Abstract
The Sp/Krüppel-like factor (Sp/Klf) family is comprised of around 25 zinc finger transcription factors that recognize CACCC boxes and GC-rich elements. We have investigated basic Krüppel-like factor (Bklf/Klf3) and show that in erythroid tissues its expression is highly dependent on another family member, erythroid Krüppel-like factor (Eklf/Klf1). We observe that Bklf mRNA is significantly reduced in erythroid tissues from Eklf-null murine embryos. We find that Bklf is driven primarily by two promoters, a ubiquitously active GC-rich upstream promoter, 1a, and an erythroid downstream promoter, 1b. Transcripts from the two promoters encode identical proteins. Interestingly, both the ubiquitous and the erythroid promoter are dependent on Eklf in erythroid cells. Eklf also activates both promoters in transient assays. Experiments utilizing an inducible form of Eklf demonstrate activation of the endogenous Bklf gene in the presence of an inhibitor of protein synthesis. The kinetics of activation are also consistent with Bklf being a direct Eklf target. Chromatin immunoprecipitation assays confirm that Eklf associates with both Bklf promoters. Eklf is typically an activator of transcription, whereas Bklf is noted as a repressor. Our results support the hypothesis that feedback cross-regulation occurs within the Sp/Klf family in vivo.
The mammalian Sp/Krüppel-like factor (Sp/Klf) family consists of 9 Sp and 17 Klf subfamily proteins (78, 88). Sp/Klfs bind GC-rich elements and related CACCC sequences in DNA by means of three tandem C2H2 zinc fingers found at or near their C termini (27, 46, 75). While this zinc finger domain is highly conserved among family members (85), other regions of the proteins are not. Accordingly, although they recognize similar DNA-binding sites, some Sp/Klfs, such as erythroid Klf (Eklf)/Klf1, serve predominantly as activators (56); others, such as basic Klf (Bklf)/Klf3, are regarded as transcriptional repressors (84). It is possible, however, that many and perhaps all Sp/Klfs function as either activators or repressors, depending on promoter and cellular context (38). Sp/Klf proteins have been found to play critical roles in a diversity of biological processes (9, 21), including erythropoiesis (8, 53, 63, 68, 87, 91), adipogenesis (7, 58, 64), and carcinogenesis (10, 29, 61, 90).
Many Sp/Klfs, such as Sp1, are broadly expressed, whereas others, such as Eklf, exhibit more-limited expression patterns. Eklf is so named because its expression is restricted mainly to erythroid cells, but it is also detectable in macrophages and mast cells (52, 56). It is a potent transcriptional activator recognized for binding CACCC boxes and related sequences that fall into the general consensus sequence 5′-NCNCNCCCN-3′ (27). These motifs are found in the regulatory regions of many erythroid genes (27, 56, 71). The adult β-globin promoter, for instance, contains an Eklf recognition site that is critical for β-globin expression (23, 27). As a result, Eklf knockout mice die at around embryonic day 15 (E15) with a severe β-globin deficiency (63, 68).
Efforts have been made to rescue the Eklf-null phenotype by the overexpression of human Aγ-globin. However, despite a correction of the globin chain imbalance, the null mice still die in utero due to hemolysis (67). This result suggests that other critical Eklf target genes exist. Recently, several genes have been shown to be underexpressed in Eklf-null fetal liver (24, 33, 70). These include the genes for α-hemoglobin stabilizing protein (Ahsp), which binds and stabilizes free α-hemoglobin and prevents its cytotoxic precipitation, and dematin, a cytoskeletal protein required for membrane integrity in erythrocytes. Numerous other Eklf target genes have been proposed from studies of differential expression in Eklf-null fetal liver. However, in many cases, it is uncertain whether these are direct targets or whether they are genes that are indirectly influenced by hypoxia, incomplete erythroid maturation, or skewed cellular heterogeneity in the Eklf-null mice.
Work with physiologically relevant cell lines has been employed to circumvent these complexities. The B1.6 cell line (17), for instance, has allowed for the validation of Ahsp and dematin as direct targets of Eklf (33). This line was generated by immortalizing E14.5 Eklf-null fetal liver erythroid progenitors using the J2 retrovirus. The cells were then rescued using a transgene encoding Eklf fused to the ligand-binding domain of the estrogen receptor Eklf-ER (17, 50). A mutant form of the ER was used, so that the fusion would respond to the synthetic steroid tamoxifen but not to estrogen, which may be present in culture medium (50). The addition of tamoxifen results in the accumulation of Eklf-ER in the nucleus, the activation of Eklf target genes, and the subsequent hemoglobinization and terminal differentiation of the cells (17).
It has long been noted that there are several apparently abundant CACCC box-binding proteins present in erythroid cells, in addition to Eklf and the ubiquitous protein Sp1 (19, 32). A cDNA-encoding Bklf was isolated using probes specific for the zinc finger domain of Eklf and Sp1 and relaxed-stringency hybridization with a mouse erythroleukemia (MEL) cDNA library (20). Bklf is found in a wide range of tissues and cell types but is particularly abundant in hematopoietic tissue (20). Like Eklf, Bklf binds CACCC motifs in preference to GC boxes (20). Bklf has been shown to bind in vitro the promoter CACCC elements of many erythroid genes, such as adult β-globin, fetal Aγ-globin, Gata-1, carbonic anhydrase I, porphobilinogen deaminase (Pbgd), and pyruvate kinase (20, 66). The CACCC-binding activity of Bklf is more readily detected in yolk sac and fetal liver than that of Eklf (20), suggestive of a potential role in hematopoiesis. The phenotype of Bklf-null mice, however, is complex, consistent with Bklf being broadly expressed. Precise target genes and biological roles for the protein have been difficult to define (unpublished results).
Functionally, Bklf was initially shown to be a transcriptional activator. However, transactivation was observed only with high levels of Bklf and was not as strong as Eklf-mediated activation (20). Bklf has since been established as a potent transcriptional repressor in many systems (66, 84, 86). The protein is highly basic, hence the name basic Krüppel-like factor, and it includes a repression domain that contains a Pro-Val-Asp-Leu-Thr motif through which Bklf interacts with the corepressor C-terminal binding protein (84).
It has previously been noted that Bklf protein levels are reduced in the fetal liver (erythroid tissue) but are unaffected in the fetal brain (nonerythroid) in Eklf−/− mice (20, 68). Whether this is a direct or indirect effect has not previously been explored. We have now found that Bklf expression is driven from two promoters. The upstream promoter, 1a, is active in a wide range of tissues, while the downstream promoter, 1b, which gives rise to a novel Bklf transcript (but encodes an identical protein), is active predominantly in erythroid tissues. We find that transcripts from both promoters are underexpressed in Eklf-null fetal liver. Activation of both promoters by Eklf is observed for heterologous cells in transient transfection experiments. Chromatin immunoprecipitation experiments confirm that Eklf-ER directly binds to both promoters in B1.6 cells. The induction of Eklf-ER activates the endogenous downstream erythroid promoter 1b in the B1.6 line but, unexpectedly, no activation of the upstream promoter is observed with this system. The kinetics of the induction of promoter 1b and the observation that induction occurs in the presence of an inhibitor of protein synthesis further suggest that Bklf is a direct Eklf target gene.
We have also examined several other Klf genes and, interestingly, find that Tieg/Klf10 is also activated by Eklf-ER in B1.6 cells in the presence of a protein synthesis inhibitor and is underexpressed in Eklf-null fetal liver. These results reveal cross-regulation of the Klf subfamily in erythroid cells.
MATERIALS AND METHODS
Genotyping.
Genotyping of the Eklf-null mice was performed as described previously (33).
RNA extraction.
E14.5 fetal livers and tissue sections (up to 140 mg) from 13- to 16-week-old female C57BL/6 mice (68) were freshly homogenized for RNA extraction after washing with phosphate-buffered saline (Sigma, St. Louis, MO). Adult mice were sacrificed by cervical dislocation to avoid indirect effects caused by terminal anesthesia. Total RNA was extracted with TRI REAGENT (Sigma) per the supplier's protocol but with an additional centrifugation step at 12,000 × g for 10 min at 4°C following homogenization to reduce contamination by genomic DNA. RNA was then cleaned up with RNeasy kits (QIAGEN Pty. Ltd., Victoria, Australia) and was subsequently subjected to DNase treatment with a DNA-free kit (Ambion, Austin, TX) as instructed by the supplier.
Real-time RT-PCR.
Up to 5 μg of RNA was used as a template for cDNA synthesis primed by random hexamers with a SuperScript III First Strand cDNA Synthesis kit (Invitrogen, Carlsbad, CA). Quantitative real-time reverse transcription (RT)-PCR was performed with SYBR Green PCR Master Mix by using an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). Approximately 10 ng of cDNA, as estimated by UV spectrophotometry, was used in each RT-PCR. Samples were normalized with respect to 18S rRNA levels. Reactions containing serial dilutions of gel-purified amplicon were included in each RT-PCR run in order to construct standard curves for relative quantification. Minus RT and no-template reactions were included as negative controls. The analysis of real-time PCR data was done with ABI Prism 7000 sequence detection system software (version 1.1).
Real-time PCR primers.
Paired primers were designed with PrimerExpress software (Version 2.0) (Applied Biosystems) to cross, where possible, exon-exon boundaries to prevent amplification of any contaminating genomic DNA. Primer sequence specificity was checked using the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/BLAST) (4). For each primer set, optimal reaction concentrations, ranging between 200 nM and 600 nM, were determined. The sequences of forward and reverse primers used are as follows: 18S, 5′-CACGGCCGGTACAGTGAAAC-3′ (forward) and 5′-AGAGGAGCGAGCGACCAA-3′ (reverse); Bklf exons 4/5, 5′-GAAATGTCACCCCCTTTAATGAAC-3′ and 5′-CACGATGACGGAAGGATGGT-3′; Bklf exons 1a/2, 5′-CGGGCCTGGGTTTCTTG-3′ and 5′-GATCAAACATGAGCATCCTTTCAG-3′; Bklf exons 1b/2, 5′-GGTGGAATTCTGTTCAGGTCAAC-3′ and 5′-CCACGCCTTCTAGGGTGTTCT-3′; Klf10, 5′-GCAGCCAACCATGCTCAAC-3′ and 5′-CCCCTCTCTGGGCTTTTCAG-3′; Klf13, 5′-CGAGAAAGTTTACGGGAAATCTTC-3′ and 5′-CAGGCGAAAGGCCTCTCA-3′; and Klf16, 5′-TCACACCTGCGGACTCACA-3′ and 5′-CAGAACGGGCGAACTTCTTG-3′.
EMSAs.
Nuclear extracts were prepared and electrophoretic mobility shift assays (EMSAs) were carried out as described previously (20). Anti-Eklf and anti-Bklf antibodies have been described previously (20). Anti-Sp1 and anti-Sp3 antibodies were supplied by Santa Cruz Biotechnology, Santa Cruz, CA.
Oligonucleotides used in the synthesis of radiolabeled probes are as follows: mouse β-major globin CACCC probe, 5′-TAGAGCCACACCCTGGTAAG-3′ and 5′-CTTACCAGGGTGTGGCTCTA-3′; Bklf promoter 1b CACCC probe, 5′-AGTACTGGGTGTGGGCAGAATCTTATCTGAAGCT-3′ and 5′-AGCTTCAGATAAGATTCTGCCCACACCCAGTACT-3′.
5′ RACE.
One μg MEL total RNA was used as a template for first-strand cDNA synthesis with a SMART rapid amplification of cDNA ends (RACE) cDNA amplification kit (BD Biosciences, Palo Alto, CA) as instructed by the supplier. RACE PCR was performed as described in the Advantage 2 Polymerase kit manual (BD Biosciences) with an additional five repetitions of the final cycle. For each reaction, cDNA synthesized from ∼20 ng total RNA was used. 5′ RACE was primed with an oligonucleotide, 5′-TCCACCGGCTCCACCTGTATCCC-3′, specific for the third exon of Bklf. Confirmatory 5′ RACE PCR was conducted with a primer, 5′-GGGACTGGATCAAACATGAGCATCCTT-3′, specific for exon 2 of Bklf. 3′ RACE primers, specific for Bklf exon 1b, were as follows: 5′-ATTGCATCCCATCTGAAGCCAAGC-3′ and 5′-GAGAGGCACAGATTCGGAAATATCCG-3′. RACE products were resolved by 2% agarose gel electrophoresis and purified with Spin-X centrifuge tube filters (Trace Biosciences, Castle Hill, New South Wales, Australia). Products were cloned into the pGEM-T Easy vector (Promega Corporation, Madison, WI) and subjected to blue-white colony selection as instructed by the supplier. Automated sequencing was performed by the Automated DNA Sequencing Service, Sydney University Prince Alfred Macromolecular Analysis Centre.
Primer extension.
Primer extension was carried out as previously described (5). RNA from MEL and NIH 3T3 cells was primed with an oligonucleotide specific for Bklf exon 1a, 5′-CCATCGATGGCGGCCAAGAAACCCAGGCCCGGCT-3′, and was extended with avian myeloblastosis virus reverse transcriptase (Promega).
Cell culture.
MEL, NIH 3T3, and COS cells were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (low glucose) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum and 1% (vol/vol) penicillin, streptomycin, and glutamine solution (Gibco-BRL Life Technologies, Grand Island, NY). Drosophila melanogaster Schneider line 2 (SL-2) cells were grown at 24°C in Schneider medium (Gibco) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum and 1% (vol/vol) penicillin, streptomycin, and glutamine solution. B1.6 erythroblast cells were cultured as described previously (17). Cells were induced by the addition of 1 mM 4-hydroxytamoxifen (in ethanol) to a final concentration of 100 nM. As a negative control, ethanol was added to a final concentration of 0.0001% (vol/vol). RNA was extracted as described above at 48 h postinduction or at 0, 2, 4, 8, and 24 h postinduction for the time course study. For translation inhibition analyses, cycloheximide (made up in ethanol) was added to subconfluent cells to a final concentration of 5 μg/ml. Thirty minutes later, cells were induced with 4-hydroxytamoxifen or with ethanol as a negative control as described above. RNA was extracted 8 h after tamoxifen induction to avoid cell death due to cycloheximide cytotoxicity.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were conducted with αEklf and preimmune serum as previously reported (33, 34, 49). Immunoprecipitated DNA was analyzed by real-time PCR using the following primer sets: Bklf promoter 1a, 5′-GCTGCCTCCACCCAAGTG-3′ and 5′-ACCAACTCTGGGCAGTCACAT-3′; Bklf promoter 1b, 5′-CTGGGTGTGGGCAGAATCTT-3′ and 5′-GCCAGGGCGAGTCCAACT-3′; negative control region approximately 10 kb upstream of Bklf, 5′-GCCTGCGGGAGGTGATTAC-3′ and 5′-TTCTTGAAGCAAAGCCAAGAATATC-3′. β-major globin and nectin control primers have been described previously (33).
Vectors and cloning of the Bklf promoters.
To clone Bklf promoter 1a, a 129SVJ mouse genomic library in Lambda FIX II vector (Stratagene, La Jolla, CA) was screened as suggested by the supplier. An approximately 500-bp sequence encompassing a portion of intron 1 and of exon 2 of Bklf was radiolabeled with [α-32P]dCTP (1.7 × 106 cpm/ml) and was used to probe the library. A 2.3-kb NotI/NotI fragment containing exon 1a plus approximately 1.8 kb of upstream sequence and 400 bp of intron 1 was obtained from a positively identified clone and then subloned into pBlueScript KS II− (Stratagene) to create pBKS-Bklfprom1a. Two fragments (−189 to +89 and −532 to +89) were amplified from pBKS-Bklfprom1a by PCR with the forward primers 5′-ATTAGGTACCGGCTCCCAGCCTCTACTTACCC-3′ and 5′-ATTAGGTACCCGGAGCGCTCAGGCG-3′, respectively, and the reverse primer 5′-ATTAAAGCTTAACCCAGGCCCGGCTC-3′. The fragments were then subcloned into KpnI/HindIII pGL4.10[luc2] (Promega), a promoterless vector, to create pGL4.10[luc2]-Bklfprom1a(−189+89) and pGL4.10[luc2]-Bklfprom1a(−532+89). To clone Bklf promoter 1b, 286-bp (−225 to +61), 190-bp (−129 to +61), 182-bp (−121 to +61) and 96-bp (−35 to +61) fragments were amplified from C57BL/6 genomic DNA (supplied by Richard Pearson, School of Molecular and Microbial Biosciences, Sydney, Australia) by PCR using the forward primers 5′-ATTAGGTACCTTGGAAAGCTGGAGAGAGTC-3′, 5′-ATTAGGTACCGGGTGTGGGCAGAATC-3′, 5′-ATTAGGTACCGCAGAATCTTATCTGAAGCTATG-3′, and 5′-ATTAGGTACCGCAGAAAAGTAAAATTGGGTG-3′, respectively, and the reverse primer 5′-ATTAAAGCTTTTCATTGCTTGGCTTCAGATG-3′. The fragments were subcloned into KpnI/HindIII pGL4.10[luc2] to form pGL4.10[luc2]-Bklfprom1b(−225+61), pGL4.10[luc2]-Bklfprom1b(−129+61), pGL4.10[luc2]- Bklfprom1b(−121+61), and pGL4.10[luc2]-Bklfprom1b(−35+61). pPac and pPac-Eklf were kindly provided by Menie Merika and Stuart Orkin (Harvard Medical School, Boston, MA). pSG5-Eklf was generously supplied by James Bieker (Mount Sinai School of Medicine, New York, NY).
Transactivation assays.
SL-2 cells were transfected in six-well plates using the calcium phosphate method (72). Zero, 10, 50 or 100 ng pPac-Eklf (supplemented to a total 100-ng vector with pPac) was transfected along with 1 μg pGL4.10[luc2], pGL4.10[luc2]-Bklfprom1a(−189+89), pGL4.10[luc2]-Bklfprom1a(−532+89), pGL4.10[luc2]-Bklfprom1b(−225+61), pGL4.10[luc2]-Bklfprom1b(−129+61), pGL4.10[luc2]-Bklfprom1b(−121+61), or pGL4.10[luc2]-Bklfprom1b(−35+61). For all transfections, 10 ng pGL4.74[hRluc/TK] (Promega) was included as a transfection control. Forty-eight hours posttransfection, cells were lysed and assayed for luciferase activity by using a dual-luciferase reporter assay system (Promega). MEL cells were transfected in six-well plates with 1 μg of the pGL4.10[luc2] constructs and 10 ng pGL4.74[hRluc/TK] and were harvested 48 h later. Transfection was achieved with Tfx-50 reagent (Promega) per the manufacturer's instructions. In all cases, reporter activity was normalized with respect to Renilla luciferase levels.
Overexpression of Eklf in COS cells.
Plates (100 mm) of COS cells, at 50% confluence, were transfected with 5 μg pSG5-Eklf (or 5 μg pMT3 as a negative control) using FuGENE6 (Roche Diagnostics) as instructed by the manufacturer. The medium was changed 48 h following transfection, and cells were harvested for nuclear extracts 24 h later as described previously (20).
Bioinformatics.
GenBank cDNA and genomic searches were conducted using the BLAST algorithm (4) at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST). Full-length Bklf cDNAs from the DDBJ and GenBank were obtained from nucleotide searches (August 2006) at the NCBI website (http://www.ncbi.nlm.nih.gov). Additional start points of transcription were identified (August 2006) from oligonucleotide-cap cDNA entries at the Database of Transcriptional Start Sites (DBTSS; http://dbtss.hgc.jp/). Vertebrate conservation analyses were performed with the University of California—Santa Cruz (UCSC) Genome Browser (February 2006; http://genome.ucsc.edu/) using the mouse (mm8) genome assembly (44).
Nucleotide sequence accession number.
A full-length Bklf cDNA entry containing exon 1b has been submitted to GenBank (accession number DQ981866).
RESULTS
Bklf mRNA is reduced in Eklf-null cells.
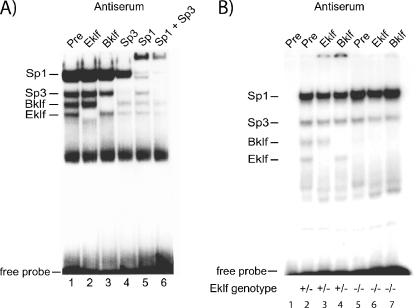
Several studies have employed a probe containing the critical CACCC box of the β-major globin promoter in EMSAs to elucidate DNA-binding proteins in erythroid cells (20, 32). The major CACCC-binding activities detected include those of Sp1, Sp3, Eklf, and Bklf (Fig. 1A). While the antibodies for Sp1 and Sp3 produce some nonspecific inhibition of binding of other proteins, the annotation of Sp1 and Sp3 in Fig. 1A is consistent with previous identifications of these prevalent CACCC-binding factors (12, 31). Interestingly, when E14.5 Eklf−/− fetal liver extracts are examined, the DNA-binding activity of Bklf, but not that of Sp1 or Sp3, is reduced (Fig. 1B). This result is in accordance with previous observations, including Western blot experiments, that show that Bklf protein levels are diminished in Eklf-null fetal liver but not in nonerythroid tissues (20). These results raise the possibility that Bklf is directly or indirectly dependent on Eklf for its expression, stability, or activity in erythroid cells.
FIG. 1.
Bklf protein is diminished in Eklf-null murine fetal liver. (A) EMSA showing the identity of proteins found in erythroid cells which recognize the β-major globin promoter CACCC box. MEL cell nuclear extracts were allowed to bind to a radiolabeled probe containing the β-major globin promoter CACCC box in the presence of either preimmune serum or an antibody specific for Eklf, Bklf, Sp1, and/or Sp3. (B) EMSA analysis of murine fetal liver nuclear extracts prepared from E14.5 Eklf-null and heterozygous (denoted below each lane) littermates. Lane 1 contains the β-major globin CACCC probe alone. Bklf and Eklf bands were confirmed by antibody supershifting. All lanes that lack either Eklf- or Bklf-specific antibody (αEklf or αBklf) contain preimmune serum as a control.
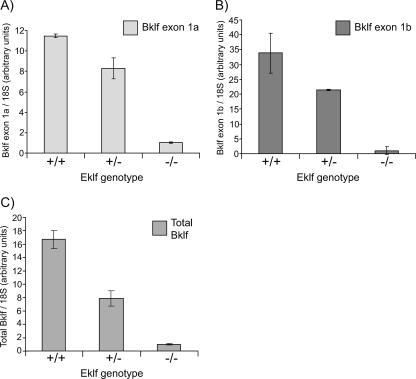
Eklf is known to be a potent activator of transcription. We first investigated whether Bklf's dependence on Eklf is observed at the mRNA level. RNA from E14.5 livers of Eklf−/−, Eklf+/−, and wild-type murine fetuses was analyzed by real-time RT-PCR using Bklf specific primers. Bklf transcripts were underrepresented in Eklf−/− fetal livers compared to the fetal livers of Eklf+/− and wild-type littermates (see Fig. 5C). In accordance with this result, a recent microarray study on genes that are differentially expressed in Eklf−/− fetal liver also identified Bklf as such a downregulated gene, although this result was not further validated (33). In addition, Bklf mRNA is diminished in Eklf+/− compared to wild-type fetal liver, suggestive of dose dependency. Taken together, these data suggest that Eklf is required for the direct or indirect activation of the Bklf gene at the transcriptional level.
FIG. 5.
Both Bklf promoters are less active in E14.5 Eklf−/− murine fetal liver. Analysis of the two Bklf transcripts in E14.5 littermate Eklf+/+, Eklf+/−, and Eklf−/− fetal livers was performed by quantitative real-time RT-PCR. (A) Bklf exon 1a transcripts. (B) Bklf exon 1b transcripts. (C) Total Bklf transcripts: Bklf-specific primers were designed to amplify a region spanning the exon 4/5 boundary. Bklf mRNA levels were normalized to 18S rRNA levels.
Identification of a novel Bklf leader exon, 1b.
Bklf's expression is highly dependent on Eklf in erythroid tissue, yet Bklf mRNA is also expressed in other tissues, for instance, the fetal head and the adult lung, where Eklf is not abundant (20). Many genes have multiple promoters that allow their expression in a variety of different biological contexts (6). We hypothesized that Bklf may utilize several promoters to achieve its broad expression profile.
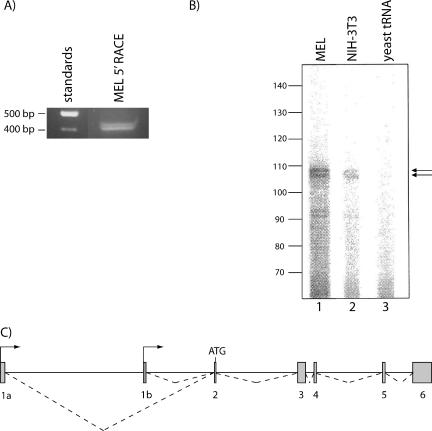
To explore this possibility, we used 5′ RACE with a primer targeted to exon 3 of Bklf. Two distinct fragments were amplified from MEL cell RNA (Fig. 2A) and were subsequently cloned into pGEM-T Easy vector and sequenced. The two fragments were identical, with the exception that one contained an additional 31 base pairs at its 5′ end (Fig. 3). As anticipated, this fragment contained a portion of exon 3 (up to the RACE primer) and exon 2, but it also contained an alternative first exon which differed from the previously published leader exon of Bklf (RefSeq accession number NM_008453; 42) (Fig. 3). A BLAST analysis of the mouse genome revealed that this novel first exon, hereby named exon 1b, lies within the first intron of Bklf (Fig. 2C). 3′ RACE confirmed that transcripts containing exon 1b extend to include all the exons of the gene (data not shown). A USCS Genome Browser analysis of the putative promoter region immediately upstream of this novel exon revealed a high level of conservation across a wide range of divergent vertebrates including the chicken (Gallus gallus) and the frog (Xenopus tropicalis) (data not shown). This result supports the view that the novel promoter (1b) is of functional importance. The two identified promoters (1a and 1b) generate transcripts that contain a single variant leader exon (exon 1a or 1b) and five common downstream exons. The AUG start codon lies within exon 2; hence, both transcripts encode the same Bklf protein. The sequence of this novel Bklf cDNA was submitted to GenBank.
FIG. 2.
Characterization of the transcriptional start sites of the alternative exons of Bklf. (A) 5′ RACE from MEL cDNA generates two distinct bands which represent transcripts containing different lengths of the novel Bklf exon 1b. (B) Primer extension of Bklf mRNA using a primer specific for Bklf exon 1a. Lanes 1 and 2 contain primer-extended total RNA from MEL cells and NIH 3T3 cells, respectively. Lane 3 contains yeast tRNA as a negative control. Arrows indicate the two major start points of transcription for Bklf exon 1a. (C) A schematic of the murine Bklf locus showing the relative positions of the two alternative first exons, 1a and 1b. Exons are denoted by gray boxes. The ATG start codon is also shown.
FIG. 3.
The sequence of Bklf exon 1a (A) and exon 1b (B) and their flanking genomic regions. Note that approximately 8.3 kb of intron 1 has been omitted. Exons are written in boldface type, and their transcriptional start sites, as determined by results shown in Fig. 2A and B, are denoted by triangles. For each exon, the 5′-most position of these start sites has been designated position +1. For exon 1b, an upstream transcriptional start site that was infrequently detected by 5′ RACE cloning is indicated with a boldface underline. For exon 1a, other putative transcription start sites are underlined. These transcription start sites were inferred from full-length cDNA entries in GenBank (DDBJ accession numbers AK007959 [42], AK157576, AK143838, and AK010713 [14, 41]; GenBank accession numbers BC116938 and BC119214 [77]) and from queries of the DBTSS (clone names BY338164, BY724103, BY707959, BY742654, BY732683, BY732370, BY188922, BY062971, and BB628801 [45, 80]). Motifs that precisely fit the Eklf binding consensus of 5′-NCNCNCCC-3′ (or its reverse complement) are boxed. The putative Gata-binding site in promoter 1b is underlined.
We carried out additional experiments to further define the start points of transcription. 5′ RACE investigations using a number of Bklf-specific primers consistently yielded fragments exhibiting the two exon 1b putative transcription start sites shown in Fig. 3 (data not shown). A single clone was identified, however, which contained an additional 64 base pairs of noncoding 5′ sequence, suggestive of a weak upstream transcriptional start site (Fig. 3).
Despite the frequent detection of 5′ RACE products containing exon 1b, no products were found to contain the previously identified first exon (1a), which is present in the majority of Bklf cDNAs lodged at the DDBJ and GenBank. Their absence is possibly due to the high GC content (approximately 77%) of exon 1a compared to that of exon 1b (approximately 46%), which the RACE polymerase may have difficulty reading through. Therefore, we employed primer extension, using a primer specific for exon 1a, to determine the start sites of transcription for this upstream promoter. In this way, two major transcription start sites were identified (Fig. 2B and Fig. 3). An analysis of cDNA entries lodged at DDBJ, GenBank, and the DBTSS revealed multiple other proximal start sites (Fig. 3). This result suggests that promoter 1a is a typical GC-rich promoter that exhibits numerous start sites of transcription.
Bklf promoter 1b is active in erythroid tissues.
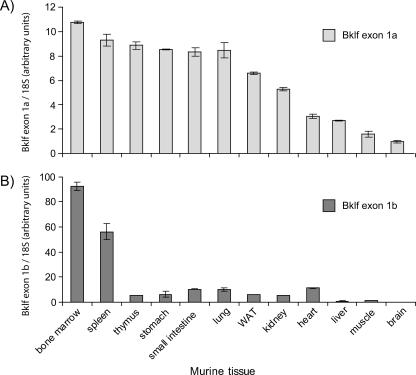
To investigate whether the two identified Bklf promoters have distinct expression patterns, we examined the levels of mRNA they produced in a range of adult murine tissues. Real-time RT-PCR primers were designed to amplify regions crossing the exon 1a/2 and exon 1b/2 boundaries, respectively. This allowed discrimination between the two different transcripts and also reduced the amplification of any trace amounts of contaminating genomic DNA. Transcripts from promoter 1a were found ubiquitously (Fig. 4A). In contrast, transcripts containing exon 1b were found at high levels in the bone marrow and spleen (Fig. 4B). Interestingly, Eklf's expression is restricted to these two hematopoietic organs (56). This result raised the possibility that promoter 1b might be dependent on Eklf and may account for the reduction in Bklf mRNA observed with the Eklf−/− fetal livers. Quantitative assays using known amounts of amplicon as template suggest that levels of the two Bklf transcripts are roughly equivalent in erythroid tissue (data not shown).
FIG. 4.
The downstream Bklf promoter is active predominantly in hematopoietic tissue. The levels of the two Bklf transcripts in murine tissues were determined by quantitative real-time RT-PCR. (A) Levels of Bklf transcripts generated from the upstream promoter and containing exon 1a. (B) Levels of Bklf mRNA arising from the downstream promoter and containing exon 1b. All values were normalized to 18S rRNA levels. WAT, white adipose tissue.
Bklf promoters 1a and 1b are dependent on Eklf in erythroid cells.
Given the restricted expression of transcripts containing exon 1b (Fig. 4B), we reasoned that promoter 1b might be highly dependent on Eklf and that its inactivity might contribute to the reduction in Bklf protein levels in the Eklf−/− fetal liver. We found exon 1b transcripts at approximately 30-fold lower levels in Eklf−/− than in wild-type fetal livers (Fig. 5B). Interestingly, transcripts containing exon 1a that arise from the ubiquitous promoter were found at approximately 10-fold lower levels in the null fetal livers (Fig. 5A). This suggests that both promoter 1b and, to a lesser extent, promoter 1a are activated by Eklf. It should be noted, however, that Eklf is clearly not crucial for expression from promoter 1a, as evidenced by its ubiquitous activity (Fig. 4A).
Eklf activates both Bklf promoters in transactivation assays.
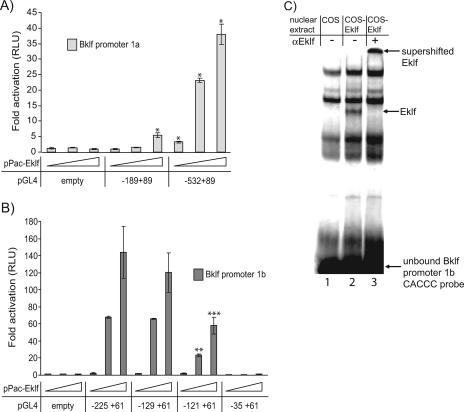
To further explore the possibility that Eklf can activate transcription from both promoters, we first performed transactivation experiments with Drosophila SL-2 cells, which are often used to examine Klf proteins since they lack CACCC-binding activity compared to mammalian cells (18). Fragments of both Bklf promoters were cloned into the promoterless reporter construct pGL4.10[luc2]. Bklf promoter 1a is GC rich and contains many elements that fit the Eklf-binding consensus sequence of 5′-NCNCNCCCN-3′ (Fig. 3). A 278-bp proximal promoter region from −189 to +89 that contains several canonical Eklf recognition sites was activated slightly in the presence of Eklf (Fig. 6A). An extended promoter element (−532 to +89) showed higher activation by Eklf, thus suggesting that CACCC boxes upstream of −189 contribute to Eklf activation (Fig. 6A). We also tested promoter 1b and found that the region −225 to +61 is potently activated by Eklf (Fig. 6B). This region contains three canonical Eklf recognition sites. A truncated fragment (−129 to +61) which retains all of these CACCC sites is activated by Eklf to a similar degree (Fig. 6B). In particular, one of the potential Eklf-binding sites (−123 to −129) is identical to the critical β-major globin promoter CACCC box, which is bound by Eklf in vivo. Deleting this motif results in a >50% reduction in promoter activity in response to Eklf (Fig. 6B, compare the −129 to +61 construct with the −121 to +61 construct). In addition, EMSA analysis demonstrated that Eklf can bind this Bklf promoter 1b CACCC box (Fig. 6C). Lastly, a further truncated promoter construct (−35 to +61), which contains only one of the three canonical Eklf-binding sites, was found not to be activated by Eklf (Fig. 6B). This implies that the 3′-most CACCC box (+3 to +9) is not sufficient for Eklf activation of Bklf promoter 1b. In addition, all promoter 1a and 1b constructs that displayed Eklf-mediated activation in SL-2 cells were also active in MEL cells (data not shown); the MEL cell line expresses Eklf (Fig. 1A). Taken together, these results corroborate the data presented here that both Bklf promoters, particularly promoter 1b, are responsive to Eklf.
FIG. 6.
Eklf transactivation assays of the Bklf promoters in SL-2 cells. Zero, 10, 50, or 100 ng pPac-Eklf (supplemented to 100 ng with pPac) was cotransfected with 1 μg pGL4.10[luc2] reporter vector containing fragments of Bklf promoter 1a (−189 to +89 or −532 to +89) (A) or varied lengths (−225 to +61, −129 to +61, −121 to +61, or −35 to +61) of Bklf promoter 1b (B). Firefly luciferase activity has been expressed as activation above that observed for cells transfected with each reporter vector and pPac alone. (A and B) Cell lysates were assayed for firefly luciferase activity 48 h posttransfection. Reporter activity has been normalized to Renilla luciferase levels. *, P < 0.05 compared to pGL4.10[luc2]-empty cotransfected with 10, 50, or 100 ng pPac-Eklf as appropriate (paired Student's t tests). **, P < 0.0005 and ***, P < 0.05 compared to pGL4.10[luc2]-Bklfprom1b(−129+61) cotransfected with 50 ng and 100 ng pPac-Eklf, respectively (paired Student's t tests). (C) EMSA analysis demonstrating that Eklf binds in vitro a CACCC box present in Bklf promoter 1b. Nuclear extracts were obtained from mock-transfected (lane 1) and Eklf-overexpressing (lanes 2 to 3) COS cells. Nuclear extracts were allowed to bind to a radiolabeled probe containing the Bklf promoter 1b CACCC box in the presence of either anti-Eklf (lane 3) or preimmune serum (lanes 1 and 2). RLU, relative light units.
Eklf-ER activates the endogenous Bklf promoter 1b in B1.6 cells.
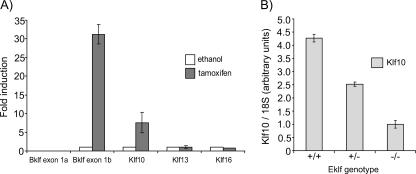
To investigate the dependence of Bklf on Eklf in a more physiological system, the B1.6 cell line was used. The B1.6 line is an erythroid cell line derived from Eklf-null knockout mice into which a transgene encoding a tamoxifen-inducible Eklf-ER protein has been introduced (17, 50). Real-time RT-PCR analysis demonstrated that, prior to activation of Eklf-ER by tamoxifen, transcripts from promoter 1a were undetectable, while mRNA from promoter 1b was at a trace level (Fig. 7A). Following induction, transcripts from promoter 1a remained undetected while promoter 1b was activated strongly. cDNA levels of readily detectable Klfs, such as Klf13 and Klf16, which were used as controls, displayed no change upon induction. This result further provides strong evidence that promoter 1b is activated, either directly or indirectly, by Eklf. Promoter 1a, on the other hand, appears to be nonresponsive to Eklf-ER in this cellular system.
FIG. 7.
Bklf exon 1b and Klf10 transcripts are upregulated in the presence of Eklf. (A) Klf mRNA levels in B1.6 cells that were harvested 48 h following induction with either 100 nM tamoxifen or with ethanol (0.0001%) as a control. (B) Klf10 transcript levels in Eklf+/+, Eklf+/−, and Eklf−/− fetal livers. All mRNA levels were determined by quantitative real-time RT-PCR and normalized to 18S rRNA levels.
We also examined other Klfs. Interestingly, Klf10 showed an approximately eightfold increase in expression upon induction (Fig. 7A). This result corroborates a previous microarray observation that Klf10 levels increase upon Eklf-ER induction (33). Klf10 transcripts were also found to be reduced approximately fourfold in Eklf-null compared to wild-type fetal liver (Fig. 7B). This result further suggests that Klf10 is also regulated by Eklf in eythroid cells, although not to the extent that is observed for Bklf (compare Fig. 5).
Eklf-ER activates Bklf promoter 1b in the presence of cycloheximide.
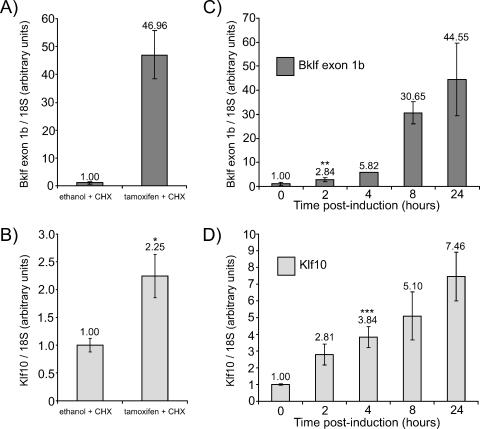
It was possible that Eklf-ER was indirectly activating Bklf via some intermediate downstream targets. We investigated this possibility by exposing B1.6 cells to cycloheximide, a translation inhibitor, prior to and during induction with tamoxifen. Due to the cytotoxicity of cycloheximide, cells were harvested 8 h (rather than 48 h) after induction. Activation of promoter 1b occurred in induced cells in the presence of cycloheximide, suggesting that the Bklf gene is a direct target of Eklf (Fig. 8A). In addition, Klf10 mRNA was also upregulated by Eklf-ER in the presence of cycloheximide, although to a lesser degree than Bklf, thus suggesting that it too is a direct target of Eklf (Fig. 8B).
FIG. 8.
Bklf and Klf10 are directly activated by Eklf-ER. (A and B) B1.6 cells were exposed to cycloheximide (CHX) 30 min before induction with tamoxifen and were harvested for RNA 8 h thereafter. Bklf exon 1b levels (A) and Klf10 mRNA levels (B) were determined by quantitative real-time RT-PCR. (C and D) B1.6 cells were harvested at numerous time points after tamoxifen induction in the absence of cycloheximide. Again, Bklf exon 1b levels (C) and Klf10 mRNA levels (D) were determined by quantitative real-time RT-PCR. In all cases, 18S rRNA levels were used for normalization. *, **, and ***, P < 0.05 (paired Student's t test, compared to ethanol control or 0-h time point as appropriate).
We also performed a time course study of Bklf and Klf10 mRNA levels following induction. It was expected that indirect target genes would exhibit a considerable lag phase before being expressed. However, statistically significant increases in Bklf exon 1b and Klf10 transcripts were detected after only 2 and 4 h postinduction, respectively, thus providing further evidence that the Bklf and Klf10 genes are direct targets of Eklf (Fig. 8C and D).
Eklf-ER binds endogenous Bklf promoters 1a and 1b in B1.6 cells.
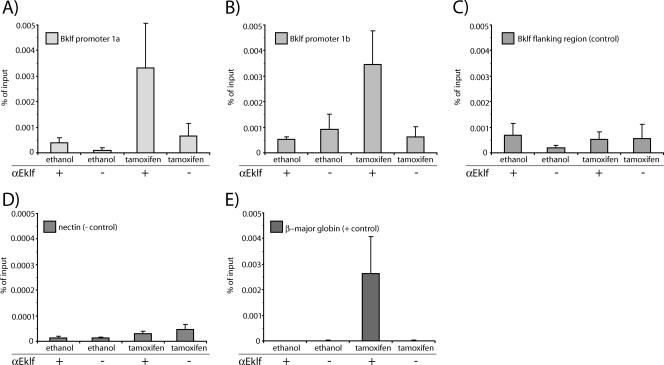
To confirm that Eklf activates Bklf by direct binding, we conducted ChIP assays using αEklf antiserum to detect the presence or absence of Eklf-ER at the Bklf promoters. ChIP material was obtained from uninduced and tamoxifen-induced B1.6 cells. As a positive control, Eklf-ER was detected at the β-major globin promoter, a known target of Eklf (Fig. 9E). Upon induction of the cells, Eklf-ER was found to associate with Bklf promoter 1b. No Eklf-ER was detected in negative control regions: the nectin gene and a region 10 kb upstream of the Bklf locus (Fig. 9B to D). Interestingly, Eklf-ER was also detected at Bklf promoter 1a following induction of the cells (Fig. 9A). This was somewhat unexpected given that no Bklf transcripts from promoter 1a were observed in induced B1.6s (Fig. 9A). Nonetheless, it is consistent with the previous observation that transcripts containing Bklf exon 1a are reduced in Eklf-null fetal liver (Fig. 5A).
FIG. 9.
Eklf ChIP at the two Bklf promoters. ChIP material was extracted from B1.6 cells that were induced with tamoxifen or ethanol (control). Chromatin was immunoprecipitated using an Eklf antibody (denoted by “+”) or preimmune serum (−). Primers used for real-time PCR quantification were targeted against Bklf promoter 1a (A), Bklf promoter 1b (B), a control region 10 kb upstream of the Bklf locus (C), the nectin promoter (D), and the β-major globin promoter (E).
DISCUSSION
Many erythroid genes utilize alternative promoters to achieve complex expression patterns (81). In particular, several genes involved in the heme biosynthesis pathway, including ALAD, PBGD, and UROS, possess an upstream housekeeping promoter that enables widespread expression and a downstream promoter that is active in erythroid cells (2, 3, 16, 43). Similarly, the genes encoding the transcription factors p45 Nf-e2 and Gata-2 each have dual promoters with housekeeping and erythroid roles (57, 59, 65). We have shown here that murine Bklf is another erythroid gene that is regulated by at least two promoters that exhibit different expression profiles but produce transcripts encoding identical proteins.
Transcripts containing exon 1a were readily detected by RT-PCR in all tissues examined, thus suggesting that the upstream promoter (1a) is widely active. In this sense, it can be considered a housekeeping promoter that allows Bklf expression in both erythroid and nonerythroid cells and tissues. Promoter 1a is GC rich, a trait which is characteristic of promoters which generate ubiquitous transcripts (15, 73, 93). In addition, oligonucleotide-cap cDNA analysis reveals that promoter 1a has multiple transcriptional start sites spread over a region of approximately 300 bp. It is well established that most genes exhibit some fluctuation in their start points of transcription (79). However, this phenomenon has been noted to be particularly pronounced for GC-rich, housekeeping promoters that lack a TATA box, as is the case for Bklf promoter 1a (13, 15).
In contrast, the novel downstream promoter 1b is active primarily in hematopoietic tissues, such as the fetal liver, adult spleen, and bone marrow and in erythroid cells, such as the tamoxifen-induced B1.6 erythroblast line and MEL cells. In contrast to transcripts containing exon 1a, mRNA containing exon 1b was barely detected in tissues such as the liver and skeletal muscle and was undetected in the brain. This result suggests that, unlike promoter 1a, promoter 1b does not exhibit a widespread basal level of activity. Consistent with its restricted activity, promoter 1b is noticeably less GC rich than promoter 1a and does not resemble a typical housekeeping promoter. It lacks a canonical TATA box, although it does have an AT-rich element (TAAAAT) at −26 to −21. In addition, promoter 1b contains a motif that perfectly fits the consensus of a downstream promoter element (+28 A/G G A/T C/T G/A/C +32) (47).
In concordance with its erythroid cell-restricted activity, we have demonstrated here that Bklf promoter 1b is directly activated by Eklf-ER in the B1.6 erythroid cell line. It is also highly dependent on Eklf in murine fetal liver. This dependence presumably accounts for the expression of transcripts containing exon 1b being restricted to hematopoietic tissues, such as the spleen and bone marrow, where Eklf is known to be present (56). A 190-bp region of promoter 1b (−129 to +61) is strongly activated by Eklf in reporter assays with SL-2 cells. This region contains three elements that fit the Eklf-binding consensus. One of these elements (centered around −126) is identical to the proximal CACCC box of the β-major globin promoter (32), albeit on the opposite strand, and is bound by Eklf in vitro. Deletion of this CACCC box leads to a significant reduction in the activation of the promoter by Eklf. Interestingly, in Bklf promoter 1b, this CACCC box is flanked by a motif that fits the Gata-binding consensus of 5′-A/T GATA A/G-3′ (25, 54, 89). Gata-1 has previously been noted to physically interact with Eklf and to synergistically potentiate transcriptional activation (55).
The ubiquitously active promoter 1a also appears to be dependent on Eklf in erythroid cells. Exon 1a transcripts are markedly reduced in the Eklf−/− fetal liver. In B1.6 cells, Eklf-ER is detected at promoter 1a by ChIP analysis; however, no resultant transcripts in B1.6 cells are detected. It is possible that other factors that are required for promoter 1a activity are absent in the cells, perhaps as a side effect of the immortalization process. While the −189 to +89 fragment of promoter 1a displays only mild activation by Eklf in SL-2 cells, an extended region (−532 to +89) is strongly activated. This result suggests that CACCC boxes between −532 and −189 contribute significantly to Eklf activation of Bklf promoter 1a.
Despite its dependence on Eklf in erythroid cells, Bklf promoter 1a is also comparably active in nonerythroid tissues where Eklf is absent, such as lung, gastrointestinal, and white adipose tissues. This implies that in nonerythroid tissues, promoter 1a may be bound by factors which are capable of substituting for Eklf and which are sufficient to activate transcription. Indeed, it is possible that other members of the Klf family may utilize Eklf's cognate binding site(s) in promoter 1a and thus drive Bklf expression in nonerythroid tissues. Within the family, lung Krüppel-like factor (Lklf/Klf2) and gut-enriched Krüppel-like factor (Gklf/Klf4) are the most highly related to Eklf (9, 21, 38, 78, 85) and, hence, these two factors are suitable candidates to assume such a compensatory role. This hypothesis is interesting and raises the possibility that promoter 1a and, by inference, other typical GC-rich housekeeping promoters, may be ubiquitously expressed by virtue of being dependent on some tissue-restricted Klfs in some tissues and other Klfs in other tissues, rather than being dependent exclusively on ubiquitous GC-box-binding proteins, such as Sp1. However, if compensation by other Klfs is occurring, the question of why promoter 1b is not similarly active in nonerythroid tissues is raised. A possible explanation for this is that promoter 1b may contain sequence elements other than its Eklf-binding site(s) which are bound exclusively by erythroid factors which, in turn, are crucial for transcriptional activation, for instance, Gata-1.
Eklf has been found to function predominantly as an activator, whereas Bklf is generally a repressor. Both proteins have also been shown to bind in vitro to an overlapping set of CACCC elements found in erythroid promoters, such as those of the β-globin, carbonic anhydrase I, Gata-1, Pbgd, and pyruvate kinase genes as well as in globin locus control region DNase hypersensitive sites (20). Therefore, given the opposing roles of the proteins in transcriptional regulation, the most likely consequence of Eklf's activation of Bklf is that it serves to temper Eklf's stimulatory effects. That is to say that Bklf upregulation inhibits the unbridled activation of Eklf's target genes. In support of this idea, reporter assays conducted with SL-2 cells have shown that Bklf can silence Eklf activation of a CACCC-containing promoter (84). There is also evidence that Bklf promoter 1b is itself subject to autorepression. In B1.6 cells, Bklf exon 1b levels are higher in the cycloheximide-treated cells than in untreated cells (compare Fig. 8A and C at the 8-h time point). This is one possible consequence of unchecked Eklf-mediated activation.
Such transcriptional antagonism has been observed among members of the Klf family in reporter assays using a number of promoters, including those of the Ap-2α (36), cytochrome p4501A1 (Cyp1a1) (35, 39, 40, 95), and cyclin D1 (48, 60, 74, 96) genes. In particular, Klf4 and Klf5 have been shown to exhibit opposing effects at a number of promoters in transient transfection experiments. Klf4 represses while Klf5 activates the smooth muscle SM α-actin (51) and SM22-α (1) promoters as well as the cyclin D1 (60, 74) and laminin-α1 (69) promoters. It is possible that, similar to Klf4 and Klf5, Eklf and Bklf may demonstrate contrasting functions in order to fine-tune the expression of a set of common target genes. Future studies will endeavor to identify target genes of Bklf which are either shared with Eklf or which are distinct, due to subtle differences in the binding preferences of the two transcription factors.
Further evidence of cross-regulation within the Klf family comes from the observation in this study that Eklf also upregulates Klf10 expression in the fetal liver and in tamoxifen-treated B1.6 erythroblasts. Klf10 is a negative regulator of cell cycle progression (11, 37, 82). Its induction is therefore consistent with the reduced proliferation of B1.6 cells observed after Eklf-ER induction. Interestingly, the human KLF10 gene contains two alternative promoters (26). The upstream promoter is widely active, while the function of the downstream promoter is unclear (26). It is possible that these alternative promoters are conserved in the mouse and that one or both may be responsive to Eklf.
The results presented here provide evidence that the Klf family is subject to internal hierarchical regulation, with Eklf activating the Bklf and Klf10 genes in erythroid cells. Other examples of cross-regulation and autoregulation have also been inferred from transient transfection experiments and work with cell lines. Klf8 binds and represses the Klf4 promoter (92). In addition, reporter assays have demonstrated that Klf4 is able to activate its own promoter, while Klf5 represses it (22). Further indirect evidence of hierarchical regulation comes from observations of concomitant changes in Klf expression during lymphocyte differentiation. For example, Klf2, Bklf, and Klf4 are expressed in resting B lymphocytes and are downregulated upon activation through the B-cell receptor (28, 30, 94). Similarly, Eklf, Klf2, and Bklf are expressed in quiescent T cells and are downregulated following activation (83, 94). Another member of the KLF family, KLF13, is also regulated during T-cell activation, albeit at a posttranscriptional level. As such, KLF13 protein, but not mRNA, is upregulated during T-cell activation (62, 76). Taken together with the results presented here, these studies allude to a complex web of hierarchical regulation which presumably functions to finely adjust the expression patterns of downstream target genes.
The widespread expression of Bklf, combined with potential redundancy by other members of the Klf family, has complicated interpretations of the Bklf-null mice data (not shown). However, although determination of the biological role of Bklf has been difficult, the results presented here provide strong in vivo evidence that Bklf is dependent on the erythroid transcriptional activator Eklf. Eklf has been shown to be crucial for erythropoiesis and directly binds to and regulates numerous erythroid promoters, including that of the adult β-globin gene. The observation that Bklf has two Eklf-dependent promoters, of which one appears to be active predominantly in erythroid tissues, strongly suggests that Bklf performs an erythroid role in vivo. Investigations on whether Bklf represses genes activated by Eklf in vivo are continuing.
Acknowledgments
This work was supported by NIH grant NHLBI HL073443 and grants from the Australian ARC and NHMRC to M.C. A.F. is supported by an Australian Postgraduate Award.
We are grateful to Menie Merika and Stuart Orkin for providing pPac and pPac-Eklf and to James Bieker for supplying pSG5-Eklf. We also thank Hannah Nicholas, Richard Pearson, and Alexis Verger for their advice in preparing the manuscript. We express our gratitude to Dale Hancock, Denise Hodge, Richard Pearson, Nancy Sue, and Jane van Vliet for experimental support.
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Adam, P. J., C. P. Regan, M. B. Hautmann, and G. K. Owens. 2000. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J. Biol. Chem. 275:37798-37806. [DOI] [PubMed] [Google Scholar]
- 2.Aizencang, G., C. Solis, D. F. Bishop, C. Warner, and R. J. Desnick. 2000. Human uroporphyrinogen-III synthase: genomic organization, alternative promoters, and erythroid-specific expression. Genomics 70:223-231. [DOI] [PubMed] [Google Scholar]
- 3.Aizencang, G. I., D. F. Bishop, D. Forrest, K. H. Astrin, and R. J. Desnick. 2000. Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. J. Biol. Chem. 275:2295-2304. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 6.Ayoubi, T. A., and W. J. Van De Ven. 1996. Regulation of gene expression by alternative promoters. FASEB J. 10:453-460. [PubMed] [Google Scholar]
- 7.Banerjee, S. S., M. W. Feinberg, M. Watanabe, S. Gray, R. L. Haspel, D. J. Denkinger, R. Kawahara, H. Hauner, and M. K. Jain. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 278:2581-2584. [DOI] [PubMed] [Google Scholar]
- 8.Basu, P., P. E. Morris, J. L. Haar, M. A. Wani, J. B. Lingrel, K. M. Gaensler, and J. A. Lloyd. 2005. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood 106:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 10.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 11.Blok, L. J., M. E. Grossmann, J. E. Perry, and D. J. Tindall. 1995. Characterization of an early growth response gene, which encodes a zinc finger transcription factor, potentially involved in cell cycle regulation. Mol. Endocrinol. 9:1610-1620. [DOI] [PubMed] [Google Scholar]
- 12.Bouwman, P., H. Gollner, H. P. Elsasser, G. Eckhoff, A. Karis, F. Grosveld, S. Philipsen, and G. Suske. 2000. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 19:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 14.Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, R. Kodzius, K. Shimokawa, V. B. Bajic, S. E. Brenner, S. Batalov, A. R. Forrest, M. Zavolan, M. J. Davis, L. G. Wilming, V. Aidinis, J. E. Allen, A. Ambesi-Impiombato, R. Apweiler, R. N. Aturaliya, T. L. Bailey, M. Bansal, L. Baxter, K. W. Beisel, T. Bersano, H. Bono, A. M. Chalk, K. P. Chiu, V. Choudhary, A. Christoffels, D. R. Clutterbuck, M. L. Crowe, E. Dalla, B. P. Dalrymple, B. de Bono, G. Della Gatta, D. di Bernardo, T. Down, P. Engstrom, M. Fagiolini, G. Faulkner, C. F. Fletcher, T. Fukushima, M. Furuno, S. Futaki, M. Gariboldi, P. Georgii-Hemming, T. R. Gingeras, T. Gojobori, R. E. Green, S. Gustincich, M. Harbers, Y. Hayashi, T. K. Hensch, N. Hirokawa, D. Hill, L. Huminiecki, M. Iacono, K. Ikeo, A. Iwama, T. Ishikawa, M. Jakt, A. Kanapin, M. Katoh, Y. Kawasawa, J. Kelso, H. Kitamura, H. Kitano, G. Kollias, S. P. Krishnan, A. Kruger, S. K. Kummerfeld, I. V. Kurochkin, L. F. Lareau, D. Lazarevic, L. Lipovich, J. Liu, S. Liuni, S. McWilliam, M. Madan Babu, M. Madera, L. Marchionni, H. Matsuda, S. Matsuzawa, H. Miki, F. Mignone, S. Miyake, K. Morris, S. Mottagui-Tabar, N. Mulder, N. Nakano, H. Nakauchi, P. Ng, R. Nilsson, S. Nishiguchi, S. Nishikawa, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559-1563. [DOI] [PubMed] [Google Scholar]
- 15.Carninci, P., A. Sandelin, B. Lenhard, S. Katayama, K. Shimokawa, J. Ponjavic, C. A. Semple, M. S. Taylor, P. G. Engstrom, M. C. Frith, A. R. Forrest, W. B. Alkema, S. L. Tan, C. Plessy, R. Kodzius, T. Ravasi, T. Kasukawa, S. Fukuda, M. Kanamori-Katayama, Y. Kitazume, H. Kawaji, C. Kai, M. Nakamura, H. Konno, K. Nakano, S. Mottagui-Tabar, P. Arner, A. Chesi, S. Gustincich, F. Persichetti, H. Suzuki, S. M. Grimmond, C. A. Wells, V. Orlando, C. Wahlestedt, E. T. Liu, M. Harbers, J. Kawai, V. B. Bajic, D. A. Hume, and Y. Hayashizaki. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38:626-635. [DOI] [PubMed] [Google Scholar]
- 16.Chretien, S., A. Dubart, D. Beaupain, N. Raich, B. Grandchamp, J. Rosa, M. Goossens, and P. H. Romeo. 1988. Alternative transcription and splicing of the human porphobilinogen deaminase gene result either in tissue-specific or in housekeeping expression. Proc. Natl. Acad. Sci. USA 85:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coghill, E., S. Eccleston, V. Fox, L. Cerruti, C. Brown, J. Cunningham, S. Jane, and A. Perkins. 2001. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 97:1861-1868. [DOI] [PubMed] [Google Scholar]
- 18.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 19.Crossley, M., and S. H. Orkin. 1993. Regulation of the beta-globin locus. Curr. Opin. Genet. Dev. 3:232-237. [DOI] [PubMed] [Google Scholar]
- 20.Crossley, M., E. Whitelaw, A. Perkins, G. Williams, Y. Fujiwara, and S. H. Orkin. 1996. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 16:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang, D. T., J. Pevsner, and V. W. Yang. 2000. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 32:1103-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang, D. T., W. Zhao, C. S. Mahatan, D. E. Geiman, and V. W. Yang. 2002. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 30:2736-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donze, D., T. M. Townes, and J. J. Bieker. 1995. Role of erythroid Kruppel-like factor in human gamma- to beta-globin gene switching. J. Biol. Chem. 270:1955-1959. [DOI] [PubMed] [Google Scholar]
- 24.Drissen, R., M. von Lindern, A. Kolbus, S. Driegen, P. Steinlein, H. Beug, F. Grosveld, and S. Philipsen. 2005. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans, T., M. Reitman, and G. Felsenfeld. 1988. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc. Natl. Acad. Sci. USA 85:5976-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fautsch, M. P., A. Vrabel, M. Subramaniam, T. E. Hefferen, T. C. Spelsberg, and E. D. Wieben. 1998. TGFbeta-inducible early gene (TIEG) also codes for early growth response alpha (EGRalpha): evidence of multiple transcripts from alternate promoters. Genomics 51:408-416. [DOI] [PubMed] [Google Scholar]
- 27.Feng, W. C., C. M. Southwood, and J. J. Bieker. 1994. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493-1500. [PubMed] [Google Scholar]
- 28.Fruman, D. A., G. Z. Ferl, S. S. An, A. C. Donahue, A. B. Satterthwaite, and O. N. Witte. 2002. Phosphoinositide 3-kinase and Bruton's tyrosine kinase regulate overlapping sets of genes in B lymphocytes. Proc. Natl. Acad. Sci. USA 99:359-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghaleb, A. M., M. O. Nandan, S. Chanchevalap, W. B. Dalton, I. M. Hisamuddin, and V. W. Yang. 2005. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 15:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glynne, R., G. Ghandour, J. Rayner, D. H. Mack, and C. C. Goodnow. 2000. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol. Rev. 176:216-246. [DOI] [PubMed] [Google Scholar]
- 31.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartzog, G. A., and R. M. Myers. 1993. Discrimination among potential activators of the beta-globin CACCC element by correlation of binding and transcriptional properties. Mol. Cell. Biol. 13:44-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodge, D., E. Coghill, J. Keys, T. Maguire, B. Hartmann, A. McDowall, M. Weiss, S. Grimmond, and A. Perkins. 2006. A global role for EKLF in definitive and primitive erythropoiesis. Blood 107:3359-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Im, H., J. A. Grass, H. M. Christensen, A. Perkins, and E. H. Bresnick. 2002. Histone deacetylase-dependent establishment and maintenance of broad low-level histone acetylation within a tissue-specific chromatin domain. Biochemistry 41:15152-15160. [DOI] [PubMed] [Google Scholar]
- 35.Imataka, H., K. Sogawa, K. Yasumoto, Y. Kikuchi, K. Sasano, A. Kobayashi, M. Hayami, and Y. Fujii-Kuriyama. 1992. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 11:3663-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imhof, A., M. Schuierer, O. Werner, M. Moser, C. Roth, R. Bauer, and R. Buettner. 1999. Transcriptional regulation of the AP-2α promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol. Cell. Biol. 19:194-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnsen, S. A., M. Subramaniam, R. Janknecht, and T. C. Spelsberg. 2002. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 21:5783-5790. [DOI] [PubMed] [Google Scholar]
- 38.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaczynski, J., J. S. Zhang, V. Ellenrieder, A. Conley, T. Duenes, H. Kester, B. van Der Burg, and R. Urrutia. 2001. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J. Biol. Chem. 276:36749-36756. [DOI] [PubMed] [Google Scholar]
- 40.Kaczynski, J. A., A. A. Conley, M. Fernandez Zapico, S. M. Delgado, J. S. Zhang, and R. Urrutia. 2002. Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem. J. 366:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564-1566. [DOI] [PubMed] [Google Scholar]
- 42.Kawai, J., A. Shinagawa, K. Shibata, M. Yoshino, M. Itoh, Y. Ishii, T. Arakawa, A. Hara, Y. Fukunishi, H. Konno, J. Adachi, S. Fukuda, K. Aizawa, M. Izawa, K. Nishi, H. Kiyosawa, S. Kondo, I. Yamanaka, T. Saito, Y. Okazaki, T. Gojobori, H. Bono, T. Kasukawa, R. Saito, K. Kadota, H. Matsuda, M. Ashburner, S. Batalov, T. Casavant, W. Fleischmann, T. Gaasterland, C. Gissi, B. King, H. Kochiwa, P. Kuehl, S. Lewis, Y. Matsuo, I. Nikaido, G. Pesole, J. Quackenbush, L. M. Schriml, F. Staubli, R. Suzuki, M. Tomita, L. Wagner, T. Washio, K. Sakai, T. Okido, M. Furuno, H. Aono, R. Baldarelli, G. Barsh, J. Blake, D. Boffelli, N. Bojunga, P. Carninci, M. F. de Bonaldo, M. J. Brownstein, C. Bult, C. Fletcher, M. Fujita, M. Gariboldi, S. Gustincich, D. Hill, M. Hofmann, D. A. Hume, M. Kamiya, N. H. Lee, P. Lyons, L. Marchionni, J. Mashima, J. Mazzarelli, P. Mombaerts, P. Nordone, B. Ring, M. Ringwald, I. Rodriguez, N. Sakamoto, H. Sasaki, K. Sato, C. Schonbach, T. Seya, Y. Shibata, K. F. Storch, H. Suzuki, K. Toyo-oka, K. H. Wang, C. Weitz, C. Whittaker, L. Wilming, A. Wynshaw-Boris, K. Yoshida, Y. Hasegawa, H. Kawaji, S. Kohtsuki, and Y. Hayashizaki. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409:685-690. [DOI] [PubMed] [Google Scholar]
- 43.Kaya, A. H., M. Plewinska, D. M. Wong, R. J. Desnick, and J. G. Wetmur. 1994. Human delta-aminolevulinate dehydratase (ALAD) gene: structure and alternative splicing of the erythroid and housekeeping mRNAs. Genomics 19:242-248. [DOI] [PubMed] [Google Scholar]
- 44.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura, K., A. Wakamatsu, Y. Suzuki, T. Ota, T. Nishikawa, R. Yamashita, J. Yamamoto, M. Sekine, K. Tsuritani, H. Wakaguri, S. Ishii, T. Sugiyama, K. Saito, Y. Isono, R. Irie, N. Kushida, T. Yoneyama, R. Otsuka, K. Kanda, T. Yokoi, H. Kondo, M. Wagatsuma, K. Murakawa, S. Ishida, T. Ishibashi, A. Takahashi-Fujii, T. Tanase, K. Nagai, H. Kikuchi, K. Nakai, T. Isogai, and S. Sugano. 2006. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klevit, R. E. 1991. Recognition of DNA by Cys2, His2 zinc fingers. Science 253:1367-1393. [DOI] [PubMed] [Google Scholar]
- 47.Kutach, A. K., and J. T. Kadonaga. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20:4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laub, F., R. Aldabe, V. Friedrich, Jr., S. Ohnishi, T. Yoshida, and F. Ramirez. 2001. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev. Biol. 233:305-318. [DOI] [PubMed] [Google Scholar]
- 49.Letting, D. L., C. Rakowski, M. J. Weiss, and G. A. Blobel. 2003. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 23:1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, Y., S. Sinha, and G. Owens. 2003. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J. Biol. Chem. 278:48004-48011. [DOI] [PubMed] [Google Scholar]
- 52.Luo, Q., X. Ma, S. M. Wahl, J. J. Bieker, M. Crossley, and L. J. Montaner. 2004. Activation and repression of interleukin-12 p40 transcription by erythroid Kruppel-like factor in macrophages. J. Biol. Chem. 279:18451-18456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumoto, N., A. Kubo, H. Liu, K. Akita, F. Laub, F. Ramirez, G. Keller, and S. L. Friedman. 2006. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 107:1357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merika, M., and S. H. Orkin. 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merika, M., and S. H. Orkin. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minegishi, N., J. Ohta, N. Suwabe, H. Nakauchi, H. Ishihara, N. Hayashi, and M. Yamamoto. 1998. Alternative promoters regulate transcription of the mouse GATA-2 gene. J. Biol. Chem. 273:3625-3634. [DOI] [PubMed] [Google Scholar]
- 58.Mori, T., H. Sakaue, H. Iguchi, H. Gomi, Y. Okada, Y. Takashima, K. Nakamura, T. Nakamura, T. Yamauchi, N. Kubota, T. Kadowaki, Y. Matsuki, W. Ogawa, R. Hiramatsu, and M. Kasuga. 2005. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 280:12867-12875. [DOI] [PubMed] [Google Scholar]
- 59.Moroni, E., T. Mastrangelo, R. Razzini, L. Cairns, P. Moi, S. Ottolenghi, and B. Giglioni. 2000. Regulation of mouse p45 NF-E2 transcription by an erythroid-specific GATA-dependent intronic alternative promoter. J. Biol. Chem. 275:10567-10576. [DOI] [PubMed] [Google Scholar]
- 60.Nandan, M. O., H. S. Yoon, W. Zhao, L. A. Ouko, S. Chanchevalap, and V. W. Yang. 2004. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene 23:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narla, G., K. E. Heath, H. L. Reeves, D. Li, L. E. Giono, A. C. Kimmelman, M. J. Glucksman, J. Narla, F. J. Eng, A. M. Chan, A. C. Ferrari, J. A. Martignetti, and S. L. Friedman. 2001. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563-2566. [DOI] [PubMed] [Google Scholar]
- 62.Nikolcheva, T., S. Pyronnet, S. Y. Chou, N. Sonenberg, A. Song, C. Clayberger, and A. M. Krensky. 2002. A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J. Clin. Investig. 110:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316-318. [DOI] [PubMed] [Google Scholar]
- 64.Oishi, Y., I. Manabe, K. Tobe, K. Tsushima, T. Shindo, K. Fujiu, G. Nishimura, K. Maemura, T. Yamauchi, N. Kubota, R. Suzuki, T. Kitamura, S. Akira, T. Kadowaki, and R. Nagai. 2005. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1:27-39. [DOI] [PubMed] [Google Scholar]
- 65.Pan, X., N. Minegishi, H. Harigae, H. Yamagiwa, M. Minegishi, Y. Akine, and M. Yamamoto. 2000. Identification of human GATA-2 gene distal IS exon and its expression in hematopoietic stem cell fractions. J. Biochem. (Tokyo) 127:105-112. [DOI] [PubMed] [Google Scholar]
- 66.Perdomo, J., A. Verger, J. Turner, and M. Crossley. 2005. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 25:1549-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perkins, A. C., K. R. Peterson, G. Stamatoyannopoulos, H. E. Witkowska, and S. H. Orkin. 2000. Fetal expression of a human Agamma globin transgene rescues globin chain imbalance but not hemolysis in EKLF null mouse embryos. Blood 95:1827-1833. [PubMed] [Google Scholar]
- 68.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318-322. [DOI] [PubMed] [Google Scholar]
- 69.Piccinni, S. A., A. L. Bolcato-Bellemin, A. Klein, V. W. Yang, M. Kedinger, P. Simon-Assmann, and O. Lefebvre. 2004. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J. Biol. Chem. 279:9103-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pilon, A. M., D. G. Nilson, D. Zhou, J. Sangerman, T. M. Townes, D. M. Bodine, and P. G. Gallagher. 2006. Alterations in expression and chromatin configuration of the alpha hemoglobin-stabilizing protein gene in erythroid Kruppel-like factor-deficient mice. Mol. Cell. Biol. 26:4368-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raich, N., and P. H. Romeo. 1993. Erythroid regulatory elements. Stem Cells 11:95-104. [DOI] [PubMed] [Google Scholar]
- 72.Sambrook, J., E. K. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 73.Schug, J., W. P. Schuller, C. Kappen, J. M. Salbaum, M. Bucan, and C. J. Stoeckert, Jr. 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shie, J. L., Z. Y. Chen, M. Fu, R. G. Pestell, and C. C. Tseng. 2000. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 28:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shields, J. M., and V. W. Yang. 1998. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 26:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song, A., Y. F. Chen, K. Thamatrakoln, T. A. Storm, and A. M. Krensky. 1999. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity 10:93-103. [DOI] [PubMed] [Google Scholar]
- 77.Strausberg, R. L., E. A. Feingold, L. H. Grouse, J. G. Derge, R. D. Klausner, F. S. Collins, L. Wagner, C. M. Shenmen, G. D. Schuler, S. F. Altschul, B. Zeeberg, K. H. Buetow, C. F. Schaefer, N. K. Bhat, R. F. Hopkins, H. Jordan, T. Moore, S. I. Max, J. Wang, F. Hsieh, L. Diatchenko, K. Marusina, A. A. Farmer, G. M. Rubin, L. Hong, M. Stapleton, M. B. Soares, M. F. Bonaldo, T. L. Casavant, T. E. Scheetz, M. J. Brownstein, T. B. Usdin, S. Toshiyuki, P. Carninci, C. Prange, S. S. Raha, N. A. Loquellano, G. J. Peters, R. D. Abramson, S. J. Mullahy, S. A. Bosak, P. J. McEwan, K. J. McKernan, J. A. Malek, P. H. Gunaratne, S. Richards, K. C. Worley, S. Hale, A. M. Garcia, L. J. Gay, S. W. Hulyk, D. K. Villalon, D. M. Muzny, E. J. Sodergren, X. Lu, R. A. Gibbs, J. Fahey, E. Helton, M. Ketteman, A. Madan, S. Rodrigues, A. Sanchez, M. Whiting, A. Madan, A. C. Young, Y. Shevchenko, G. G. Bouffard, R. W. Blakesley, J. W. Touchman, E. D. Green, M. C. Dickson, A. C. Rodriguez, J. Grimwood, J. Schmutz, R. M. Myers, Y. S. Butterfield, M. I. Krzywinski, U. Skalska, D. E. Smailus, A. Schnerch, J. E. Schein, S. J. Jones, and M. A. Marra. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 99:16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suske, G., E. Bruford, and S. Philipsen. 2005. Mammalian SP/KLF transcription factors: bring in the family. Genomics 85:551-556. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki, Y., H. Taira, T. Tsunoda, J. Mizushima-Sugano, J. Sese, H. Hata, T. Ota, T. Isogai, T. Tanaka, S. Morishita, K. Okubo, Y. Sakaki, Y. Nakamura, A. Suyama, and S. Sugano. 2001. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki, Y., R. Yamashita, S. Sugano, and K. Nakai. 2004. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 32:D78-D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan, J. S., N. Mohandas, and J. G. Conboy. 2006. High frequency of alternative first exons in erythroid genes suggests a critical role in regulating gene function. Blood 107:2557-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tau, K. R., T. E. Hefferan, K. M. Waters, J. A. Robinson, M. Subramaniam, B. L. Riggs, and T. C. Spelsberg. 1998. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology 139:1346-1353. [DOI] [PubMed] [Google Scholar]
- 83.Teague, T. K., D. Hildeman, R. M. Kedl, T. Mitchell, W. Rees, B. C. Schaefer, J. Bender, J. Kappler, and P. Marrack. 1999. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc. Natl. Acad. Sci. USA 96:12691-12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turner, J., and M. Crossley. 1999. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci. 24:236-240. [DOI] [PubMed] [Google Scholar]
- 86.Turner, J., H. Nicholas, D. Bishop, J. M. Matthews, and M. Crossley. 2003. The LIM protein FHL3 binds basic Kruppel-like factor/Kruppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J. Biol. Chem. 278:12786-12795. [DOI] [PubMed] [Google Scholar]
- 87.Van Loo, P. F., P. Bouwman, K. W. Ling, S. Middendorp, G. Suske, F. Grosveld, E. Dzierzak, S. Philipsen, and R. W. Hendriks. 2003. Impaired hematopoiesis in mice lacking the transcription factor Sp3. Blood 102:858-866. [DOI] [PubMed] [Google Scholar]
- 88.van Vliet, J., L. A. Crofts, K. G. Quinlan, R. Czolij, A. C. Perkins, and M. Crossley. 2006. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics 87:474-482. [DOI] [PubMed] [Google Scholar]
- 89.Wall, L., E. deBoer, and F. Grosveld. 1988. The human beta-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev. 2:1089-1100. [DOI] [PubMed] [Google Scholar]
- 90.Wang, X., and J. Zhao. 2007. KLF8 transcription factor participates in oncogenic transformation. Oncogene 26:456-461. [DOI] [PubMed] [Google Scholar]
- 91.Wani, M. A., R. T. Means, Jr., and J. B. Lingrel. 1998. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 7:229-238. [DOI] [PubMed] [Google Scholar]
- 92.Wei, H., X. Wang, B. Gan, A. M. Urvalek, Z. K. Melkoumian, J. L. Guan, and J. Zhao. 2006. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J. Biol. Chem. 281:16664-16671. [DOI] [PubMed] [Google Scholar]
- 93.Yamashita, R., Y. Suzuki, S. Sugano, and K. Nakai. 2005. Genome-wide analysis reveals strong correlation between CpG islands with nearby transcription start sites of genes and their tissue specificity. Gene 350:129-136. [DOI] [PubMed] [Google Scholar]
- 94.Yusuf, I., and D. A. Fruman. 2003. Regulation of quiescence in lymphocytes. Trends Immunol. 24:380-386. [DOI] [PubMed] [Google Scholar]
- 95.Zhang, W., J. M. Shields, K. Sogawa, Y. Fujii-Kuriyama, and V. W. Yang. 1998. The gut-enriched Kruppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J. Biol. Chem. 273:17917-17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao, J., Z. C. Bian, K. Yee, B. P. Chen, S. Chien, and J. L. Guan. 2003. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol. Cell 11:1503-1515. [DOI] [PubMed] [Google Scholar]