the identification of previously unknown components of viral particles (Fig. 1).
FIG. 1.
Analysis of virion composition by mass spectrometry. Typical approaches for analyzing virion protein composition by MALDI-TOF mass spectrometry (MS) and LC-MS/MS are shown. 1D, one dimensional.
Nonenveloped Viruses
There have been few proteomic studies on virions from nonenveloped eukaryotic viruses, but one has been reported for adenovirus. Chelius et al. (19) used the well-characterized adenovirus type 5 virions as a test system, using LC-MS/MS with both one-dimensional and two-dimensional (2D) chromatographic separations to characterize the protein complement of virions. This method identified all of the expected viral structural proteins, although the 23-kDa viral protease was detected only after 2D separation. Since this protein is known to be present at only 10 copies per virion, those authors concluded that LC-MS/MS was a useful method for identifying proteins in whole virions that are present at copy numbers of 10 or more.
Enveloped RNA Viruses
Virions from two RNA viruses, severe acute respiratory syndrome (SARS) coronavirus and human immunodeficiency virus type 1 (HIV-1), have been subjected to proteomic analysis. SARS coronavirus virions were analyzed by 2D LC-MS/MS, confirming the presence of S (spike), M (membrane), and N (nucleocapsid) proteins (122). This method failed to detect the E (envelope) protein, but a single E peptide was identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by electrospray ionization-MS/MS, suggesting that it may be present in low abundance. Possible cellular components of these virions were not investigated.
Two groups used LC-MS/MS to identify viral and host proteins in HIV-1 virions. Saphire et al. (89) compared tryptic digests of whole virions produced in Jurkat and 293T cells. In both cell types, the expected HIV proteins along with several human proteins that were previously reported to be incorporated into HIV-1 were identified. The human CD48 protein and histones H1, H2A, H3, and H4 were also identified as virion components. Chertova et al. (20) analyzed HIV-1 virions derived from monocyte-derived macrophages by SDS-PAGE followed by tryptic digestion of proteins in individual gel slices. They found 253 different human proteins, 33 of which were already known to be virion components. The numerous newly identified human proteins included histones, in keeping with the results described by Saphire et al., and several proteins of the endosomal compartment that may reflect the use of the late endosomal pathway for virion budding from monocytes.
The large discrepancy in the numbers of host proteins identified by Saphire et al. and Chertova et al. likely reflects a combination of the different sensitivities of the methods used for sample preparation for mass spectrometry as well as genuine differences in virions produced in the different cell types. It has been reported that preparations of HIV-1 virions can also contain cellular vesicles, which would complicate any analysis of cellular proteins incorporated into the virions (10, 39). However, Chertova et al. used CD45 immunoaffinity depletion to remove these vesicles from their virion preparations prior to density gradient purification, thereby limiting the number of cellular proteins that would be present in the virion preparations but not actually incorporated into the virions. While many of the 253 cellular proteins may be incorporated into the HIV-1 virions, it seems unlikely that all of these proteins are of functional significance, and some may be packaged accidentally due to their abundance and cellular localization. Detailed functional studies will be necessary to determine which of the packaged cellular proteins actually contribute to HIV-1 infection. It is also worth noting that despite the very high number of human proteins identified in HIV-1 virions, some human proteins reported to be HIV-1 virion components by other methods, such as Staufen and tRNA synthetase, were not found (for a review of previously identified HIV virion components, see reference 16). Thus, mass spectrometry-based analyses are not completely comprehensive and, like most methods, will have some false-positive and false-negative results.
Herpesviruses
Virion proteomics have been performed for several herpesviruses. These include human cytomegalovirus (CMV) (HCMV) and murine cytomegalovirus of the betaherpesviruses and EBV, Kaposi's sarcoma-associated herpesvirus (KSHV), rhesus monkey rhadinovirus (RRV), and murine gammaherpesvirus 68 (MHV68) of the gammaherpesviruses (5, 9, 13, 51, 55, 74, 113, 123). To date, there are no reports of virion proteomic studies of any alphaherpesviruses, including the intensively studied herpes simplex virus type 1 (HSV-1). All infectious herpesvirus virions are composed of an icosahedral nucleocapsid surrounded by a structural matrix called the tegument, which is in turn surrounded by a lipid envelope. The multiple structural components and large size (150 to 200 nm in diameter) of the virions give the potential for packaging numerous viral and host proteins.
A proteomic analysis of purified HCMV virions was initially conducted by Baldick and Shenk in 1996 (5), where protein components were identified by SDS-PAGE followed by N-terminal sequencing. In this way, six viral proteins and one actin-like cellular protein (not previously known to be packaged), in addition to known virion components, were identified. These proteins were also found in noninfectious enveloped particles and dense bodies, two types of aberrant viral particles generated in HCMV infection. Recently, Varnum et al. (113) used the more sensitive LC-MS/MS and LC-Fourier transform ion cyclotron resonance mass spectrometry techniques to determine the composition of the HCMV proteome. As expected, they identified the set of proteins described previously by Baldick and Shenk as well as less abundant viral and cellular proteins, totaling 71 HCMV and 71 host proteins. Twenty-four of the viral proteins were not previously identified in virions, and 12 of these proteins were from open reading frames (ORFs) not previously shown to express proteins. The LC-Fourier transform ion cyclotron resonance-mass spectrometry approach allowed for the relative abundance of the virion proteins to be determined, indicating that the pp65 (UL83) tegument protein was the most prevalent viral component and that gM was the most abundant glycoprotein. Approximately 50% of the viral proteins in the virion were found to be tegument proteins, compared to 30% capsid proteins, 13% envelope proteins, and 7% proteins of undefined localization. Dense HCMV bodies were also analyzed as part of this study and, consistent with previous reports, were found to contain considerably fewer proteins than did the virions, with 29 viral and 13 cellular proteins identified. It is important that the HCMV virions were extensively purified from culture media on both sedimentation and density gradients and verified to be free of cellular organelles by electron microscopy. Therefore, the cellular proteins in the HCMV virion preparations are likely to be inside the virion. While the localization of the cellular proteins within the virion was not determined, the numerous cytoplasmic proteins identified are expected to be in the tegument compartment, picked up as the capsids move through the cytoplasm from the endoplasmic reticulum (ER) to the Golgi apparatus.
Highly purified murine CMV virions have also been analyzed by LC-MS/MS, leading to the identification of 58 viral proteins, 20 of which were uncharacterized, and 7 cellular proteins (actin, annexin, cofilin, histone H2A, elongation factor 1α, glyceraldehyde-3-phosphate dehydrogenase, and rho GDP dissociation inhibitor) (55). All of the cellular proteins except histone H2A were also found by Varnum et al. in HCMV virions.
The virion compositions of several gammaherpesviruses have been determined. For EBV and KSHV, this involved treatments to induce lytic infection (since these viruses are usually in a latent state) followed by extensive gradient purifications of the virions. Using LC-MS/MS analyses, Johannsen et al. (51) identified 35 viral proteins that largely comprised homologues of proteins previously found in other herpesvirus virions, while none of the EBV latency proteins were detected. Similar to results obtained for CMV, the most numerous viral proteins were tegument components; however, for EBV, these included five proteins unique to gammaherpesviruses. The six most prevalent cellular proteins identified (actin, hsp70, hsp90, cofilin, β-tubulin, and enolase) were found in the tegument, and these six proteins were also identified in HCMV virions.
Purified virions from KSHV, the human virus most closely related to EBV, have also been the subject of proteomic analyses by two groups. Zhu et al. (123) identified 24 viral and multiple cellular proteins using LC-MS/MS, while Bechtel et al. (9) identified 10 viral and nine cellular proteins using MALDI. All but one of the viral proteins identified by Bechtel et al. were also found by Zhu et al., and 19 out of the 24 viral proteins identified by Zhu et al. were found by Johannsen et al. to have corresponding proteins in EBV virions. As for EBV, actin, hsp70, hsp90, tubulin, and enolase were identified as being cellular components of the KSHV virions in both analyses. In addition, Zhu et al. further characterized the form of actin identified as nonmuscle β-actin and showed that the actin and myosin proteins in their virion preparation were resistant to trypsin digestion, indicating that they were indeed inside the virions. Similarly, Bechtel et al. showed that six of the cellular proteins that they identified (enolase, Hsc70, tubulin, moesin, ezrin, and EF-2b) were trypsin resistant in virion preparations and therefore likely to be packaged inside the virion (the remaining four cellular proteins were not tested).
The virion compositions of gammaherpesviruses that infect mice (MHV68) and rhesus monkeys (RRV) have also been studied. Bortz et al. (13) identified 15 viral proteins in MHV68; these overlap considerably but not entirely with those identified in KSHV. O'Connor and Kedes (74) used two different mass spectrometry approaches to identify 33 viral proteins in RRV; MS/MS of protein bands excised from polyacrylamide gels and multidimensional protein identification technology in which proteolytic peptides from the entire virion were analyzed en masse by 2D LC-MS/MS. The identified proteins included three proteins not previously found in any herpesvirus virions. As for other herpesviruses, the majority of the virion components were from the tegument.
Poxviruses
Poxviruses produce several types of infectious particles, the first of which are intracellular mature virions (IMV), which contain a single membrane (25). IMVs are wrapped by two Golgi membranes to form intracellular enveloped virions, which are then released at the cell surface, losing one membrane to form cell-associated extracellular virions and extracellular enveloped virions. The composition of IMVs from vaccinia virus has been the subject of study by several groups. Initially, Takahashi et al. (101) identified 17 virally encoded components by N-terminal sequencing. Jensen et al. (49) used 2D gel electrophoresis coupled with MALDI-TOF mass spectrometry to identify 13 viral and five cellular proteins in IMVs, the latter of which were predominantly mitochondrial proteins. Since the cellular proteins in the IMV preparations were accessible to proteases, they are likely derived from mitochondrial fragments and not internal to the virions. Yoder et al. (119) used multiple mass spectrometry approaches to analyze vaccinia virus IMVs, resulting in the identification of 63 virally encoded proteins, 2 of which were from ORFs not previously known to be expressed. Around the same time, Chung et al. (22) analyzed the IMV proteome by LC-MS/MS, identifying 75 viral and 23 cellular protein components. Ten of the viral proteins were not previously known to be IMV components (A6L, A15L, A16L, A31R, C6L, E6R, G3L, G9R, K4L, and L3L), and six of these were also described by Yoder et al. Most of the cellular proteins in the IMVs have also been found in virions from other viruses (including actin, tubulin, ubiquitin, elongation factor 1α, hsp70, and hsp90), but a few abundant cellular proteins found in virions from other viruses were not detected (e.g., myosin, moesin, and cofilin). Since IMVs are prepared from cell lysates, it is possible that some of the cellular proteins copurifying with these particles bind to the outside of the virions as opposed to being incorporated into the virion. This important point remains to be addressed.
In addition to vaccinia virus, IMV particles from the myxoma leporipoxvirus have been subjected to proteomic analysis (120), leading to the identification of 17 viral proteins. These proteins are homologues of the viral proteins found in vaccinia virus IMVs, with the exception of M151R (B13R in vaccinia virus), which has not been found in vaccinia virus virions, and M093L, which lacks a vaccinia virus homologue.
Other Enveloped DNA Viruses
Singapore grouper iridovirus contains a 140-kb genome that encodes 162 proteins and produces large enveloped virions. Virion proteins were initially analyzed by SDS-PAGE followed by MALDI-TOF mass spectrometry, leading to the identification of 26 virally encoded proteins (96). Later, that same group used a shotgun proteomic approach in which whole virions were digested with trypsin and the resulting peptides were separated by LC and then analyzed by MALDI (95). The additional viral proteins identified brought the total number of components in the virion to 44, 20 of which were found by both methods. Fourteen of the virion proteins had no apparent homologues in other iridoviruses.
White spot syndrome virus (WSSV) of the Nimaviridae is another large enveloped DNA virus that infects shrimp and other crustaceans. Its 300-kb circular DNA genome encodes about 180 proteins, and the rod-shaped virions have been studied by several groups. In 2002, Huang et al. (45) analyzed WSSV virions by SDS-PAGE and identified 18 viral proteins from excised bands by MALDI-TOF and MS/MS. All but one of those proteins were shown to be late proteins. A similar approach described by Tsai et al. (109) led to the identification of 33 viral proteins, 20 of which had not been previously identified. In a subsequent paper, that same group used detergent and salt extractions to fractionate purified WSSV virions in order to categorize the localization of the major virion proteins as nucleocapsid (four proteins reported), tegument (four proteins reported), or envelope (seven proteins reported) components (110). Xie et al. (117) conducted a similar analysis of fractionated WSSV virions; of the 30 viral proteins identified, they classified 23 as being envelope proteins and 8 as being nucleocapsids. These proteins included the envelope and nucleocapsid proteins described by Tsai et al. (110), with the exception of two envelope proteins reported by Tsai et al. (VP36B and VP53A), which were not identified.
A proteomic analysis of the virions from the gigantic mimivirus has recently been conducted, resulting in the identification of 114 viral proteins (83). In addition to major structural proteins, mimivirus virions were found to contain 12 proteins predicted to be involved in transcription, 9 proteins from oxidative pathways, 3 DNA repair proteins, 2 topoisomerases, and 45 proteins with no sequence similarity to other proteins in the database. It is also remarkable that no amoeba host proteins were found despite the fact that two amoebal genome sequences are present in the database. This suggests that mimivirus packages few or no host proteins, in marked contrast to HIV, vaccinia virus, and herpesvirus virions.
Bacteriophages
Four recent studies have examined the structural proteomes of bacteriophages. Roberts et al. (85) analyzed the proteome of coliphage T1 using isoelectric focusing to separate the components of the virion followed by MALDI-TOF. T1 was previously estimated to contain 13 to 15 structural proteins in the mature virion (66, 108, 116), and the 2D gel profile of the T1 virion performed in that study resolved 15 proteins. Mass spectrometry identified four abundant proteins: the major tail protein (gp41), a major capsid component (gp49), the major head subunit (gp47), and a protein that is associated with the major head subunit (gp48).
The bacteriophage Φ11b, which infects Flavobacterium, was also examined in a study reported by Borriss et al. (12). SDS-PAGE analysis of the phage particle resolved 11 proteins, and 5 were identified through LC-mass spectrometry, including the major capsid protein (gp21), minor head protein (gp18), portal protein (gp16), and two hypothetical proteins. Lavigne et al. (60) studied the T7-like psuedomonal bacteriophage pKMV using two approaches: separation of the phage particle proteins by SDS-PAGE coupled with LC-electrospray ionization-MS/MS and a whole-phage shotgun analysis using trypsin-digested phage particles separated by reversed-phase high-performance liquid chromatography followed by MS/MS. The data gathered from these two approaches allowed the identification of 12 virion proteins. Six of these proteins were previously annotated as being structural proteins due to sequence similarity with known phage structural proteins. Five additional proteins that had little or no sequence similarity to known phage proteins were successfully classified as structural proteins.
Naryshkina et al. (73) examined the proteome of pYS40, a lytic tailed bacteriophage of Thermus thermophilus. This phage has a large genome, with a total of 170 potential ORFs. Mass spectrometry was performed on proteins resolved by SDS-PAGE, and two major virion components were identified. They are believed to correspond to the major head and tail proteins. Their functions could not have been predicted by sequence comparisons because of a lack of homologues in the database. Purified phage was also examined by a shotgun proteomic approach using multidimensional protein identification technology. Peptides matching 33 pYS40 proteins were detected, including the tail sheath protein (gp69), baseplate assembly protein (gp150), fibritin neck whisker (gp152), and a DEAD box helicase (gp79). The remaining proteins identified were novel proteins with no database homologues, including a group of 13 proteins detected at high levels that may correspond to the late gene cluster.
Studies examining the composition of phage particles provide valuable information that can be used to aid in the correct annotation of phage genomes and allow comparative genomics studies. Phage genomes are highly mosaic as a result of horizontal gene exchange through both homologous and nonhomologous recombination. Although it is often difficult to detect sequence similarity between genes that are likely homologous due to the extreme divergence of phage genomes, strong conservation of gene order can facilitate identification. Thus, the identification of major capsid and tail proteins in phages with no identifiable sequence similarity to phages already in the genome database can allow the annotation of the morphogenetic region.
Summary
In summary, several different mass spectrometric approaches have been used successfully to analyze the composition of a variety of virions. These methods have proven to be complementary and together have led to a more complete picture of the viral particle. While the data sets obtained by different methods largely agree, each method has its own strengths and weaknesses, and this can result in missed or additional protein identifications. The enveloped DNA viruses in general are composed of a large number of viral proteins, including many that are not strictly structural but are known or predicted to function once released into the host cell. In herpesviruses, the tegument is the major site of packaging of these proteins. Numerous host proteins are also packed in HIV, vaccinia virus, and herpesvirus particles. There is some overlap in the nature of the host proteins packaged in all of these virions; in particular, several highly abundant host proteins have been reported for many types of virions. For example, actin, β-tubulin, elongation factor 1α, elongation factor 2, hsp70, and hsp90 have been found in HIV, vaccinia virus, and two or more types of herpesvirus virions. It is not yet known whether these cellular proteins play a conserved functional role in these virions, are passenger proteins that are accidentally packed due to their location and high abundance, or are contaminants that copurify with the virions. The latter possibility seems unlikely, since the different types of virions analyzed were prepared using different methods and, in each case, checked for purity prior to analysis. Whether or not the above-described host proteins are purposefully packaged in the virions will require more extensive investigation. However, it is not hard to imagine that host chaperone proteins like hsp70 and hsp90 may be required to chaperone various viral proteins during packaging and/or release from the virions and hence are packaged along with their viral partners.
STRUCTURAL PROTEOMICS
High-resolution structures of viral proteins are extremely valuable for elucidating protein function, for mechanisms of action, and for structure-aided design of inhibitors that could be used in antiviral therapies. However, structural studies using nuclear magnetic resonance spectroscopy and X-ray crystallography have been successful for only a small fraction of viral proteins. One of the challenges in generating these structures is in producing large quantities of viral proteins in soluble form, since proteins from eukaryotic viruses are notoriously difficult to express in Escherichia coli. There are numerous examples of individual viral proteins that are produced poorly in E. coli but that are expressed in soluble form in insect cells, suggesting that baculovirus expression is generally superior for viral protein expression.
A systematic comparison of the expression and solubility of a large number of viral proteins in E. coli and insect cells was conducted by Gao et al. (37). This involved the use of a high-throughput multiwell plate system for isolating bacmids, generating baculoviruses, and screening the expression, solubility, and recovery of hexahistidine-tagged proteins on nickel resin. A total of 337 proteins from HSV-1, EBV, CMV, and vaccinia viruses were scored for expression level and solubility in insect cells. A subset of 152 of these proteins was also tested for expression and solubility in an E. coli strain augmented with tRNA genes for codons rarely used in E. coli but common in mammalian proteins. For all three herpesviruses, expression in insect cells was superior to that in E. coli, with only 27 to 29% of the proteins scored as being expressed in E. coli and 90% of HSV and EBV proteins and 50% of CMV proteins expressed in insect cells. For vaccinia virus, however, expression rates were similar, at about 80%. For all four viruses, solubility of the expressed proteins in either system was a major issue, with approximately 20% of proteins expressed in soluble form. Similarly, the Structural Proteomics in Europe (SPINE) group reported problems with the expression and solubility of herpesvirus proteins in E. coli, with improved results using baculovirus (35). Like Gao et al., they had higher success rates with poxvirus proteins in E. coli expression. Tarbouriech et al. (102) monitored the expression of 23 EBV proteins in E. coli and found only 8 to be produced in soluble form. This outcome was not improved by modifying the constructs by N- or C-terminal truncations, changing the E. coli strain, or changing the expression temperature. The refolding of insoluble proteins also failed to result in soluble protein for the 12 EBV proteins tested (102) but was more successful (four out of nine proteins) for secreted vaccinia virus immunomodulators (35).
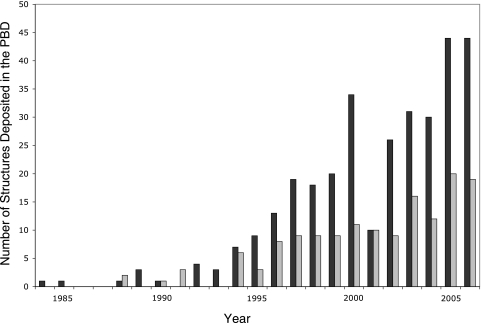
Despite the difficulties mentioned above, structures have been determined for numerous eukaryotic viral proteins or protein domains, totaling approximately 300 different structures at the time of writing of this review. As expected, the number of viral protein structures solved has been rapidly increasing in recent years (Fig. 2). For example, less than 10 viral protein structures were determined in 1995, including the first herpesvirus structure (11), compared to 44 unique structures deposited in the Protein Data Bank (PDB) in 2005. The number of structures determined varies considerably for different viruses. This likely represents a combination of the amount of effort a virus receives as well as the ease of production of soluble proteins and the tendency of these proteins to crystallize, which may be inherently different for different classes of viruses. An example of a virus that has moved relatively quickly in structural terms is SARS coronavirus. Since its genome sequence was determined in 2003 (65, 86), structures for 13 different proteins/domains have been deposited in the PDB from the 28 encoded proteins (see http://sars.scripps.edu/ProtSummary1.htm for a summary of these structures). This surpasses the number of unique structures for any single herpesvirus (currently 12 for HSV-1 and 11 for EBV) or for vaccinia virus (nine structures), each of which encodes four to nine times more proteins than SARS. Another example of a virus that has proven to be very amenable to structural studies is hepatitis C virus (HCV). Protein structures corresponding to most of the HCV coding sequences have been determined, providing insight into the mechanisms of action of the nonstructural proteins and facilitating the development of inhibitors for these proteins (57, 61, 63, 64, 106, 118).
FIG. 2.
Viral protein structures in the PDB. The numbers of protein structures from eukaryotic viruses (black) and phages (gray) are shown according to the year deposited in the PDB.
Presently, there are approximately 150 unique structures of bacteriophage proteins in the PBD. These proteins include representatives from a total of 43 different bacteriophages and six prophages, many of which have only a single solved structure. More than 45% of the total number of phage structures in the PDB belong to four intensively studied phages: T4, with 30 structures; lambda, with 18 structures; and T7 and P22, with 10 structures each. The deposition rates of phage proteins have mirrored those of viral structures, with early structures, including the repressor and Cro proteins from phage 434, deposited in the PDB in 1988. These structures were preceded by a few phage structures, including the lambda Cro and repressor proteins (2, 77). In the 10-year period between 1988 and 1998, a total of 42 phage structures were deposited, while in the 2-year period spanning 2005 to 2006, there were 40 structures deposited.
Viral protein structures have come largely from individual laboratories with interests in specific viral proteins or subsets of viral proteins, a complete discussion of which is beyond the scope of this review. However, in recent years, organized groups have used high-throughput approaches to tackle multiple viral protein targets. For example, the Consortium for Functional and Structural Proteomics of SARS Coronavirus-Related Proteins (http://sars.scripps.edu) studies the structure and function of many SARS proteins, which has led to four high-resolution structures (53, 79, 82, 87). The European Vizier group (http://www.vizier-europe.org/) studies the replication proteins of several RNA viruses, including SARS, measles virus, influenza virus, HCV, dengue virus, and viruses that cause gastroenteritis and encephalitis, with the aim of identifying new drug targets against these viruses. To date, they have reported structures for the VP9 outer coat protein of Banna virus (46) and the ORF-9b SARS protein (72), both in conjunction with SPINE. SPINE focuses on the development and implementation of methods suitable for high-throughput structure determination of proteins that impact human health, including proteins from vaccinia virus (nine proteins targeted by the Oxford group), EBV (studied by the Grenoble group), and SARS (targeted by groups at Oxford and Marseille) (http://www.spineurope.org) (35). To date, they reported one measles virus protein structure (52), four SARS protein structures (although only two for NSP9 have been published) (32, 99), four vaccinia virus protein structures, including A41L (none have been made publicly available), and four EBV protein structures (102). Structures were determined for EBV BARF1, dUTPase, uracil DNA glycosylase, and the EBV protease, with the latter three in complex with inhibitors (15, 103, 104). Finally, the Ontario Center for Structural Proteomics, a center affiliated with the Midwest Center for Structural Genomics, has implemented a systematic approach to solving bacteriophage proteins using both nuclear magnetic resonance spectroscopy and X-ray crystallography. To date, they have published five structures from bacteriophage lambda (gpU, CII, NinB, FII, and W) (30, 48, 67-69) as well as one from the related phage HK97 (PDB accession number 2OB9).
PROTEIN INTERACTIONS
Among the most important applications of proteomic approaches is in providing an unbiased account of the interactions of viral proteins with other viral proteins or with the host cellular proteins. The large number of sequenced viral genomes enables the use of proteomic methods to study virus-virus and virus-host interactions, providing valuable insight into the protein interactions that allow viruses to infect and replicate within the host cell.
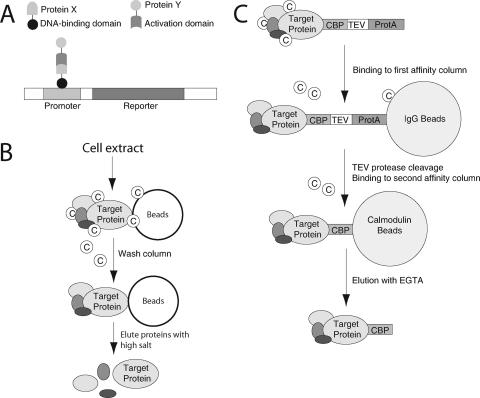
The most common method used to screen for protein interactions is a yeast two-hybrid screen using GAL4 and/or LexA fusion proteins (33) (Fig. 3A). Results with this method are very dependent on the quality of the library being screened and the relative representation of each cDNA. They also depend on the expression level of individual proteins and their ability to localize to the yeast nuclei, where the reporter constructs are located. As a result, many bona fide protein interactions are missed by this method (31). Tandem affinity purification (TAP) tagging approaches are becoming increasingly common for the detection of protein complexes (84, 121). This involves expressing the target protein fused to two affinity tags in host cells and purifying the tagged protein on two affinity columns under conditions that do not disrupt protein interactions (Fig. 3C). Copurifying proteins are then identified by mass spectrometry. While this method is excellent for identifying stable complexes, it tends to miss more transient interactions, which dissociate during the multistep purification. A more comprehensive method for detecting protein interactions of various strengths is an affinity column approach, where the purified target protein is immobilized on resin (97) (Fig. 3B). Cell extracts are passed through the column, and the profile of retained proteins is determined by mass spectrometry and compared to those proteins retained on negative control columns. The ability to detect specific interactions by this method depends on the concentration of individual proteins in the soluble lysate and hence is also not completely comprehensive. In addition, this approach requires a highly purified target protein, making it less popular than other methods.
FIG. 3.
Proteomic methods for protein-protein interactions. Schematic representation of the yeast-based two-hybrid screen (A), affinity column profiling (B), and TAP tagging (C) are shown. IgG, immunoglobulin G; TEV, tobacco etch virus protease site; CBP, calmodulin binding protein.
In addition to false-negative results, each of the above-described methods is expected to have some degree of false-positive results. Various controls must be performed to assess the specificity of the interactions and to determine whether the interaction is mediated by nucleic acid, which can indirectly tether two proteins together. The latter possibility can be addressed by TAP tagging and affinity column approaches by treating extracts with DNase I and RNase A, ethidium bromide, or benzonase. Any protein-protein interactions identified would then require additional experiments to determine if the interaction was direct (as opposed to being mediated by another protein) and of functional significance.
Each of the above-described proteomic methods has the potential to detect unanticipated interactions, making them suitable for exploring poorly understood systems. In the long term, the information gained from these types of studies will increase our understanding of viral infections and may lead to improved strategies for treating them. In this review, we will focus on studies using unbiased screening approaches to identify binding partners, as opposed to approaches designed to verify already known or suspected interactions.
Virus-Virus Interactions
Screens with individual viral proteins.
There have been several studies in which individual viral proteins have been screened for interactions with other viral proteins. For example, Mayer et al. (70) used a TAP tagging approach coupled with mass spectrometry to determine the composition of ribonucleoprotein complexes from Borna disease virus in persistently infected cells. They showed that TAP-tagged nucleoprotein (N) copurified with four viral proteins identified as being two forms of nucleoprotein (p38 and p39), the matrix protein (M), and the phosphoprotein (P).
Several different proteins from herpesviruses have been screened for viral protein interactions. Taylor and Knipe (105) isolated protein complexes formed with the ICP8 single-stranded DNA binding protein of HSV-1 by immunoprecipitation and mass spectrometry, identifying six viral proteins in addition to many cellular proteins. Vittone et al. (115) screened 13 HSV-1 tegument proteins for interactions with each other in a LexA-based two-hybrid system. This led to the identification of nine interactions, including three self-interactions, five of which were not previously known. Schierling et al. (91) generated a random genomic library of HCMV fused to the GAL4 activation domain and screened it for interactions with HCMV protein UL82 (pp71), involved in the activation of immediate-early transcription. The screen identified interactions with UL32 (pp150) and the UL35 tegument protein, the latter of which was verified by other assays. Lake and Hutt-Fletcher (59) also used a two-hybrid approach to screen an EBV cDNA library for interactions with the EBV BFRF1 protein (a membrane protein homologous to the UL34 alphaherpesvirus protein), identifying an interaction with the UL31 homologue BFLF2.
The viral protein interactions of a few different vaccinia virus proteins have also been determined. Szajner et al. (100) investigated the viral protein interactions of the vaccinia virus F10 kinase. Epitope-tagged F10 was isolated from vaccinia virus-infected cells by immunoaffinity chromatography, and interacting proteins were analyzed by SDS-PAGE and mass spectrometry. Interactions were seen for the previously identified F10 targets A30 and G7 as well as for A15, D2, D3, and J1. The same method was used by this laboratory to investigate the viral protein interactions of four vaccinia virus membrane proteins, A21, A28, H2, and L5. The results revealed that these four proteins form a complex along with four additional uncharacterized proteins (93).
Genome-wide screens.
A small number of genome-wide screens for viral protein-viral protein interactions have been conducted. The first genome-wide protein-protein interaction study was carried out on bacteriophage T7 using a yeast two-hybrid system (8). A random library of T7 DNA fragments was cloned into DNA binding domain and activation domain vectors, and the fragments were tested against one another. There were 25 interactions detected, including 9 interactions for which no previous genetic or biochemical evidence existed. These interactions suggested cross-connections between pathways and provided information about the potential functions of these proteins. For example, the interactions of proteins 4.7 and 6.5 with DNA polymerase suggested that these previously unannotated proteins affect some aspect of DNA replication.
Viruses in which mature proteins are generated from the proteolytic processing of polyproteins have also been subjected to genome-wide screens for viral protein interactions. Flajolet et al. (34) conducted a two-hybrid screen of a random genomic HCV library against itself, which proved to be more successful in detecting interactions than screening with mature HCV proteins. The screen of 200 bait clones identified NS3-E2, NS5A-E1, NS4A-NS2, and NS4A-NS3 interactions, the latter of which was previously known. A similar two-hybrid screen was performed with random genomic fragments of wheat streak mosaic virus, which, like HCV, uses the proteolytic processing of polyproteins to generate mature viral proteins (21). The screen identified multiple interactions between protein domains that were not detected in two-hybrid assays with the mature proteins. However, the lack of two-hybrid interactions of mature proteins generated from processed polyproteins is not a general rule, as interactions were detected between mature proteins from the potyviruses potato virus A and pea seed-borne mosaic virus in two-hybrid screens (41).
There have been two publications that reported genome-wide screens of large DNA viruses. McCraith et al. (71) conducted a two-hybrid screen of 266 predicted vaccinia virus proteins. This resulted in the identification of 37 protein-protein interactions, 28 of which were previously known. Uetz et al. (111) performed genome-wide two-hybrid screens for two herpesviruses, KSHV and varicella-zoster virus (VZV). For VZV, approximately 10,000 potential interactions were tested for the 69 proteins encoded by this virus as well as protein fragments, resulting in 173 nonredundant protein pairs. Tests of 12,000 potential interactions of the 89 KSHV full-length proteins, and fragments thereof, identified 123 nonredundant protein pairs, including five of the seven previously known KSHV interactions. Fifty percent of the newly identified interactions were confirmed by coimmunoprecipitation, and several others have known orthologous interactions in other herpesviruses, suggesting that many of these interactions may be biologically significant. The KSHV data set was used to predict orthologous protein interactions in other herpesviruses (HSV-1, VZV, EBV, and CMV), and of the 112 predicted interactions, 72 were confirmed by coimmunoprecipitation. However, only 5 of the 19 interactions predicted for VZV were actually detected in the two-hybrid screen, which is in keeping with other reports of a high degree of false-negative results in two-hybrid assays (31).
Virus-Host Interactions
Like viral protein-viral protein interactions, data for virus-host interactions allow the thorough characterization of the life cycles of viruses and have the potential to reveal important interactions that could be targeted for drug therapy. Screens designed to identify cellular protein interactions with specific viral proteins have been conducted for many individual viral proteins but have yet to be performed on a viral genome-wide scale. By far, the most common screening method used to date is the yeast two-hybrid screen. While two-hybrid approaches have identified many functionally important virus-host interactions, reports of two-hybrid screens conducted with individual proteins from eukaryotic viruses are too numerous to be covered here.
A small but increasing number of studies have used proteomics methods to reveal additional virus-host interactions. Affinity column approaches, coupled to either mass spectrometry or N-terminal sequencing, have been used by several groups to identify proteins from cell extracts that are retained by an immobilized viral protein. Using an HIV-1 Nef affinity column and N-terminal sequencing, Cohen et al. (24) identified human thioesterase II as a Nef binding partner, an interaction which had also been observed in a two-hybrid screen. Kang et al. (54) analyzed hepatocyte proteins retained on an HCV core protein column by two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry of individual spots, leading to the identification of four different intermediate filament proteins as core binding partners. Holowaty et al. (44) identified several human proteins retained on an Epstein-Barr nuclear antigen 1 (EBNA1) column by MALDI-TOF mass spectrometry of protein bands after SDS-PAGE. The strength and specificity of these interactions were further assessed by repeated passage of the eluted proteins through the EBNA1 column and by performing the same experiment with a charge-matched protein affinity column. A TAP tagging approach was also used to assess stable host cell interactions formed by EBNA1 when expressed in human cells (44). This identified a subset of cellular protein interactions found by the affinity column approach, including one with USP7, which was later shown to enable EBNA1 to modulate the p53 pathway (90).
In general, simple immunoprecipitation approaches are not amenable to proteomic analyses due to the high degree of background proteins in the samples and interference from the antibody bands. However, this approach can been used successfully for proteomic analyses if the antibodies are of high specificity and affinity and particularly if they are covalently coupled to beads, reducing the amount of antibody in the final sample. For example, immunoprecipitation has been used to identify interactions with HSV-1 proteins ICP8 and ICP27 (36, 105). Immunoprecipitation of ICP8 from HSV-1-infected cells led to the identification of over 50 cellular proteins by LC-MS/MS in addition to the viral proteins mentioned above (105). The large number of proteins associated with DNA processes is consistent with the known function of ICP0 as a single-stranded DNA binding protein; however, at least some of these interactions are likely to be indirect, mediated by nucleic acid or other proteins. Similar studies with the multifunctional ICP27 protein identified several cellular partners, including three translation initiation factors (36). Finally, the host interactions of the nsP3 protein of Sindbis alphavirus during the course of infection have been explored using a different immunoaffinity purification approach, which involved expressing nsP3 fused to a green fluorescent protein tag and recovering the protein on magnetic beads coupled to green fluorescent protein antibody (27, 28). Recovered proteins were then identified by SDS-PAGE and MALDI-TOF mass spectrometry, revealing an interaction of nsP3 with host G3BP throughout the course of infection and an interaction with 14-3-3 late in infection.
VIRUS-INDUCED CHANGES IN THE CELLULAR PROTEOME
Another important area of study examines how viral infection or expression of a specific viral protein affects the expression of the cellular proteome. There have been many studies using microarrays to profile cellular changes at the transcription level in response to viral infection or individual viral protein expression (for reviews, see references 29, 56, and 80), and these studies will not be discussed here. However, there is also a need to determine changes at the protein level, in part because changes observed in mRNA abundance do not always correspond to changes at the protein level (43, 107). In addition, many viral proteins affect protein turnover without affecting the transcription rate of the protein, for example, by promoting or interfering with polyubiquitination (reviewed in references 6, 62, 94, and 98). Virus-induced changes in the cellular proteome have been assessed by comparing protein profiles before and after viral infection by two general approaches: 2D gel electrophoresis and quantitative mass spectrometry techniques. 2D gel methods also enable the detection of altered forms of a protein, including posttranslational modifications, which could have major effects on the function of the cellular protein.
2D gel electrophoresis has been used to determine changes in the cellular proteome upon infection by several different viruses, where protein spots that differ before and after infection are excised and identified by mass spectrometry. The host interactions of three different plant viruses have been studied by this approach. Ventelon-Debout et al. (114) used this method to study infection by rice yellow mottle virus and found changes in 24 to 40 cellular proteins depending on the cultivar of rice infected. These proteins included several stress response proteins such as salt-induced protein, heat shock proteins, and superoxide dismutase (SOD). A similar study compared changes in tomato protein expression after infection with tobacco mosaic virus and also found changes in antioxidant enzymes (including SOD) as well as peptidases, chitinases, and proteins in the ascorbate-glutathione cycle (17). In a somewhat different application, Cooper et al. (26) conducted 2D gel analyses on tobacco plants infected with an unknown virus and identified the proteins expressed after infection as a means of identifying the viral proteins and hence the nature of the virus itself (identified as potato virus X).
The host interactions of a small number of animal viruses have also been studied by 2D gel approaches. Alfonso et al. (1) used 2D electrophoresis to examine changes in Vero cells after infection with African swine fever virus and identified 12 induced cellular proteins, which included SOD and two heat shock proteins (hsp70 and hsp27). Brasier et al. (14) reported changes in 24 nuclear proteins upon infection with respiratory syncytial virus, and, like the above-described viruses, these proteins included heat shock proteins and SOD. 2D gel analysis was also used to compare changes in cellular protein levels before and after EBV infection as well as before and after the expression of a single EBV protein, EBNA2 (92). Changes were identified in 20 cellular proteins upon EBNA2 expression, several of which showed similar changes upon EBV infection, suggesting that EBNA2 has a major role in affecting the cellular proteome in an EBV infection.
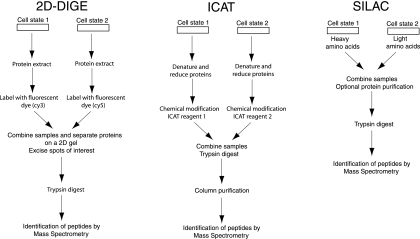
Jiang et al. (50) studied the cellular response to SARS infection by both a 2D gel approach and mass spectrometry utilizing isotope-coded affinity tag (ICAT) technology. The 2D gel approach used in that study differed from those described above in using different fluorescent dyes to uniformly label the protein samples, which could then be mixed together and analyzed on a single 2D gel, as opposed to analyzing the two samples on separate 2D gels (Fig. 4). This approach, called difference gel electrophoresis (DIGE) (112), has greatly facilitated the comparison of two (or three) samples by removing the gel-to-gel variability factor and by using dyes with a greater dynamic range than the traditionally used silver or Coomassie stains (reviewed in reference 78). However, like all 2D gel electrophoresis methods, DIGE is limited in its ability to detect very large proteins or proteins with extreme isoelectric points and is not useful for studying membrane proteins. The ICAT adaptation of mass spectrometry used by Jiang et al. (50) involves the labeling of two protein samples with isotopically light and heavy ICAT reagents, which react with the thiol group in the cysteine residues of proteins (42, 78) (Fig. 4). The heavy- and light-labeled samples are then combined, trypsinized, and analyzed by LC-mass spectrometry, enabling the quantification of the relative abundance of each protein in the two samples. This method is more sensitive than DIGE for the detection of low-abundant proteins but cannot detect proteins lacking cysteine or changes in posttranslational modifications. Using these two methods, Jiang et al. (50) identified quantitative changes of 1.5-fold or more upon SARS infection for 186 cellular proteins (out of 355 identified proteins), but only 15 proteins were identified as being differentially expressed by both methods. DIGE identified 48 unique proteins, 30 of which were altered in abundance upon SARS infection, while ICAT identified 322 proteins, 167 of which were altered. This reflected inherent differences of the two methods to detect specific proteins as discussed above, where ICAT was more sensitive in general and better for the identification of higher-molecular-weight proteins, while 2D electrophoresis favored lower-molecular-weight proteins.
FIG. 4.
Methods for comparative proteomics. Schematic representations for the 2D DIGE, ICAT, and SILAC methods used to compare the relative abundance of individual proteins in two samples are shown.
Several groups used mass spectrometry-based approaches to identify differences in the abundances of host proteins after viral infection. Go et al. (40) used ICAT as well as 18O labeling coupled with LC-MS/MS to monitor changes in Drosophila melanogaster cells infected with flock house virus (FHV) and found 150 proteins that were increased and 66 proteins that were decreased in abundance out of 1,500 identified proteins. Baas et al. (3) used MS/MS after various chromatographic separations to identify changes induced by influenza virus infection of macaques and reported several changes for the 3,548 proteins identified. Jacobs et al. (47) used multidimensional liquid chromatographic separations coupled to mass spectrometry to identify proteomic changes after HCV infection of highly permissive Huh-7.5 cells. The multiple proteins showing altered abundance included many that are involved in lipid metabolism.
Finally, Bartee et al. (7) examined the effects of expressing the KSHV membrane-associated K5 ubiquitin ligase protein in human cells using stable isotope labeling with amino acids in cell culture (SILAC) to differentially label samples with light or heavy isotopes of C and N before and after K5 expression, respectively (Fig. 4). This method is similar to ICAT in allowing quantitative comparisons of individual proteins in combined samples but gives more uniform labeling of proteins (provided that long labeling times are used) and, unlike ICAT, does not require the purification of the tryptic peptides (to remove excess ICAT reagent) prior to mass spectrometry (75, 76). As the name implies, SILAC can be used only in tissue culture cells and not on primary cells or tissues. Bartee et al. (7) compared the ratios of heavy and light proteins in plasma, Golgi apparatus, and ER membrane fractions in order to assess the effect of K5 expression on human membrane proteins. Over 100 cellular proteins were repeatedly identified for each of these compartments. While the only consistent change in protein expression in the Golgi apparatus and ER was the K5 protein itself, four cellular proteins were found to be consistently down-regulated in the plasma membrane (including the expected major histocompatibility complex class I), and one (leukocyte adhesion molecule) was confirmed by other methods to be a target of K5 ubiquitination.
CONCLUDING REMARKS
Recent advances in mass spectrometry methods coupled with the development of proteomic approaches have greatly facilitated the detection and identification of proteins. In addition, detection of virus-induced changes in the cellular proteome has been greatly facilitated by DIGE and ICAT and SILAC mass spectrometry methods. These advances have enabled a more comprehensive characterization of virions and of virus-virus and virus-host interactions involved in infection and pathogenesis. However, it is important to stress that proteomic analyses are not an end point but rather are a starting point for functional studies; discoveries made through proteomics will inevitably require more traditional methods to determine their functional significance. To date, proteomic approaches have been applied to only a small number of viruses and individual viral proteins, and high-resolution structures are available for only a small percentage of the viral proteome. Challenges in the expression of eukaryotic viral proteins will need to be overcome to facilitate structure determinations for many of these proteins. Viral proteomics is clearly in its infancy, and huge amounts of information remain untapped, particularly from the large number of viruses that are completely uncharacterized. More extensive applications of proteomic tools to viruses will inevitably provide valuable information on viral pathogenesis and life cycles as well as reveal new insights into cellular functions.
Acknowledgments
We thank Aled Edwards for helpful comments and critical reading of the manuscript.
L.F. is a Canada Research Chair in Molecular Virology.
REFERENCES
- 1.Alfonso, P., J. Rivera, B. Hernaez, C. Alonso, and J. M. Escribano. 2004. Identification of cellular proteins modified in response to African swine fever virus infection by proteomics. Proteomics 4:2037-2046. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, W. F., D. H. Ohlendorf, Y. Takeda, and B. W. Matthews. 1981. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature 290:754-758. [DOI] [PubMed] [Google Scholar]
- 3.Baas, T., C. R. Baskin, D. L. Diamond, A. Garcia-Sastre, H. Bielefeldt-Ohmann, T. M. Tumpey, M. J. Thomas, V. S. Carter, T. H. Teal, N. Van Hoeven, S. Proll, J. M. Jacobs, Z. R. Caldwell, M. A. Gritsenko, R. R. Hukkanen, D. G. Camp II, R. D. Smith, and M. G. Katze. 2006. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 80:10813-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 5.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry, M., and K. Fruh. 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 2006:pe21. [DOI] [PubMed] [Google Scholar]
- 7.Bartee, E., A. L. McCormack, and K. Fruh. 2006. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. [DOI] [PubMed] [Google Scholar]
- 9.Bechtel, J. T., R. C. Winant, and D. Ganem. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 11.Bochkarev, A., J. Barwell, R. Pfuetzner, W. Furey, A. Edwards, and L. Frappier. 1995. Crystal structure of the DNA binding domain of the Epstein-Barr virus origin binding protein EBNA1. Cell 83:39-46. [DOI] [PubMed] [Google Scholar]
- 12.Borriss, M., T. Lombardot, F. O. Glockner, D. Becher, D. Albrecht, and T. Schweder. 2007. Genome and proteome characterization of the psychrophilic Flavobacterium bacteriophage 11b. Extremophiles 11:95-104. [DOI] [PubMed] [Google Scholar]
- 13.Bortz, E., J. P. Whitelegge, Q. Jia, Z. H. Zhou, J. P. Stewart, T. T. Wu, and R. Sun. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 77:13425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasier, A. R., H. Spratt, Z. Wu, I. Boldogh, Y. Zhang, R. P. Garofalo, A. Casola, J. Pashmi, A. Haag, B. Luxon, and A. Kurosky. 2004. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78:11461-11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buisson, M., J. F. Hernandez, D. Lascoux, G. Schoehn, E. Forest, G. Arlaud, J. M. Seigneurin, R. W. Ruigrok, and W. P. Burmeister. 2002. The crystal structure of the Epstein-Barr virus protease shows rearrangement of the processed C terminus. J. Mol. Biol. 324:89-103. [DOI] [PubMed] [Google Scholar]
- 16.Cantin, R., S. Methot, and M. J. Tremblay. 2005. Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J. Virol. 79:6577-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casado-Vela, J., S. Selles, and R. B. Martinez. 2006. Proteomic analysis of tobacco mosaic virus-infected tomato (Lycopersicon esculentum M.) fruits and detection of viral coat protein. Proteomics 6(Suppl. 1):S196-S206. [DOI] [PubMed] [Google Scholar]
- 18.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 19.Chelius, D., A. F. Huhmer, C. H. Shieh, E. Lehmberg, J. A. Traina, T. K. Slattery, and E. Pungor, Jr. 2002. Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J. Proteome Res. 1:501-513. [DOI] [PubMed] [Google Scholar]
- 20.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80:9039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi, I. R., D. C. Stenger, and R. French. 2000. Multiple interactions among proteins encoded by the mite-transmitted wheat streak mosaic tritimovirus. Virology 267:185-198. [DOI] [PubMed] [Google Scholar]
- 22.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claverie, J. M., H. Ogata, S. Audic, C. Abergel, K. Suhre, and P. E. Fournier. 2006. Mimivirus and the emerging concept of “giant” virus. Virus Res. 117:133-144. [DOI] [PubMed] [Google Scholar]
- 24.Cohen, G. B., V. S. Rangan, B. K. Chen, S. Smith, and D. Baltimore. 2000. The human thioesterase II protein binds to a site on HIV-1 Nef critical for CD4 down-regulation. J. Biol. Chem. 275:23097-23105. [DOI] [PubMed] [Google Scholar]
- 25.Condit, R. C., N. Moussatche, and P. Traktman. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 66:31-124. [DOI] [PubMed] [Google Scholar]
- 26.Cooper, B., D. Eckert, N. L. Andon, J. R. Yates, and P. A. Haynes. 2003. Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J. Am. Soc. Mass Spectrom. 14:736-741. [DOI] [PubMed] [Google Scholar]
- 27.Cristea, I. M., J. W. Carroll, M. P. Rout, C. M. Rice, B. T. Chait, and M. R. MacDonald. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269-30278. [DOI] [PubMed] [Google Scholar]
- 28.Cristea, I. M., R. Williams, B. T. Chait, and M. P. Rout. 2005. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4:1933-1941. [DOI] [PubMed] [Google Scholar]
- 29.DeFilippis, V., C. Raggo, A. Moses, and K. Fruh. 2003. Functional genomics in virology and antiviral drug discovery. Trends Biotechnol. 21:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edmonds, L., A. Liu, J. J. Kwan, A. Avanessy, M. Caracoglia, I. Yang, K. L. Maxwell, J. Rubenstein, A. R. Davidson, and L. W. Donaldson. 2007. The NMR structure of the gpU tail-terminator protein from bacteriophage lambda: identification of sites contributing to Mg(II)-mediated oligomerization and biological function. J. Mol. Biol. 365:175-186. [DOI] [PubMed] [Google Scholar]
- 31.Edwards, A. M., B. Kus, R. Jansen, D. Greenbaum, J. Greenblatt, and M. Gerstein. 2002. Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 18:529-536. [DOI] [PubMed] [Google Scholar]
- 32.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245. [DOI] [PubMed] [Google Scholar]
- 34.Flajolet, M., G. Rotondo, L. Daviet, F. Bergametti, G. Inchauspe, P. Tiollais, C. Transy, and P. Legrain. 2000. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242:369-379. [DOI] [PubMed] [Google Scholar]
- 35.Fogg, M. J., P. Alzari, M. Bahar, I. Bertini, J. M. Betton, W. P. Burmeister, C. Cambillau, B. Canard, M. Carrondo, M. Coll, S. Daenke, O. Dym, M. P. Egloff, F. J. Enguita, A. Geerlof, A. Haouz, T. A. Jones, Q. Ma, S. N. Manicka, M. Migliardi, P. Nordlund, R. J. Owens, Y. Peleg, G. Schneider, R. Schnell, D. I. Stuart, N. Tarbouriech, T. Unge, A. J. Wilkinson, M. Wilmanns, K. S. Wilson, O. Zimhony, and J. M. Grimes. 2006. Application of the use of high-throughput technologies to the determination of protein structures of bacterial and viral pathogens. Acta Crystallogr. D Biol. Crystallogr. 62:1196-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontaine-Rodriguez, E. C., T. J. Taylor, M. Olesky, and D. M. Knipe. 2004. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330:487-492. [DOI] [PubMed] [Google Scholar]
- 37.Gao, M., N. Brufatto, T. Chen, L. L. Murley, R. Thalakada, M. Domagala, B. Beattie, D. Mamelak, V. Athanasopoulos, D. Johnson, G. McFadden, C. Burks, and L. Frappier. 2005. Expression profiling of herpesvirus and vaccinia virus proteins using a high-throughput baculovirus screening system. J. Proteome Res. 4:2225-2235. [DOI] [PubMed] [Google Scholar]
- 38.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 39.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 40.Go, E. P., W. R. Wikoff, Z. Shen, G. O'Maille, H. Morita, T. P. Conrads, A. Nordstrom, S. A. Trauger, W. Uritboonthai, D. A. Lucas, K. C. Chan, T. D. Veenstra, H. Lewicki, M. B. Oldstone, A. Schneemann, and G. Siuzdak. 2006. Mass spectrometry reveals specific and global molecular transformations during viral infection. J. Proteome Res. 5:2405-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo, D., M. L. Rajamaki, M. Saarma, and J. P. Valkonen. 2001. Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 82:935-939. [DOI] [PubMed] [Google Scholar]
- 42.Gygi, S. P., B. Rist, S. A. Gerber, F. Turecek, M. H. Gelb, and R. Aebersold. 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17:994-999. [DOI] [PubMed] [Google Scholar]
- 43.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen 1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 45.Huang, C., X. Zhang, Q. Lin, X. Xu, Z. Hu, and C. L. Hew. 2002. Proteomic analysis of shrimp white spot syndrome viral proteins and characterization of a novel envelope protein VP466. Mol. Cell. Proteomics 1:223-231. [DOI] [PubMed] [Google Scholar]
- 46.Jaafar, F. M., H. Attoui, M. W. Bahar, C. Siebold, G. Sutton, P. P. Mertens, P. De Micco, D. I. Stuart, J. M. Grimes, and X. De Lamballerie. 2005. The structure and function of the outer coat protein VP9 of Banna virus. Structure 13:17-28. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs, J. M., D. L. Diamond, E. Y. Chan, M. A. Gritsenko, W. Qian, M. Stastna, T. Baas, D. G. Camp II, R. L. Carithers, Jr., R. D. Smith, and M. G. Katze. 2005. Proteome analysis of liver cells expressing a full-length hepatitis C virus (HCV) replicon and biopsy specimens of posttransplantation liver from HCV-infected patients. J. Virol. 79:7558-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain, D., Y. Kim, K. L. Maxwell, S. Beasley, R. Zhang, G. N. Gussin, A. M. Edwards, and S. A. Darst. 2005. Crystal structure of bacteriophage lambda cII and its DNA complex. Mol. Cell 19:259-269. [DOI] [PubMed] [Google Scholar]
- 49.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. K. Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang, X. S., L. Y. Tang, J. Dai, H. Zhou, S. J. Li, Q. C. Xia, J. R. Wu, and R. Zeng. 2005. Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol. Cell. Proteomics 4:902-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson, K., J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard, and S. Longhi. 2003. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278:44567-44573. [DOI] [PubMed] [Google Scholar]
- 53.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, A. Brooun, M. Griffith, K. Moy, M. K. Yadav, J. Velasquez, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2006. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 80:7894-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang, S. M., M. J. Shin, J. H. Kim, and J. W. Oh. 2005. Proteomic profiling of cellular proteins interacting with the hepatitis C virus core protein. Proteomics 5:2227-2237. [DOI] [PubMed] [Google Scholar]
- 55.Kattenhorn, L. M., R. Mills, M. Wagner, A. Lomsadze, V. Makeev, M. Borodovsky, H. L. Ploegh, and B. M. Kessler. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187-11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kellam, P. 2001. Post-genomic virology: the impact of bioinformatics, microarrays and proteomics on investigating host and pathogen interactions. Rev. Med. Virol. 11:313-329. [DOI] [PubMed] [Google Scholar]
- 57.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 58.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St. Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. [DOI] [PubMed] [Google Scholar]
- 59.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99-106. [DOI] [PubMed] [Google Scholar]
- 60.Lavigne, R., J. P. Noben, K. Hertveldt, P. J. Ceyssens, Y. Briers, D. Dumont, B. Roucourt, V. N. Krylov, V. V. Mesyanzhinov, J. Robben, and G. Volckaert. 2006. The structural proteome of Pseudomonas aeruginosa bacteriophage phiKMV. Microbiology 152:529-534. [DOI] [PubMed] [Google Scholar]
- 61.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 62.Lindner, H. A. 7 February 2007. Deubiquitination in virus infection. Virology. doi: 10.1016/j.virol.2006.12.035. [DOI] [PMC free article] [PubMed]
- 63.Lorenz, I. C., J. Marcotrigiano, T. G. Dentzer, and C. M. Rice. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831-835. [DOI] [PubMed] [Google Scholar]
- 64.Love, R. A., H. E. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331-342. [DOI] [PubMed] [Google Scholar]
- 65.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The Genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 66.Martin, D. T., C. A. Adair, and D. A. Ritchie. 1976. Polypeptides specified by bacteriophage T1. J. Gen. Virol. 33:309-319. [DOI] [PubMed] [Google Scholar]
- 67.Maxwell, K. L., P. Reed, R. G. Zhang, S. Beasley, A. R. Walmsley, F. A. Curtis, A. Joachimiak, A. M. Edwards, and G. J. Sharples. 2005. Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange. Proc. Natl. Acad. Sci. USA 102:11260-11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maxwell, K. L., A. A. Yee, C. H. Arrowsmith, M. Gold, and A. R. Davidson. 2002. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J. Mol. Biol. 318:1395-1404. [DOI] [PubMed] [Google Scholar]
- 69.Maxwell, K. L., A. A. Yee, V. Booth, C. H. Arrowsmith, M. Gold, and A. R. Davidson. 2001. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J. Mol. Biol. 308:9-14. [DOI] [PubMed] [Google Scholar]
- 70.Mayer, D., S. Baginsky, and M. Schwemmle. 2005. Isolation of viral ribonucleoprotein complexes from infected cells by tandem affinity purification. Proteomics 5:4483-4487. [DOI] [PubMed] [Google Scholar]
- 71.McCraith, S., T. Holtzman, B. Moss, and S. Fields. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meier, C., A. R. Aricescu, R. Assenberg, R. T. Aplin, R. J. Gilbert, J. M. Grimes, and D. I. Stuart. 2006. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure 14:1157-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naryshkina, T., J. Liu, L. Florens, S. K. Swanson, A. R. Pavlov, N. V. Pavlova, R. Inman, L. Minakhin, S. A. Kozyavkin, M. Washburn, A. Mushegian, and K. Severinov. 2006. Thermus thermophilus bacteriophage phiYS40 genome and proteomic characterization of virions. J. Mol. Biol. 364:667-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ong, S. E., B. Blagoev, I. Kratchmarova, D. B. Kristensen, H. Steen, A. Pandey, and M. Mann. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1:376-386. [DOI] [PubMed] [Google Scholar]
- 76.Ong, S. E., L. J. Foster, and M. Mann. 2003. Mass spectrometric-based approaches in quantitative proteomics. Methods 29:124-130. [DOI] [PubMed] [Google Scholar]
- 77.Pabo, C. O., and M. Lewis. 1982. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature 298:443-447. [DOI] [PubMed] [Google Scholar]
- 78.Patton, W. F. 2002. Detection technologies in proteome analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 771:3-31. [DOI] [PubMed] [Google Scholar]
- 79.Peti, W., M. A. Johnson, T. Herrmann, B. W. Neuman, M. J. Buchmeier, M. Nelson, J. Joseph, R. Page, R. C. Stevens, P. Kuhn, and K. Wuthrich. 2005. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 79:12905-12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piersanti, S., Y. Martina, G. Cherubini, D. Avitabile, and I. Saggio. 2004. Use of DNA microarrays to monitor host response to virus and virus-derived gene therapy vectors. Am. J. Pharmacogenom. 4:345-356. [DOI] [PubMed] [Google Scholar]
- 81.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 82.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Renesto, P., C. Abergel, P. Decloquement, D. Moinier, S. Azza, H. Ogata, P. Fourquet, J. P. Gorvel, and J. M. Claverie. 2006. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 80:11678-11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 85.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318:245-266. [DOI] [PubMed] [Google Scholar]
- 86.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 87.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure (Cambridge) 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanger, F., G. M. Air, B. G. Barrell, N. L. Brown, A. R. Coulson, C. A. Fiddes, C. A. Hutchison, P. M. Slocombe, and M. Smith. 1977. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265:687-695. [DOI] [PubMed] [Google Scholar]
- 89.Saphire, A. C., P. A. Gallay, and S. J. Bark. 2006. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J. Proteome Res. 5:530-538. [DOI] [PubMed] [Google Scholar]
- 90.Saridakis, V., Y. Sheng, F. Sarkari, M. N. Holowaty, K. Shire, T. Nguyen, R. G. Zhang, J. Liao, W. Lee, A. M. Edwards, C. H. Arrowsmith, and L. Frappier. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1: implications for EBV-mediated immortalization. Mol. Cell 18:25-36. [DOI] [PubMed] [Google Scholar]
- 91.Schierling, K., T. Stamminger, T. Mertens, and M. Winkler. 2004. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J. Virol. 78:9512-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schlee, M., T. Krug, O. Gires, R. Zeidler, W. Hammerschmidt, R. Mailhammer, G. Laux, G. Sauer, J. Lovric, and G. W. Bornkamm. 2004. Identification of Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) target proteins by proteome analysis: activation of EBNA2 in conditionally immortalized B cells reflects early events after infection of primary B cells by EBV. J. Virol. 78:3941-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Senkevich, T. G., S. Ojeda, A. Townsley, G. E. Nelson, and B. Moss. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. USA 102:18572-18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41:139-156. [DOI] [PubMed] [Google Scholar]
- 95.Song, W., Q. Lin, S. B. Joshi, T. K. Lim, and C. L. Hew. 2006. Proteomic studies of the Singapore grouper iridovirus. Mol. Cell. Proteomics 5:256-264. [DOI] [PubMed] [Google Scholar]
- 96.Song, W. J., Q. W. Qin, J. Qiu, C. H. Huang, F. Wang, and C. L. Hew. 2004. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. J. Virol. 78:12576-12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sopta, M., R. W. Carthew, and J. Greenblatt. 1985. Isolation of three proteins that bind to mammalian RNA polymerase II*. J. Biol. Chem. 260:10353-10360. [PubMed] [Google Scholar]
- 98.Sulea, T., H. A. Lindner, and R. Menard. 2006. Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 387:853-862. [DOI] [PubMed] [Google Scholar]
- 99.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szajner, P., H. Jaffe, A. S. Weisberg, and B. Moss. 2004. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology 330:447-459. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi, T., M. Oie, and Y. Ichihashi. 1994. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology 202:844-852. [DOI] [PubMed] [Google Scholar]
- 102.Tarbouriech, N., M. Buisson, T. Geoui, S. Daenke, S. Cusack, and W. P. Burmeister. 2006. Structural genomics of the Epstein-Barr virus. Acta Crystallogr. D Biol. Crystallogr. 62:1276-1285. [DOI] [PubMed] [Google Scholar]
- 103.Tarbouriech, N., M. Buisson, J. M. Seigneurin, S. Cusack, and W. P. Burmeister. 2005. The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure 13:1299-1310. [DOI] [PubMed] [Google Scholar]
- 104.Tarbouriech, N., F. Ruggiero, M. de Turenne-Tessier, T. Ooka, and W. P. Burmeister. 2006. Structure of the Epstein-Barr virus oncogene BARF1. J. Mol. Biol. 359:667-678. [DOI] [PubMed] [Google Scholar]
- 105.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian, Q., S. B. Stepaniants, M. Mao, L. Weng, M. C. Feetham, M. J. Doyle, E. C. Yi, H. Dai, V. Thorsson, J. Eng, D. Goodlett, J. P. Berger, B. Gunter, P. S. Linseley, R. B. Stoughton, R. Aebersold, S. J. Collins, W. A. Hanlon, and L. E. Hood. 2004. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3:960-969. [DOI] [PubMed] [Google Scholar]
- 108.Toni, M., G. Conti, and G. C. Schito. 1976. Viral protein synthesis during replication of bacteriophage T1. Biochem. Biophys. Res. Commun. 68:545-552. [DOI] [PubMed] [Google Scholar]
- 109.Tsai, J.-M., H.-C. Wang, J.-H. Leu, H.-H. Hsiao, A. H.-J. Wang, G.-H. Kou, and C.-F. Lo. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78:11360-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsai, J.-M., H.-C. Wang, J.-H. Leu, A. H.-J. Wang, Y. Zhuang, P. J. Walker, G.-H. Kou, and C.-F. Lo. 2006. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 80:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 112.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18:2071-2077. [DOI] [PubMed] [Google Scholar]
- 113.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ventelon-Debout, M., F. Delalande, J. P. Brizard, H. Diemer, A. Van Dorsselaer, and C. Brugidou. 2004. Proteome analysis of cultivar-specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing rice yellow mottle virus infection. Proteomics 4:216-225. [DOI] [PubMed] [Google Scholar]
- 115.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wagner, E. F., H. Ponta, and M. Schweiger. 1977. Development of E. coli virus T1: the pattern of gene expression. Mol. Gen. Genet. 150:21-28. [DOI] [PubMed] [Google Scholar]
- 117.Xie, X., L. Xu, and F. Yang. 2006. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 80:10615-10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463-467. [DOI] [PubMed] [Google Scholar]
- 119.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J. 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zachertowska, A., D. Brewer, and D. H. Evans. 2006. Characterization of the major capsid proteins of myxoma virus particles using MALDI-TOF mass spectrometry. J. Virol. Methods 132:1-12. [DOI] [PubMed] [Google Scholar]
- 121.Zeghouf, M., J. Li, G. Butland, A. Borowska, V. Canadien, D. Richards, B. Beattie, A. Emili, and J. F. Greenblatt. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463-468. [DOI] [PubMed] [Google Scholar]
- 122.Zeng, R., H. Q. Ruan, X. S. Jiang, H. Zhou, L. Shi, L. Zhang, Q. H. Sheng, Q. Tu, Q. C. Xia, and J. R. Wu. 2004. Proteomic analysis of SARS associated coronavirus using two-dimensional liquid chromatography mass spectrometry and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by mass spectrometric analysis. J. Proteome Res. 3:549-555. [DOI] [PubMed] [Google Scholar]
- 123.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]