Abstract
Interleukin-12 (IL-12) and IL-23 are heterodimeric cytokines that serve as critical regulators of T helper cell development. The Il12b gene, which encodes the p40 subunit of both IL-12 and IL-23, is expressed in macrophages and dendritic cells following induction by bacterial products. Although the Il12b promoter, like the promoters of most proinflammatory genes, can support transcriptional induction in typical transfection assays, we show that it is not sufficient for transcription in an insulated chromatin environment. Using a DNase I hypersensitivity assay, two potential distal control regions were identified. One region, DNase I-hypersensitive site 1 (HSS1), located 10 kb upstream of the transcription start site, exhibited hypersensitivity only in stimulated macrophages. In an insulated environment, a 105-bp fragment spanning HSS1 was sufficient for transcription when combined with the Il12b promoter. Although several elements are likely to contribute to activity of the endogenous HSS1 enhancer, including an evolutionarily conserved binding site for C/EBP proteins, the only element required for activity in transient- and stable-transfection assays bound Oct-1 and Oct-2, both of which are expressed constitutively in macrophages. Oct-1 and Oct-2 were recruited to the enhancer upon macrophage stimulation, and the Oct site appeared important for nucleosome remodeling at HSS1. These results suggest that the HSS1 enhancer and Oct proteins play central roles in Il12b induction upon macrophage activation.
Interleukin-12 (IL-12) is representative of a large number of proinflammatory cytokines and other mediators of inflammation, in that it is produced by monocytes/macrophages, dendritic cells, and neutrophils following interaction with microbial products. A central function of IL-12 is to provide a critical bridge between innate and adaptive immunity (13, 44). IL-12 is important for the differentiation of naïve T helper cells into T helper 1 cells, which combat infection by intracellular pathogens (14, 29). Although beneficial for normal immune responses, overproduction of IL-12 can contribute to autoimmune and chronic inflammatory diseases (44). Thus, an understanding of the regulated expression of IL-12 may provide insight into the control of infectious and inflammatory diseases.
IL-12 consists of p35 and p40 subunits, which are covalently linked to form a biologically functional p70 heterodimer (44). In macrophages and dendritic cells, the p35 and p40 genes (Il12a and Il12b, respectively) are transcribed upon cell activation by bacterial products, including lipopolysaccharide (LPS) (24, 44). IL-12 p40 also forms heterodimers with another protein, p19. The p19/p40 cytokine, IL-23, contributes unique functions in regulating immune responses (13, 44).
The murine Il12b promoter contains several transcription factor binding sites that contribute to gene induction in LPS-stimulated macrophages (6, 23, 31, 33, 53, 54). The DNA elements that have been characterized most extensively bind NF-κB, C/EBP, AP-1, and NFAT family members. An analysis of mice lacking specific NF-κB subunits revealed that c-Rel is selectively required for Il12b induction in LPS-stimulated macrophages (35, 36). Transcription of Il12b is also regulated by chromatin, as a positioned nucleosome at the promoter overlaps the transcription factor binding sites, and remodeling of this nucleosome by SWI/SNF complexes appears to be essential for transcriptional activation (34, 49). LPS-induced remodeling is independent of c-Rel but requires protein synthesis and Toll-like receptor 4 (TLR4) signaling (48). Recent evidence suggests that SWI/SNF complexes are broadly required for the induction of secondary response genes (i.e., genes that require new protein synthesis for induction) following LPS stimulation (34).
Previous studies of inducible genes have focused primarily on events occurring at the promoter. For example, early studies of the FOS promoter revealed several cis-acting elements that contribute to inducible transcription, including the serum response element and the cyclic AMP response element (4, 43). Studies of the human IFNB1 promoter provide another paradigm for understanding the mechanisms by which cis-acting elements at an inducible promoter function to control transcription (1, 25, 27, 30).
Studies of rapidly induced genes have focused primarily on their promoters because the promoters typically contain elements that support rapid induction in transfection experiments. However, for efficient induction in a chromatin environment, distal control regions are thought to be required for most mammalian genes. Although some distant control regions appear to be constitutively active, others play critical and direct roles in the rapid induction of transcription. Examples of inducible genes in the immune system for which distant control regions have been identified and characterized include Il4, Il2, and Il2ra (2, 3, 15, 16, 20, 21, 38, 41, 50, 51). At the Il4 locus, multiple distant control regions are activated during Th2 differentiation, but most appear to be constitutively active in mature resting Th2 cells (2, 41). However, one region (VA) directly contributes to inducible Il4 transcription during Th2 activation and functions as an inducible enhancer (2, 3). Interestingly, several of the Il4 control regions are found in close proximity to one another in T lymphocytes (39). Although the precise mechanism by which enhancers contribute to inducible transcription remains unknown, it is clear that a complete understanding of proinflammatory gene activation in response to microbial stimuli cannot be attained by focusing solely on the promoters for proinflammatory genes.
We report here that, although the Il12b promoter is sufficient for LPS-induced transcription in typical transfection assays, by itself it is completely inactive in an insulated chromatin environment. We therefore performed a systematic search for distal control elements 27 kb upstream and 20 kb downstream of the Il12b transcription start site by using a DNase I hypersensitivity assay. Two DNase I-hypersensitive sites (HSS1 and HSS2) were identified, and a detailed examination of one of these sites (HSS1) suggests a central role of Oct proteins in Il12b induction.
MATERIALS AND METHODS
Plasmids.
The murine Il12b promoter containing sequences from positions −356 to +55 was amplified by PCR and inserted into the pGL2B vector (Promega) cleaved with MluI and BglII. Il12b promoter sequences (−356 to +55) in pGL2B were then excised with KpnI and BglII and subcloned into a chloramphenicol acetyltransferase (CAT) reporter vector (33). DNA fragments derived from the HSS1 region were amplified by PCR by using an upstream primer containing a SacI site and a downstream primer containing an MluI site. These fragments were inserted upstream of the Il12b promoter in the CAT reporter plasmid. Deletion mutants (Del A to Del H) were generated by PCR with an upstream primer containing a SacI site. Substitution mutants (Sub A to Sub K) were derived from the deletion mutants by inserting a second PCR product extending from the starting nucleotide of fragment C to the deletion end point, using an upstream primer containing a KpnI site and a downstream primer containing a SacI site and the sequence adjacent to the deletion end point. Fragments C and C1 in the Il12b promoter-CAT vector were digested with HindIII and MluI and subcloned into the TATA/Inr-CAT vector (33). The insulator green fluorescence protein (GFP) reporter plasmid was a gift from Mark Schlissel (28). The murine Il12b promoter from positions −356 to +55, with or without the DNA sequences from the HSS1 site, was amplified by PCR from CAT reporter constructs by using upstream and downstream primers containing a NotI site. The fragments were cloned into a NotI site in the insulator GFP reporter plasmids. All of the DNA constructs were verified by restriction mapping and sequencing.
Cell lines and reagents.
The RAW264.7 and J774 murine macrophage cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) supplemented with 10% low-endotoxin fetal bovine serum (Omega Scientific) and penicillin/streptomycin (Omega Scientific). The B6.129/J2 macrophage line was a gift from Jill Suttles and Peter Tontonoz and was maintained in RPMI 1640 (Cellgro) supplemented with 10% low-endotoxin fetal bovine serum (Omega Scientific) and penicillin/streptomycin (Omega Scientific). This line was generated by transformation of bone marrow from wild-type mice in a mixed 129-C57BL/6 background with the murine J2 retrovirus. Superfect (QIAGEN) was used for transient transfection of RAW264.7 cells. J774 cells were stably transfected by electroporation. The parameters for electroporation were described previously (33). Stable lines were selected and maintained in DMEM supplemented with 10% fetal bovine serum, penicillin/streptomycin, and 364 μg G418 (GibcoBRL)/ml.
Bone marrow- and fetal liver-derived primary macrophages were prepared from wild-type and mutant mice as described previously (35). Macrophages were stimulated with 10 μg LPS (Sigma)/ml with or without 10 U recombinant murine gamma interferon (IFN-γ) (PharMingen)/ml. c-Rel−/−, Oct-1−/−, and Oct-2−/− mice were described previously (9, 19, 46).
Reporter assays.
For CAT assays, 0.5 × 106 cells/well were plated in six-well plates. The following day, the cells were washed with phosphate-buffered saline and transfected with 2 μg DNA. DNA was incubated in DMEM without serum or antibiotics in a total volume of 100 μl with Superfect at a 1:3 ratio (μg of DNA to μl of Superfect) for 5 to 10 min at room temperature. A total of 600 μl of complete DMEM supplemented with 10% fetal bovine serum (Omega Scientific) and penicillin/streptomycin (Omega Scientific) was added to the DNA/Superfect mix and then added to the cells, and the cells were incubated at 37°C for 2.5 to 3 h. Cells were washed with phosphate-buffered saline and split into two wells in 2.5 ml of complete DMEM. The cells in one of the wells were activated with 10 μg LPS (Sigma)/ml 6 h posttransfection and incubated for 24 h. Cells were then harvested and used in reporter assays as described previously (33).
Restriction enzyme accessibility and DNase I hypersensitivity assays.
DNase I hypersensitivity assays were performed as described previously (8, 52). Primers for generating probe 1 were 5′-CTCAATGCGTAAAGTGAGCAGGATTGC-3′ and 5′-TAGCTCAGGCTAGCCTCAACTCTTC-3′. Primers for generating probe 2 were 5′-GGCCAACCACCCTTGTTAATATATTG-3′ and 5′-GTCCATAGTGCTACTATATTTGGTTGAAAGG-3′. Primers for generating probe 3 were 5′-GGCTAGGTGTACATGTATGTGCATATATC-3′ and 5′-GAAAGAAATGAATGAGTTCCCACC-3′. Primers for generating probe 4 were 5′-TTGGGGCAAGTCCTTCCTTTTTCTGC-3′ and 5′-AGTGTCAAAACATTCTGGGGG-3′. The probes were labeled, and the Southern blot-based restriction enzyme accessibility assay was performed as described previously (52).
Gel shifts, Western blots, and enzyme-linked immunosorbent assays.
Nuclear extracts were prepared as described previously (33). Gel shift probes for Oct-1, Oct-2, and C/EBPβ contain sequences derived from the HSS1 site. Oct-1 (SC-232X), Oct-2 (SC-233X), and C/EBPβ (SC-150X) antibodies were from Santa Cruz Biotechnology. Gel shifts, Western blots, and enzyme-linked immunosorbent assays were performed as described previously (52).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays analyzed by semiquantitative PCR and real-time PCR were performed as described previously (34, 52). Some primer pairs used for semiquantitative analysis were described previously (52). Primer pairs for semiquantitative analysis of C/EBPβ binding to the HSS1 site were as follows: for fragment I, 5′-AAAAGAATATACCTTCTCCTCATC-3′ and 5′-TGGTACTAAGGAAATGCTCTATTG-3′; for fragment II, 5′-ATGTCTCTCACATTGGTCATCTGCAAG-3′ and 5′-AACTTTTTCTTTCTGTGTGACATAATTTATG-3′; for fragment III, 5′-GCTTTCTCACCCTCTTCTTCTCC-3′ and 5′-ACCTGCTGTTGTAAACCATCTTAG-3′; for fragment IV, 5′-GGCTAGGTGTACATGTATGTGCATATATC-3′ and 5′-GAAAGAAATGAATGAGTTCCCACC-3′; for fragment V, 5′-TCAGGTCACAAGTACCTATTACAGG-3′ and 5′-TGAGGCAACTGGAAGAGTCAGAGC-3′; and for fragment VI, 5′-AACTGTTACGGTCTTAGGCATGGTCTGG-3′ and 5′-TAGCCATGGGCAGGTGATTTAAAC-3′. Primer pairs for monitoring Oct-1 and Oct-2 binding to the HSS1 site were as follows: for HSS1, 5′-GCATGATCAGAGCATTGTCTTTGTG-3′ and 5′-TTTATGCAAATCCTCCTGGCTGC-3′; for the promoter, 5′-CCTGGGATTTCGACGTCTATATTCCCTCTGT-3 and 5′-GAGTTAGCGACAGGGAAGAGGAGAG-3′. Primer sequences for real-time PCR analysis are available upon request.
RESULTS
Identification of DNase I-hypersensitive sites within the murine Il12b locus.
Il12b, located on mouse chromosome 11, contains eight exons that span 14 kb (42). The adjacent genes, located approximately 563 kb upstream and 55 kb downstream of the Il12b transcription start site, are Adra1b and Ublcp1, respectively.
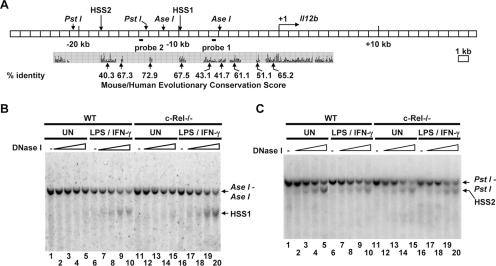
To identify distant control regions in the murine Il12b locus, a systematic search 27 kb upstream and 20 kb downstream of the transcription start site was performed using a DNase I hypersensitivity assay. Briefly, nuclei from unactivated or LPS-plus-IFN-γ-activated bone marrow-derived macrophages were incubated with increasing amounts of DNase I. Genomic DNA was purified from each sample and cleaved to completion with the restriction enzyme AseI or PstI. Southern blots were then performed with probes hybridizing to various regions spanning the Il12b locus. With this assay, two DNase I-hypersensitive sites were identified (Fig. 1). HSS1, located approximately 10 kb upstream of the transcription start site, is undetectable in unstimulated macrophages but is strongly induced upon macrophage activation (Fig. 1B, lanes 1 to 10). DNase I cleavage at this hypersensitive site yields a 4-kb band that is apparent below the 5.6-kb AseI-AseI band (Fig. 1B). HSS1 was briefly described in a previous analysis of the mechanism by which the anti-inflammatory cytokine IL-10 suppresses Il12b transcription (52). A second hypersensitive site, HSS2, was detected further upstream (kb −17 to −19) of the transcription start site (Fig. 1C, lanes 1 to 10). DNase I cleavage at this site yields a band that migrates further than the 7.6-kb PstI-PstI band (Fig. 1C). In contrast to HSS1, HSS2 was observed in both unstimulated and stimulated macrophages.
FIG. 1.
Identification of two c-Rel-independent DNase I-hypersensitive sites upstream of the murine Il12b gene. (A) The murine Il12b locus is shown, along with the positions of DNase I-hypersensitive sites (HSS1 and HSS2), restriction enzyme recognition sites, and two probes used for the DNase I hypersensitivity Southern blots. Evolutionary conservation of DNA sequences within the mouse and human Il12b loci, determined using the University of California, Santa Cruz Genome Browser, is shown below the blot. (B) Aliquots of nuclei from wild-type (WT) and c-Rel−/− bone marrow-derived macrophages that were left unstimulated (UN) or were stimulated for 4 h with LPS (10 μg/ml) and IFN-γ (10 U/ml) (LPS/IFN-γ) were incubated with increasing amounts of DNase I (0, 0.2, 0.4, 0.8, and 1.2 μg). The DNA samples were then purified and digested with AseI. Cleavage products were visualized by Southern blotting with probe 1. The positions of the AseI-AseI fragment and the fragment extending from HSS1 to the proximal AseI site are indicated. (C) The procedures described above for panel B were followed except the purified DNA samples were digested with PstI. Cleavage products were visualized by Southern blotting with probe 2. The positions of the PstI-PstI fragment and the fragment extending from HSS2 to the proximal PstI site are indicated.
An analysis of the evolutionary conservation of the hypersensitive regions using the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway) suggested that HSS1 resides within a 545-bp region (from bp −9437 to −9981) of high conservation between the mouse, rat, human, and dog genomes (67.5% identity between the mouse and human Il12b loci [Fig. 1A]). A 1,364-bp region (from bp −16572 to −17936) in close proximity to HSS2 shares high conservation between the four genomes (40.3% identity between the mouse and human Il12b loci [Fig. 1A]). However, we have not conclusively determined whether HSS2 resides within or is adjacent to this conserved region. Interestingly, several other regions within the Il12b locus, including the Il12b promoter, did not exhibit significant DNase I hypersensitivity (Fig. 1A and data not shown).
c-Rel is not required for HSS1 and HSS2 formation.
As described above, the NF-κB family member c-Rel is critical for Il12b induction in macrophages stimulated with LPS plus IFN-γ (35, 36). However, we previously showed that nucleosome remodeling at the endogenous Il12b promoter is only slightly reduced in c-Rel−/− macrophages, leading to the hypothesis that c-Rel acts downstream of, or parallel to, nucleosome remodeling in the Il12b activation pathway (48). One possibility was that c-Rel is required for the activation of a distant enhancer. To determine whether c-Rel is required for the formation of HSS1 and HSS2, DNase I hypersensitivity assays were performed using bone marrow-derived macrophages from c-Rel−/− mice (19, 36, 52). Interestingly, both the inducible and constitutive hypersensitive sites were readily detected in the c-Rel-deficient macrophages (Fig. 1B, lanes 11 to 20; Fig. 1C, lanes 11 to 20). These results demonstrate that c-Rel, although critical for Il12b transcription, is not required for the chromatin changes that are responsible for the appearance of HSS1 and HSS2.
Defining the boundaries of the inducible chromatin changes surrounding HSS1.
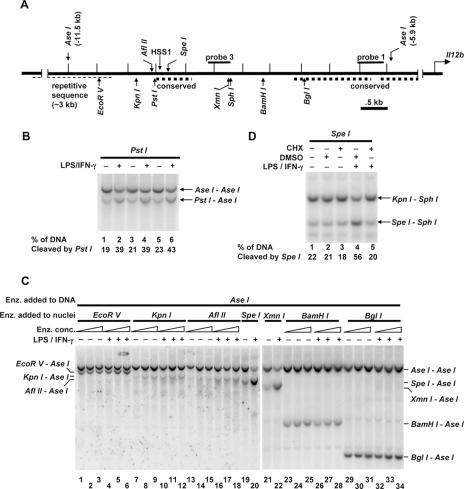
The DNase I hypersensitivity results suggested that the chromatin structure approximately 10 kb upstream of the Il12b start site is altered following stimulation with LPS plus IFN-γ. To analyze more precisely the locations and boundaries of these chromatin changes, a restriction enzyme accessibility assay was employed. First, to determine whether this assay can be used to monitor chromatin changes at HSS1 and to assess the reproducibility of this assay, three independent aliquots of B6.129/J2 macrophages were left unstimulated, with three additional aliquots stimulated with LPS plus IFN-γ. The B6.129/J2 line was used for this analysis because Il12b transcription is strongly and reproducibly activated in this line and HSS1 DNase I hypersensitivity was comparable to that observed in primary bone marrow-derived macrophages (data not shown). Nuclei from each of the six samples were incubated with restriction enzyme PstI, which has the potential to cleave at a recognition site located at bp −10061 (Fig. 2A). Genomic DNA was then purified and cleaved to completion with the restriction enzyme AseI, followed by gel electrophoresis and Southern blotting.
FIG. 2.
Defining the boundaries of LPS-induced chromatin changes in the vicinity of HSS1. (A) The positions of HSS1, relevant restriction enzyme recognition sites, and the Southern blot probes are indicated on the Il12b 5′ flanking region extending from kb −5.5 to −12 relative to the transcription start site. The thick dashed lines represent evolutionarily conserved regions located from bp −9437 to −9981 (containing HSS1), from bp −7521 to −6186, and from bp −6012 to −5327. The thin dashed line represents repetitive sequence. (B) LPS-induced chromatin changes in the vicinity of HSS1 were monitored using a restriction enzyme accessibility/Southern blot assay with nuclei from B6.129/J2 cells that were left unstimulated (−) or were stimulated with LPS plus IFN-γ (LPS/IFN-γ) for 4 h (+). Nuclei were incubated with 40 (lanes 1 and 2), 80 (lanes 3 and 4), or 160 (lanes 5 and 6) units of PstI to monitor accessibility at HSS1. Genomic DNA was then purified and cleaved with AseI. The digested DNA fragments were visualized by Southern blotting with probe 3. The PstI-AseI fragment reveals the efficiency of PstI cleavage in the isolated nuclei. Quantification of cleavage percentage was determined by phosphorimager analysis. (C) LPS-induced chromatin changes at different locations between kb −5.9 and −11.5 were monitored using the restriction enzyme accessibility/Southern blot assay. Aliquots of nuclei were incubated with different amounts of EcoRV (lanes 1 to 6), KpnI (lanes 7 to 12), AflII (lanes 13 to 18), SpeI (lanes 19 and 20), XmnI (lanes 21 and 22), BamHI (lanes 23 to 28), or BglI (lanes 29 to 34) enzyme (Enz.). Purified genomic DNA was cleaved with AseI. The digested DNA fragments were visualized by Southern blotting with probe 1. The EcoRV-AseI, KpnI-AseI, AflII-AseI, SpeI-AseI, XmnI-AseI, BamHI-AseI, and BglI-AseI fragments reveal the efficiency of nuclear cleavage with EcoRV, KpnI, AflII, SpeI, XmnI, BamHI, and BglI, respectively. (D) B6.129/J2 cells were pretreated for 15 min with cycloheximide (CHX) (+) dissolved in dimethyl sulfoxide (DMSO) (10 μg/ml) (lanes 3 and 5), with DMSO alone (lanes 2 and 4), or were left untreated (−) (lane 1). Cells were then stimulated with LPS plus IFN-γ for 1 h (lanes 4 and 5) or were left unstimulated (lanes 1 to 3). Nuclei were incubated with SpeI. Purified SpeI-digested DNA was digested with KpnI and SphI. The digested DNA fragments were visualized by Southern blotting with probe 3. The SpeI-SphI fragment reveals the efficiency of SpeI cleavage in the isolated nuclei. All data in panels B, C, and D are representative of at least three independent experiments.
When a radiolabeled probe derived from the Il12b locus was used (Fig. 2A, probe 3), cleavage at the two AseI sites flanking HSS1 yielded a 5,600-bp product, whereas an additional cleavage at the PstI site yielded a fragment of 4,075 bp. Approximately 20% of the genomic DNA was cleaved by PstI in nuclei from unstimulated cells (Fig. 2B, lanes 1, 3, and 5), with approximately 40% cleaved in nuclei from stimulated cells (Fig. 2B, lanes 2, 4, and 6).
Stimulus-dependent restriction enzyme cleavage was also apparent when the nuclei were incubated with AflII (which cleaves at bp −10121), SpeI (bp −9869), and XmnI (bp −8725) (Fig. 2C, lanes 13 to 22). In contrast, stimulus-dependent cleavages were not observed with two enzymes that cleave at positions farther from the transcription start site, EcoRV (bp −10916) and KpnI (bp −10256), or with two enzymes that cleave closer to the start site, BamHI (bp −8088) and BglI (bp −7369) (Fig. 2C, lanes 1 to 12 and lanes 23 to 34). These results suggest that the region in which LPS-induced changes in chromatin structure take place extends from at least bp −10121 (AflII site) at the distal end to at least bp −8725 (XmnI site) at the proximal end, but not beyond bp −10256 (KpnI site) at the distal end or bp −8088 (BamHI site) at the proximal end. This region includes the 545-bp region that exhibits high evolutionary conservation (−9981 to −9437) plus 700 to 1,300 bp proximal to these conserved sequences. It is important to note, however, that no suitable restriction enzyme recognition sequence was available between the SpeI site at bp −9869 and the XmnI site at bp −8725. Thus, we cannot rule out the possibility that the XmnI site is within a control region that is independent of the control region containing the clustered AflII, PstI, and SpeI sites.
To further analyze LPS-induced nucleosome remodeling at the HSS1 site, its requirement for new protein synthesis was monitored. When the B6.129/J2 line was pretreated for 15 min with the protein synthesis inhibitor cycloheximide, the increase in restriction enzyme cleavage typically observed following stimulation with LPS plus IFN-γ was eliminated (Fig. 2D). Similar results were recently reported in J774 macrophages (34). New protein synthesis is similarly required for Il12b transcription and for LPS-induced increases in restriction enzyme accessibility at the Il12b promoter (34, 49).
The HSS1 region functions as a transcriptional enhancer in transient-transfection experiments.
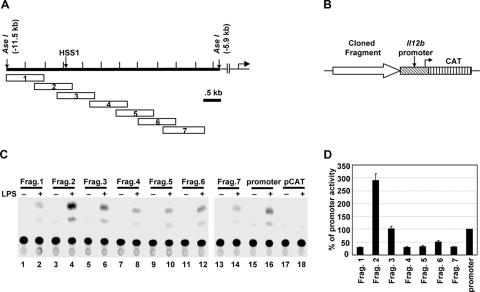
The inducible DNase I hypersensitivity and restriction enzyme accessibility at HSS1 suggest that this region might function as an inducible Il12b enhancer. To determine whether enhancer activity could be detected, seven overlapping DNA fragments of approximately 1 kb each were fused upstream of a 400-bp Il12b promoter fragment in a CAT reporter plasmid (Fig. 3A and B). Transient-transfection experiments were then performed with the RAW264.7 macrophage line, with the transfected cells left unstimulated or stimulated with LPS for 24 h. RAW264.7 cells were used for the transient-transfection experiments because the J774 and B6.129/J2 macrophage lines do not transfect with sufficiently high efficiency for detectable reporter activity (data not shown). In our hands, RAW264.7 cells express the endogenous Il12b gene poorly in response to either LPS or LPS plus IFN-γ, but they express many other LPS-induced genes and therefore support TLR4 signaling pathways (33). We do not know why the endogenous Il12b locus is not expressed in RAW264.7 cells, but we previously found that transfected Il12b promoter-reporter plasmids were efficiently induced in these cells, and we therefore suspected that the line would be equally appropriate for a basic characterization of the HSS1 enhancer. The CAT reporter gene was used for these experiments because we previously found that luciferase reporter activity is strongly induced in RAW264.7 cells, even when luciferase transcripts are driven by constitutively active promoters (33). This nonspecific induction of luciferase activity has the potential to yield misleading results. In contrast, CAT activity was only slightly induced when constitutive promoters were used to drive CAT transcription in these cells (33).
FIG. 3.
The DNA region surrounding HSS1 exhibits enhancer activity in transient-transfection experiments. (A) Individual Il12b DNA fragments (fragments 1 through 7) that were subcloned into the CAT reporter vector are shown. (B) Critical regions of the CAT reporter plasmids are diagrammed, including the fragment being tested for enhancer activity, the 400-bp Il12b promoter fragment, and the CAT reporter gene. (C) RAW264.7 cells were transfected with plasmids containing each of the seven fragments (fragments 1 to 7 [Frag. 1 to Frag. 7]) diagrammed in panel A (lanes 1 to 14), as well as a plasmid containing only the Il12b promoter (lanes 15 and 16), and the pCAT vector lacking both a promoter and enhancer (lanes 17 and 18). Transfected cells were left unstimulated (−) or were stimulated with LPS (10 μg/ml) for 24 h (+). IFN-γ was not included during stimulation because this cytokine has no effect on promoter activity upon transient transfection of this line (33). CAT enzyme activities were measured as described previously (7). (D) The means plus standard errors of the means (error bars) from four independent experiments (after phosphorimager quantification) are shown. The activity of the Il12b promoter was set at 100%.
LPS-induced CAT activity dependent on the Il12b promoter was observed with all plasmids tested, but only fragment 2 substantially enhanced the CAT activity (Fig. 3C and D). This fragment spans the DNase I cleavage site and three of the restriction enzyme recognition sites that showed LPS-induced increases in cleavage (AflII, PstI, and SpeI) (Fig. 3C). Fragment 3, which overlaps fragment 2 and also includes the DNase I cleavage site and the same three restriction enzyme recognition sites, supported CAT activity at an enhanced level relative to that observed with the other five 1-kb fragments. However, the activity obtained with this plasmid was comparable to that obtained with the Il12b promoter in the absence of a 1-kb insert. Figure 3D shows the average signals with standard deviations obtained in three independent transfection experiments.
Localization and analysis of the core HSS1 enhancer.
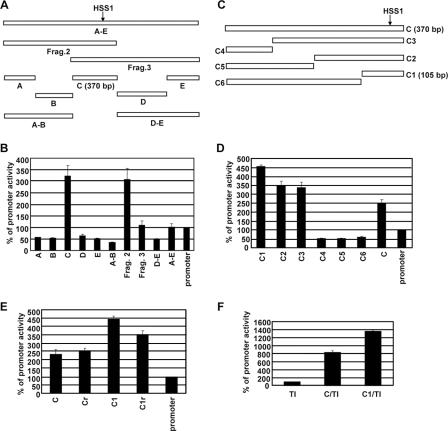
To localize the core enhancer within fragments 2 and 3, a series of smaller DNA fragments, as well as a larger fragment encompassing both fragments 2 and 3, were analyzed in the context of the Il12b promoter-CAT reporter plasmid (Fig. 4A). Only one small fragment, a 370-bp fragment called fragment C, supported enhanced CAT activity (Fig. 4B). Of further note, fragment A-E, which encompasses both fragments 2 and 3, yielded activity comparable to that of fragment 3 (Fig. 4B). These results suggest that all sequences responsible for the HSS1 enhancer activity detectable in a transient-transfection assay reside within the 370-bp fragment C. Furthermore, fusion of these sequences to the Il12b promoter in the context of fragment 2 or fragment C is preferable to fusion in the context of fragment 3, which places the critical sequences farther from the promoter. Importantly, the 370-bp fragment C encompasses the DNase I-hypersensitive site and restriction sites that showed LPS-induced increases.
FIG. 4.
Identification and characterization of a 105-bp core enhancer region. (A) The transient-transfection assay in RAW264.7 cells was used to analyze a variety of DNA fragments to more precisely localize the sequences responsible for enhancer activity. Fragment A-E contains sequences spanning both fragments 2 and 3 (Frag.2 and Frag.3) from Fig. 3A. Fragment C, which exhibited the strongest enhancer activity, is 370 bp long. (B) Transfected cells were stimulated with LPS (10 μg/ml) for 24 h. (No significant activity was observed with any of the plasmids in the absence of stimulation [not shown]). CAT reporter activities were then determined. The means plus standard errors of the means (error bars) from four independent experiments (after phosphorimager quantification) are shown. The activity of the Il12b promoter was set at 100%. (C) The sequences within the 370-bp fragment C that were responsible for enhancer activity were further localized by analysis of six subfragments (C1 through C6). Fragment C1 is 105 bp long. (D) CAT reporter activities of plasmids containing the fragments diagrammed in panel C were determined. The means plus standard errors of the means (error bars) from four independent experiments are shown. (No significant activity was observed with any of the plasmids in the absence of stimulation [not shown]). (E) The transient-transfection assay was used to monitor the enhancer activities of fragments C and C1 in the forward (C and C1) and reverse (Cr and C1r) orientations in the context of the Il12b promoter-CAT reporter plasmid. The Il12b promoter-CAT reporter plasmid in the absence of an inserted enhancer was analyzed as a control (promoter). Transfected cells were stimulated with LPS (10 μg/ml) for 24 h. The means plus standard errors of the means (error bars) from four independent experiments are shown. (F) The enhancer activities of fragments C and C1 in the context of a minimal heterologous core promoter containing consensus TATA and Inr elements (C/TI and C1/TI) were examined. The TI promoter-CAT reporter plasmid in the absence of an inserted enhancer fragment was tested as a control (TI). The means plus standard errors of the means (error bars) from three independent experiments are shown.
To localize more precisely the sequences within fragment C that are responsible for enhancer activity, smaller fragments were analyzed in the context of the Il12b promoter-CAT reporter plasmid (Fig. 4C). The results revealed that enhancer activity is contained entirely within a 105-bp region (−9994 to −9890) termed fragment C1 (Fig. 4D). Both fragments C and C1 were capable of supporting weak enhancer activity in either orientation in the context of the Il12b promoter-CAT reporter plasmid (Fig. 4E). Furthermore, both of these fragments supported LPS-induced CAT activity when inserted into a CAT reporter plasmid containing only a minimal consensus TATA box and Inr element (Fig. 4F) (7).
Identification of a DNA element required for HSS1 enhancer activity.
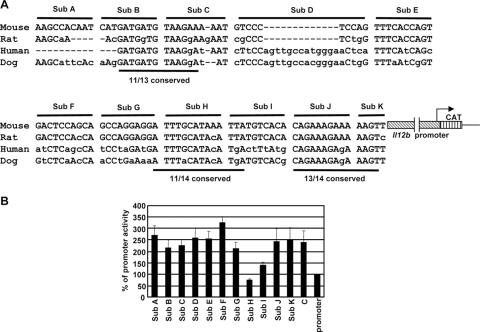
The 105-bp C1 fragment found to be required for enhancer activity (−9994 to −9890) is located at the distal edge of the 545-bp region of evolutionary conservation between DNA sequences encoding the mouse, rat, human, and dog C1 fragments (−9981 to −9437). It is interesting that a careful examination of the sequence within this 105-bp region revealed three segments that exhibit particularly strong conservation (Fig. 5A). To determine which DNA elements are important for enhancer activity in the transient-transfection assay, 11 different substitution mutants were prepared in the context of the Il12b promoter-CAT reporter plasmid (Sub A through Sub K [Fig. 5A]). In these plasmids, the mutations were introduced into the 370-bp fragment C. Analysis of the mutants by transient transfection in RAW264.7 cells revealed that enhancer activity was abolished by only one mutation, Sub H, with a substantial decrease in activity observed with an adjacent mutation, Sub I (Fig. 5B). The Sub H and Sub I mutations disrupt one of the three regions of evolutionary conservation. In a stable-transfection assay in J774 cells (see below), the Sub H mutation also abolished enhancer activity, whereas the Sub I mutation only slightly reduced enhancer activity (data not shown). These data suggest that the DNA sequence altered by the Sub H mutation represents a critical enhancer element. Importantly, a small DNA fragment encompassing only the Sub H and Sub I regions was unable to support enhancer activity (data not shown), demonstrating that additional sequences within the 105-bp fragment are essential for activity. However, these additional sequences must function in a redundant manner, at least in the transfection assay, as none of the other individual mutations significantly decreased enhancer activity.
FIG. 5.
Localization of a DNA element required for the LPS-induced enhancer activity. (A) The DNA sequence of mouse Il12b fragment C1 (−9,994 to −9,890 bp) is shown, along with the homologous sequences from the rat, human, and dog genomes. The nucleotides altered in each of the 11 substitution mutants (Sub A through Sub K) are indicated, as are three regions that are particularly well-conserved between the species. The 11 mutations were analyzed in the context of the 370-bp fragment C in the Il12b promoter-CAT reporter plasmid. Gaps introduced to maximize alignment are indicated by dashes. (B) RAW264.7 cells were transfected with enhancer-promoter-CAT reporter plasmids containing wild-type fragment C and the 11 mutant enhancers. The Il12b promoter alone (promoter) was analyzed as a control. Transfected cells were stimulated with LPS (10 μg/ml) for 24 h. (No significant activity was observed with any of the plasmids in the absence of stimulation [not shown]). The means plus standard errors of the means (error bars) from four independent experiments are shown.
Oct proteins bind the DNA sequence altered by the Sub H mutation.
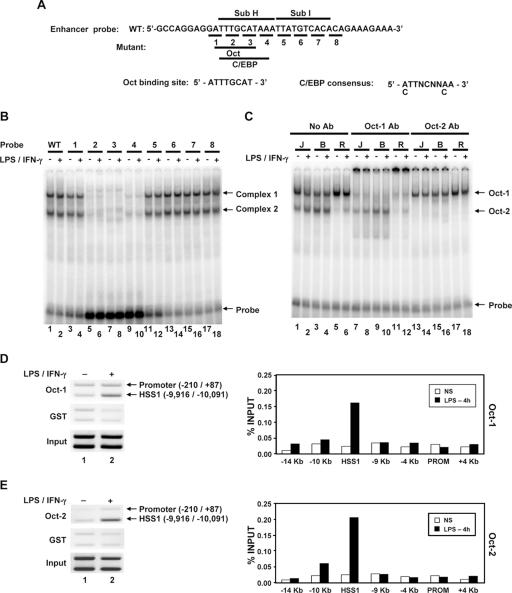
To identify transcription factors capable of binding the DNA sequence altered by the Sub H mutation, gel shift assays were performed with nuclear extracts prepared from the B6.129/J2 line and a 40-bp DNA probe (Fig. 6A). Two major protein-DNA complexes were detected with the wild-type probe, with no differences between unstimulated and stimulated macrophages (Fig. 6B, lanes 1 and 2). Analysis of a series of mutant probes, each of which alters 3 bp, revealed that both complexes were eliminated by three different mutations within the Sub H region (Fig. 6B).
FIG. 6.
Oct-1 and Oct-2 bind the critical Sub H region of the HSS1 enhancer. (A) The DNA sequence of the probe used for gel shift assays is shown. Nucleotides altered in each of the eight mutant probes are underlined. The predicted Oct and C/EBP binding sites within the probe are shown. (B) 32P-labeled wild-type (WT) (lanes 1 and 2) and mutant (lanes 3 to 18) probes (probes 1 to 8) were used for gel shift assays with nuclear extracts from unstimulated B6.129/J2 cells (−) or B6.129/J2 cells stimulated for 4 h with LPS (+). (C) The identities of the two protein-DNA complexes were determined by adding Oct-1 (lanes 7 to 12) and Oct-2 (lanes 13 to 18) antibodies (Ab) to gel shift reaction mixtures. Nuclear extracts were made from J744 (J), B6.129/J2 (B), and RAW264.7 (R) cells. (D) Oct-1 binding to the endogenous HSS1 region was monitored using a ChIP assay. (Left) Formaldehyde-cross-linked chromatin samples prepared from unstimulated peritoneal macrophages (lane 1) or peritoneal macrophages stimulated for 4 h with LPS plus IFN-γ (lane 2) were immunoprecipitated with Oct-1 antibodies (top blot) or control GST antibodies (middle blot). The presence of the HSS1 region and the Il12b promoter in the immunoprecipitates was determined by semiquantitative PCR. Input samples are shown in the bottom blot. (Right) ChIP experiments were performed with bone marrow-derived macrophages, with the Oct-1 antibody immunoprecipitates analyzed by real-time PCR, using a variety of primer pairs that amplify regions spanning the Il12b locus. Similar results were obtained in a second independent experiment (not shown). NS, not stimulated; PROM, promoter. (E) ChIP assays analyzed by both semiquantitative PCR (left) and real-time PCR (right) were performed with Oct-2 or GST antibodies. For both semiquantitative and real-time PCR analysis, chromatin samples were prepared from bone marrow-derived macrophages.
An examination of the DNA sequence within the Sub H region revealed the presence of potential binding sites for Oct and C/EBP proteins (Fig. 6A). Antibodies against C/EBP family members had no effect on the complexes observed in the gel shift experiments (data not shown). However, the lower-mobility complex was eliminated by antibodies directed against Oct-1, and the higher-mobility complex was eliminated by antibodies against Oct-2 (Fig. 6C). This experiment was performed with extracts derived from three different macrophage lines (J774, B6.129/J2, and RAW264.7), which revealed that Oct-1 is readily detectable in all three lines, whereas Oct-2 is readily detectable only in the J774 and B6.129/J2 lines (Fig. 6C). Because OCA-B (also known as OBF1 and Bob-1) has been shown to interact with Oct-1 and Oct-2 in B lymphocytes (11, 22, 37, 40), the presence of OCA-B in complex 1 or 2 was examined by gel shift analysis. OCA-B antibodies failed to supershift or disrupt either complex (data not shown). Furthermore, by Western blot analysis, OCA-B was undetectable in macrophage extracts, whereas it was readily detected in extracts from B-cell lines (data not shown).
To monitor the recruitment of Oct proteins to the endogenous HSS1 enhancer in primary murine macrophages, ChIP assays were performed with peritoneal or bone marrow-derived macrophages left unstimulated or stimulated with LPS plus IFN-γ. Sheared chromatin from formaldehyde-treated cells was precipitated with antibodies directed against either Oct-1 or Oct-2 and was first analyzed by PCR with a mixture of two primer pairs. One primer pair amplified the HSS1 region, and the second pair amplified the Il12b promoter. In the DNA samples prepared from macrophages stimulated with LPS plus IFN-γ, the HSS1 PCR product was enriched relative to the promoter product when either the Oct-1 or Oct-2 antibodies were used (Fig. 6D and E, top blots). In contrast, the HSS1 product was not enriched when a glutathione S-transferase (GST) control antiserum was used (Fig. 6D and E, middle blots). The immunoprecipitated chromatin was further evaluated by real-time PCR, using PCR primer pairs that amplify seven different regions spanning the Il12b locus. The HSS1 region was selectively enriched in Oct-1 and Oct-2 antibody immunoprecipitates prepared from stimulated macrophages (Fig. 6D and E, bar graphs). These results suggest that Oct-1 and Oct-2 associate with the endogenous HSS1 enhancer upon macrophage activation. Interestingly, interactions between Oct-1 and Oct-2 and the Il12b promoter were not detected. These data contrast with evidence that some transcription factors can interact with enhancers and promoters simultaneously by DNA looping, although this possibility cannot be ruled out (12).
Il12b expression in Oct-1- and Oct-2-deficient macrophages.
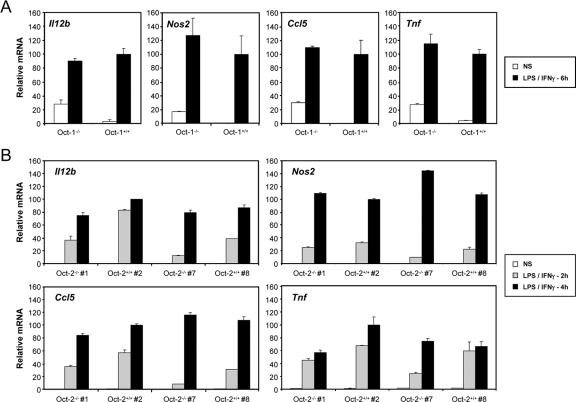
Although the functions of Oct-1 and Oct-2 have been studied extensively in B cells and other cell types (26, 45, 47), little is known about the functions of these proteins in macrophages. To examine their relevance for Il12b expression, macrophages were derived from the fetal livers of one Oct-1−/− mouse and two Oct-2−/− mice, as well as their wild-type littermates. Il12b mRNA was then monitored by real-time RT-PCR, with mRNAs from three other LPS-induced genes (Nos2, Ccl5, and Tnf) monitored as controls. Although variable results were obtained, no significant reduction in expression of any of the four genes was observed in the mutant samples in comparison to the wild-type samples (Fig. 7). One explanation for these negative results is that Oct-1 and Oct-2 act redundantly to stimulate Il12b transcription. Unfortunately, we were unable to obtain Oct-1−/− Oct-2−/− double mutant fetuses to test this possibility.
FIG. 7.
Analysis of Il12b expression in fetal liver macrophages from Oct-1−/− and Oct-2−/− mice. Fetal liver macrophages from one Oct-1−/− mouse and two Oct-2−/− mice, as well as their wild-type littermates, were left unstimulated or were stimulated with LPS plus IFN-γ (LPS/IFN-γ) for 6 h (for Oct-1−/− mice) or for 2 and 4 h (for Oct-2−/− mice). Real-time RT-PCR was then used to monitor Il12b mRNA levels, with Nos2, Ccl5, and Tnf mRNA levels monitored as controls. NS, not stimulated; Oct-2−/− #1, Oct-2−/− mouse 1.
C/EBPβ associates with the HSS1 enhancer.
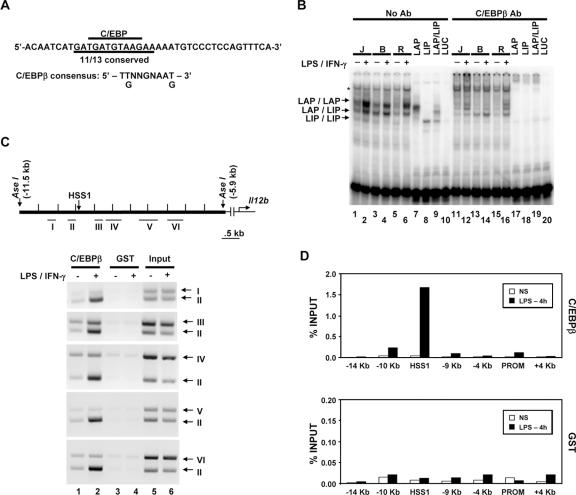
As shown in Fig. 5A, the 105-bp HSS1 enhancer fragment contains three segments that exhibit particularly strong evolutionary conservation. Although only the Sub H region was important for enhancer activity in the transient-transfection assay, we suspect that the other two regions, and possibly additional regions that exhibit significant evolutionary conservation both within and adjacent to the 105-bp fragment, are important for activity of the endogenous enhancer. Interestingly, the highly conserved DNA element spanning the Sub B/Sub C region of the enhancer contains a DNA sequence that perfectly matches the consensus recognition sequence for C/EBP proteins (Fig. 5A and 8A). In addition, gel shift experiments performed with extracts from the three macrophage lines and a probe encompassing the Sub B/Sub C region yielded complexes that comigrated with complexes formed by in vitro-translated LAP, a C/EBPβ isoform containing the bZIP DNA binding domain and N-terminal transactivation domains (Fig. 8B, lanes 1 to 7) (6). This complex is thought to consist of LAP homodimers. A second abundant complex in the macrophage extracts comigrates with a complex consisting of a heterodimer between LAP and LIP, a smaller C/EBPβ isoform that lacks the transactivation domains (Fig. 8B, lane 9) (6). The levels of these complexes were greatly diminished by C/EBPβ antibodies (Fig. 8B, lanes 11 to 19).
FIG. 8.
C/EBPβ binds the endogenous HSS1 enhancer. (A) The DNA sequence of the probe used for gel shift assays is shown, along with the C/EBP consensus recognition sequence. (B) Gel shift (lanes 1 to 10) and antibody (Ab) supershift (lanes 11 to 20) assays were performed as described in the legend to Fig. 6. Nuclear extracts from J774 (J), B6.129/J2 (B), and RAW264.7 (R) cells were examined, as well as in vitro-translated LAP (C/EBPβ large isoform; lanes 7 and 17), LIP (C/EBPβ small isoform; lanes 8 and 18), and cotranslated LAP and LIP (LAP/LIP) (lanes 9 and 19). In vitro-translated luciferase (LUC) (lanes 10 and 20) was tested as a negative control. The positions of protein-DNA complexes corresponding to LAP homodimers, LAP/LIP heterodimers, and LIP homodimers are indicated. A protein-DNA complex that does not interact with C/EBPβ antibodies is also apparent (*). (C) The Il12b genomic region examined for C/EBPβ binding by semiquantitative ChIP is shown. The locations of DNA fragments amplified by PCR (fragments I through VI) are indicated. The binding of C/EBPβ to the endogenous Il12b locus was monitored by ChIP using C/EBPβ or GST antibodies and chromatin samples prepared from peritoneal macrophages. Semiquantitative PCR was used to analyze the DNA fragments present in the immunoprecipitates. (D) ChIP experiments were performed with real-time PCR as described in the legend to Fig. 6, using chromatin prepared from bone marrow-derived macrophages. Similar results were obtained in a second independent experiment (not shown).
ChIP experiments provided further support for the hypothesis that C/EBPβ contributes to HSS1 enhancer activity, as DNA fragments in close proximity to HSS1 were enriched following immunoprecipitation of cross-linked chromatin from stimulated macrophages with C/EBPβ antibodies (Fig. 8C). The immunoprecipitates were analyzed with various combinations of primer pairs from the Il12b locus to determine the approximate locations of the C/EBPβ contact sites. DNA amplified by primer pair II, which overlaps HSS1, was enriched to a much greater extent than the other five DNA fragments analyzed (Fig. 8C). This finding was confirmed by real-time PCR analysis of C/EBPβ antibody immunoprecipitates from an independent set of unstimulated and stimulated macrophages (Fig. 8D). Together, these results provide strong evidence that C/EBPβ associates with the endogenous Il12b locus within or in close proximity to the HSS1 enhancer.
Although mutations in the consensus C/EBP binding site at the Sub B/Sub C region had no effect on enhancer activity in transient- and stable-transfection assays (Fig. 5B and data not shown), these results suggest that C/EBP proteins may indeed contribute to enhancer activity. C/EBP proteins may not be important in the transfection experiments because of the artificial nature of these assays, or the functions of C/EBP proteins may be redundant with the functions of other enhancer binding proteins. Unfortunately, in the absence of functional data, we cannot exclude the possibility that the binding of C/EBPβ to the endogenous enhancer is fortuitous and without functional relevance.
HSS1 enhancer activity in an insulated chromatin environment.
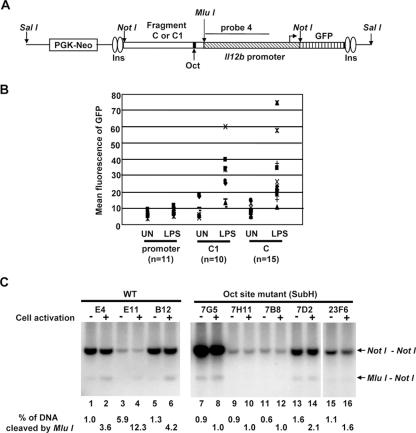
The transient-transfection results, combined with the DNase I hypersensitivity and restriction enzyme accessibility results, support the hypothesis that DNA sequences in the vicinity of HSS1 function as an LPS-induced enhancer. However, one weakness of this hypothesis is that fragments 2, C, and C1 stimulate Il12b promoter activity by only three- to fivefold in transient-transfection experiments. To determine whether this weak activity might be due to the artificial nature of the transient-transfection assay, the activity of the HSS1 region was tested in a chromatin environment by using a stable-transfection assay. For this assay, fragments C and C1 were fused upstream of the Il12b promoter in a vector containing a GFP reporter gene (28). In this vector, the enhancer-promoter-GFP cassette is flanked on each side by two chicken β-globin insulators (Fig. 9A). A phosphoglycerate kinase (PGK)-neomycin (Neo) drug selection cassette is located on the plasmid on the side opposite that of the insulators. The resulting plasmids (Il12b promoter alone, fragment C1 plus promoter, and fragment C plus promoter) were linearized using the restriction enzyme SalI, followed by transfection into the J774 macrophage line. (Although the J774 line, which produces large quantities of IL-12 p40, is not useful for transient-transfection experiments in our hands, it transfects with sufficient efficiency for selection of clones containing stably integrated plasmids.) Transfected cells were immediately plated on 96-well plates and were selected in growth medium containing G418, yielding monoclonal or oligoclonal colonies. After LPS stimulation for 24 h, the mean GFP fluorescence was determined for each colony (Fig. 9B).
FIG. 9.
Enhancer activity and nucleosome remodeling in an insulated chromatin environment. (A) The insulator vector used for the generation of stable cell lines is diagrammed. Two copies of the 1.2-kb chicken β-globin insulator (Ins) are present at each end of the reporter cassette. A NotI cloning site was used for insertion of the enhancer fragments (fragment C or C1) and Il12b promoter, with a downstream GFP reporter gene. A PGK-Neo drug resistance cassette is located outside of the insulated reporter cassette. The plasmids were linearized with SalI before transfection. The location of probe 4 used for the Southern blot assay is indicated. (B) Insulator plasmids containing the Il12b promoter alone or the Il12b promoter with upstream fragment C1 or fragment C were stably transfected into J774 cells. Several monoclonal or oligoclonal drug-resistant colonies were left unstimulated (UN) or were stimulated with LPS for 24 h. The mean GFP fluorescence was then determine by flow cytometry. Eleven colonies containing the Il12b promoter alone, 10 colonies containing fragment C1, and 15 colonies containing fragment C were analyzed. (C) Three monoclonal or oligoclonal J774 lines containing stably integrated plasmids with wild-type (WT) fragment C and five lines containing integrated plasmids with the Sub H mutant version of fragment C were analyzed using the restriction enzyme accessibility assay. Cells were left unstimulated (odd-numbered lanes) or were stimulated with LPS for 4 h (even-numbered lanes) prior to preparation of nuclei. Nuclei were incubated with MluI (40 units), and genomic DNA was then purified and cleaved to completion with NotI. The digested DNA fragments were visualized by Southern blotting with probe 4. The NotI-NotI fragment corresponds to the in vitro cleavage product. The MluI-NotI fragment reveals the efficiency of MluI cleavage in the isolated nuclei.
In these experiments, the activity of the isolated Il12b promoter was very low in unstimulated cells and was stimulated only slightly by LPS (Fig. 9B), in striking contrast to the results obtained in transient-transfection experiments and in previous stable-transfection experiments performed with promoter-GFP reporter plasmids in the absence of insulators (48). However, in the presence of fragments C1 and C, strong LPS-induced promoter activity was observed (Fig. 9B). Although considerable clonal variability was observed, there was a consistent trend toward strong LPS-induced GFP fluorescence. We suspect that the magnitude of the enhancer effect is greater in these stable-transfection experiments than in the transient-transfection experiments because the insulator elements allow the promoter to assemble into an inactive chromatin structure that cannot be converted to an active state in the absence of the enhancer. These results greatly strengthen the hypothesis that the HSS1 region contains a biologically important enhancer that is a target of LPS signaling pathways.
The Oct protein binding site contributes to inducible chromatin remodeling at HSS1.
In contrast to the results shown in Fig. 9B, stable-transfection experiments performed in the absence of the insulator elements suggested that the Il12b promoter is sufficient for strong, inducible transcription in J774 cells (48). We now suspect that inducible transcription was observed in the earlier experiments only when the plasmid integrated into the chromosome adjacent to control regions that were capable of functioning as enhancers. Consistent with this hypothesis, inducible transcription from the promoter-reporter plasmids lacking insulator elements was observed only in a small percentage of clones analyzed (48; also data not shown). In contrast, in the insulator-containing plasmids containing the HSS1 enhancer upstream of the Il12b promoter, inducible reporter gene transcription was observed in a high percentage of clones (Fig. 9B).
We also previously observed highly efficient restriction enzyme cleavage at the stably integrated Il12b promoter when insulator elements were omitted, and this high-efficiency cleavage was observed both before and after LPS stimulation (48). We now suspect that endogenous enhancers in the vicinity of the integration site or the enhancer associated with the drug resistance gene were responsible for this constitutive accessibility.
An analysis of restriction enzyme accessibility at the integrated insulator-enhancer-promoter-reporter plasmids revealed very low cleavage efficiency at an MluI site located 28 bp downstream of the Oct site within the Il12b enhancer, with cleavage efficiency consistently enhanced following LPS stimulation (MluI does not cleave within the endogenous Il12b enhancer). The results obtained with three representative clonal lines with variable numbers of copies of integrated plasmid are shown in Fig. 9C (lanes 1 to 6). Similar results were obtained when a cleavage site within the promoter was analyzed (data not shown). These results provide evidence that the insulator elements allow the integrated reporter cassette to be assembled into a relatively “closed” chromatin structure and that factors bound to the HSS1 enhancer can support chromatin remodeling events in this context upon LPS stimulation.
To determine whether the Oct binding site within HSS1 is required for the LPS-induced increases in restriction enzyme cleavage observed in this stable-transfection assay, stably transfected lines containing the wild-type HSS1 enhancer (fragment C) were compared to lines containing the Sub H mutation that alters the Oct site. Although strong LPS-induced GFP fluorescence was observed with most colonies containing wild-type fragment C (Fig. 9B), none of the colonies containing the Sub H mutation exhibited significant GFP fluorescence (data not shown). Next, several clones were analyzed by restriction enzyme accessibility. In contrast to the results obtained with the wild-type enhancer, LPS-induced restriction enzyme cleavage was not observed with colonies containing the Sub H-mutant enhancer. Figure 9C (lanes 7 to 16) shows the results obtained with five independent clones containing the mutant enhancer. These results suggest that an intact Oct binding site is required for LPS-induced restriction enzyme cleavage at HSS1. Further experiments are needed to determine whether Oct1 alone or in combination with other enhancer binding proteins is directly responsible for recruitment of remodeling complexes to the HSS1 enhancer.
DISCUSSION
A systematic search for distant control regions at the murine Il12b locus using a DNase I hypersensitivity assay led to the identification of one region, termed HSS1, that is likely to make important contributions to inducible Il12b transcription. A second region, which was not examined further, exhibited DNase I hypersensitivity both before and after macrophage stimulation, raising the possibility that it contributes to Il12b expression but is not a target of TLR signaling pathways. HSS1 is located within a 545-bp region that is conserved through evolution, and LPS-induced changes in chromatin structure apparently occur throughout this region and extend even farther toward the transcription start site. Nevertheless, in an extensive analysis of enhancer mutants using a transient-transfection assay, only one DNA element was essential for enhancer activity. This site was recognized by Oct-1 and Oct-2, which are present in both unstimulated and stimulated macrophages. However, the Oct proteins associated with the endogenous enhancer only in stimulated cells. Although the conservation of 545 bp surrounding HSS1 and our accumulated knowledge of transcriptional enhancers strongly suggest that other transcription factors make important contributions to HSS1 enhancer activity, Oct proteins appears to be critical nonredundant activators of this enhancer.
With the availability of genome sequence information for several different vertebrates, it is now possible to identify conserved noncoding sequences in the vicinity of almost all genes. Like most genes, the Il12b gene is flanked by several regions that exhibit evolutionary conservation, including the Il12b promoter, the HSS1 enhancer, a region in close proximity to HSS2, and several other regions. It is currently unclear why only HSS1 and HSS2 exhibit detectable DNase I hypersensitivity. Clearly, DNase I hypersensitivity is not essential for functional activity, as the Il12b promoter is fully functional yet does not exhibit DNase I hypersensitivity. The absence of DNase I hypersensitivity at the promoter is consistent with our analysis of restriction enzyme accessibility. Although potent LPS-induced increases in restriction enzyme accessibility at the promoter were observed using a ligation-mediated PCR-based assay (44), more recent studies using a Southern blot assay (as used in this study) revealed that restriction enzyme cleavage at the promoter occurs at a much lower efficiency than at the enhancer, even when the same restriction enzymes are used for nuclear cleavage (34). The reason for this difference remains unknown, but one possibility is that LPS stimulation is accompanied by the removal of nucleosomes from the HSS1 enhancer, with nucleosomes at the promoter merely altered through the action of SWI/SNF remodeling complexes. Because the promoter is undoubtedly important for transcription in the absence of DNase I hypersensitivity, we can speculate that other conserved noncoding sequences in the vicinity of the Il12b gene may be equally important for inducible transcription, despite the absence of DNase I hypersensitivity. To resolve this issue, it will be necessary to systematically delete these conserved regions from the Il12b locus by homologous recombination in embryonic stem cells or by analysis of bacterial artificial chromosome transgenes.
Although Oct-1 and Oct-2 have been widely studied as critical activators of transcription in B cells and other cell types (26, 45, 47), there have been surprisingly few studies of these proteins in macrophages. Oct-1 and Oct-2 are known to be expressed in macrophages, and Oct proteins have been implicated in activation of the CD36 and inducible nitric oxide synthetase genes in macrophages (10, 17, 18, 32). However, to our knowledge, no Oct-1 or Oct-2 loss-of-function studies have been performed in macrophages, other than the study that led to the identification of CD36 as an Oct-2 target (18). Furthermore, Oct binding sites have not been commonly identified in functional studies of other proinflammatory genes.
We do not know why the HSS1 enhancer contains only one nonredundant site in transfection experiments. This may reflect a peculiarity of transfection experiments and may have no special significance to the mechanism of action of the endogenous enhancer. Alternatively, all other HSS1 enhancer elements may simply act redundantly with other elements in the enhancer, with the Oct site serving as the only nonredundant element. However, we must also consider the possibility that the important role of the Oct site in transfection assays is due to a special mechanistic contribution to enhancer activity, even at the endogenous locus. Perhaps, Oct proteins carry out the most important interactions with coactivators responsible for communicating with specific factors bound at the promoter or with the general transcription machinery. Indeed, critical interactions between enhancer-bound Oct proteins and the general transcription machinery have previously been documented (5). Alternatively, Oct proteins may be responsible for stimulating the chromatin remodeling events required for full enhancer function. Although our restriction enzyme accessibility experiments suggest that the Oct site is necessary for LPS-induced accessibility, the importance of the Oct site in transiently transfected plasmids that are unlikely to assemble into inaccessible chromatin structures suggests that its unique requirement in these assays is unrelated to specific nucleosome remodeling events. It is noteworthy that our systematic analysis of the Il12b promoter revealed that a binding site for C/EBPβ is far more important than binding sites for other factors, including NF-κB, AP-1, and NFAT, in transient-transfection assays (33). Moreover, in a systematic analysis of the murine IL-10 promoter, only an Sp1 binding site was found to be critical for LPS-induced promoter function, although other important sites must also exist (7).
The results of our gel shift analyses suggest that Oct-1 and Oct-2 are the Oct family members that are most abundantly expressed in murine macrophages. The two proteins are equally expressed in unstimulated and stimulated macrophages, yet binding to the endogenous HSS1 enhancer was observed only in stimulated cells. These results suggest that enhancer binding by other inducible transcription factors that we failed to detect is required for Oct binding. Alternatively, enhancer binding by Oct-1 and Oct-2 in macrophages may require an inducible posttranslational modification or association with a specific cofactor. If a cofactor is indeed involved, it does not appear to be OBF-1, as this factor does not appear to be expressed in primary or transformed murine macrophages and Il12b induction occurred normally in OBF-1−/− macrophages.
Finally, it is worth noting that the identification and characterization of the HSS1 enhancer should be of considerable value for our ongoing attempts to use the Il12b gene as a model for understanding the molecular mechanisms by which inducible mammalian genes in general and proinflammatory cytokine genes in particular are activated. We have previously characterized the nucleosome organization and remodeling events required for activation of the Il12b promoter, and we have recently classified LPS-induced genes according to their remodeling requirements, kinetics of activation, and protein synthesis requirements (34, 48, 49). With the identification of the HSS1 enhancer, we can now begin to explore how the enhancer for a proinflammatory gene contributes to events occurring at the promoter, including nucleosome remodeling, transcription factor binding, assembly of the preinitiation complex, and transcription initiation.
Acknowledgments
We thank Gary Felsenfeld and Mark Schlissel for providing the insulator vector used for the stable-transfection experiments and Jill Suttles and Peter Tontonoz for the B6.129/J2 murine macrophage line.
S.T.S. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S., and A. Rao. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765-775. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal, S., O. Avni, and A. Rao. 2000. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12:643-652. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz, L. A., K. T. Riabowol, and M. Z. Gilman. 1989. Multiple sequence elements of a single functional class are required for cyclic AMP responsiveness of the mouse c-fos promoter. Mol. Cell. Biol. 9:4272-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolino, E., and H. Singh. 2002. POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol. Cell 10:397-407. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, M. N., L. Zhou, and S. T. Smale. 2003. C/EBPβ regulation in lipopolysaccharide-stimulated macrophages. Mol. Cell. Biol. 23:4841-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brightbill, H. D., S. E. Plevy, R. L. Modlin, and S. T. Smale. 2000. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J. Immunol. 164:1940-1951. [DOI] [PubMed] [Google Scholar]
- 8.Carey, M., and S. T. Smale. 2000. Transcriptional regulation in eukaryotes. Concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Corcoran, L. M., M. Karvelas, G. J. V. Nossal, Z. S. Ye, T. Jacks, and D. Baltimore. 1993. Oct-2, although not required for early B cell development, is critical for later B cell maturation and for postnatal survival. Genes Dev. 7:570-582. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, T. L., I. L. Ross, and D. A. Hume. 1996. Transcription factor Oct-2 is expressed in primary murine macrophages. Blood 88:4072. [PubMed] [Google Scholar]
- 11.Gstaiger, M., L. Knoepfel, O. Georgiev, W. Schaffner, and C. M. Hovens. 1995. A B-cell coactivator of octamer-binding transcription factors. Nature 373:360-362. [DOI] [PubMed] [Google Scholar]
- 12.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, C. A. 2005. New IL-12 family members: IL-23 and IL-27, cytokines with divergent functions. Nat. Rev. Immunol. 5:521-531. [DOI] [PubMed] [Google Scholar]
- 14.Jankovic, D., M. C. Kullberg, S. Hieny, P. Caspar, C. M. Collazo, and A. Sher. 2002. In the absence of IL-12, CD4+ T cell responses to intracellular pathogens fail to default to a Th2 pattern and are host protective in an IL-10−/− setting. Immunity 16:429-439. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. P., and W. J. Leonard. 2002. The basis for TCR-mediated regulation of the IL-2 receptor alpha chain gene: role of widely separated regulatory elements. EMBO J. 21:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. P., J. Kelly, and W. J. Leonard. 2001. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity 15:159-172. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. M., C. B. Ko, Y. P. Park, Y. J. Kim, and S. G. Paik. 1999. Octamer motif is required for the NF-kappaB-mediated induction of the inducible nitric oxide synthase gene expression in RAW 264.7 macrophages. Mol. Cells 9:99-109. [PubMed] [Google Scholar]
- 18.Konig, H., P. Pfisterer, L. M. Corcoran, and T. Wirth. 1995. Identification of CD36 as the first gene dependent on the B-cell differentiation factor Oct-2. Genes Dev. 9:1598-1607. [DOI] [PubMed] [Google Scholar]
- 19.Kontgen, F., R. J. Grumont, A. Strasser, D. Metcalf, R. Li, D. Tarlinton, and S. Gerondakis. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965-1977. [DOI] [PubMed] [Google Scholar]
- 20.Lécine, P., M. Algarté, P. Rameil, C. Beadling, P. Bucher, M. Nabholz, and J. Imbert. 1996. Elf-1 and Stat5 bind to a critical element in a new enhancer of the human interleukin-2 receptor alpha gene. Mol. Cell. Biol. 16:6829-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, B. B., S. L. Cross, N. F. Halden, D. G. Roman, M. B. Toledano, and W. J. Leonard. 1990. Delineation of an enhancerlike positive regulatory element in the interleukin-2 receptor alpha-chain gene. Mol. Cell. Biol. 10:850-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo, Y., H. Fujii, T. Gerster, and R. G. Roeder. 1992. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell 71:231-241. [DOI] [PubMed] [Google Scholar]
- 23.Ma, X., J. M. Chow, G. Gri, G. Carra, F. Gerosa, S. F. Wolf, R. Dzialo, and G. Trinchieri. 1996. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J. Exp. Med. 183:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, X., and G. Trinchieri. 2001. Regulation of interleukin-12 production in antigen-presenting cells. Adv. Immunol. 79:55-92. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 26.Matthias, P. 1998. Lymphoid-specific transcription mediated by the conserved octamer site: who is doing what? Semin. Immunol. 10:155-163. [DOI] [PubMed] [Google Scholar]
- 27.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 28.Muljo, S. A., and M. S. Schlissel. 2002. The variable, C(H)1, C(H)2, and C(H)3 domains of Ig heavy chain are dispensable for pre-BCR function in transgenic mice. Int. Immunol. 14:577-584. [DOI] [PubMed] [Google Scholar]
- 29.Mullen, A. C., F. A. High, A. S. Hutchins, H. W. Lee, A. V. Villarino, D. M. Livingston, A. L. Kung, N. Cereb, T. P. Yao, S. Y. Yang, and S. L. Reiner. 2001. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science 292:1907-1910. [DOI] [PubMed] [Google Scholar]
- 30.Munshi, N., Y. Yie, M. Merika, K. Senger, S. Lomvardas, T. Agalioti, and D. Thanos. 1999. The IFN-beta enhancer: a paradigm for understanding activation and repression of inducible gene expression. Cold Spring Harbor Symp. Quant. Biol. 64:149-159. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, T. L., M. G. Cleveland, P. Kulesza, J. Magram, and K. M. Murphy. 1995. Regulation of interleukin 12 p40 expression through an NF-κB half-site. Mol. Cell. Biol. 15:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann, M., H. Fries, C. Scheicher, P. Keikavoussi, A. Kolb-Maurer, E. Brocker, E. Serfling, and E. Kampgen. 2000. Differential expression of Rel/NF-kappaB and octamer factors is a hallmark of the generation and maturation of dendritic cells. Blood 95:277-285. [PubMed] [Google Scholar]
- 33.Plevy, S. E., J. H. Gemberling, S. Hsu, A. J. Dorner, and S. T. Smale. 1997. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 17:4572-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Carrozzi, V. R., A. A. Nazarian, C. C. Li, S. L Gore, R. Sridharan, A. N. Imbalzano, and S. T. Smale. 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20:282-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjabi, S., A. Hoffmann, H. C. Liou, D. Baltimore, and S. T. Smale. 2000. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl. Acad. Sci. USA 97:12705-12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjabi, S., K. J. Williams, S. Saccani, L. Zhou, A. Hoffmann, G. Ghosh, S. Gerondakis, G. Natoli, and S. T. Smale. 2005. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 19:2138-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schubart, D. B., A. Rolink, M. H. Kosco-Vilbois, F. Botteri, and P. Matthias. 1996. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature 383:538-542. [DOI] [PubMed] [Google Scholar]
- 38.Smale, S. T., and A. G. Fisher. 2002. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 20:427-462. [DOI] [PubMed] [Google Scholar]
- 39.Spilianakis, C. G., and R. A. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5:1017-1027. [DOI] [PubMed] [Google Scholar]
- 40.Strubin, M., J. W. Newell, and P. Matthias. 1995. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 80:497-506. [DOI] [PubMed] [Google Scholar]
- 41.Takemoto, N., N. Koyano-Nakagawa, T. Yokota, N. Arai, S. Miyatake, and K. Arai. 1998. Th2-specific DNase I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int. Immunol. 10:1981-1985. [DOI] [PubMed] [Google Scholar]
- 42.Tone, Y., S. A. Thompson, J. M. Babik, K. F. Nolan, M. Tone, C. Raven, and H. Waldmann. 1996. Structure and chromosomal location of the mouse interleukin-12 p35 and p40 subunit genes. Eur. J. Immunol. 26:1222-1227. [DOI] [PubMed] [Google Scholar]
- 43.Treisman, R. 1992. The serum response element. Trends Biochem. Sci. 17:423-426. [DOI] [PubMed] [Google Scholar]
- 44.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133-146. [DOI] [PubMed] [Google Scholar]
- 45.Verrijzer, C. P., and P. C. Van der Vliet. 1993. POU domain transcription factors. Biochim. Biophys. Acta 1173:1-21. [DOI] [PubMed] [Google Scholar]
- 46.Wang, V. E., T. Schmidt, J. Chen, P. A. Sharp, and D. Tantin. 2004. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 24:1022-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wegner, M., D. W. Drolet, and M. G. Rosenfeld. 1993. POU-domain proteins: structure and function of developmental regulators. Curr. Opin. Cell Biol. 5:488-498. [DOI] [PubMed] [Google Scholar]
- 48.Weinmann, A. S., D. M. Mitchell, S. Sanjabi, M. N. Bradley, A. Hoffmann, H. C. Liou, and S. T. Smale. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2:51-57. [DOI] [PubMed] [Google Scholar]
- 49.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 50.Yeh, J. H., P. Lecine, J. A. Nunes, S. Spicuglia, P. Ferrier, D. Olive, and J. Imbert. 2001. Novel CD28-responsive enhancer activated by CREB/ATF and AP-1 families in the human interleukin-2 receptor alpha-chain locus. Mol. Cell. Biol. 21:4515-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yui, M. A., G. Hernandez-Hoyos, and E. V. Rothenberg. 2001. A new regulatory region of the IL-2 locus that confers position-independent transgene expression. J. Immunol. 166:1730-1739. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, L., A. A. Nazarian, and S. T. Smale. 2004. Interleukin-10 inhibits interleukin-12 p40 gene transcription by targeting a late event in the activation pathway. Mol. Cell. Biol. 24:2385-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, C., K. Gagnidze, J. H. Gemberling, and S. E. Plevy. 2001. Characterization of an activation protein-1-binding site in the murine interleukin-12 p40 promoter. Demonstration of novel functional elements by a reductionist approach. J. Biol. Chem. 276:18519-18528. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, C., K. Rao, H. Xiong, K. Gagnidze, F. Li, C. Horvath, and S. Plevy. 2003. Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J. Biol. Chem. 278:39372-39382. [DOI] [PubMed] [Google Scholar]