Abstract
TFIID is a general transcription factor required for transcription of most protein-coding genes by RNA polymerase II. TAF7L is an X-linked germ cell-specific paralogue of TAF7, which is a generally expressed component of TFIID. Here, we report the generation of Taf7l mutant mice by homologous recombination in embryonic stem cells by using the Cre-loxP strategy. While spermatogenesis was completed in Taf7l−/Y mice, the weight of Taf7l−/Y testis decreased and the amount of sperm in the epididymides was sharply reduced. Mutant epididymal sperm exhibited abnormal morphology, including folded tails. Sperm motility was significantly reduced, and Taf7l−/Y males were fertile with reduced litter size. Microarray profiling revealed that the abundance of six gene transcripts (including Fscn1) in Taf7l−/Y testes decreased more than twofold. In particular, FSCN1 is an F-action-bundling protein and thus may be critical for normal sperm morphology and sperm motility. Although deficiency of Taf7l may be compensated in part by Taf7, Taf7l has apparently evolved new specialized functions in the gene-selective transcription in male germ cell differentiation. Our mouse studies suggest that mutations in the human TAF7L gene might be implicated in X-linked oligozoospermia in men.
TFIID, a general transcription factor, plays a central role in transcription initiation of most protein-coding genes by RNA polymerase II. TFIID is a multiprotein complex consisting of TATA-binding protein (TBP) and 12 to 15 TBP-associated factors (TAFs) (20, 35). The assembly of TFIID at the promoter region recruits other basal transcription factors and RNA polymerase II (2, 31). TAFs play important roles in transcriptional regulation. Some TAFs directly interact with transcriptional activators and thus serve as coactivators. In addition, interactions between TAFs are critical for promoter recognition and selectivity by RNA polymerase II (17, 36).
Strikingly, studies of a number of tissue-specific TAFs in Drosophila melanogaster and mouse have identified cell-type-specific transcription programs. In Drosophila melanogaster, five testis-specific homologues of widely expressed TAFs have been reported: Can (homologue of dTAF5), Nht (homologue of dTAF4), Mia (homologue of dTAF6), Sa (homologue of dTAF8), and Rye (homologue of dTAF12) (18, 19). Null mutations in can, nht, mia, and sa result in the same male sterile phenotype, and all four genes are required for meiotic cell cycle progression and onset of spermatid differentiation (27). In addition, Rye interacts with Nht, suggesting that these five testis-specific TAFs in Drosophila function in the same transcription regulatory pathway (18). Mechanistically, these TAFs may counteract transcriptional repression by Polycomb group (PcG) proteins in spermatocytes (8). In mice, TAF4B (homologue of TAF4) is highly expressed in the testis and the granulosa cells of the ovary, where it is required for follicular development (14). Testes of TAF4B-deficient males are initially normal but undergo progressive germ cell loss, resulting in male sterility by 3 months of age (12). In addition to tissue-restricted TAFs, TRF2 is a testis-specific homologue of TBP and is essential for spermiogenesis in mouse (28, 44). These results, together with other studies, support the presence of tissue-specific transcription programs in regulating germ cell differentiation (23, 33).
We have identified a testis-specific homologue of the generally expressed TAF7 in mouse, named TAF7L (30, 39). TAF7 interacts with multiple transcription activators (9). TAF7 also interacts with other TAFs, including TAF1 (the largest subunit of TFIID), but not with TAF10 or TBP (9, 24). Binding of TAF7 to TAF1 inhibits the acetyltransferase activity of TAF1, which is important for the transcription of major histocompatibility complex class I genes (16). Like TAF7 in somatic tissues, TAF7L interacts with TAF1 and is associated with TBP in testes, indicating that TAF7L is a bona fide TAF (30). Subcellular localization of TAF7L in male germ cells is dynamic. TAF7L is cytoplasmic in spermatogonia and early spermatocytes (preleptotene, leptotene, and zygotene); however, TAF7L translocates into the nuclei of pachytene spermatocytes and round spermatids. In contrast, TAF7 is nuclear from spermatogonia to pachytene spermatocytes and appears to be absent in round spermatids. Biochemical studies indicate that TAF7L might replace TAF7 in the TFIID complex to modulate the transcription program in spermatogenesis (30). To assess the role of TAF7L in spermatogenesis, we generated mice lacking TAF7L by gene targeting in embryonic stem (ES) cells. Here, we describe the effects of this mutation on gene transcription and production, morphology, and motility of spermatozoa.
MATERIALS AND METHODS
Disruption of the Taf7l gene.
In the Taf7l targeting construct, we introduced loxP sites into the first and sixth introns (see Fig. 2A). Using a Taf7l-containing bacterial artificial chromosome clone (RP22-415C9) as template, three DNA fragments (2 kb, 3.4 kb, and 3 kb) were amplified by high-fidelity PCR. The CMV-HyTK double-selection cassette is flanked by loxP sites and enables hygromycin-positive selection and thymidine kinase-negative selection. Three loxP sites in the final targeting construct are in the same orientation (see Fig. 2A). The construct was sequenced, except for the HyTK cassette, and no mutations were found.
FIG. 2.
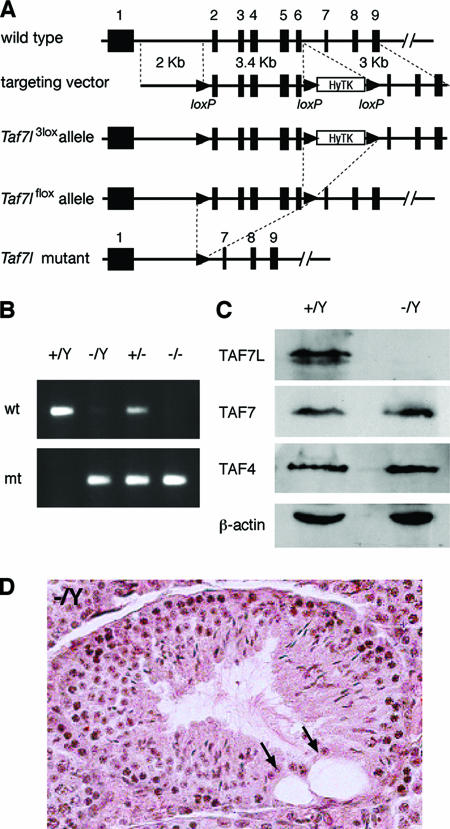
Targeted inactivation of Taf7l in mice. (A) Schematic presentation of the Taf7l gene, the targeting construct, and various alleles. Exons 1 to 9 are shown as rectangles. Exons 10 to 13 are not shown. Deletion of exons 2 to 6 (aa 96 to 263) in the Taf7l mutant allele is expected to cause a frameshift. (B) Genotyping of Taf7l alleles. Genotypes are indicated. wt, wild-type allele; mt, Taf7l mutant allele. (C) Western blot of Taf7l−/Y testes. Equal amounts (20 μg) of testis protein extracts were loaded. TAF7L was absent in Taf7l−/Y testes. The abundances of TAF7 and TAF4 did not differ in Taf7l−/Y and wild-type testes. (D) Histological analysis of 7-month-old Taf7l−/Y testes. Two large vacuoles (arrows) were present in the seminiferous tubule, despite the presence of a full spectrum of germ cells.
Hybrid V6.5 ES cells (C57BL/6 × 129) were used for gene targeting (11). ES cells were electroporated with the linearized Taf7l targeting construct and selected for integration in the presence of hygromycin B (120 μg/ml; Invitrogen). Subsequently, 384 hygromycin-resistant ES cell colonies were screened for homologous recombination by PCR. Of the hygromycin-resistant clones, 9.4% contained the Taf7l3lox allele. Twelve clones were subjected to Southern blot analysis and confirmed (data not shown).
Two Taf7l3lox-positive ES cell clones were then electroporated with the pOG231 plasmid that transiently expresses Cre recombinase. Two days after electroporation, cells were passaged and then subjected to selection with ganciclovir (2 μM; Sigma) for removal of the HyTK cassette. Viable colonies were picked and screened by PCR. Recombination between the HyTK-flanking loxP sites resulted in the Taf7lflox allele at a frequency of 12.5% (see Fig. 2A).
Generation and genotyping of mice.
ES cells harboring the Taf7lflox allele were injected into BALB/c blastocysts that were subsequently transferred to uteri of pseudopregnant Swiss Webster females. The resulting male chimeras were bred with BALB/c females to obtain germ line transmission of injected ES cells. Agouti females were genotyped by PCR. Taf7lflox mice were crossed with ACTB-Cre transgenic mice to obtain the Taf7l− (mutant) allele (26). The ACTB-Cre transgene was subsequently removed from Taf7l mutant mice by breeding. We backcrossed Taf7l+/− mice to C57BL/6J (B6) and 129 strains for more than five generations (strain 129X1/SvJ, stock no. 000691; The Jackson Laboratory). Mice from either B6 or 129 backgrounds were used in this study. All offspring were genotyped by PCR. Wild-type (300-bp) and flox (490-bp) alleles were assayed by PCR with the primers CCATTCTTCTAAATCCCTAGC and TCGCTTGGGAACTCATCAATT. The PCR product (218 bp) of the mutant allele was amplified by PCR with the primers CCATTCTTCTAAATCCCTAGC and CATCGTGTAATTTGGGTTGAC.
Coimmunoprecipitation and Western blot analysis.
Nuclear extracts from testes were prepared as previously described (30). Extracted proteins (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the presence of TBP, TAF7, and TAF7L was revealed using monoclonal antibodies 3G3, 19TA, and 46TA as previously described (6, 30). The filters were then reprobed with antibodies against TAF5 and TAF6 (4, 10). Immunoprecipitations were performed as previously described (30). Briefly, 200 μg of extract was immunoprecipitated with anti-TBP antibody (3G3) overnight at 4°C with 100 μl of protein G-Sepharose. Beads were washed three times for 10 min at room temperature with buffer A (20% glycerol, 50 mM Tris-HCl, pH 7.9, 1 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40) containing 0.5 M KCl and once with buffer A containing 0.1 M KCl. Precipitated material was eluted using a peptide against the 3G3 epitope as described previously (6). Eluted proteins were probed on immunoblots using the antibodies described above.
EM and histology.
Electron microscopy (EM) was performed at the Biomedical Imaging Core Facility at the University of Pennsylvania as previously described (42). Cauda epididymides from 8-week-old wild-type and Taf7l−/Y mice were fixed and processed for EM. For histological analysis, testes were fixed in Bouin's solution, embedded in paraffin, sectioned, and stained with hematoxylin and eosin as described previously (32).
Sperm count.
Cauda epididymides were dissected and minced in phosphate-buffered saline solution. Sperm were squeezed out with fine forceps and allowed to disperse in phosphate-buffered saline at room temperature for 10 min, followed by repeated pipetting. Samples were fixed in 4% paraformaldehyde. Sperm were counted using a hematocytometer. Sperm counting was performed four times for each sample.
Sperm motility assay.
Uncapacitated cauda epididymal sperm from wild-type and Taf7l−/Y mice were collected by placing minced cauda epididymides in Krebs-Ringer bicarbonate medium (HM) without Ca2+, bovine serum albumin, and NaHCO3 as previously described (25). The working “complete” medium was prepared by adding CaCl2 (1.7 mM), pyruvate (1 mM), NaHCO3 (25 mM), and bovine serum albumin (3 mg/ml), followed by gassing with 5% CO2 and 95% O2 to pH 7.3. One drop of the sperm suspension was transferred to the incubation chamber at 37°C. The incubation time for capacitation was 1 hour at 37°C in a 5% CO2-95% O2 incubator. Aliquots of each sperm suspension were loaded into a 100-μm-deep chamber prewarmed at 37°C (Conception Technologies). Sperm motility parameters were quantified using a computer-assisted semen analysis system running IVOS (version 12.2L; Hamilton Thorne Research). At least 400 sperm per sample were analyzed. For statistical analysis, eight motion parameters, motility, average path velocity (VAP), straight-line velocity (VSL), curvilinear velocity (VCL), amplitude of lateral head displacement, beat-cross frequency (BCF), straightness, and linearity, were examined (see Table 2). For statistical testing, sperm motility measurements of each parameter were pooled for each genotype and for time of observation. Considering the log-normal distribution, Student's t test for independent observations was applied to define differences between the wild type and the mutant in VAP, VSL, VCL, and BCF means (normalized by natural logarithms). For the same purpose, the nonparametric amplitude of lateral head displacement and STR distributions were tested by Friedman's analysis of variance. Statistical analyses were performed using the InStat program (GraphPad software).
TABLE 2.
Motility of sperm from Taf7l+/Y and Taf7l−/Y micea
| Genotype and condition | Motility (%) | VAP (μm/s) | VSL (μm/s) | VCL (μm/s) | ALH (μm) | BCF (Hz) | STR (%) | LIN (%) |
|---|---|---|---|---|---|---|---|---|
| +/Y | ||||||||
| Uncapacitated | 72.7 ± 4.2 | 166.9 ± 13.0 | 123.9 ± 11.4 | 270.6 ± 28.1 | 12.3 ± 0.8 | 10.5 ± 1.5 | 73.3 ± 1.5 | 47.3 ± 3.1 |
| Capacitated | 70.0 ± 15.6 | 160.3 ± 13.1 | 121.5 ± 9.2 | 261.0 ± 34.6 | 12.8 ± 3.2 | 15.0 ± 8.1 | 73.5 ± 2.1 | 48.0 ± 2.8 |
| −/Y | ||||||||
| Uncapacitated | 56.3 ± 2.1* | 93.8 ± 5.1* | 66.3 ± 3.5* | 161.6 ± 14.6* | 9.4 ± 1.4 | 19.6 ± 0.8* | 69.3 ± 6.0 | 43.3 ± 5.0 |
| Capacitated | 45.5 ± 9.2* | 90.0 ± 23.3* | 63.3 ± 13.8* | 154.3 ± 39.2* | 9.3 ± 1.8 | 20.5 ± 2.5* | 69.0 ± 4.2 | 45.5 ± 9.2 |
Values represent means ± standard deviations. Three mice of each genotype were analyzed. Asterisks indicate statistically significant differences (P < 0.05). ALH, amplitude of lateral head displacement; STR, straightness; LIN, linearity.
Microarray analysis.
Total RNA was prepared from 8-week-old testes by using TRIzol reagent (Invitrogen) and subsequently purified using an RNeasy minikit (QIAGEN). Both wild-type and Taf7l−/Y testes were analyzed in duplicate. Five micrograms of total RNA from each sample was used for the generation of biotinylated cRNA. The cRNA samples were hybridized to Mouse Genome 430 2.0 GeneChips (Affymetrix) at the University of Pennsylvania Microarray Core Facility according to the manufacturer's expression analysis technical manual (Affymetrix). Microarray Analysis Suite 5.0 (Affymetrix) was used to quantify microarray signals. The Microarray Analysis Suite 5.0 metrics output was imported into GeneSpring v7 (Agilent Technologies) with normalization to the median of the Affymetrix spike-in controls. SAM (Statistical Analysis of Microarrays) two-paired analysis was applied to identify genes with significant differences at a 13% false discovery rate.
Quantitative and semiquantitative reverse transcription (RT)-PCR analyses.
Total RNA was isolated from 8-week-old wild-type and Taf7l−/Y testes by using TRIzol reagent. One microgram of total RNA for each sample was converted into cDNA by reverse transcription with oligo(dT)18V primers and was diluted to a final volume of 200 μl, 5 μl of which was used in each PCR. Three replicates were used for each real-time PCR. Reverse transcriptase-negative templates served as controls. Real-time PCR was run on a LightCycler (Roche). Quantification was normalized to Actb within the log phase of the amplification curve.
Spermatogenic cell populations enriched for specific germ cell types were the same as previously used, and their preparations have been described previously (41). Each enriched population contained a small amount of developmentally adjacent germ cells. A semiquantitative RT-PCR technique was used as previously described (41). Briefly, 70 ng of poly(A)+ RNA was used for reverse transcription primed with oligo(dT)18V in a 25-μl reaction mixture. Each RT reaction was diluted to a total volume of 200 μl, and 5 μl was used for each PCR. To avoid saturation of PCR, products were taken after various cycles (20-30) and analyzed by gel electrophoresis. One microgram of total RNA from adult wild-type and XXY* testes was used for reverse transcription. Controls without reverse transcriptase were negative (data not shown). The following gene-specific primers were used: Actb, AGAAGAGCTATGAGCTGCCT and TCATCGTACTCCTGCTTGCT; Fscn1, ACCGATCAGGAGACCTTCCA and GAGTCTTTGATGTTGTAGGCG; 4732473B16Rik, TGAGCTGGCCACAGGTGAA and ACTTTGACCAGCTTCTGCAC; Cpa6, GAACCAGAAGTGAAGGCTGT and CTTTAGCAGGTGCATTGTGAT; Adc, GGGGTCTTCAACTCAGTCCT and ACAAGGTGTCTGTGATCTCC; D1Ertd622e, AACTTGCACAGTGACATCATC and AGTCCCGTGTCCAGCTGTTT; and Sfmbt2, GACGGATGTGGTACGATTCA and GTGCTCCTTCCGTGTGCTTT. Primers for Pgk2 and Prm1 have been described previously (43).
Microarray data accession number.
Microarray tabular data have been deposited in the Gene Expression Omnibus database under accession no. GSE5510.
RESULTS
Taf7l is the ancestral gene of Taf7 and has evolved to be testis specific.
While the Taf7l coding region is interrupted by 12 introns, the Taf7 coding region completely lacks introns (Fig. 1A). This suggests that Taf7l is the ancestral gene and that Taf7 is a retroposed paralogue of Taf7l. Interestingly, the Slc25a2 gene, adjacent to Taf7, is also a retroposed gene (of Slc25a) (7). These two functional retroposed genes are located between the protocadherin alpha and beta gene clusters on mouse chromosome 18 and human chromosome 5. Thus, the Taf7-Slc25a2 retroposition occurred prior to the radiation of eutherians (at least 80 million years ago). One intron is present in the 5′ untranslated region of the mouse Taf7 gene (Fig. 1A). However, the Taf7 orthologue in human and rat does not appear to have an intron. The most parsimonious explanation is that the acquisition of the single intron by Taf7 is specific to the mouse lineage.
FIG. 1.
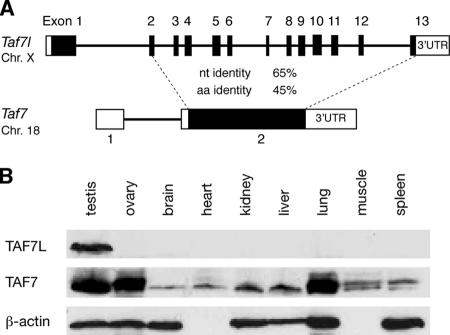
Contrasting exon/intron structures and expression patterns of the mouse Taf7l and Taf7 genes. (A) Taf7 is a retroposed derivative of Taf7l. The gene structures were determined by alignment of Taf7l (accession no. AK017109) and Taf7 (NM_011901) cDNA sequences with their genomic sequences. Coding regions are shown in black. Percent identities in the coding regions for nucleotide (nt) and aa sequences are indicated. Compared with TAF7, TAF7L contains ∼100 additional residues at its amino terminus. No significant nt identity is present in the untranslated regions (UTR). (B) Western blot of TAF7L and TAF7 in adult mouse tissues. Equal amounts (20 μg) of protein extracts for each tissue were loaded. β-Actin served as a control. Chr., chromosome.
We then examined the expression of Taf7 and Taf7l by Western blot analysis. While the TAF7 protein was present in all tissues examined at various abundances, TAF7L was testis specific (Fig. 1B). These data are in agreement with those of previous studies (30, 39, 45). There exist a number of precedents in which widely expressed intron-bearing ancestral genes gave rise to tissue-specific retroposed genes, particularly testis-specific genes (38). However, it is highly unusual for an ancestral gene such as Taf7l, initially widely expressed, to evolve tissue specificity.
Inactivation of the Taf7l gene.
To elucidate the role of Taf7l in spermatogenesis, we generated a floxed Taf7l conditional allele (Taf7lflox) in mice (Fig. 2A). Taf7lflox/Y males and Taf7lflox/flox females displayed normal fertility. By crossing with ACTB-Cre transgenic mice, we obtained Taf7l−/Y mice that lack exons 2 to 6 (26). Deletion of exons 2 to 6 (amino acids [aa] 96 to 263) resulted in a frameshift. In ACTB-Cre mice, Cre recombinase is under the control of the human β-actin promoter and is widely expressed. In the hybrid genetic backgrounds, both Taf7l−/Y males and Taf7l−/− females were fertile and the litter sizes were similar to that of wild-type controls. We backcrossed this knockout allele to C57BL/6 and 129 backgrounds (at least five backcrosses). All subsequent analyses were performed on both C57BL/6 and 129 mice, and no difference was observed. Western blot analysis showed that the TAF7L protein is absent in Taf7l−/Y testes (Fig. 2C). In contrast, the abundance of TAF7 is not affected in Taf7l−/Y testes.
Expression of TFIID components in Taf7l−/Y testes.
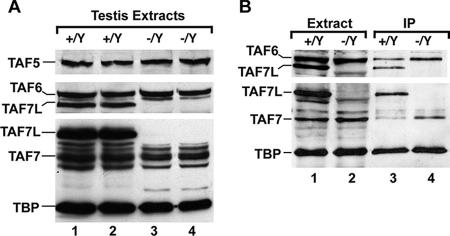
Two independent nuclear extracts made from wild-type or mutant testes were analyzed for the expression of TFIID components (Fig. 3). Our results showed that no significant change in expression of TBP or the other TAFs (TAF5 and TAF6) was observed in Taf7l−/Y testes (Fig. 3A). Nuclear extracts were immunoprecipitated with antibodies against TBP. As expected, TAF7L was coimmunoprecipitated with TBP from wild-type but not Taf7l−/Y testes (Fig. 3B). As previously described, little TAF7 was precipitated with TBP in the wild type, whereas a significantly larger amount was found in the immunoprecipitated fraction from Taf7l−/Y testes (Fig. 3B) (30). These results suggest that there is competition between TAF7 and TAF7L for integration into TFIID so that in the presence of TAF7L, TAF7 is excluded, whereas in its absence, TAF7 can more readily associate with TBP. However, TAF7 did not appear to be upregulated in Taf7l-deficient spermatids (see Fig. S1 in the supplemental material).
FIG. 3.
Competition between TAF7 and TAF7L for association with TBP. (A) Expression analysis of TBP and TBP-associated factors in Taf7l−/Y testes. Ten micrograms of two independent extracts from testes of wild-type (Taf7l+/Y) or Taf7l−/Y animals was probed with antibodies against the indicated proteins. The filter was then reprobed with antibodies against TAF5 and TAF6. (B) Results of coimmunoprecipitation assays. Lanes 1 and 2 show starting extracts from wild-type and mutant testes used for immunoprecipitation (IP) with anti-TBP antibody. Lanes 3 and 4 show peptide-eluted material from immunoprecipitations. The filter was probed with antibodies against TBP, TAF7, and TAF7L and then reprobed with antibodies against TAF6. To avoid masking TAF7 with a signal from the heavy chain of anti-TBP antibody used in immunoprecipitation, the blot was revealed using conjugated goat anti-mouse κ chain antibody as previously described (24).
Reduced sperm production and fertility in Taf7l−/Y mice.
Both Taf7l−/Y males and Taf7l−/− females appeared to be grossly healthy and produced healthy offspring. However, we found that although Taf7l−/Y males were fertile, they produced smaller litters. The litter size (average ± standard deviation) sired by Taf7l−/Y males (3.8 ± 2.1) was sharply reduced in comparison to that of wild-type littermate controls (8.3 ± 2.5) (P < 0.0004). The body weight of Taf7l−/Y males was similar to that of wild-type littermates. Histological analysis of 8-week-old Taf7l−/Y testes revealed no spermatogenic arrest, as evidenced by the presence of a full range of spermatogenic cells, including spermatogonia, spermatocytes, and spermatids (data not shown). However, aged Taf7l−/Y testes (>7 months old) developed large vacuoles in seminiferous tubules (15.2% ± 4.6% of tubules with at least one vacuole) (Fig. 2D); such vacuoles were rarely present in aged wild-type testes. Interestingly, the weights of testes from 8-week-old Taf7l−/Y mice were 10% less than those of wild-type males (P < 0.0039). The sperm counts of Taf7l−/Y mice were reduced by >50% (P < 0.0007) (Table 1).
TABLE 1.
Sperm production in Taf7l+/Y and Taf7l−/Y micea
| Parameter | Value for genotype (mean ± SD)b
|
Ratio of −/Y value to +/Y value | P value | |
|---|---|---|---|---|
| +/Y | −/Y | |||
| Body wt (g) | 25.2 ± 2.6 | 25.8 ± 2.3 | 1.02 | <0.55 |
| Testicular wt (mg) | 172 ± 17 | 150 ± 12 | 0.87 | <0.0039c |
| No. of sperm/ cauda (106) | 7.70 ± 3.45 | 3.30 ± 0.81 | 0.43 | <0.0007c |
Mice from the C57BL/6J background were used at 8 weeks of age.
Twelve mice of each genotype were used.
Values were statistically significant (Student's t test).
Structural defects and impaired motility in sperm from Taf7l−/Y mice.
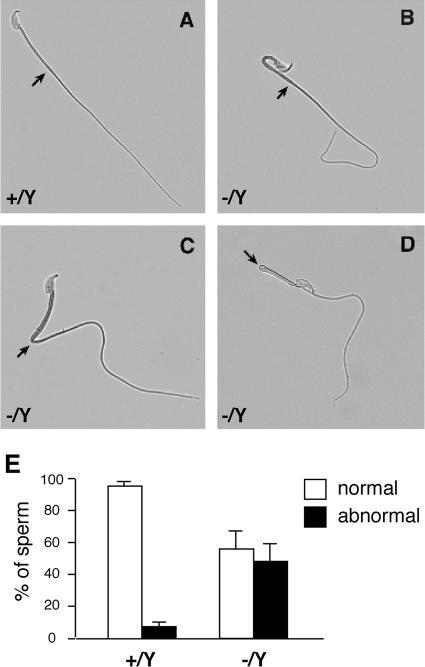
During the sperm count analysis, we observed that a high percentage of sperm tails from Taf7l−/Y mice were folded (Fig. 4). Sperm tails were frequently bent by 180° within the middle piece (Fig. 4B) or at the junction between the middle and principal pieces (Fig. 4D). Sharp angulation of sperm tails also occurred at the junction of the middle and principal pieces (Fig. 4C). Overall, 46% of Taf7l−/Y sperm tails were folded or angulated, compared with 5.8% in the wild type (Fig. 4E).
FIG. 4.
Morphological defects in Taf7l-deficient sperm. Sperm from adult cauda epididymides were analyzed. (A) Wild-type sperm. (B) Taf7l mutant sperm folded at the proximal middle piece. The sperm head is bent back on the tail. (C) Flagellar angulation of Taf7l mutant sperm at the distal middle piece. (D) The middle piece is bent over the principal piece. In addition, the principal piece of Taf7l mutant sperm is abnormally curved. Arrows indicate a junction between the middle and principal pieces. (E) Percentage of sperm with normal and angulated tails. Three 8-week-old mice of each genotype (Taf7l+/Y or Taf7l−/Y) were analyzed. Two hundred sperm from cauda epididymides were counted for each animal.
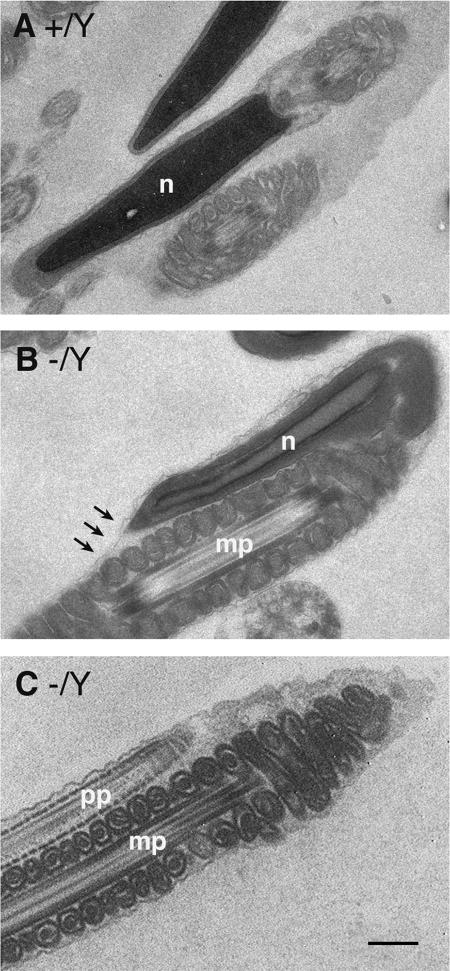
We further examined the sperm tail defects by electron microscopy (Fig. 5). When the sperm head was folded back on the middle piece, the cytoplasmic membrane appeared to be retracted from the region of folding and thus extended continuously from the apex of the sperm head onto the middle piece (Fig. 5B). We confirmed that a high percentage of sperm tails folded around the annulus (the junction of the middle and principal pieces) (Fig. 5C).
FIG. 5.
Ultrastructural defects in Taf7l-deficient sperm. (A) Wild-type sperm. (B) Taf7l mutant sperm. The sperm head (n) is folded over the middle piece (mp). Arrows indicate the continuity of the cytoplasmic membrane from the apex of the sperm head to the middle piece. (C) Taf7l mutant sperm. The principal piece (pp) is bent 180° over the middle piece around the annulus. Bar, 500 nm.
We analyzed sperm motility by computer-assisted sperm analysis under both noncapacitating and capacitating conditions. The percentage of motile sperm was significantly reduced in Taf7l−/Y mice in comparison with that in the wild type (Table 2). Consistently, the values of three velocity parameters (VAP, VSL, and VCL) were significantly lower for Taf7l−/Y mice. However, the BCF of Taf7l mutant sperm was significantly higher, which is consistent with the observation that sperm with folded tails beat more rapidly but fail to generate forward motion. Surprisingly, the motility of mutant sperm was further reduced from 56.3% to 45.5% upon capacitation (Table 2).
Altered gene expression in Taf7l−/Y testes.
To identify genes with altered expression in Taf7l−/Y testes systematically, we performed transcript profiling of testes from 8-week-old mice using Affymetrix Mouse Genome 430 2.0 GeneChips, representing more than 39,000 transcripts. With an expression cutoff of twofold change or greater, our microarray analysis identified 16 genes that were downregulated in Taf7l−/Y testes. Quantitative PCR analysis validated the downregulation of six genes (out of 16) in Taf7l−/Y testes, including four genes with known functions or motifs (Cpa6, 2.7-fold; Adc, 2.4-fold; Sfmbt2, 2.3-fold; and Fscn1, 3.4-fold) and two genes of unknown function (Table 3). CPA6 and ADC are metabolic enzymes. SFMBT2 (sex comb-like with four mbt domains 2) is a PcG protein and thus a putative transcription factor. Interestingly, FSCN1 is a widely expressed actin-bundling protein involved in cell motility (1). FSCN3, a testis-specific paralogue of FSCN1, localizes specifically to the elongating spermatid head (34). However, the expression of Fscn3 is not altered in Taf7l−/Y testes, as assayed by both microarray and quantitative PCR analyses.
TABLE 3.
Downregulation of six genes in Taf7l−/Y testes
| Gene symbol | Accession no. | Description | Fold decrease
|
|
|---|---|---|---|---|
| Microarray | Real-time PCR | |||
| Fscn1 | NM_007984 | Fascin homolog 1, actin-bundling protein | 4.6 | 3.4 |
| 4732473B16Rik | BB407092 | RIKEN cDNA 4732473B16 gene | 2.4 | 3.3 |
| Cpa6 | NM_177834 | Carboxypeptidase A6 | 2.1 | 2.7 |
| Adc | AV282351 | Arginine decarboxylase | 2.2 | 2.4 |
| D1Ertd622e | NM_133825 | DNA segment, chromosome 1, ERATO Doi 622, expressed | 2.1 | 2.3 |
| Sfmbt2 | BM200222 | Scm-like with four mbt domains 2 | 2.7 | 2.3 |
We then examined the expression of these six genes during spermatogenesis (Table 3). Even though they were all expressed in spermatids, the expression profiles of these genes during spermatogenesis were quite diverse (Fig. 6). Interestingly, Cpa6 was specifically expressed in round spermatids. The expression level of Adc was relatively low in spermatogonia and early spermatocytes but increased in pachytene spermatocytes and peaked in round spermatids. The transcript level of 4732473B16Rik was highest in early spermatocytes. Fscn1, Sfmbt2, and D1Ertd622e appeared to be expressed throughout spermatogenesis. We then asked whether these genes are germ cell specific by examining their expression in wild-type and germ cell-deficient (XXY*) testes (21). This analysis showed that two genes (4732473B16Rik and Cpa6) were germ cell specific, three genes (Fscn1, Adc, and D1Ertd622e) were expressed in germ cells at a higher level than in somatic cells of testes, and Sfmbt2 was expressed at similar levels between germ cells and somatic cells in the testis (Fig. 6).
FIG. 6.
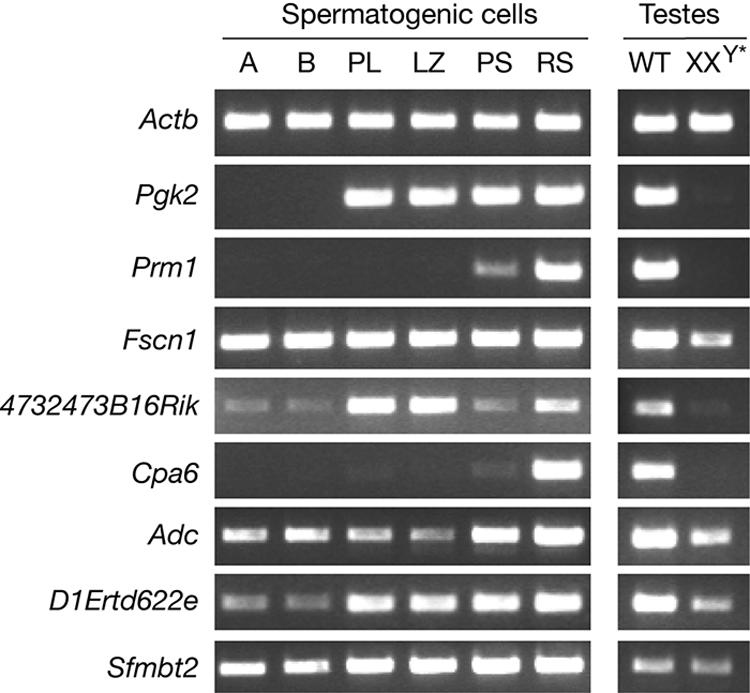
Expression analysis during spermatogenesis. Relative transcript levels among different spermatogenic cell populations were assayed by RT-PCR. Actb served as a ubiquitous expression control. Pgk2 is transcribed at the onset of meiosis. Prm1 is expressed in postmeiotic germ cells. A, type A spermatogonia; B, type B spermatogonia; PL, preleptotene spermatocytes; LZ, mixed leptotene and zygotene spermatocytes; PS, pachytene spermatocytes; RS, round spermatids; WT, wild-type adult testes; XXY*, germ cell-deficient adult testes.
DISCUSSION
Although Taf7l is testis specific, it appears to be the ancestral gene from which Taf7 originated by retroposition. A number of known retrogenes are derived from generally expressed X-linked ancestral genes, and most of these retrogenes are testis specific (22, 38). Therefore, the evolution of Taf7 as a retrogene is an exception in that it is widely expressed. One possibility is that Taf7l was initially widely expressed (as the ancestral housekeeping gene). When Taf7 was produced by retroposition during evolution and was widely expressed in somatic tissues, Taf7l evolved to be specialized in the testis with no selection pressure from the soma. This explanation is further supported by the presence of only one Taf7 homologue in the chicken (Gallus gallus) genome. The chicken Taf7 gene, comprising 11 introns, is present on the region of chromosome 4 that is syntenic with the Taf7l-containing region of the mammalian X chromosome, indicating that the chicken Taf7 gene is orthologous to the mammalian Taf7l gene. Strikingly, the only other two X-linked TAF genes (TAF1 and TAF9B) in mammals both have autosomal homologues (TAF1L and TAF9) (15, 40). Interestingly, the TAF1L gene is testis specific, arose by retroposition relatively recently in the old-world monkey lineage, including humans, and can functionally substitute the somatic TAF1 (40). These studies point to the unique selection pressure on X-linked TAFs and the continuous creation of testis-specific TAFs during evolution.
Recent studies support the notion that special transcription mechanisms operate in germ cells (23, 33). Apart from the presence of testis-specific TAFs, TRF2/TLF is a testis-specific paralogue of TBP. Disruption of Trf2 in mice causes spermiogenic arrest and thus male sterility (28, 44). In comparison, the loss of TAF7L results in reduced fertility but not sterility. Although TAF7L is missing from TFIID, only a small number of genes are affected. The relatively mild phenotypes caused by the loss of TAF7L are surprising. However, the enhanced reproductive fitness (increased sperm count, increased sperm motility, and increased litter size) conferred by Taf7l in one generation has probably had dramatic cumulative effects on an evolutionary time scale.
The transcript levels of six genes are reduced in Taf7l−/Y testis more than twofold. The deficiency of Taf7l might be compensated in part but not fully by Taf7, suggesting that Taf7l has evolved specialized functions in transcription regulation in spermatogenesis. It is possible that developmental stage-specific transcripts might be missed by the microarray profiling of whole testes. However, the altered transcript levels of these six genes might be implicated in the sperm defects (amount, morphology, and motility) in Taf7l−/Y mice. The most strongly downregulated gene is Fscn1 (Table 3). FSCN1 is a filamentous actin (F actin)-bundling protein involved in cytoplasmic protrusion and cell motility (1). In mammalian spermatozoa, actin filaments are found primarily in the subacrosomal space along the nucleus (37). The actin cytoskeleton undergoes dynamic changes in sperm capacitation and the acrosome reaction (5). Globular actin polymerizes to form F actin during capacitation. Prior to the acrosome reaction, F actin undergoes depolymerization. Actin is also detected in the outer dense fibers of the sperm tail, suggesting that actin may regulate sperm motility (3, 13, 29). We postulate that altered expression of Fscn1 contributes to reduced sperm motility and folding of sperm tails in Taf7l−/Y testes. In addition, mutant sperm motility is further reduced upon capacitation (Table 2). These results suggest that bundling of F actin is important for the regulation of sperm motility and capacitation. However, the mechanism underlying FSCN1 function in spermatozoa remains to be determined. In addition, the expression of two metabolic enzymes (CPA6 and ADC) in spermatids might also be related to sperm function (Fig. 6).
In Drosophila, testis-specific TAFs oppose transcriptional repression mediated by PcG proteins, and both groups of proteins localize to the nucleolus in spermatocytes (8). Interestingly, Sfmbt2 encodes a PcG protein of unknown function and is downregulated in Taf7l−/Y testes. In addition to nuclear localization in pachytene spermatocytes and round spermatids, TAF7L localizes to specific chromatin domains in meiotically dividing spermatocytes (30). These results suggest that cross talk between testis-specific TAFs and PcG proteins in the regulation of gene-selective transcription in male germ cell differentiation might be conserved between Drosophila species and mice.
Here, we show that targeted disruption of Taf7l in mice results in reduced sperm count and motility. Taf7l is an X-linked, single-copy testis-specific gene in both mice and humans (39). Thus, the human TAF7L gene may also play an important role in spermatogenesis. Because of the hemizygous state of the X chromosome in men, mutations in TAF7L might cause oligozoospermia (reduced sperm count) in humans.
Supplementary Material
Acknowledgments
We thank J. R. McCarrey for germ cell preparations, J. Tobias for microarray data analysis, Q. C. Yu for electron microscopy, and D. Yu for help in real-time PCR analysis. We thank F. Yang and J. Pan for technical contributions and comments on the manuscript.
This study was supported by the University of Pennsylvania Research Foundation and NIH/NICHD grant HD 045866 (P.J.W.), NIH grant 1-R01-HD41552 (G.L.G.), Fogarty International Center grant NIH 5-D43-TW 00671 (M.G.B.), and the Howard Hughes Medical Institute (D.C.P). Work at the IGBMC (I.D) was supported by grants from the CNRS, the INSERM, the Fondation pour la Recherche Médicale, the Ministère de la Recherche et de la Technologie, the European Union RTN-00026, the Association pour la Recherche contre le Cancer, and the Ligue Nationale contre le Cancer. M.K. was a recipient of a fellowship from the Fondation pour la Recherche Médicale.
Footnotes
Published ahead of print on 22 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, J. C. 2004. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16:590-596. [DOI] [PubMed] [Google Scholar]
- 2.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 3.Aumüller, G., and J. Seitz. 1988. Immunocytochemical localization of actin and tubulin in rat testis and spermatozoa. Histochemistry 89:261-267. [DOI] [PubMed] [Google Scholar]
- 4.Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8:591-600. [DOI] [PubMed] [Google Scholar]
- 5.Breitbart, H., G. Cohen, and S. Rubinstein. 2005. Role of actin cytoskeleton in mammalian sperm capacitation and the acrosome reaction. Reproduction 129:263-268. [DOI] [PubMed] [Google Scholar]
- 6.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho, J. A., N. Rioseco-Camacho, D. Andrade, J. Porter, and J. Kong. 2003. Cloning and characterization of human ORNT2: a second mitochondrial ornithine transporter that can rescue a defective ORNT1 in patients with the hyperornithinemia-hyperammonemia-homocitrullinuria syndrome, a urea cycle disorder. Mol. Genet. Metab. 79:257-271. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., M. Hiller, Y. Sancak, and M. T. Fuller. 2005. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310:869-872. [DOI] [PubMed] [Google Scholar]
- 9.Chiang, C. M., and R. G. Roeder. 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267:531-536. [DOI] [PubMed] [Google Scholar]
- 10.Dubrovskaya, V., A. C. Lavigne, I. Davidson, J. Acker, A. Staub, and L. Tora. 1996. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 15:3702-3712. [PMC free article] [PubMed] [Google Scholar]
- 11.Eggan, K., H. Akutsu, J. Loring, L. Jackson-Grusby, M. Klemm, W. M. Rideout III, R. Yanagimachi, and R. Jaenisch. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA 98:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falender, A. E., R. N. Freiman, K. G. Geles, K. C. Lo, K. Hwang, D. J. Lamb, P. L. Morris, R. Tjian, and J. S. Richards. 2005. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 19:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouquet, J. P., M. L. Kann, and J. P. Dadoune. 1990. Immunoelectron microscopic distribution of actin in hamster spermatids and epididymal, capacitated and acrosome-reacted spermatozoa. Tissue Cell 22:291-300. [DOI] [PubMed] [Google Scholar]
- 14.Freiman, R. N., S. R. Albright, S. Zheng, W. C. Sha, R. E. Hammer, and R. Tjian. 2001. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293:2084-2087. [DOI] [PubMed] [Google Scholar]
- 15.Frontini, M., E. Soutoglou, M. Argentini, C. Bole-Feysot, B. Jost, E. Scheer, and L. Tora. 2005. TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol. Cell. Biol. 25:4638-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gegonne, A., J. D. Weissman, and D. S. Singer. 2001. TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc. Natl. Acad. Sci. USA 98:12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 18.Hiller, M., X. Chen, M. J. Pringle, M. Suchorolski, Y. Sancak, S. Viswanathan, B. Bolival, T. Y. Lin, S. Marino, and M. T. Fuller. 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131:5297-5308. [DOI] [PubMed] [Google Scholar]
- 19.Hiller, M. A., T. Y. Lin, C. Wood, and M. T. Fuller. 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 21.Hunt, P. A., and E. M. Eicher. 1991. Fertile male mice with three sex chromosomes: evidence that infertility in XYY male mice is an effect of two Y chromosomes. Chromosoma 100:293-299. [DOI] [PubMed] [Google Scholar]
- 22.Khil, P. P., B. Oliver, and R. D. Camerini-Otero. 2005. X for intersection: retrotransposition both on and off the X chromosome is more frequent. Trends Genet. 21:3-7. [DOI] [PubMed] [Google Scholar]
- 23.Kimmins, S., N. Kotaja, I. Davidson, and P. Sassone-Corsi. 2004. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction 128:5-12. [DOI] [PubMed] [Google Scholar]
- 24.Lavigne, A. C., G. Mengus, M. May, V. Dubrovskaya, L. Tora, P. Chambon, and I. Davidson. 1996. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J. Biol. Chem. 271:19774-19780. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. A., and B. T. Storey. 1986. Bicarbonate is essential for fertilization of mouse eggs: mouse sperm require it to undergo the acrosome reaction. Biol. Reprod. 34:349-356. [DOI] [PubMed] [Google Scholar]
- 26.Lewandoski, M., E. N. Meyers, and G. R. Martin. 1997. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harbor Symp. Quant. Biol. 62:159-168. [PubMed] [Google Scholar]
- 27.Lin, T. Y., S. Viswanathan, C. Wood, P. G. Wilson, N. Wolf, and M. T. Fuller. 1996. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development 122:1331-1341. [DOI] [PubMed] [Google Scholar]
- 28.Martianov, I., G. M. Fimia, A. Dierich, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2001. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7:509-515. [DOI] [PubMed] [Google Scholar]
- 29.Paranko, J., A. Yagi, and M. Kuusisto. 1994. Immunocytochemical detection of actin and 53 kDa polypeptide in the epididymal spermatozoa of rat and mouse. Anat. Rec. 240:516-527. [DOI] [PubMed] [Google Scholar]
- 30.Pointud, J. C., G. Mengus, S. Brancorsini, L. Monaco, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2003. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116:1847-1858. [DOI] [PubMed] [Google Scholar]
- 31.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 32.Russell, L. D., R. A. Ettlin, A. P. Sinha Hikim, and E. D. Clegg. 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater, FL.
- 33.Sassone-Corsi, P. 2002. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296:2176-2178. [DOI] [PubMed] [Google Scholar]
- 34.Tubb, B., D. J. Mulholland, W. Vogl, Z. J. Lan, C. Niederberger, A. Cooney, and J. Bryan. 2002. Testis fascin (FSCN3): a novel paralog of the actin-bundling protein fascin expressed specifically in the elongate spermatid head. Exp. Cell Res. 275:92-109. [DOI] [PubMed] [Google Scholar]
- 35.Veenstra, G. J., and A. P. Wolffe. 2001. Gene-selective developmental roles of general transcription factors. Trends Biochem. Sci. 26:665-671. [DOI] [PubMed] [Google Scholar]
- 36.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 37.Vogl, A. W., K. Genereux, and D. C. Pfeiffer. 1993. Filamentous actin detected in rat spermatozoa. Tissue Cell 25:33-48. [DOI] [PubMed] [Google Scholar]
- 38.Wang, P. J. 2004. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol. Metab. 15:79-83. [DOI] [PubMed] [Google Scholar]
- 39.Wang, P. J., J. R. McCarrey, F. Yang, and D. C. Page. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27:422-426. [DOI] [PubMed] [Google Scholar]
- 40.Wang, P. J., and D. C. Page. 2002. Functional substitution for TAFII250 by a retroposed homolog that is expressed in human spermatogenesis. Hum. Mol. Genet. 11:2341-2346. [DOI] [PubMed] [Google Scholar]
- 41.Wang, P. J., D. C. Page, and J. R. McCarrey. 2005. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 14:2911-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, F., R. Fuente De La, N. A. Leu, C. Baumann, K. J. McLaughlin, and P. J. Wang. 2006. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 173:497-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, F., H. Skaletsky, and P. J. Wang. 2007. Ubl4b, an X-derived retrogene, is specifically expressed in post-meiotic germ cells in mammals. Gene Expr. Patterns 7:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, D., T. L. Penttila, P. L. Morris, M. Teichmann, and R. G. Roeder. 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292:1153-1155. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, T., and C. M. Chiang. 2001. The intronless and TATA-less human TAF(II)55 gene contains a functional initiator and a downstream promoter element. J. Biol. Chem. 276:25503-25511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.