Abstract
Phenotypic modulation of vascular smooth muscle cells (SMCs) in the blood vessel wall from a differentiated to a proliferative state during vascular injury and inflammation plays an important role in restenosis and atherosclerosis. Matrix metalloproteinase 9 (MMP9) is a member of the MMP family of proteases, which participate in extracellular matrix degradation and turnover. MMP9 is upregulated and required for SMC migration during the development of restenotic and atherosclerotic lesions. In this study, we show that FoxO4 activates transcription of the MMP9 gene in response to tumor necrosis factor alpha (TNF-α) signaling. Inhibition of FoxO4 expression by small interfering RNA or gene knockout reduces the abilities of SMCs to migrate in vitro and inhibit neointimal formation and MMP9 expression in vivo. We further show that both the N-terminal, Sp1-interactive domain and the C-terminal transactivation domain of FoxO4 are required for FoxO4-activated MMP9 transcription. TNF-α signaling upregulates nuclear FoxO4. Our studies place FoxO4 in the center of a transcriptional regulatory network that links gene transcription required for SMC remodeling to upstream cytokine signals and implicate FoxO4 as a potential therapeutic target for combating proliferative arterial diseases.
Phenotypic modulation of vascular smooth muscle cells (SMCs) from a quiescent, contractile phenotype to a proliferative one in response to physiological and pathological stimuli plays an important role in vascular development and remodeling during disease (15, 16, 23). This form of phenotypic change involves migration of SMCs from the medial layer of the blood vessel wall to the intimal layer and requires a family of matrix metalloproteinases (MMPs) (20).
There are several MMPs, including MMP2 (gelatinase A), MMP3 (stromelysin-1), and MMP9 (gelatinase B), as well as tissue inhibitors of MMPs (TIMPs) present in human vasculature (reviewed in reference 20). In normal human and experimental animal arteries, MMP2, TIMP1, and TIMP2 are constitutively expressed at levels providing a stable balance between endogenous matrix production and matrix degradation. Under pathological conditions, such as in restenosis and atherosclerosis, the expression of MMP3 and MMP9 is upregulated. MMP9 is primarily produced by SMCs and macrophages in vascular lesions and has multiple functions during phenotypic modulation of SMCs. MMP9 and MMP2 degrade basement membrane components, including type IV collagen, laminin, and elastin, allowing SMCs to migrate from the medial layer to the intimal layer (reviewed in reference 20). Degradation of extracellular matrix by MMP9 can also release and activate latent growth factors and cytokines bound to extracellular matrix components (17), which in turn further promote phenotypic changes of SMCs. MMP9-deficient mice have reduced neointima formation in an animal model of restenosis due to a defect in SMC migration (10). Atherosclerotic apoE-null mice lacking MMP9 have smaller atherosclerotic lesions containing fewer macrophages and less collagen than plaques from wild-type apoE-null mice (14). MMP9 is also implicated in destabilizing late atherosclerotic plaques (20).
Upregulation of MMP9 is triggered by inflammatory cytokines and growth factors released at sites of tissue damage and inflammation. Tumor necrosis factor alpha (TNF-α) is expressed in atherosclerotic lesions and is a known migration factor for SMCs and a potent activator of MMP9 transcription (18, 26, 32). The signaling pathways and transcriptional mechanisms that regulate MMP9 expression in response to TNF-α in cardiovascular diseases are the subject of intense research but remain elusive.
FoxO4 (AFX) is a member of the transcription factor Forkhead box O (FoxO) family, which also includes FoxO1 (FKHR), FoxO3 (FKHRL1), and FoxO6 in mice (2, 19, 29, 30). FoxO proteins are emerging as critical transcriptional integrators among pathways regulating differentiation, proliferation, metabolism, cancer, survival, and life span. FoxO proteins share common functions that require their DNA binding activity. These include induction of cell cycle arrest and apoptosis, promotion of long-term survival of quiescent cells, and activation of genes involved in DNA damage repair and detoxification in response to stress stimuli. FoxO proteins also regulate expression of downstream targets by binding to other transcription factors, acting as either coactivators or corepressors (13, 29).
Previously, we have found that FoxO4 inhibits SMC differentiation through interaction with the transcription factor myocardin (13). In this paper, we show that FoxO4 plays an additional role in promoting SMC migration. We have identified a novel transcriptional target of FoxO4, MMP9. FoxO4 activates MMP9 transcription in response to TNF-α through a mechanism that requires an Sp1 DNA binding site in the promoter of the MMP9 gene. We show that inactivation of Foxo4 inhibits the abilities of vascular SMCs to migrate in vitro and reduces neointimal formation in an animal model of restenosis. TNF-α signaling upregulates nuclear FoxO4. Our studies place FoxO4 in the center of a transcriptional regulatory network linking cytokine signals to changes in gene expression required for SMC remodeling. Since MMP9 is a key mediator of extracellular matrix remodeling during the development of restenotic and atherosclerotic lesions, wound healing after myocardial infarction, and cancer metastasis, our results suggest a potential role for FoxO4 as a therapeutic target for combating proliferative arterial diseases and cancer.
MATERIALS AND METHODS
Plasmids.
The mammalian expression vectors of FoxO4, FoxO1, and various deletion mutants were described previously (13). The MMP9-luciferase reporter construct was made by subcloning PCR-amplified inserts corresponding to the MMP9 promoter sequence from rat genomic DNA into the pGL3-Basic vector (Promega). More-detailed information about the plasmids used in this study is available upon request.
siRNA.
The Foxo4-specific small interfering RNA (siRNA) and control green fluorescent protein (GFP) siRNA were described previously (13). SMART pool Foxo4 siRNA was purchased from Dharmacon (Dharmacon, Chicago, IL). SMCs were transfected with siRNA duplex at a concentration of 50 nM, using DharmaFECT 3, following the manufacturer's protocols. COS cells were transfected with various concentrations of siRNA, using Lipofectamine 2000.
SMC migration assays in culture.
Two-dimensional cell migration was analyzed with rat aortic SMCs transfected with control GFP siRNA or Foxo4 siRNA duplex for 24 h, using a scratch wound assay. Cells were fixed and stained with Hoechst (Sigma) 19 h after the wounding. The furthest distance that cells migrated from the wound edge was measured (with an average of five independent microscope fields used for each of the three independent experiments). For mouse primary aortic cells, the scratch wound assay was performed as described above and cells were kept in culture in the presence or absence of TNF-α (12 ng/ml) and human recombinant MMP9 (50 ng/ml; Anaspec). Nineteen hours after the wounding, cells were fixed and photographed using light microscopy.
Three-dimensional cell migration was determined using transwells with a gelatin-coated membrane, following the manufacturer's procedure (Corning Life Science). The lower chamber contained either no chemoattractant (control) or 100 ng/ml of TNF-α in Dulbecco's modified Eagle's medium (DMEM)-F-12 medium. After 6 h, cells were scraped from the upper surface, the membrane was fixed with formalin, and cells were stained with hematoxylin and eosin stain and analyzed using light microscopy to count cell numbers (with an average of four randomly chosen fields used for each of the three independent experiments).
Nuclear and cytosolic fractionation of the cell lysates.
Cultured SMCs (5 × 106 cells) were washed, scraped, and pelleted in phosphate-buffered saline after 1 h stimulation with or without either TNF-α or insulin-like growth factor-I (IGF-I). Subsequent cytosolic and nuclear extraction of the lysate was carried out with a ProteoExtract subcellular-proteome-extraction kit, following the manufacturer's protocol with modification (Calbiochem). After cytosolic fractionation (500 μl), the cell pellet containing nuclear proteins was resuspended in 50 μl nuclear lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, 10 mM MgCl, 2 mM dithiothreitol, 50 mM β-glycerophosphate, 2 mM sodium orthovanadate, 50 mM NaF, 5 mM EGTA, and 1× protease inhibitor cocktail [Roche]).
Zymography.
Zymography was performed with aliquots of either conditioned medium or protein extracts from carotid arteries as indicated for the experiments, following published protocols (Chemicon). SDS-polyacrylamide gel electrophoresis (PAGE) gel (7.5%) containing 0.1% gelatin A was used.
Cell culture, transfection, and luciferase assays.
Rat aortic SMCs were a gift from Gary Owens (University of Virginia). SMCs were cultured in DMEM-F-12 medium supplemented with 10% fetal bovine serum and penicillin-streptomycin and used in passages 13 through 23. COS and C2C12 (ATCC) cells were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, and penicillin-streptomycin (Invitrogen). For experiments under serum-free conditions, SMCs were cultured in DMEM-F-12 medium with a combination of vehicle, TNF-α, and IGF-I as indicated for the experiments. Mouse primary aortic SMCs were isolated from aorta of postnatal day 1 mice, following published protocols (25). The purity of the SMC culture was above 95%, as judged by smooth muscle α-actin staining.
COS or SMC cells were transfected with combinations of plasmids indicated for each experiment, using Fugene 6 reagent according to the manufacturer's instructions (Roche). After transfection, cells were cultured as described for each experiment. Cell extracts were assayed for luciferase expression, using a luciferase assay kit (Promega). Relative promoter activities were expressed as luminescence relative units normalized for cotransfected β-galactosidase expression in the cell extracts.
ChIP and real-time PCR analysis.
Chromatin immunoprecipitation (ChIP) assays were carried out as described previously (13). Immunoprecipitated chromatin fragments were quantified by real-time PCR using SYBR green and normalized against total-input genomic DNA. Primer sequences used for PCR analysis are available upon request.
Immunoprecipitation and Western blotting.
COS cells were transfected with a combination of Myc-tagged FoxO1 or FoxO4 and Flag-Sp1 as indicated for each experiment. Twenty-four hours after transfection, cells were used for immunoprecipitation, following a procedure described previously (13), using appropriate antibodies as indicated for the experiments. Western blotting was performed according to standard protocols. Antibodies were purchased from Sigma (Flag), Santa Cruz [AFX1(FoxO4), A14(myc), Hsp90, poly(ADP-ribose) polymerase, and tubulin], Upstate [FKHRL1(FoxO3)], and Chemicon (MMP9, MMP2).
RNA and real-time PCR analysis.
Total RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer's protocols. The single-stranded cDNA was synthesized using Superscript III (Invotrogen). Semiquantitative reverse transcription-PCRs were performed for Foxo4 and MMP9 before the real-time PCR analysis. All the PCR-generated fragments were sequenced to confirm their authenticities. The real-time PCR was performed using TaqMan on an ABI-PE prism 7000 sequence detection system (Applied Biosystems) according to the protocols provided by the manufacturer. The primers and probe sequences were obtained from Applied Biosystems and are available upon request. The relative quantities of mRNA were determined using comparative cycle threshold methods and normalized against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA.
Carotid artery ligation, immunohistochemistry, and morphometric analysis.
The Foxo4-null mutation was generated via a gene-targeting strategy using 129SvJ embryonic stem cells (23a). Mice were backcrossed to wild-type FVB animals (more than five generations). Mice homozygous for the Foxo4 mutant are viable and fertile. Male mice (8 to 10 weeks old) were used in accordance with the guidelines of the National Institutes of Health and the American Heart Association for the care and use of laboratory animals. The procedure in this study was approved by the University of Texas Southwestern Medical Center Animal Care Committee. Carotid artery ligation and immunohistochemistry experiments were performed as described previously (11, 13). Morphometric analysis was performed using NIH ImageJ software. For the zymography study, ligated left common carotid arteries (LCCAs) and unligated right common carotid arteries (RCCAs) were minced in extraction buffer (20 mM triethanolamine, 0.1% Brij 35) and mixed at 4°C for 4 h. Aliquots of protein lysate (15 μg) were loaded on SDS-PAGE gels for Coomassie blue staining and for gelatin zymography.
TUNEL staining.
Transferase-mediated dUTP nick end labeling (TUNEL) staining was used to examine apoptosis, following the manufacturer's protocol (Roche). Positive-control slides were treated with DNase I, and negative-control slides were stained in the absence of terminal deoxynucleotidyl transferase enzyme.
Statistical analysis was performed using functions from Microsoft Word Excel. Student's t test was used to assay the statistical significance.
RESULTS
Inactivation of Foxo4 reduces the abilities of SMCs to migrate in vitro.
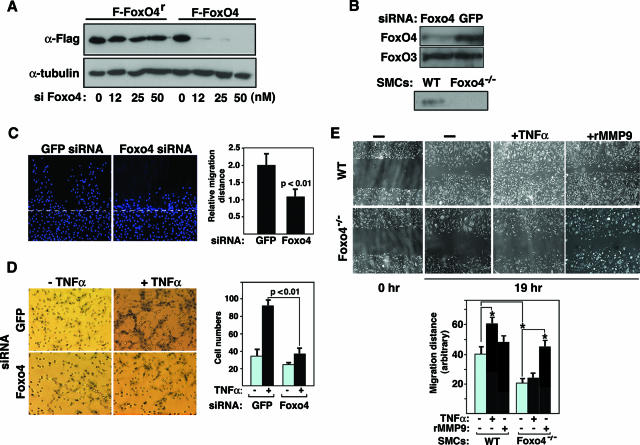
Previously, we have found that FoxO4 expression is upregulated in proliferating SMCs of the neointima upon vessel injury, suggesting that FoxO4 may promote phenotypic modulation of SMCs (13). As phenotypic switching of SMCs from a differentiated state to a proliferative one involves dedifferentiation, proliferation, and migration, we sought to investigate whether FoxO4 has potential roles in SMC proliferation and migration. We transfected rat aortic vascular SMCs with Foxo4-specific siRNA (13) or control GFP siRNA. Foxo4 siRNA effectively suppressed the expression of both transfected Flag-FoxO4 and endogenous FoxO4, without affecting a transfected siRNA-resistant Flag-FoxO4 and endogenous FoxO3 (Fig. 1A and B). No significant difference in proliferative rate was observed between the wild-type and Foxo4-knocked-down SMCs as assayed by a bromodeoxyuridine incorporation assay (data not shown). However, in a scratch migration assay, Foxo4-knocked-down SMC monolayers showed a significant decrease in the average distance migrated at 19 h after the wounding (Fig. 1C) (P < 0.01). The impact of Foxo4 inactivation on SMC migration was reinforced further by a gelatin invasion assay using a modified Boyden chamber. We found that Foxo4 inactivation resulted in a significant reduction of the number of SMCs that migrated through a gelatin-coated membrane in response to TNF-α (Fig. 1D) (92 ± 7 versus 36 ± 7 per microscopic field for control GFP siRNA-transfected SMCs versus Foxo4 siRNA-transfected cells, respectively; P < 0.01). Finally, to confirm the relevance of FoxO4 in SMC migration, we assayed migration activity in primary SMCs derived from the aortas of wild-type and Foxo4-null mice. By use of a scratch migration assay, Foxo4 deficiency was associated with significantly reduced migration of SMCs under basal conditions (Fig. 1E) (P < 0.05). Furthermore, the difference in migration between wild-type and Foxo4-null SMCs was more pronounced under treatment of cultures with TNF-α (Fig. 1E).
FIG. 1.
Inactivation of Foxo4 reduces the abilities of SMCs to migrate in vitro. (A) COS cells were transfected with either Flag-FoxO4 or siRNA-resistant Flag-FoxO4r in the presence of various concentrations of Foxo4 siRNA. Cells were harvested 48 h after transfection for Western blot analysis with anti-Flag antibody. (B) Top panel, rat aortic SMCs were transfected with Foxo4 siRNA and control GFP siRNA. Sixty hours after transfection, aliquots of cell lysates were subjected to Western blot analysis using antibodies against FoxO4 and FoxO3. Bottom panel, mouse primary aortic SMCs were isolated from wild-type (WT) and Foxo4-null mice and cultured. Cell lysates were subjected to Western blot analysis using anti-FoxO4 antibody. (C) Confluent SMC monolayers transfected with Foxo4 siRNA and control GFP siRNA were scratch wounded 24 h after transfection, and cells were kept in culture for another 19 h before being fixed and stained with Hoechst (left panel). The average distances migrated by SMCs are quantified in the right panel. Data shown are averages ± standard deviations for three independent experiments (P < 0.01). (D) Migration of vascular SMCs transfected with Foxo4 siRNA and GFP siRNA through a gelatin-coated membrane was assayed using a modified Boyden chamber in the presence and absence of the chemoattractant TNF-α (100 ng/ml, at 6 h). SMCs that migrated across the gelatin barrier were stained with hematoxylin (left panel), and the number of migrated cells per microscopic field was quantified (right panel) (with an average of four randomly chosen fields used for each of the three independent experiments). Data shown are averages ± standard deviations for three independent experiments (P < 0.01). (E) Confluent aortic SMC monolayers isolated from wild-type and Foxo4-null mice were scratch wounded, and cells were kept in culture in the absence or presence of either TNF-α or recombinant human MMP9 for another 19 h before being fixed. The average distances migrated are quantified in the lower panel. Data shown are averages ± standard deviations for three independent experiments. *, P < 0.05.
Inactivation of Foxo4 reduces TNF-α-activated MMP9 induction.
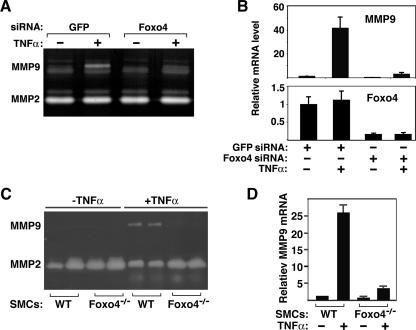
TNF-α is a potent MMP9 inducer and migration factor for SMCs (32). MMP9 is required for SMC migration (10). We hypothesized that FoxO4 may mediate the TNF-α-induced MMP9 expression. To test this hypothesis, we transfected rat aortic SMCs with Foxo4-specific siRNA (13) or control GFP siRNA. Twenty-four hours after transfection, cell culture medium was changed to serum-free medium with or without TNF-α. TNF-α-induced MMP9 expression was assayed 16 h later, using gelatin zymography to detect the enzymatic activity of secreted MMP9 (Fig. 2A) and real-time PCR analysis to detect MMP9 transcripts (Fig. 2B). TNF-α activated both the enzymatic activity and the transcription of MMP9 in SMCs transfected with control GFP siRNA. Inactivation of Foxo4 significantly attenuated the upregulation of MMP9 by TNF-α, suggesting that FoxO4 mediates MMP9 transcription in response to TNF-α in vivo. To rule out a potential off-target effect of a single Foxo4 siRNA on the TNF-α-induced MMP9 expression, we employed an independent pool of Foxo4 siRNAs and observed similar outcomes (data not shown). This strong link between FoxO4 and TNF-α-induced MMP9 induction was also observed in primary aortic SMCs isolated from wild-type and Foxo4-null mice. As shown in Fig. 2C and D, both the enzymatic activity and the transcription level of MMP9 were found to be upregulated in response to TNF-α stimulation in wild-type SMCs, whereas the TNF-α-mediated upregulation of MMP9 expression was significantly inhibited in Foxo4-deficient SMCs. To test whether the migration defect of Foxo4-null SMCs is due to the lack of MMP9, we added purified recombinant human MMP9 back to the culture medium. Addition of exogenous MMP9 is able to rescue the migration defect of Foxo4-null SMCs (Fig. 1E), suggesting that FoxO4-regulated SMC migration is MMP9 dependent.
FIG. 2.
FoxO4 mediates TNF-α-activated MMP9 expression. (A) Rat SMCs were transfected with Foxo4 siRNA and control GFP siRNA. Twenty-four hours after transfection, cell culture media were changed to serum-free media in the presence or absence of TNF-α and remained in culture for another 16 h. Aliquots of the culture media were taken for zymography assays of the gelatinolytic activities of secreted MMP9 and MMP2. (B) Cell lysates were used for analysis of mRNA transcripts of MMP9 and FoxO4 by real-time PCR. Data shown are averages ± standard deviations for three independent experiments. (C) Gelatin zymography assay with conditioned medium of primary aortic SMCs isolated from wild-type (WT) and Foxo4-null mice. Two independent primary SMC preparations are shown. (D) Real-time PCR analysis of relative mRNA levels of MMP9 in wild-type and Foxo4-deficient SMCs in the absence or presence of TNF-α stimulation. Data shown are averages ± standard deviations for three independent experiments.
FoxO4 activates the MMP9 promoter by binding to the transcription factor Sp1.
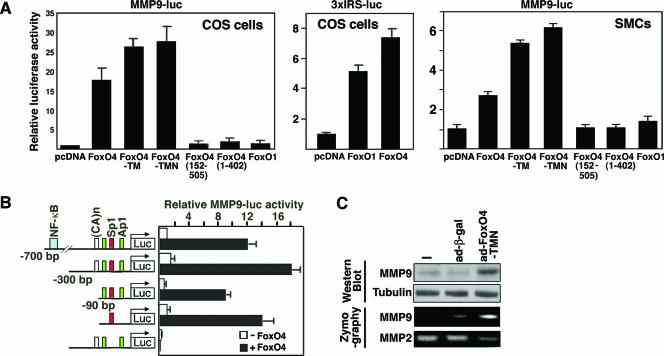
To investigate whether the MMP9 gene is the direct transcriptional target of FoxO4, we tested the effect of FoxO4 on the expression of a luciferase reporter linked to the 700-bp MMP9 promoter (Fig. 3A). Previous studies showed that this 700-bp promoter region contains highly conserved DNA binding sites for the transcription factors Ap-1, NF-κB, PEA3, and Sp1; a TATA box; and a CA repeat microsatellite sequence (8) (Fig. 3B). As shown in Fig. 3A (left panel), FoxO4 activated MMP9-luciferase in COS cells. The N-terminal amino acid sequence (residues 1 to 152) and C-terminal transactivation domain (TAD) (residues 403 to 505) of FoxO4 were required for its activity since deletion of either region led to loss of the ability of FoxO4 to activate the MMP9-luciferase reporter. We also tested whether the DNA binding activity of FoxO4 is required for activation of the MMP9-luc reporter. For this purpose, we used a FoxO4 mutant, FoxO4-TM, which has three Akt phosphorylation sites replaced with Ala, and another mutant, FoxO4-TMN, which has a conserved amino acid (Asn152) involved in DNA binding replaced with Ala, in addition to the triple mutation in the Akt phosphorylation sites. FoxO4-TMN is constitutively nucleus localized due to the triple mutation and is unable to bind a canonical Foxo binding element (13). Both FoxO4-TM and FoxO4-TMN also activated the MMP9 promoter. Similarly, FoxO4 also activated the MMP9-luc reporter in SMCs (Fig. 3A, right panel), although the activation is less than that in COS cells due to the high basal activity of the MMP9-luc reporter in SMCs. Interestingly, while FoxO1 is able to activate a control promoter-driven luciferase (3xIRS-luc) reporter containing canonical insulin-responsive sequences, it failed to activate the MMP9-luc reporter (Fig. 3A, left and middle panels).
FIG. 3.
FoxO4 activates MMP9 transcription through an Sp1 DNA binding site. (A) MMP9-luc reporter activity from COS cells (left panel) and SMCs (right panel) transfected with the MMP9-luc reporter in combination with the indicated plasmids. Luciferase activities were normalized against cotransfected β-galactosidase activities. Data shown are averages ± standard deviations for three independent experiments. Middle panel, multimerized insulin-responsive element (3xIRS)-luc reporter activity from COS cells transfected with the indicated plasmids. (B) Responsiveness of the deletion and site-specific mutants of the MMP9-luc reporter to FoxO4 in COS cells. Transcription factor binding sites of NF-κB, Sp1, and Ap1 are highlighted in color (left panel). Mutation of the Sp1 binding site impairs the responsiveness of the promoter to FoxO4. Data shown are averages ± standard deviations for three independent experiments. (C) SMCs were infected with nothing, adenoviruses expressing β-galactosidase, and FoxO4-TMN, respectively. Twenty-four hours after infection, equal amounts of culture media were used for determination of the gelatinolytic activity of MMP9 by zymography (bottom panel), and cell lysates were used for analysis of MMP9 protein level by Western blotting (top panel). Tubulin was used as a loading control in Western blot analysis.
We further mapped the FoxO4-responsive region to an Sp1 DNA binding site in the MMP9 promoter, using a series of deletion constructs and site-directed mutations that abolish individual binding sites in the MMP9 promoter (NF-κB, Ap1, and Sp1) (Fig. 3B). Deletion of either the NF-κB or the CA repeat microsatellite sequence or mutation of two Ap1 DNA binding sites had no significant effect on the responsiveness of the MMP9 promoter to FoxO4 in COS cells (Fig. 3B). However, mutation of the Sp1 DNA binding site resulted in complete loss of the responsiveness of the MMP9 promoter to FoxO4. Consistent with the observation that FoxO4 activates MMP9 transcription through the Sp1 DNA binding site, we observed that the DNA binding-defective FoxO4 mutant FoxO4-TMN activated the MMP9-luc reporter (Fig. 3A). FoxO4-TMN also upregulated the protein level and enzymatic activity of MMP9 when ectopically expressed in SMCs (Fig. 3C).
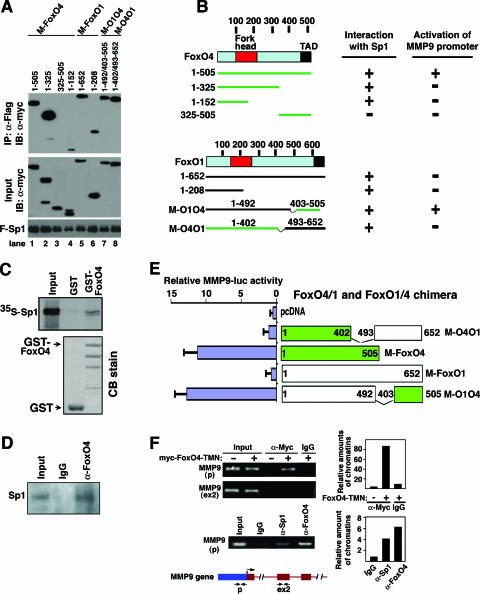
To test whether FoxO4 binds to Sp1, we carried out coimmunoprecipitation assays with Flag-Sp1 and myc-tagged full-length and deletion mutant forms of FoxO4. These experiments showed that Flag-Sp1 coimmunoprecipitated with myc-FoxO4 (Fig. 4A, lane 1, and Fig. 4B). Myc-FoxO4 (residues 1 to 325) but not myc-FoxO4 (residues 325 to 505) interacted with Flag-Sp1 (Fig. 4A, lanes 2 and 3, respectively). A smaller fragment of FoxO4 (residues 1 to 152) was also sufficient to interact with Sp1 (Fig. 4A, lane 4). Glutathione S-transferase (GST) pulldown experiments with GST-FoxO4 and in vitro-translated, 35S-labeled Sp1 indicated that FoxO4 interacted with Sp1 directly (Fig. 4C). The direct interaction between Sp1 and FoxO4 in GST pulldown experiments appears to be weaker than that observed in COS cells (Fig. 4A), suggesting that an additional binding partner(s) may be involved in stabilizing the Sp1/FoxO4 immunocomplex in COS cells. A bona fide interaction between endogenous Sp1 and FoxO4 was also observed in myoblast C2C12 cells (Fig. 4D). These experiments show that FoxO4 interacts with Sp1 and that the N-terminal region of FoxO4 (residues 1 to 152) mediates this interaction.
FIG. 4.
FoxO4 interacts with the transcription factor Sp1. (A) Coimmunoprecipitation of Flag-Sp1 with myc-tagged FoxO4 and FoxO1. COS cells were transfected with expression plasmids encoding Flag-Sp1, myc-tagged full-length and deletion mutant forms of FoxO4 and FoxO1, and chimeric constructs of FoxO4 and FoxO1 as indicated. Cell lysates were used for immunoprecipitation (IP) with anti-Flag antibody, and the immunoprecipitates were analyzed by immunoblot (IB) analysis with anti-myc antibody. Four percent of inputs is shown. (B) Schematic diagrams of plasmids used for panel A and summary of the results obtained from the coimmunoprecipitation experiments whose results are shown in panel A and from the reporter assays whose results are shown in panel E and Fig. 3A. (C) GST pulldown experiments. 35S-labeled Sp1 protein was translated in vitro using TNT reticulocyte lysate and incubated overnight with GST and GST-FoxO4 proteins conjugated to glutathione agarose beads. After being washed, the bound Sp1 proteins were separated by SDS-PAGE and analyzed by autoradiography. Ten percent input of Sp1 was loaded on the SDS-PAGE gel. Coomassie blue (CB) stains of the GST and GST-FoxO4 proteins are shown below. (D) Myoblast C2C12 cell lysates were immunoprecipitated with either control IgG or anti-FoxO4 antibody. The immunoprecipitates were subjected to Western blot analysis with anti-Sp1 antibody. (E) Specificity of FoxO4-activated MMP9 transcription. cDNA plasmids expressing chimeric FoxO4/1 or FoxO1/4 were transfected to COS cells along with an MMP9-luc reporter construct. The chimeric constructs are illustrated in the right panel. The abilities of chimeric proteins to activate MMP9 transcription were assayed by MMP9-luc reporter activity (left panel). Data shown are averages ± standard deviations for three independent experiments. (F) Chromatin immunoprecipitation of MMP9 promoter complexes. Top two panels on the left, in vivo cross-linked chromatin was prepared from rat SMCs infected with and without adenovirus expressing myc-FoxO4-TMN and immunoprecipitated with anti-myc or control IgG, followed by PCR amplification using the primer pair specific for the MMP9 promoter (p) and the coding region (ex2). Bottom panel on the left, a ChIP assay with endogenous Sp1 and FoxO4 was performed with anti-Sp1 and anti-FoxO4 antibodies. The amounts of chromatin immunoprecipitated by various antibodies were quantified by real-time PCR and normalized against input and are shown on the right (averages for two independent experiments).
As FoxO1 failed to activate the MMP9 promoter (Fig. 3A), we tested whether FoxO1 interacts with Sp1. Figure 4A shows that FoxO1 interacts with Sp1, and its FoxO4-homologous region (residues 1 to 208) is sufficient to mediate this interaction (Fig. 4A, lanes 5 and 6, and Fig. 4B). Our data suggest that the specificity of FoxO4-activated MMP9 transcription may reside in the C-terminal region of FoxO4. To test this hypothesis, we performed coimmunoprecipitation and MMP9-luc reporter assays with two chimeric FoxO4/1 and FoxO1/4 constructs: M-O4O1 contains the N-terminal region of FoxO4 (residues 1 to 402) and the C terminus of FoxO1 (residues 493 to 652), and the reverse chimera (M-O1O4) contains the N-terminal region of FoxO1 (residues 1 to 492) and the C terminus of FoxO4 (residues 403 to 505). Deletion of the C-terminal region of FoxO4 (residues 403 to 505) abolished its ability to activate MMP9 transcription, and incorporation of the FoxO1 C terminus into this construct (resulting in the M-O4O1 chimera) did not restore its ability to activate MMP9 transcription (Fig. 4E), even though this chimera binds to Sp1 (Fig. 4A, lane 8). On the other hand, whereas FoxO1 failed to activate MMP9 transcription, replacement of the FoxO1 C terminus with that of FoxO4 (resulting in the M-O1O4 chimera) restored its ability to activate the MMP9 transcription to a level similar to that of FoxO4. Binding of M-O1O4 is also comparable to that of FoxO4 (Fig. 4A, lane 7). These results suggest that binding of FoxO4 to Sp1 is necessary but not sufficient to activate MMP9 transcription and that the C-terminal region (residues 403 to 505) of FoxO4 is required for the specificity of FoxO4-activated MMP9 transcription.
To investigate whether FoxO4 regulates MMP9 transcription by associating with the chromatin region containing the Sp1 DNA binding site in the promoter of the MMP9 gene, we carried out ChIP assays. Endogenous chromatin fragments of SMCs infected with adenovirus expressing myc-FoxO4-TMN were precipitated with anti-myc antibody or control immunoglobulin G (IgG) (Fig. 4F). DNA from the immunoprecipitates was subjected to PCR analysis using the primer pair encompassing the Sp1 site in the MMP9 promoter. As shown in Fig. 4F, chromatin fragments containing the Sp1 DNA binding site from the MMP9 promoter region were specifically immunoprecipitated by anti-myc antibody in cells expressing myc-FoxO4-TMN. No chromatin fragments were precipitated from myc-FoxO4-TMN-expressing cells by control IgG or by anti-myc antibody in the absence of myc-FoxO4-TMN. Binding of FoxO4 to the promoter region of MMP9 was specific, as no chromatin fragment in the coding region of MMP9 (ex2) was immunoprecipitated with anti-myc antibody in myc-FoxO4-TMN-expressing cells. Finally, to test whether endogenous FoxO4 binds to the MMP9 promoter, we performed ChIP assays with anti-FoxO4 antibody with chromatin fragments from proliferating SMCs. Chromatin fragments containing the Sp1 DNA binding sites were observed specifically in the immunoprecipitates with anti-FoxO4 and Sp1 antibodies (Fig. 4F, third panel on the left).
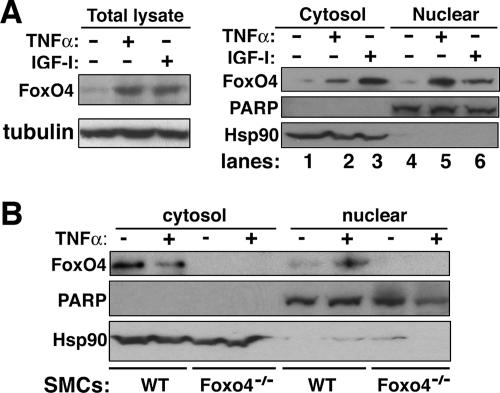
TNF-α signaling upregulates nuclear FoxO4.
FoxO proteins shuttle between the cytoplasm and nucleus in response to growth and oxidative stress signals (5, 6). IGF-I has been shown to activate the phosphatidylinositol 3-kinase (PI3K)-Akt growth signaling pathway and to promote nuclear export of FoxO proteins through 14-3-3 proteins (5; reviewed in references 29 and 30). TNF-α has been shown to activate the ROS-Jun N-terminal protein kinase (JNK) stress signaling pathway and promote nuclear import of FoxO4 (6). We thus tested whether TNF-α induces MMP9 transcription by promoting FoxO4 nuclear translocation in aortic SMCs. We performed cellular fractionations of SMCs treated with or without TNF-α and IGF-I. As shown in Fig. 5A, the total FoxO4 protein is upregulated in TNF-α- and IGF-I-stimulated rat aortic SMCs (Fig. 5A, left panel). Moreover, upregulation of FoxO4 upon treatment by TNF-α is more pronounced in the nucleus than in the cytoplasm (Fig. 5A, right panel, lanes 2 and 5). On the other hand, IGF-I resulted in a higher protein level of FoxO4 in the cytoplasm than in the nucleus (Fig. 5A, right panel, lanes 3 and 6), suggesting that TNF-α promotes FoxO4 nuclear accumulation whereas IGF-I promotes FoxO4 nuclear export. A similar effect of TNF-α on the nuclear translocation of FoxO4 was also observed in mouse primary aortic SMCs, albeit with no upregulation of total FoxO4 protein by TNF-α (Fig. 5B). As a control for the specificity of the anti-FoxO4 antibody, we used lysates from mouse Foxo4−/− SMCs in the Western blot analysis. No FoxO4 proteins in Foxo4-null SMCs were detected using anti-FoxO4 antibody.
FIG. 5.
TNF-α upregulates nuclear FoxO4. (A) Subconfluent rat aortic SMCs in serum-free medium were treated with either TNF-α (50 ng/ml) or IGF-I (100 ng/ml) for 1 h. Total, cytosolic, and nuclear fractions of the cell lysates were used for Western blot analysis using antibodies against FoxO4. Poly(ADP-ribose) polymerase (PARP) and Hsp90 were used as loading controls for nuclear and cytoplasmic proteins, respectively. (B) Primary aortic SMCs from wild-type (WT) and Foxo4-null mice were treated with or without TNF-α. Cytosolic and nuclear fractions of the cell lysates were subjected to Western blot analysis as described for panel A.
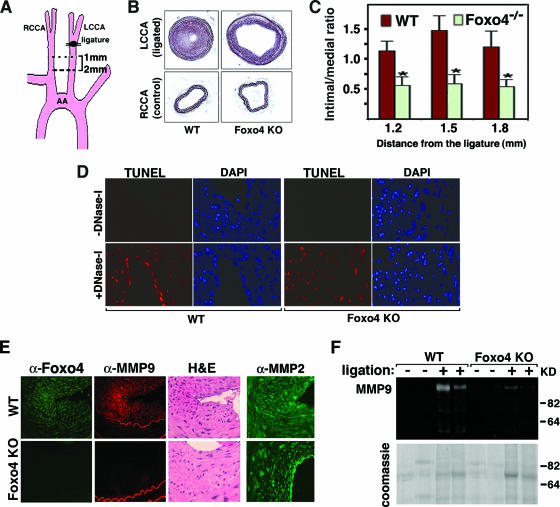
Foxo4 deficiency reduces the extent of intimal formation in vivo.
To examine the involvement of endogenous FoxO4 in the modulation of SMC phenotypes in vivo, we subjected Foxo4 knockout (KO) mice to a flow cessation injury model (Fig. 6A). We chose this model because it has been shown that intimal SMC hyperplasia due to the phenotypic change of medial SMCs and their production of extracellular matrix are the predominant contributors to intimal thickening, whereas very few inflammatory cells were observed in the neointima (11). Foxo4-null mice are viable and fertile and show no obvious developmental defects (9, 23a). However, 4 weeks after ligation injury, Foxo4-null mice displayed reduced neointimal formation in contrast to the robust growth of the neointima in wild-type mice (Fig. 6B and C). To test whether the reduced neointimae in Foxo4-null mice is due to increased apoptosis, we performed TUNEL assays on sections of ligated carotid arteries of both wild-type and Foxo4-null mice. Little apoptosis was observed in the carotid sections of either wild-type or Foxo4-null mice (Fig. 6D), suggesting that the inhibition of neointima formation in Foxo4 KO mice is not due to elevated apoptosis of Foxo4-null SMCs.
FIG. 6.
Foxo4 deficiency reduces the extent of intimal hyperplasia. (A) Flow cessation model. The LCCAs of 8- to 10-week-old mice were ligated near the bifurcation. Four weeks after ligation, the carotids were harvested and fixed. Sections between 1 and 2 mm below the ligature were used for the subsequent analyses performed for panels B to D. AA, aortic arch. (B) Hart elastin staining of cross sections of representative ligated LCCAs and unligated RCCAs from wild-type (WT) and Foxo4 KO mice. (C) Intimal areas of cross sections of LCCAs of wild-type and Foxo4-null mice were calculated and normalized against the medial areas. Data shown are averages ± standard errors of the means for 10 wild-type mice and 16 Foxo4-null mice. *, P < 0.05. Neointimal formation is significantly reduced in Foxo4-deficient mice. (D) TUNEL staining of sections of carotid arteries of wild-type and Foxo4 KO mice 4 weeks after ligation. The positive-control slides were treated with DNase I prior to the staining. DAPI, 4′,6′-diamidino-2-phenylindole. (E) Coimmunofluorescence staining of FoxO4 and MMP9 with anti-FoxO4 and anti-MMP9 antibodies on cross sections of ligated LCCAs of wild-type and Foxo4 KO mice, respectively. MMP2 immunostaining and hematoxylin and eosin staining (H&E) were performed on separate, adjacent sections. (F) Gelatin zymography with protein extracts of unligated and ligated carotid arteries of wild-type and Foxo4-null mice (upper panel). Aliquots of the lysates were loaded on SDS-PAGE gel and stained with Coomassie blue for the loading control (lower panel). The amounts of protein in ligated wild-type and Foxo4-null carotid arteries were similar.
Expression of MMP9 is low in normal blood vessels and dramatically upregulated upon vessel injury. MMP9 has been shown to be required for SMC migration in vivo in the formation of intimal hyperplasia. MMP9-null mice have reduced neointimal formation upon carotid artery ligation (10). To test whether MMP9 is a downstream effector of FoxO4, we examined the expression of MMP9 in the intimae of injured carotid arteries of Foxo4-null mice and compared it with that in the wild-type littermates. As shown in Fig. 6E, MMP9 expression is upregulated in intimae of wild-type mice compared to that in the medial area. The upregulation of MMP9 expression is significantly attenuated in Foxo4-null mice, suggesting that the reduction of intimal hyperplasia in Foxo4-null mice may be mediated through MMP9. In contrast, there was minimal change in MMP2 expression in injured carotids of wild-type and Foxo4-null mice, a finding consistent with regional differences in FoxO4 activity in different vascular beds.
To further confirm that MMP9 expression is reduced in the neointimae of Foxo4-null mice, we performed a zymography assay with protein extracts from ligated and unligated carotid arteries of wild-type and Foxo4-null mice 28 days postligation. MMP9 activity in the ligated carotid arteries of Foxo4-null mice is significantly reduced compared to that in wild-type mice (Fig. 6F). Taken together, these data suggest that the inhibition of neointimae in Foxo4-null mice is due to the migration defect of Foxo4-null SMCs caused by reduction of MMP9 production.
DISCUSSION
Phenotypic modulation of vascular SMCs in response to injury is one of the key events in development of restenotic and atherosclerotic lesions. In this paper, we show that FoxO4 is required for SMC migration. Inhibition of FoxO4 expression inhibits SMC migration in vitro and reduces intimal hyperplasia in vivo (Fig. 1 and 6). Inactivation of Foxo4 by either siRNA or gene knockout inhibits upregulation of MMP9 expression in vitro after TNF-α stimulation and in vivo after vessel injury (Fig. 2 and 6). The reduced neointimal phenotype of Foxo4-null mice after carotid artery ligation is similar to that of MMP9-null mice (10), suggesting a genetic interaction between Foxo4 and MMP9. Furthermore, we show that FoxO4 activates MMP9 transcription and upregulates the protein level and enzymatic activity of MMP9 (Fig. 3). The FoxO4-regulated SMC migration is MMP9 dependent, as the migration defect of Foxo4-null SMCs can be rescued by exogenous recombinant MMP9 protein (Fig. 1E). Altogether, these data strongly indicate that FoxO4 promotes phenotypic modulation of SMC through activation of MMP9.
Our results also suggest that FoxO4 activates MMP9 transcription by acting as a coactivator of the transcription factor Sp1 (Fig. 3 and 4). The mechanism by which FoxO4 activates MMP9 transcription does not require FoxO4 DNA binding activity but does require its N-terminal Sp1-binding domain (residues 1 to 152) and its C-terminal sequence (residues 403 to 505). Activation of MMP9 transcription by FoxO4 is specific. We observed that the C-terminal sequence (residues 403 to 505) of FoxO4 is required for the specific activation of MMP9 by FoxO4. Transfer of this region of FoxO4 to the corresponding region of FoxO1 confers responsiveness of MMP9 to FoxO1. It is possible that this sequence interacts with an additional, yet-to-be-identified transcription factor(s) that is present in the transcription complex in the promoter of MMP9.
The promoter of MMP9 contains several highly conserved functional binding sites for transcription factors, including Ap1, NF-κB, and Sp1. It has been shown that NF-κB and Ap-1 are involved in TNF-α-activated MMP9 expression in SMCs in culture (4). The relative importance of Sp1 and NF-κB in mediating TNF-α-activated gene expression has been explored (3, 24). Ainbinder et al. showed that Sp1 is required for priming the transcriptional apparatus to ensure a rapid induction of A20 protein by NF-κB after stimulation by TNF-α (3). Furthermore, Pazdrak et al. showed that while both Sp1 and NF-κB are required for the initial induction of intercellular adhesion molecule 1 transcription, only Sp1 is required for sustained transcription after an initial period of 1 to 3 h of TNF-α stimulation (24). Our studies suggest that Sp1 could sustain TNF-α-induced MMP9 transcription by recruiting FoxO4 as a coactivator.
The effect of TNF-α on the cellular localizations and protein levels of FoxOs remains elusive. Abid et al. have shown that TNF-α induces phosphorylation of FoxO proteins on conserved Akt phosphorylation sites [FoxO1(S256) and FoxO4(S193)] in human coronary artery SMCs (CASMCs) and induces nuclear exclusion of FoxO1 and FoxO3 (1). It was not studied whether phosphorylation of FoxO4 on Ser193 will induce nuclear exclusion of FoxO4 in coronary artery SMCs. Essers et al. have shown that TNF-α promotes nuclear import of human FoxO4 in fibroblasts through phosphorylation of FoxO4 on Thr447 and Thr451 by JNK (6). The JNK signaling pathway has also been shown to phosphorylate and promote nuclear translocation of Caenorhabditis elegans Foxo (DAF-16) and Drosophila Foxo (Dfoxo) (21, 31). Our studies suggest that TNF-α promotes Foxo4 nuclear accumulation in aortic SMCs (Fig. 5).
The mechanism by which TNF-α promotes FoxO4 nuclear localization remains to be determined. As Thr447/Thr451 are not conserved among different FoxO proteins and species, TNF-α may promote nuclear import of mouse FoxO4 through an alternative mechanism. It has been shown that stress-activated JNK can phosphorylate 14-3-3 and disrupt its interaction with FoxO3, therefore promoting nuclear localization of FoxO3 (28). Recently, Lehtinen et al. have identified a conserved phosphorylation site of FoxO3 on Ser207 (corresponding to Ser212 in FoxO1 and Ser153 in FoxO4) that can be phosphorylated by protein kinase MST1 in response to oxidative stress signals (12). Phosphorylation of FoxO3 on Ser207 disrupts its interaction with 14-3-3 proteins and promotes its nuclear translocation (12). It is possible that TNF-α promotes FoxO4 nuclear accumulation in aortic SMCs through activated JNK, which could phosphorylate 14-3-3 and disrupt its binding to FoxO4. Alternatively, TNF-α could activate MST1, which in turn phosphorylates FoxO4 at S153 and disrupts its interaction with 14-3-3. TNF-α is a cytokine that has been shown to increase intracellular H2O2 level, which in turn produces cellular oxidative stress (6, 7). MST1 is expressed in vascular SMCs and upregulated in balloon-injured rat carotid arteries (22).
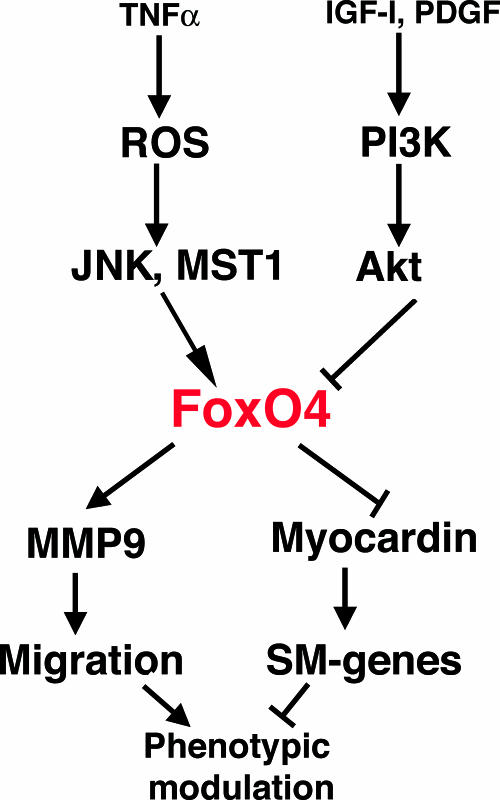
In addition to promoting nuclear translocation of FoxO4, TNF-α also increases the total amount of FoxO4 protein in the rat aortic SMCs (Fig. 5A). The mechanism by which TNF-α upregulates FoxO4 remains to be determined. We have observed a small but consistent upregulation of Foxo4 transcription in TNF-α-stimulated cells even though the difference is not statistically significant (Fig. 2B), suggesting that TNF-α may activate Foxo4 transcription. Alternatively, a TNF-α-activated signaling pathway may stabilize FoxO4 protein. Taken together, our data propose a mechanistic model by which FoxO4 regulates phenotypic modulation of SMCs in response to cytokine and growth factor signaling (Fig. 7). Upon vascular injury, the inflammatory cytokine TNF-α upregulates nuclear FoxO4 expression through the ROS-JNK/ROS-MST1-activated stress signaling pathway. FoxO4 activates MMP9 transcription and inhibits expression of contractile smooth muscle differentiation genes by acting as a transcriptional coactivator of Sp1 and a corepressor of myocardin (13), respectively. Consequently, FoxO4 promotes phenotypic modulation of SMCs from a differentiated state to a proliferative one. The transcriptional activity of FoxO4 is either negatively regulated through its subcellular localization by Akt kinase through the PI3K-Akt signaling pathway, which promotes nuclear export, or positively regulated through the stress signal-activated JNK/MST1 pathway, which promotes nuclear import.
FIG. 7.
A model of the role of FoxO4 in phenotypic modulation of SMCs in response to cytokine and growth factor signals. During phenotypic modulation of SMCs in restenosis and atherosclerosis, nuclear FoxO4 is upregulated in response to TNF-α, which leads to suppression of myocardin-mediated smooth muscle (SM) differentiation, activation of MMP9 transcription, and promotion of SMC migration. The activity of FoxO4 is negatively regulated by Akt kinases through the PI3K-Akt signaling pathway.
Because MMP9 plays important roles in atherosclerosis and cancer metastasis, there has been an intense effort to find MMP9 inhibitors. The use of synthetic MMP9 inhibitors in clinical trials has not been proven to be efficacious (27), due to lack of specificity. Understanding the signaling pathways that regulate MMP9 expression in specific cell types and disease conditions may provide opportunities for future therapeutic intervention. We have shown that FoxO4 specifically activates MMP9 expression in response to the proinflammatory cytokine TNF-α. Since FoxO4 is the convergence point of cytokine and growth factor signaling pathways and it regulates the expression of specific sets of genes, inhibition of FoxO4 could provide a therapeutic opportunity to combat inflammatory arterial diseases and cancer.
Acknowledgments
This study was supported by a scientist development grant from the American Heart Association and NIH RO1 HL085749-01 to Z.-P.L. and grants from the NIH and the Donald W. Reynolds Clinical Cardiovascular Research Center to E.N.O. R.A.D. is an American Cancer Society Research Professor and an Ellison Medical Foundation Scholar and is supported by the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science and the NIH (grants PO1 CA095616 and MMHCC U01).
We thank G. Owens and T. Kitamura for reagents, H. Yanagisawa for fruitful discussions, and S. Hacker and J. Shelton for technical assistance.
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Abid, M. R., K. Yano, S. Guo, V. I. Patel, S. Shrikhande, K. C. Spokes, C. Ferran, and W. C. Aird. 2005. Forkhead transcription factors inhibit vascular smooth muscle cell proliferation and neointimal hyperplasia. J. Biol. Chem. 280:29864-29873. [DOI] [PubMed] [Google Scholar]
- 2.Accili, D., and K. C. Arden. 2004. Foxos at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117:421-426. [DOI] [PubMed] [Google Scholar]
- 3.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, N. Diamant, and R. Dikstein. 2002. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 22:6354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, M., R. P. Fabunmi, A. H. Baker, and A. C. Newby. 1998. Synergistic upregulation of metalloproteinase-9 by growth factors and inflammatory cytokines: an absolute requirement for transcription factor NF-kappa B. FEBS Lett. 435:29-34. [DOI] [PubMed] [Google Scholar]
- 5.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 6.Essers, M. A., S. Weijzen, A. M. de Vries-Smits, I. Saarloos, N. D. de Ruiter, J. L. Bos, and B. M. Burgering. 2004. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23:4802-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goossens, V., J. Grooten, K. De Vos, and W. Fiers. 1995. Direct evidence for tumor necrosis factor-induced mitochondrial reactive oxygen intermediates and their involvement in cytotoxicity. Proc. Natl. Acad. Sci. USA 92:8115-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, C. 1996. Molecular mechanism of transcriptional activation of human gelatinase B by proximal promoter. Cancer Lett. 106:185. [DOI] [PubMed] [Google Scholar]
- 9.Hosaka, T., W. H. Biggs III, D. Tieu, A. D. Boyer, N. M. Varki, W. K. Cavenee, and K. C. Arden. 2004. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA 101:2975-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, C., and Z. S. Galis. 2004. Matrix metalloproteinase-2 and -9 differentially regulates smooth muscle cell migration and cell-mediated collagen organization. Arterioscler. Thromb. Vasc. Biol. 24:54-60. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, A., and V. Linder. 1997. Remodeling with neointima formation in the mouse carotid artery after cessation of blood flow. Arterioscler. Thromb. Vasc. Biol. 17:2238-2244. [DOI] [PubMed] [Google Scholar]
- 12.Lehtinen, M. K., Z. Yuan, P. R. Boag, Y. Yang, J. Villen, E. B. Becker, S. DiBacco, N. de la Iglesia, S. Gygi, T. K. Blackwell, and A. Bonni. 2006. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125:987-1001. [DOI] [PubMed] [Google Scholar]
- 13.Liu, Z. P., Z. Wang, H. Yanagisawa, and E. N. Olson. 2005. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell 9:261-270. [DOI] [PubMed] [Google Scholar]
- 14.Luttun, A., E. Lutgens, A. Manderveld, K. Maris, D. Collen, P. Carmeliet, and L. Moons. 2004. Loss of matrix metalloproteinase-9 or matrix metalloproteinase-12 protects apolipoprotein E-deficient mice against atherosclerotic media destruction but differentially affects plaque growth. Circulation 109:1408-1414. [DOI] [PubMed] [Google Scholar]
- 15.Miano, J. M. 2002. Mammalian smooth muscle differentiation: origins, markers and transcriptional control. Results Probl. Cell Differ. 38:39-59. [DOI] [PubMed] [Google Scholar]
- 16.Majesky, M. W. 2003. Vascular smooth muscle diversity: insights from developmental biology. Curr. Atheroscler. Rep. 5:208-213. [DOI] [PubMed] [Google Scholar]
- 17.McCawley, L. J., and L. M. Matrisian. 2001. Matrix metalloproteinases: they're not just for matrix anymore! Curr. Opin. Cell Biol. 13:534-540. [DOI] [PubMed] [Google Scholar]
- 18.Moon, S. K., H. M. Kim, Y. C. Lee, and C. H. Kim. 2004. Disialoganglioside (GD3) synthase gene expression suppresses vascular smooth muscle cell responses via the inhibition of ERK1/2 phosphorylation, cell cycle progression, and matrix metalloproteinase-9 expression. J. Biol. Chem. 279:33063-33070. [DOI] [PubMed] [Google Scholar]
- 19.Morris, B. J. 2005. A forkhead in the road to longevity: the molecular basis of lifespan becomes clearer. J. Hypertens. 23:1285-1309. [DOI] [PubMed] [Google Scholar]
- 20.Newby, A. C. 2005. Dual role of matrix metalloproteinases (matrixins) in intimal thickening and atherosclerotic plaque rupture. Physiol. Rev. 85:1-31. [DOI] [PubMed] [Google Scholar]
- 21.Oh, S. W., A. Mukhopadhyay, N. Svrzikapa, F. Jiang, R. J. Davis, and H. A. Tissenbaum. 2005. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA 102:4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono, H., T. Ichiki, H. Ohtsubo, K. Fukuyama, I. Imayama, Y. Hashiguchi, J. Sadoshima, and K. Sunagawa. 2005. Critical role of Mst1 in vascular remodeling after injury. Arterioscler. Thromb. Vasc. Biol. 25:1871-1876. [DOI] [PubMed] [Google Scholar]
- 23.Owens, G. K., M. S. Kumar, and B. R. Wamhoff. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84:767-801. [DOI] [PubMed] [Google Scholar]
- 23a.Paik, J.-H., R. Kollipara, G. Chu, H. Ji, Y. Xiao, Z. Ding, L. Miao, Z. Tothova, J. W. Horner, D. R. Carrasco, S. Jiang, D. G. Gilliland, L. Chin, W. H. Wong, D. H. Castrillon, and R. A. DePinho. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128:309-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pazdrak, K., X. Z. Shi, and S. K. Sarna. 2004. TNFalpha suppresses human colonic circular smooth muscle cell contractility by SP1- and NF-kappaB-mediated induction of ICAM-1. Gastroenterology 127:1096-1109. [DOI] [PubMed] [Google Scholar]
- 25.Ray, J. L., R. Leach, J. M. Herbert, and M. Benson. 2001. Isolation of vascular smooth muscle cells from a single murine aorta. Methods Cell Sci. 23:185-188. [DOI] [PubMed] [Google Scholar]
- 26.Rectenwald, J. E., L. L. Moldawer, T. S. Huber, J. M. Seeger, and C. K. Ozaki. 2000. Direct evidence for cytokine involvement in neointimal hyperplasia. Circulation 102:1697-1702. [DOI] [PubMed] [Google Scholar]
- 27.St-Pierre, Y., J. Couillard, and C. Themsche. 2004. Regulation of MMP-9 gene expression for the development of novel molecular targets against cancer and inflammatory diseases. Expert Opin. Ther. Targets 8:473-489. [DOI] [PubMed] [Google Scholar]
- 28.Sunayama, J., F. Tsuruta, N. Masuyama, and Y. Gotoh. 2005. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 170:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Heide, L. P., M. F. Hoekman, and M. P. Smidt. 2004. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 380:297-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt, P. K., H. Jiang, and M. Aoki. 2005. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle 4:7. [DOI] [PubMed] [Google Scholar]
- 31.Wang, M. C., D. Bohmann, and J. Jasper. 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121:115-125. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Z., M. R. Castresana, and W. H. Newman. 2001. NF-kappaB is required for TNF-alpha-directed smooth muscle cell migration. FEBS Lett. 508:360-364. [DOI] [PubMed] [Google Scholar]