Abstract
The Forkhead box f1 (Foxf1) transcription factor is expressed in mesenchymal cells of the lung, liver, and gallbladder. Although Foxf1 deficiency causes severe abnormalities in the development of these organs, the molecular mechanisms underlying Foxf1 function remain uncharacterized. In this study we inactivated Foxf1 function in lung mesenchymal cells and mouse embryonic fibroblasts (MEFs) by use of either short interfering RNA duplexes or a membrane-transducing Foxf1 dominant negative (DN) mutant protein (Foxf1 DN), the latter of which is fused to the human immunodeficiency virus TAT protein transduction domain. Although Foxf1 did not influence DNA replication or cell survival, Foxf1 depletion severely diminished mesenchyme migration. Foxf1 deficiency in mesenchymal cells was associated with reduced expression of the integrin-beta3 (Itgβ3) subunit. Furthermore, we generated transgenic mice containing a tetracycline-inducible Foxf1 DN transgene. Adenovirus-mediated activation of Foxf1 DN in transgenic MEFs caused diminished cell migration and reduced Itgβ3 expression. A chromatin immunoprecipitation assay demonstrated that Foxf1 protein binds to the bp −871 to −815 region of the mouse Itgβ3 promoter. Deletion of the −871 to −815 Itgβ3 promoter region completely abolished the ability of Foxf1 to activate transcription of the Itgβ3 promoter in cotransfection experiments, indicating that the mouse Itgβ3 is a direct transcriptional target of Foxf1 protein. Foxf1 plays an essential role in mesenchyme migration by transcriptionally regulating Itgβ3.
The embryonic development of gut-derived organs is dependent on extensive cellular proliferation and migration. These processes are required to establish the appropriate positioning of epithelial cells with developing mesenchyme (11, 52, 55). The migration of mesenchymal cells is regulated by distinct signaling pathways which stimulate the expression of mesenchyme-specific transcription factors (7, 8). These in turn bind cooperatively to distinct promoter regions and activate the expression of mesenchyme-specific target genes essential for cell migration, including those encoding extracellular matrix proteins (ECM) and their receptors, integrins (40, 43). Understanding the transcriptional regulation of these genes by mesenchyme-specific transcription factors will provide insight regarding mesenchyme migration and organ morphogenesis during embryonic development.
Integrin αVβ3 is one of the most important receptors for ECM proteins, which can bind several different ligands, such as fibronectin, vitronectin, thrombospondin, collagen, tenascin-C, fibrin, von Willebrand factor, and osteopontin (13, 40). The integrin αVβ3 is expressed in a variety of mesenchymal cells, including endothelium, smooth muscle cells, osteoclasts (14, 39, 40), and cells that migrate through different tissues, such as neutrophils, monocytes, and lymphocytes (14). αVβ3 expression is increased during tissue remodeling, inflammation, and cancer, where αVβ3 plays a key role in mediating the migration and adhesion of endothelial cells (14, 48).
The Forkhead box (Fox) proteins belong to an extensive family of transcription factors which share homology in the winged helix/Forkhead DNA binding domain (6, 25). Fox proteins play important roles in regulating the transcription of genes involved in cellular proliferation (12, 23, 44, 51, 54), differentiation (3, 7, 8, 11, 56), metabolic homeostasis (15, 35), and development of cancer (2, 19, 22, 49). The expression of the mesenchyme-specific Foxf1 (HFH-8 or Freac-1) gene initiates during gastrulation at 6.5 days postcoitum (dpc) in extraembryonic mesoderm, allantois, and lateral mesoderm (29, 37). Foxf1 expression continues in splanchnic (visceral) mesoderm and septum transversum mesenchyme, both of which are critical for the mesenchymal/epithelial induction of gut-derived organs, such as the liver, gallbladder, lung, stomach, and intestine (20, 30, 37). Foxf1−/− embryos die by 8 dpc due to defects in extraembryonic mesoderm development (17, 29). Foxf1 haploinsufficiency in Foxf1+/− mice is associated with a variety of developmental abnormalities in mesenchyme of the lung, gallbladder, esophagus, and trachea, suggesting that Foxf1 is an important transcriptional regulator in mesenchymal cells during organ morphogenesis (17, 20, 26, 28). Furthermore, Foxf1 is expressed in hepatic stellate cells and capillary endothelial cells of the adult liver and lung (16, 17, 24). Foxf1+/− mice displayed defective stellate cell activation during liver regeneration (16) as well as severe pulmonary hemorrhage in response to lung injury (21), suggesting that Foxf1 is essential for organ repair.
Although reduced Foxf1 levels in Foxf1+/− mice were associated with severe defects in mesenchyme of the lung, liver, and gallbladder (16, 17, 20), molecular mechanisms underlying Foxf1 function remain uncharacterized. In this study, we used either short interfering RNA (siRNA) duplexes or the Foxf1 dominant negative (DN) mutant protein (Foxf1 DN) to inhibit Foxf1 function in mouse fetal lung mesenchymal cell line MFLM-91U and mesenchyme-derived mouse embryonic fibroblasts (MEFs). Our results demonstrate that Foxf1 induces cell migration without detectable changes in cell proliferation or survival. Diminished mesenchyme migration in Foxf1-deficient cells was associated with reduced mRNA and protein levels of integrin-beta3 (Itgβ3), which plays a key role in cell migration. Foxf1 protein binds to the bp −871 to −815 region of the mouse Itgβ3 promoter. Deletion of the −871 to −815 Itgβ3 promoter region completely abolished the ability of Foxf1 to activate transcription of the Itgβ3 promoter in cotransfection experiments, indicating that the mouse Itgβ3 is a direct transcriptional target of Foxf1 protein. Altogether, these studies demonstrated that the Foxf1 transcription factor plays a key role in mesenchyme migration and the transcriptional activation of Itgβ3.
MATERIALS AND METHODS
Generation of TetO-Foxf1 DN transgenic mice.
The Foxf1 DN transgene corresponds to the cytomegalovirus-Tet operator (CMV-TetO) promoter-driven T7-tagged Engrailed transcriptional repression domain, which was cloned in frame with the N terminus of the Foxf1 winged helix DNA binding domain. This Foxf1 DN fusion protein binds to the Foxf1 DNA binding sites and recruits the Engrailed transcriptional repression domain, used previously to create a Gata6 DN inhibitor (4, 27). To generate transgenic mice, the Foxf1 DN construct was injected into pronuclei of CD-1 mouse eggs (24). The fertilized mouse eggs were transferred to surrogate mothers by the University of Chicago Transgenic Mouse Facility. Transgenic mice were identified by PCR analysis of the mouse tail DNA with primers specific to Engrail. MEFs were prepared from 12.5-dpc mouse embryos as described previously (50).
Western blot analysis.
Total or nuclear protein extracts were prepared from either MFLM-91U cells or MEFs and then subjected to Western blot analysis (17) using rabbit polyclonal antibody against Foxf1 (1:200 dilution [19]), Foxf2 (1:200 dilution; Affinity BioReagents, Golden, CO), or Itgβ3 (1:1,000 dilution; Cell Signaling). We also used a mouse monoclonal antibody against β-actin (clone AC-15, 1:5,000 dilution; Sigma) or T7 (1:10,000 dilution; Novagen). Detection of the immune complex was accomplished by using secondary antibodies directly conjugated with horseradish peroxidase followed by chemiluminescence (Supersignal; Pierce, Rockford, IL).
siRNA transfection and immunofluorescent staining.
MFLM-91U cells were obtained from Ann Akeson (Children's Hospital, Cincinnati, OH [1]) and cultured in serum-free UltraCULTURE medium (BioWhittaker). In order to inhibit Foxf1 expression in MFLM-91U cells, we used a 21-nucleotide siRNA duplex (Dharmacon) specific to the nucleotide +584 to +602 region of the mouse Foxf1 cDNA (siFoxf1, 5′-CCATTTGGCTGGCAACGTGTT). We transfected 100 nM of either siFoxf1 or control siRNA duplexes against a nontargeting sequence (Dharmacon) into MFLM-91U cells or MEFs by use of Lipofectamine 2000 reagent (Invitrogen) in serum-free culture medium as described previously (22, 50). Cells were harvested at 48 or 72 h after transfection and used for preparation of total RNA and protein. For immunofluorescent staining, MFLM-91U cells or MEFs were fixed with 10% paraformaldehyde and then stained with rabbit polyclonal antibody against cleaved caspase-3 (1:200 dilution; Cell Signaling) followed by anti-rabbit antibody conjugated with tetramethyl rhodamine isocyanate (TRITC). MFLM-91U cells were also pulse-labeled with bromodeoxyuridine (BrdU) for 15 min and then fixed with ethanol and immunostained for BrdU incorporation using the BrdU labeling and detection kit I (Roche Diagnostics) as described previously (22). MEFs were labeled with BrdU for 60 min.
Immunohistochemical staining.
Wild-type (WT), high Foxf1+/−, and low Foxf1+/− 15.5-dpc embryos were harvested, fixed overnight with 10% buffered formalin, and then embedded into paraffin blocks. Paraffin 5-μm sections were used for immunostaining with mouse monoclonal antibody against alpha smooth muscle actin (αSM) (clone 1A4, 1:1,000; Sigma). Antibody-antigen complexes were detected using secondary antibody directly conjugated with alkaline phosphatase (AP) followed by avidin-AP complex (Vector Labs). 5-Bromo-4-chloro-3-indolylphosphate (BCIP)-Nitro Blue Tetrazolium was used as an AP substrate as described previously (18). Sections were counterstained with nuclear fast red (Vector Labs, Burlingame, CA). We also used a rabbit polyclonal antibody against Itgβ3 (1:500; Cell Signaling). Antibody-antigen complexes were detected using biotinylated secondary antibody, avidin-horseradish peroxidase complex, and 3,3′-diaminobenzidine substrate (all from Vector Labs) as described previously (20, 24). Itgβ3-stained sections were counterstained with hematoxylin.
RT-PCR analysis.
RNA-STAT-60 (Tel-Test “B” Inc., Friendswood, TX) was used to prepare total RNA from MFLM-91U cells, MEFs, or lung tissue of Foxf1+/− 18.5-dpc embryos. After digestion of RNA with DNase I, reverse transcriptase PCR (RT-PCR) analysis was performed as described previously (20, 22). The following sense and antisense primers were used for amplification: for mouse Foxf1, 5′-CCTGGAGCAGCCATACCTT and 5′-TAAGATCCTCCGCCTGTTGT; for mouse Foxf2, 5′-ATCATCCTCTGCCTCCTGTG and 5′-AGCGCGATGTACGAGTAAGG; for mouse Foxm1b, 5′-GGATCCTGCCACCCCAGACCTTGTTC and 5′-GTCGACTCCCTGATGCTTTTCGCTGTC; for mouse cyclophilin, 5′-AGCTCTGAGCACTGGAGAGAAA and 5′-TCCTGAGCTACAGAAGGAATGG; for mouse Itgβ3, 5′-GACGCATCCCATTTGCTAGT and 5′-ACTGTGGTCCCAGGAATGAG; for human Foxf1, 5′-GGAGCGGGAAGTGACAAGA and 5′-AGCGAAGGAAGAGGAGGAAC; and for human cyclophilin, 5′-TGTGGATGCAGAAGATGGAT and 5′-AAACATGGCAGTGACACCAC. Two different cDNA concentrations were used for RT-PCRs to ensure that RT-PCR conditions were in the linear range.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed using in situ cross-linked MFLM-91U cells as described previously (50, 53). The resulting extract was subsequently sonicated and used for immunoprecipitation (IP) with Foxf1, Foxf2, or Foxm1b rabbit antibodies as described previously (38). IP with P-selectin rabbit antibody (BD Biosciences, Palo Alto, CA) was used as a control. Cross-links were reversed by the addition of 100 μl Tris-EDTA (TE) buffer containing 10 μg of RNase A. Proteinase K (10 μg) was then used to digest IP samples for 16 h at 65°C as described previously (50). DNA was extracted from the digested samples by use of PCR purification columns according to the manufacturer's instructions (QIAGEN, MD). ChIP DNA samples were used for PCR with primers specific to the bp −988 to −867 Itgβ3 promoter region (sense, 5′-CTGCCTCCTTAGGCTGGAAT; antisense, 5′-TAAAACTAGGGCAGGCGATG).
Expression and purification of recombinant TAT-Foxf1 DN and TAT-Engrail proteins.
T7-Foxf1 DN or T7-Engrail constructs were cloned into the pTAT vector, which was used to transform BL21 Star cells (Invitrogen). Since these fusion proteins contain six-His tags, Ni-nitrilotriacetic acid agarose (QIAGEN) was used for their purification. Purification was performed in native conditions following the manufacturer's protocol (QIAGEN) as described previously (33). Elution fractions were analyzed using GelCode blue stain reagent (Pierce) or by Western blotting using T7 antibody.
Cotransfection studies and infection of MEFs with recombinant adenoviruses.
MEFs or U2OS cells were transiently transfected with CMV-Foxf1, CMV-Foxm1b, or CMV-hepatocyte nuclear factor 6 (HNF-6) expression plasmid and the corresponding luciferase (LUC) reporter construct by use of Fugene 6 reagent (Roche) as described previously (23). CMV-Renilla was used as an internal control to normalize transfection efficiency. Purified TAT-T7-Foxf1 DN or TAT-T7-Engrail proteins (10 μg/ml) were added to the cell cultures at 24 and 48 h after transfection. A dual luciferase assay (Promega) was performed 24 h after the last protein treatment as described previously (23, 24). In separate experiments, transgenic TetO-Foxf1 DN MEFs were infected at a multiplicity of infection of 100 inclusion-forming units per cell with adenovirus containing tetracycline activator (Ad-TA) (Tet-off system) as described previously (24, 45) and 48 h later used for a luciferase assay. Adenovirus containing LacZ (Ad-LacZ) was used as a control.
Integrin-beta3 promoter constructs (−900 Itgβ3-LUC, −793 Itgβ3-LUC) were generated by PCR amplification using the following primers: sense, 5′-GGTACCTGCCGATTGCACCACATCGCC and 5′-GGTACCCCCTTCTTCGTAAGG ATG; antisense, 5′-CTCGAGTTCACGCCGCGCGCGCGCCAC. HEK-293 cells were transiently transfected with 300 ng of −900 Itgβ3-LUC or −793 Itgβ3-LUC plasmid and 100 ng of CMV-Foxf1 or CMV-empty vector and then used for dual luciferase assays (Promega) as described previously (23).
Migration and adhesion.
Adhesion assays were performed as described previously (5). Briefly, 24-well plates were coated with fibronectin (20 μg/ml in water for 1 h at 37°C). After the fibronectin solution was removed and the dishes were washed, the dishes were blocked with 1% bovine serum albumin for 1 h at 37°C. MFLM-91U cells (1 × 105 per ml) were added to the wells and then incubated for 30 min at 37°C. After three washings with phosphate-buffered saline, attached cells were fixed and then adhesion was quantified by counting cell number. All adhesion experiments were done in duplicate and repeated at least three times.
Cell migration was performed using a modification of the method of Varani (agarose drop assay) as described previously (32, 47). Briefly, MFLM-91U cells or MEFs (1 × 107 per ml) in serum-free medium were mixed with low-melting-point agarose solution and then plated as small drops onto tissue culture plastic. Agarose drops were allowed to solidify for 15 min at 4°C and then covered with serum-free medium containing fibronectin (20 μg/ml). Cell migration was assayed after 12 or 24 h by counting the cells which migrated from the agarose drop. The means ± standard deviations (SD) were calculated using all four sides of the drop in three individual agarose drops.
Statistical analysis.
Student's t test was used to determine statistical significance. P values of less than 0.05 were considered significant. Values for all measurements were expressed as the means ± SD.
RESULTS
Foxf1 induces the migration of MFLM-91U cells.
To determine whether Foxf1 transcription factor is expressed in mesenchymal cell lines, we examined Foxf1 expression by RT-PCR using primers specific to Foxf1. Abundant Foxf1 mRNA levels were detected in mesenchyme-derived MEFs and mouse fetal lung mesenchymal MFLM-91U cells (Fig. 1A), the latter of which display cellular characteristics of lung endothelial precursor cells (1). Low Foxf1 levels were observed in human U2OS osteosarcoma cells, whereas mouse MFLM-4 cells lacked detectable Foxf1 expression (Fig. 1A).
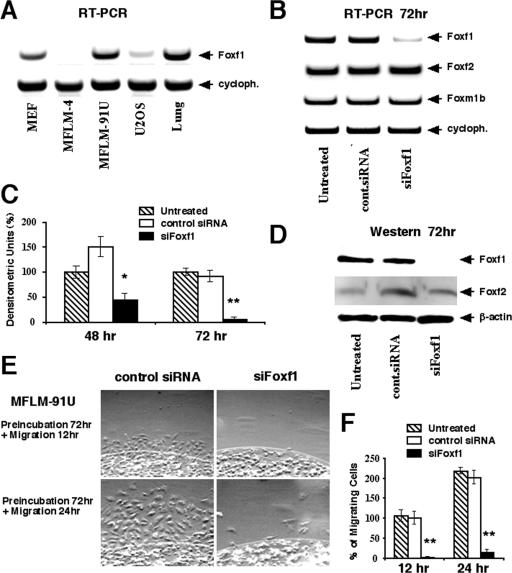
FIG. 1.
Depletion of Foxf1 levels by siRNA causes diminished mesenchyme migration. (A) Foxf1 is expressed in mesenchymal cell lines. Total RNA was prepared from mouse fetal lung mesenchymal cells (MFLM-4 and MFLM-91U), human osteosarcoma U2OS cells, or MEFs and then examined for Foxf1 and cyclophilin (cycloph.) levels by RT-PCR. (B) Transfection of Foxf1 siRNA into MFLM-91U cells effectively diminishes expression of endogenous Foxf1. Total RNA was prepared 72 h after transfection with either Foxf1-specific siRNA (siFoxf1) or control (cont.) siRNA and then analyzed for expression levels of Foxf1, Foxf2, Foxm1b, and cyclophilin by RT-PCR. (C and D) Transfection of siFoxf1 reduces Foxf1 protein levels in MFLM-91U cells. Nuclear protein extracts were prepared 48 or 72 h after transfection with either siFoxf1 or control siRNA and then analyzed for Foxf1 or Foxf2 by Western blot analysis. Levels of β-actin were determined using total protein extract. Foxf1 expression was normalized to its corresponding β-actin level and expressed relative to untreated cells. Values are means ± SD. (E and F) Foxf1 depletion reduces cell migration. (E) MFLM-91U cells were transfected for 72 h with either siFoxf1 or control siRNA and then placed into an agarose drop. Cell migration was assayed in the next 12 or 24 h using a phase-contrast microscope. (F) Numbers of cells migrating from the agarose drop were counted in six independent experiments and expressed as means ± SD. P values of <0.05 are shown with double asterisks.
Since previous reports have demonstrated that diminished Foxf1 levels in Foxf1+/− mice are associated with reduced numbers of mesenchymal cells in the lung, liver, and gallbladder (17, 20, 26, 28), we wanted to determine the role of Foxf1 in mesenchyme proliferation, survival, and migration. Mesenchymal MFLM-91U cells were transiently transfected with siRNA duplex specific to mouse Foxf1 cDNA (siFoxf1) or with control siRNA duplex. Total RNA was prepared 72 h after siRNA transfection and then analyzed for Foxf1 levels by RT-PCR (Fig. 1B). These transfection studies revealed that siFoxf1 efficiently reduced the expression of mouse Foxf1, whereas the transfection of control siRNA duplex did not influence the expression levels of this gene (Fig. 1B). Furthermore, we used Western blotting to demonstrate that Foxf1 protein levels were reduced by 50% or 95% in MFLM-91U cells transfected with siFoxf1 for 48 or 72 h, respectively (Fig. 1C and D). Interestingly, siFoxf1 transfection did not influence the expression levels of Foxf2 and Foxm1b genes (Fig. 1B and D), demonstrating a specificity of Foxf1 depletion by siFoxf1.
We next performed an agarose drop migration assay to investigate the consequences of Foxf1 depletion on the migration of mesenchymal cells. MFLM-91U cells were transfected for 72 h with either siFoxf1 or control siRNA and then placed into agarose drops. Transfected cells were allowed to migrate from the agarose for 12 or 24 h in a tissue culture dish coated with fibronectin, an extracellular matrix protein promoting cell migration (40). In comparison with either untransfected cells or cells transfected with control siRNA, depletion of Foxf1 levels by siFoxf1 caused a 95% decrease in numbers of migrating MFLM-91U cells (Fig. 1E and F), suggesting that Foxf1 is essential for mesenchyme migration.
Foxf1 depletion does not influence cell survival and DNA replication in vitro.
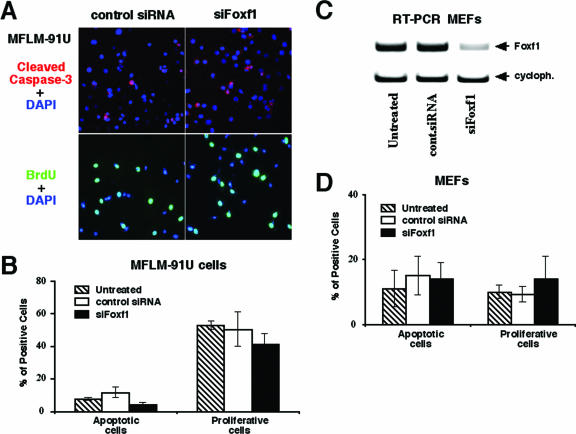
To determine the role of Foxf1 in DNA replication, MFLM-91U cells were transfected with siFoxf1 or control siRNA duplexes or were left untransfected. Seventy-two hours later, cells were pulse-labeled for 15 min with BrdU and then fixed and used for immunofluorescent staining with BrdU antibody. Depletion of Foxf1 did not influence the number of MFLM-91U cells undergoing DNA replication compared to that for either untransfected MFLM-91U cells or cells transfected with control siRNA duplex (Fig. 2A and B). Furthermore, no difference in the numbers of cells undergoing apoptosis was found for siFoxf1-transfected MFLM-91U cells, as determined by immunofluorescent staining with antibody against cleaved caspase-3 (Fig. 2A and B). Finally, siFoxf1 transfection caused a 70% reduction of Foxf1 mRNA levels in MEFs, which was insufficient to influence either DNA replication or cell survival in these cells (Fig. 2C and D). Altogether, these results suggest that depletion of Foxf1 levels does not influence cell survival or DNA replication in vitro.
FIG. 2.
Foxf1 depletion does not influence DNA replication and cell survival in vitro. (A and B) siFoxf1 transfection does not alter DNA replication and apoptosis in MFLM-91U cells. (A) MFLM-91U cells were transfected for 72 h with either siFoxf1 or control siRNA and then fixed and used for immunostaining with antibodies against cleaved caspase-3 (top panels) followed by TRITC-conjugated secondary antibodies (red). siRNA-transfected cells were treated with BrdU for 15 min and then fixed and immunostained with mouse monoclonal antibodies specific for BrdU (bottom panels) followed by anti-mouse antibody conjugated with fluorescein isothiocyanate (green). Cell nuclei were counterstained with 4′-6′-diamidino-2-phenylindole (DAPI) (blue). The numbers of caspase-3- and BrdU-positive cells were counted in five random microscope fields (magnification, ×400). (B) Three distinct siRNA transfections were used to determine the mean percentages of cells ± SD. (C) Transfection of Foxf1 siRNA into MEFs effectively diminishes expression of endogenous Foxf1. Total RNA was prepared 72 h after transfection with either Foxf1-specific siRNA (siFoxf1) or control siRNA and then analyzed for Foxf1 expression by RT-PCR. (D) Foxf1 depletion does not influence numbers of caspase-3- and BrdU-positive MEFs.
Foxf1 depletion causes reduced expression of integrin-beta3 subunit.
Previous reports have demonstrated that the Itgβ3 subunit is essential for fibronectin-mediated cell migration in a variety of normal and tumor cell lines (14, 40, 48). Therefore, we performed Western blotting and RT-PCR analysis to examine the temporal expression of Itgβ3 in MFLM-91U cells transfected with siFoxf1. In comparison with both untransfected cells and cells transfected with control siRNA, the depletion of Foxf1 levels by siFoxf1 caused significant decreases in the expression of Itgβ3 protein (Fig. 3B and C) and mRNA (Fig. 3A), a result consistent with the reduced migration of Foxf1-depleted MFLM-91U cells (Fig. 1E and F).
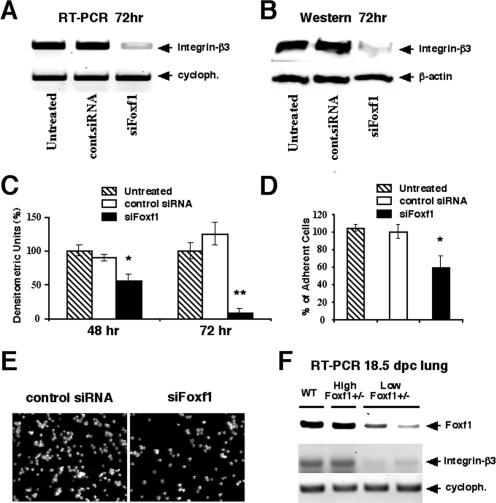
FIG. 3.
Foxf1 deficiency is associated with reduced integrin-beta3 expression and decreased cell adhesion. (A to C) siFoxf1 transfection causes reduced expression of the Itgβ3 subunit. MFLM-91U cells were transfected for 72 h with either siFoxf1 or control siRNA duplexes and then either fixed or used for preparation of total protein and RNA. (A) Total RNA was analyzed for expression levels of Itgβ3 and cyclophilin by RT-PCR analysis. (B and C) Western blotting showed significantly decreased levels of Itgβ3 protein in siFoxf1-transfected cells. Each individual sample was normalized to its corresponding β-actin level. Three independent experiments were used to determine means ± SD. A P value of <0.05 is shown with an asterisk, and a P value of <0.01 is shown with a double asterisk. (D and E) Foxf1 depletion inhibits adhesion of MFLM-91U cells. (D) MFLM-91U cells were transfected for 72 h with either siFoxf1 or control siRNA. Cells were counted and then added for 30 min to fibronectin-coated plates. After being washed, cells were fixed and the adhesion was quantified by counting all attached cells under a phase-contrast microscope. Results of four independent experiments were used to determine means ± SD. A P value of <0.05 is shown with an asterisk. (E) Microscopic images of cells transfected with either control siRNA or siFoxf1. (F) Decreased Foxf1 expression in low Foxf1+/− lungs is associated with reduced Itgβ3 levels. Total RNA was prepared from 18.5-dpc lungs of WT, high Foxf1+/−, and low Foxf1+/− embryos and then analyzed for expression levels of Foxf1, Itgβ3, and cyclophilin (cycloph.) by RT-PCR analysis.
Since Itgβ3 induces cell adhesion in mesenchymal cell lines (41), we examined the cell adhesion in Foxf1-depleted MFLM-91U cells. MFLM-91U cells were transfected for 72 h with either siFoxf1 or control siRNA and then used for the cell adhesion assay described in Materials and Methods. The depletion of Foxf1 levels by siFoxf1 caused a significant decrease in the number of adherent MFLM-91U cells compared to that for either untransfected MFLM-91U cells or cells transfected with control siRNA duplex (Fig. 3D and E). Taken together, our data suggest that Foxf1 depletion in mesenchymal MFLM-91U cells causes diminished cell migration and adhesion and reduces the expression of Itgβ3.
We have previously demonstrated that approximately half of Foxf1+/− embryos display severe mesenchymal defects and exhibit an 80% reduction in pulmonary Foxf1 levels (low Foxf1+/− embryos), whereas a subset of Foxf1+/− embryos exhibited normal lung development and compensatory increases of lung Foxf1 mRNA to wild-type levels (high Foxf1+/− embryos) (17). Therefore, we compared Itgβ3 levels in high Foxf1+/− and low Foxf1+/− 15.5-dpc embryos by use of immunohistochemistry with Itgβ3 antibody. Although the Itgβ3 protein was detected in both mesenchymal and epithelial cells of high Foxf1+/− and WT lungs (Fig. 4A and data not shown), Itgβ3 expression was selectively reduced in mesenchymal cells of low Foxf1+/− lungs (Fig. 4B). Reduced Itgβ3 staining was also observed in mesenchyme of low Foxf1+/− liver (Fig. 4C and D). Furthermore, RT-PCR analysis demonstrated that Itgβ3 mRNA levels were significantly diminished in total lung RNA prepared from low Foxf1+/− 18.5-dpc embryos, compared to either WT or high Foxf1+/− lungs (Fig. 3F). These results suggest that decreased Foxf1 expression in low Foxf1+/− embryos is associated with reduced Itgβ3 levels.
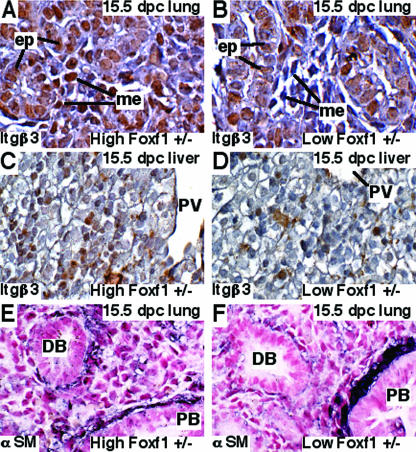
FIG. 4.
Embryonic low Foxf1+/− lungs and livers displayed reduced Itgβ3 expression. (A to D) Mesenchymal cells of low Foxf1+/− embryos displayed reduced Itgβ3 expression. Paraffin sections were prepared from high Foxf1+/− and low Foxf1+/− 15.5-dpc embryos and then immunostained with antibodies against Itgβ3. Sections were counterstained with hematoxylin (blue). (A) Itgβ3 protein (brown staining) was detected in both mesenchymal (me) and epithelial (ep) cells of high Foxf1+/− lungs. (B) Decreased Itgβ3 staining was observed in mesenchyme of low Foxf1+/− lungs. Compared to high Foxf1+/− livers (C), livers of low Foxf1+/− 15.5-dpc embryos displayed reduced Itgβ3 staining (D). PV, portal vein. (E and F) Low Foxf1+/− embryos displayed decreased αSM staining in the distal lung bronchioles. Paraffin sections of high Foxf1+/− (E) and low Foxf1+/− (F) 15.5-dpc embryos were stained with αSM antibody (dark purple) and counterstained with nuclear fast red (red). Abundant αSM expression was observed in proximal regions (PB) of low Foxf1+/− airways, while αSM staining was significantly diminished in the distal bronchioles (DB) of low Foxf1+/− lungs. Magnification, ×400.
We next used immunostaining with antibodies against αSM to visualize mesenchyme-derived myofibroblasts in high Foxf1+/− and low Foxf1+/− 15.5-dpc lungs. High Foxf1+/− lungs displayed normal αSM staining in both proximal and distal regions of pulmonary airways (Fig. 4E). In contrast, elevated αSM expression was observed in proximal regions of low Foxf1+/− airways, whereas the αSM staining was diminished in distal bronchioles of these mice (Fig. 4F). These results demonstrate that myofibroblasts accumulate in proximal lung regions of low Foxf1+/− mice, suggesting that Foxf1 deficiency causes diminished mesenchyme migration to distal lung regions.
A membrane-transducing TAT-T7-Foxf1 dominant negative protein significantly reduces Foxf1 transcriptional activity and Itgβ3 expression and inhibits migration of mesenchymal cells.
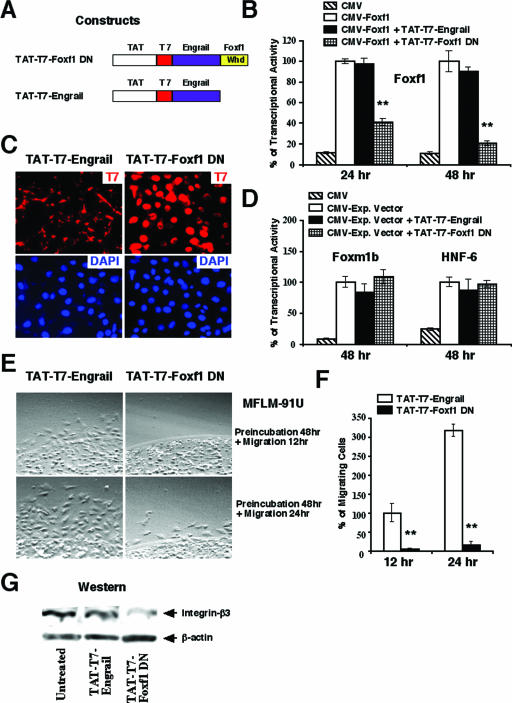
Previous reports have demonstrated that the addition of membrane transduction/nuclear localization domain from the human immunodeficiency virus TAT protein to the N termini of fusion proteins promotes their cellular uptake (33, 34). We therefore created the Foxf1 dominant negative fusion protein (TAT-T7-Foxf1 DN) consisting of the TAT polypeptide and the T7-tagged Engrailed transcriptional repression domain, which was cloned in frame with the N terminus of the Foxf1 winged helix DNA binding domain (Fig. 5A). Similar to the Gata6 DN mutant protein (4, 27), the Engrailed transcriptional repression domain was used in the TAT-T7-Foxf1 DN construct to provide an effective transcriptional repression on Foxf1 DNA binding sites. We also created a control TAT-T7-Engrail protein which lacked the Foxf1 winged helix DNA binding domain (Fig. 5A).
FIG. 5.
Inhibition of Foxf1 by TAT-T7-Foxf1 DN protein causes decreased migration of MFLM-91U cells and reduces Itgβ3 expression. (A) Schematic drawing of TAT-T7-Foxf1 DN and control TAT-T7-Engrail constructs. Whd, winged helix domain. (B) The TAT-T7-Foxf1 DN fusion protein reduces Foxf1 transcriptional activity. U2OS cells were transiently transfected with a TATA-luciferase reporter construct and a CMV-Foxf1 expression plasmid and then treated with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein. Cells were harvested 24 or 48 h later and processed for a dual luciferase assay to determine transcriptional activity. A P value of ≤0.05 is shown with double asterisks. (C) TAT-T7-Foxf1 DN protein localizes into cell nuclei. MFLM-91U cells were treated with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein for 24 h and then fixed and stained with mouse monoclonal T7 antibodies followed by anti-mouse antibodies conjugated with TRITC (red). Slides were counterstained with DAPI (blue). (D) The TAT-T7-Foxf1 DN fusion protein does not alter the transcriptional activity of either Foxm1b or HNF-6 protein in cotransfection experiments. (E and F) TAT-T7-Foxf1 DN fusion protein inhibits the migration of MFLM-91U cells. (E) MFLM-91U cells were treated for 48 h with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein. An agarose drop migration assay was performed as described in Materials and Methods. (F) Numbers of cells migrating from agarose were counted in six independent experiments and are presented as means ± SD. A P value of ≤0.05 is shown with double asterisks. (G) The TAT-T7-Foxf1 DN fusion protein reduces the expression of integrin-beta3. Total protein extracts were prepared from MFLM-91U cells treated for 48 h with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein. Western blotting was used to determine expression levels of Itgβ3 and β-actin.
Treatment of MFLM-91U cells with 10 μg/ml of either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein for 30 min demonstrated that these proteins were effectively transduced into all of the cells, as determined by immunofluorescent staining with T7 antibody (data not shown). Furthermore, MFLM-91U cells treated for 24 h with TAT-T7-Foxf1 DN protein displayed nuclear T7 staining (Fig. 4C), which is consistent with the presence of the nuclear localization sequence in the Foxf1 winged helix domain (36). Moreover, mesenchymal cells transfected with CMV-Foxf1 expression vector and the 6× Foxf1-TATA-luciferase plasmid and then treated with 10 μg/ml of the TAT-T7-Foxf1 DN protein displayed a significant reduction in Foxf1 transcriptional activity (Fig. 5B). Interestingly, the TAT-T7-Foxf1 DN protein was unable to reduce transcriptional activities of either Foxm1b or HNF-6 transcription factors in cotransfection experiments (Fig. 5D).
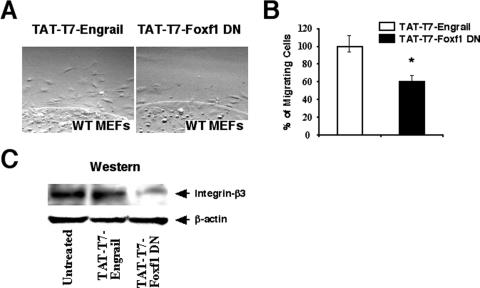
We next wanted to determine whether the inhibition of endogenous Foxf1 function by the TAT-T7-Foxf1 DN fusion protein could reduce fibronectin-mediated migration in MFLM-91U cells. MFLM-91U cells were treated for 48 h with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein and then used for an agarose drop migration assay. Cell migration was determined by counting numbers of cells migrating from agarose in the next 12 or 24 h. In comparison with what was seen for cells treated with control TAT-T7-Engrail protein, the inhibition of Foxf1 activity by the TAT-T7-Foxf1 DN protein caused a 90 to 95% decrease in the number of migrating MFLM-91U cells (Fig. 5E and F). Furthermore, a significant reduction in Itgβ3 protein was observed for MFLM-91U cells treated with TAT-T7-Foxf1 DN (Fig. 5G), a result consistent with diminished Itgβ3 expression in siFoxf1-transfected MFLM-91U cells (Fig. 3A to C). Finally, since Foxf1 is expressed in mesenchyme-derived MEFs (Fig. 1A), we wanted to determine whether Foxf1 regulates cell migration and Itgβ3 expression in these primary mouse cells. Treatment of MEFs with 10 μg/ml of TAT-T7-Foxf1 DN for 48 h significantly reduced fibronectin-mediated cell migration (Fig. 6A and B) and protein levels of Itgβ3 (Fig. 6C) compared to MEFs treated with control TAT-T7-Engrail protein. These results suggest that the TAT-T7-Foxf1 DN fusion protein effectively reduces the expression of Itgβ3 and inhibits the migration of mesenchymal cells.
FIG. 6.
The TAT-T7-Foxf1 DN protein inhibits the migration of mouse embryonic fibroblasts and reduces Itgβ3 expression in these primary cells. (A and B) The TAT-T7-Foxf1 DN protein reduces MEF migration. (A) MEFs were treated for 48 h with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail protein and then used for an agarose drop migration assay. (B) Cell migration was assayed after 12 h by counting the number of cells migrating from the agarose (± SD). A P value of ≤0.05 is shown with an asterisk. Six distinct agarose drops were used for each condition. (C) Decreased level of Itgβ3 in Foxf1-deficient MEFs. Total protein extracts were prepared from MEFs treated for 48 h with either TAT-T7-Foxf1 DN or control TAT-T7-Engrail fusion protein. Western blotting was used to determine expression levels of Itgβ3 and β-actin.
Conditional expression of Foxf1 DN protein in transgenic MEFs decreases cell migration and reduces expression of integrin-beta3 subunit.
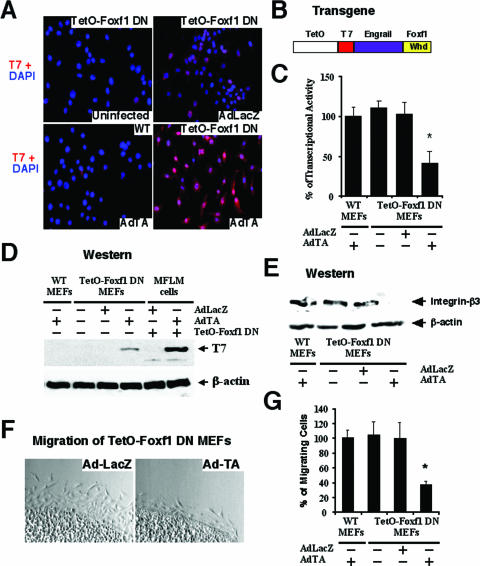
To determine whether Foxf1 induces mesenchyme migration in vivo, we generated a transgenic mouse line which used the CMV-TetO promoter to drive expression of the Foxf1 DN fusion protein (Fig. 7B). To activate the TetO-Foxf1 DN transgene, transgenic MEFs were prepared from TetO-Foxf1 DN 12.5-dpc mouse embryos and then infected in cell culture conditions for 48 h with either Ad-TA or Ad-LacZ. Western blotting with T7 antibody demonstrated that the Foxf1 DN protein is induced by Ad-TA infection (Tet-off system) in transgenic TetO-Foxf1 DN MEFs, but it was not detected either in Ad-TA-infected wild-type MEFs or in transgenic TetO-Foxf1 DN MEFs infected with Ad-LacZ (Fig. 7D). The Foxf1 DN protein was localized in nuclei of Ad-TA-infected transgenic MEFs, as demonstrated by immunofluorescent staining with T7 antibody (Fig. 7A). Furthermore, cotransfection studies with the CMV-Foxf1 expression vector and the 6× Foxf1-TATA-luciferase reporter plasmid showed that Foxf1 DN protein effectively inhibited Foxf1 transcriptional activity in transgenic TetO-Foxf1 DN MEFs when they were infected with Ad-TA (Fig. 7C). In contrast, both Ad-LacZ-infected transgenic MEFs and Ad-TA-infected WT MEFs displayed normal Foxf1 transcriptional activity (Fig. 7C). These results demonstrate that conditional expression of the Foxf1 DN transgene in transgenic MEFs effectively reduces Foxf1 transcriptional activity. Furthermore, reduced Foxf1 transcriptional activity in Ad-TA-infected transgenic TetO-Foxf1 DN MEFs was associated with diminished cell migration (Fig. 7F and G) and reduced expression of Itgβ3 protein (Fig. 7E), which is consistent with our studies using either Foxf1-specific siRNA (Fig. 1 and 3) or TAT-T7-Foxf1 dominant negative fusion protein (Fig. 5 and 6).
FIG. 7.
Conditional expression of Foxf1 DN protein in transgenic MEFs decreases cell migration and reduces expression of integrin-beta3. (A) Nuclear localization of Foxf1 DN protein in transgenic MEFs infected with Ad-TA. Transgenic TetO-Foxf1 DN or WT MEFs were infected with Ad-TA (Tet-off system) or control Ad-LacZ. Cells were then fixed and used for immunofluorescent staining with T7 antibody (red) followed by DAPI counterstaining (blue) to visualize cell nuclei. (B) Schematic drawing of the TetO-Foxf1 DN transgene, containing the CMV-TetO promoter and the Foxf1 DNA binding winged helix domain (Whd) fused with Engrail transcriptional repressor domain and T7 tag sequences. (C) Foxf1 transcriptional activity is reduced in transgenic TetO-Foxf1 DN MEFs after infection with Ad-TA. Transgenic or WT MEFs were transiently transfected with 6× Foxf1 TATA-luciferase reporter plasmid and CMV-Foxf1 expression vector and then infected with either Ad-TA or control Ad-LacZ. Cells were harvested 48 h later and processed for a dual luciferase assay to determine luciferase activity. Transcriptional activity levels are presented as means ± SD. A P value of ≤0.05 is shown with an asterisk. (D) Western blotting shows increased expression of Foxf1 DN protein in transgenic TetO-Foxf1 DN MEFs infected with Ad-TA. Total protein extracts were prepared from transgenic or WT MEFs infected with either Ad-TA or control Ad-LacZ. The positive control was MFLM-91U cells that were transiently transfected with TetO-Foxf1 DN plasmid and then infected with Ad-TA. (E) Western blotting shows decreased expression of Itgβ3 in transgenic TetO-Foxf1 DN MEFs infected with Ad-TA. Protein levels of β-actin were used as loading controls. (F and G) Activation of the Foxf1 DN transgene reduces cell migration in transgenic TetO-Foxf1 DN MEFs. Transgenic or WT MEFs were infected for 48 h with either Ad-TA or Ad-LacZ and then used for an agarose drop migration assay. Twelve hours later, numbers of migrating cells were counted in six independent agarose drops. A P value of ≤0.05 is shown with an asterisk.
Foxm1 directly regulates mouse Itgβ3 promoter.
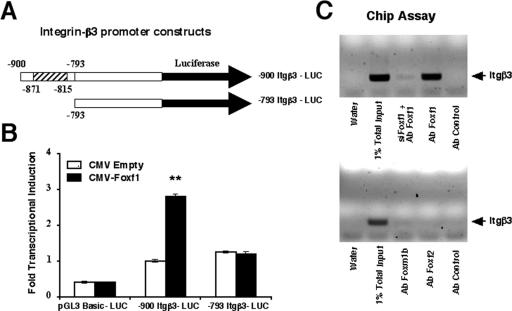
Because inhibition of Foxf1 function by either siRNA or Foxf1 dominant negative protein caused significant reduction in both Itgβ3 mRNA and protein levels, we investigated whether Foxf1 transcriptionally regulates mouse Itgβ3 promoter. One potential Foxf1 DNA binding site was identified in the bp −900 promoter region of the mouse Itgβ3 gene (−871 to −815), which contains 15 overlapping binding motifs for Foxf1 protein (Fig. 8A). Cotransfection experiments were performed with HEK-293 cells (which do not express endogenous Foxf1) by use of the CMV-Foxf1 expression vector and the LUC reporter driven by the bp −900 Itgβ3 promoter region. Cotransfection of the CMV-Foxf1 expression vector increased expression of the bp −900 Itgβ3-LUC reporter plasmid compared to that for CMV-empty vector (Fig. 8B), suggesting that Foxf1 is a transcriptional activator of the Itgβ3 gene. Furthermore, deletion of the bp −900 to −793 Itgβ3 region was sufficient to reduce the ability of Foxf1 to activate transcription of the Itgβ3 promoter in cotransfection experiments (Fig. 8B), indicating that the bp −900 to −793 Itgβ3 region is essential for transcriptional activation of the Itgβ3 promoter by Foxf1.
FIG. 8.
Foxf1 directly regulates the mouse Itgβ3 promoter through the bp −871 to −815 Itgβ3 promoter region. (A) Schematic drawing of Itgβ3 promoter constructs. Schematically shown are LUC reporter constructs that use the bp −900 Itgβ3 promoter region (includes a Foxf1 binding site) and the bp −793 Itgβ3 promoter region (without the Foxf1 binding site) to drive expression of the LUC reporter. (B) Foxf1 induces Itgβ3 promoter activity in cotransfection assays. We transiently transfected HEK-293 cells (which do not express endogenous Foxf1) with CMV-Foxf1 expression vector and the bp −900 Itgβ3-LUC or bp −793 Itgβ3-LUC reporter plasmid. Cells were harvested at 48 h after transfection and processed for dual luciferase assays to determine luciferase activity. Transcriptional induction is shown as the increase (n-fold) relative to that for CMV-empty vector (± SD). A P value of ≤0.05 is shown with double asterisks. (C) ChIP assays show that Foxf1 protein binds to the Itgβ3 promoter region in the context of endogenous DNA. Cross-linked chromatin was prepared from untreated MFLM-91U cells or MFLM-91U cells transfected with siFoxf1 for 72 h. The cross-linked and sonicated chromatin was then immunoprecipitated with antibodies (Ab) specific to Foxf1, Foxf2, Foxm1b, or P-selectin (control). The IP genomic DNA was analyzed for the amount of mouse Itgβ3 promoter DNA by use of PCR analysis with primers specific to the mouse Itgβ3 promoter region (bp −988 to −867).
We next used ChIP assays to determine whether the Foxf1 protein binds to the bp −871 to −815 Itgβ3 promoter region in the context of endogenous mouse DNA. The cross-linked and sonicated chromatin from mesenchymal MFLM-91U cells was immunoprecipitated with antibodies specific to either Foxf1 or immunoglobulin G control antibody. The binding of Itgβ3 promoter DNA with IP chromatin was determined by PCR with primers specific to the mouse bp −988 to −867 Itgβ3 promoter region. The ChIP assay demonstrated that Foxf1 specifically binds to the endogenous mouse Itgβ3 promoter, whereas depleting Foxf1 levels by siFoxf1 significantly diminished the association of Foxf1 protein with this endogenous Itgβ3 promoter region (Fig. 8C). Although Foxf2 protein was unable to bind the mouse bp −988 to −867 Itgβ3 promoter region, the Foxm1b protein showed a very weak association with the Itgβ3 promoter region (Fig. 8C). Taken together, these results demonstrate that the mouse Itgβ3 gene is a direct transcriptional target of Foxf1.
DISCUSSION
Recent studies have demonstrated that the Foxf1 protein is an important transcriptional regulator in developing mesenchyme during mouse embryonic development. Foxf1−/− embryos die by 8 dpc due to severe mesenchymal defects in the yoke sac and allantois (17, 29). Approximately half of the newborn Foxf1+/− mice die from pulmonary hemorrhage and exhibit an 80% reduction in pulmonary Foxf1 levels (low Foxf1+/−) (17). These low Foxf1+/− mice display a pulmonary hypoplasia, a fusion of lung lobes, an increased apoptosis of mesenchymal cells, and a reduced number of alveolar capillaries (17, 26). Interestingly, approximately 40% of the newborn Foxf1+/− mice exhibited normal development of alveolar capillaries (17), which was associated with compensatory increases of lung Foxf1 mRNA to wild-type levels (high Foxf1+/− mice). Although these mouse genetic studies suggest that WT levels of Foxf1 are critical for the development of lung mesenchyme in vivo, molecular mechanisms underlying Foxf1 function remain largely unknown. In this study, we used siRNA duplexes and the Foxf1 dominant negative mutant protein (Foxf1 DN) to inhibit Foxf1 function in mouse fetal lung mesenchymal cells (MFLM-91U cells) and MEFs. Our results demonstrated that Foxf1 induces cell migration in these mesenchymal cells but plays no role in DNA replication. Therefore, diminished mesenchyme migration most likely causes reduced numbers of mesenchymal cells in low Foxf1+/− embryonic lungs. Since pulmonary mesenchyme gives a rise to capillary endothelial cells (1), decreased mesenchyme numbers may also contribute to reduced vascular development in low Foxf1+/− lungs. Interestingly, Foxf1 depletion in MFLM-91U cells caused decreased cell adhesion and reduced migration, raising a possibility that Foxf1 may regulate many mesenchymal genes which are essential for both cell migration and cell adhesion.
Foxf1 is not expressed in all mesenchymal cells of developing lung, including endothelial and smooth muscle cells of large pulmonary vessels (17, 18). In this study, we also found that mouse fetal lung mesenchymal MFLM-4 cells do not express Foxf1, suggesting a functional heterogeneity of mesenchymal cells during lung development. However, it is not clear whether Foxf1 expression represents a lineage commitment toward specific mesenchymal cell populations or marks selective stages of mesenchyme differentiation.
Low Foxf1+/− embryos displayed significant increases in the number of apoptotic cells in the developing lung (17). Furthermore, adult high Foxf1+/− mice died from severe lung hemorrhage after butylated hydroxytoluene lung injury (21). This phenotype was associated with a 10-fold decrease in pulmonary Foxf1 expression and increased alveolar endothelial cell apoptosis (21), suggesting that Foxf1 may play a direct role in cell survival. However, our results demonstrate that the inhibition of Foxf1 function by siRNA transfection causes insignificant changes in the number of apoptotic cells. These results suggest that Foxf1 is not essential for cell survival and therefore that increased apoptosis in Foxf1+/− lungs can be explained by indirect effects of Foxf1 deficiency on pulmonary homeostasis and/or metabolism. Previous studies reported diminished cellular proliferation in the lateral plate mesoderm of Foxf1-deficient embryos (29, 46), suggesting that Foxf1 positively regulates mesenchyme proliferation during embryonic development. In this study, we demonstrated that a 70% reduction in Foxf1 levels does not influence DNA replication in either MEFs or MFLM-91U cells (Fig. 2). Therefore, it is possible that proliferation defects in Foxf1-deficient embryos are indirect and represent consequences of reduced BMP-4 and Shh signaling, as was reported by these studies' authors (29, 46). Furthermore, reduced vasculogenesis in the yolk sac and allantois can contribute to diminished proliferation in the lateral plate mesoderm of Foxf1−/− embryos (29). Alternatively, a 70% reduction in Foxf1 levels may be insufficient to diminish DNA replication in MEFs and MFLM-91U cells in cell culture experiments.
In this study, we demonstrated that diminished mesenchyme migration in Foxf1-deficient cells is associated with reduced mRNA and protein levels of Itgβ3, which is known to interact with the integrin-αV subunit to form αVβ3 integrin, which functions as a receptor for several ECM proteins (13, 40). The αVβ3 protein complex is regulated by expression of the Itgβ3 subunit during mesenchyme migration and differentiation (14, 40). Although little is known regarding the transcriptional mechanisms mediating Itgβ3 expression, several transcription factors can directly bind to the Itgβ3 promoter region, where they serve as transcriptional activators. These include the transcription factor Stat-6 (31), the nuclear factor of activated T cells c1 (NFATc1) (9), the homeobox HoxA10 (10), and the nuclear factor kappaB (NF-κB) (42) proteins. Interestingly, DNA binding sites for all these transcription factors are located in the Kb −2 Itgβ3 promoter region, which displays up to 80% sequence homology between mouse and human Itgβ3 genes (9). In this study, we demonstrated that this conserved Itgβ3 promoter region is also regulated by the Forkhead box Foxf1 transcription factor, which binds the Itgβ3 promoter in the −871 to −815 region. Deletion of this region completely abolished the transcriptional activity of the Itgβ3 promoter, indicating that the mouse Itgβ3 is a direct transcriptional target of Foxf1 protein. Since depletion of Foxf1 levels in lung mesenchymal cells almost completely inhibits cell migration, Foxf1 protein may represent an attractive target to inhibit cell migration during cancer invasion and tumor angiogenesis.
In summary, Foxf1 induces the migration of mesenchymal cells without detectable changes in cell proliferation and cell survival. Diminished mesenchyme migration in Foxf1-deficient cells was associated with reduced mRNA and protein levels of Itgβ3, a key player in cell migration (14, 48). Foxf1 protein physically recognized the −871 to −815 Itgβ3 promoter sequences in context of endogenous DNA. Deletion of this Foxf1 binding site completely abolished the ability of Foxf1 to activate transcription of the Itgβ3 promoter, indicating that the mouse Itgβ3 is a direct transcriptional target of Foxf1 protein.
Acknowledgments
This work was supported by American Heart Association Scientist Development Grant 0335036N (to V.V.K.), research grant 6-FY2005-325 from the March of Dimes Birth Defects Foundation (to V.V.K.), and U.S. Public Health Service grant HL 84151-01 (to V.V.K.).
We thank Ann Akeson for providing MFLM-91U and MFLM-4 cell lines. We also thank Robert Costa for critically reviewing the manuscript.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Akeson, A. L., B. Wetzel, F. Y. Thompson, S. K. Brooks, H. Paradis, R. L. Gendron, and J. M. Greenberg. 2000. Embryonic vasculogenesis by endothelial precursor cells derived from lung mesenchyme. Dev. Dyn. 217:11-23. [DOI] [PubMed] [Google Scholar]
- 2.Barr, F. G. 2001. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 20:5736-5746. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, R. M., and J. L. Brissette. 2002. Role of the nude gene in epithelial terminal differentiation. J. Investig. Dermatol. 118:303-309. [DOI] [PubMed] [Google Scholar]
- 4.Bruno, M. D., T. R. Korfhagen, C. Liu, E. E. Morrisey, and J. A. Whitsett. 2000. GATA-6 activates transcription of surfactant protein A. J. Biol. Chem. 275:1043-1049. [DOI] [PubMed] [Google Scholar]
- 5.Chernousov, M. A., R. C. Stahl, and D. J. Carey. 2001. Schwann cell type V collagen inhibits axonal outgrowth and promotes Schwann cell migration via distinct adhesive activities of the collagen and noncollagen domains. J. Neurosci. 21:6125-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-420. [DOI] [PubMed] [Google Scholar]
- 7.Costa, R. H., V. V. Kalinichenko, A. X. Holterman, and X. Wang. 2003. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331-1347. [DOI] [PubMed] [Google Scholar]
- 8.Costa, R. H., V. V. Kalinichenko, and L. Lim. 2001. Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L823-L838. [DOI] [PubMed] [Google Scholar]
- 9.Crotti, T. N., M. Flannery, N. C. Walsh, J. D. Fleming, S. R. Goldring, and K. P. McHugh. 2006. NFATc1 regulation of the human beta3 integrin promoter in osteoclast differentiation. Gene 372:92-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daftary, G. S., P. J. Troy, C. N. Bagot, S. L. Young, and H. S. Taylor. 2002. Direct regulation of beta3-integrin subunit gene expression by HOXA10 in endometrial cells. Mol. Endocrinol. 16:571-579. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, S. A. 2003. Mechanisms controlling early development of the liver. Mech. Dev. 120:19-33. [DOI] [PubMed] [Google Scholar]
- 12.Garry, D. J., Q. Yang, R. Bassel-Duby, and R. S. Williams. 1997. Persistent expression of MNF identifies myogenic stem cells in postnatal muscles. Dev. Biol. 188:280-294. [DOI] [PubMed] [Google Scholar]
- 13.Hynes, R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673-687. [DOI] [PubMed] [Google Scholar]
- 14.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 15.Kaestner, K. H. 2000. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol. Metab. 11:281-285. [DOI] [PubMed] [Google Scholar]
- 16.Kalinichenko, V. V., D. Bhattacharyya, Y. Zhou, G. A. Gusarova, W. Kim, B. Shin, and R. H. Costa. 2003. Foxf1 +/− mice exhibit defective stellate cell activation and abnormal liver regeneration following CCl4 injury. Hepatology 37:107-117. [DOI] [PubMed] [Google Scholar]
- 17.Kalinichenko, V. V., L. Lim, D. Beer-Stoltz, B. Shin, F. M. Rausa, J. Clark, J. A. Whitsett, S. C. Watkins, and R. H. Costa. 2001. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the forkhead box f1 transcription factor. Dev. Biol. 235:489-506. [DOI] [PubMed] [Google Scholar]
- 18.Kalinichenko, V. V., L. Lim, B. Shin, and R. H. Costa. 2001. Differential expression of forkhead box transcription factors following butylated hydroxytoluene lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L695-L704. [DOI] [PubMed] [Google Scholar]
- 19.Kalinichenko, V. V., M. Major, X. Wang, V. Petrovic, J. Kuechle, H. M. Yoder, B. Shin, A. Datta, P. Raychaudhuri, and R. H. Costa. 2004. Forkhead box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18:830-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalinichenko, V. V., Y. Zhou, D. Bhattacharyya, W. Kim, B. Shin, K. Bambal, and R. H. Costa. 2002. Haploinsufficiency of the mouse forkhead box f1 gene causes defects in gall bladder development. J. Biol. Chem. 277:12369-12374. [DOI] [PubMed] [Google Scholar]
- 21.Kalinichenko, V. V., Y. Zhou, B. Shin, D. Beer-Stoltz, S. C. Watkins, J. A. Whitsett, and R. H. Costa. 2002. Wild type levels of the mouse forkhead box f1 gene are essential for lung repair. Am. J. Physiol. Lung Cell. Mol. Physiol. 282:L1253-L1265. [DOI] [PubMed] [Google Scholar]
- 22.Kim, I. M., T. Ackerson, S. Ramakrishna, M. Tretiakova, I. C. Wang, T. V. Kalin, M. L. Major, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2006. The forkhead box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 66:2153-2161. [DOI] [PubMed] [Google Scholar]
- 23.Kim, I. M., S. Ramakrishna, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2005. The forkhead box M1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280:22278-22286. [DOI] [PubMed] [Google Scholar]
- 24.Kim, I. M., Y. Zhou, S. Ramakrishna, D. E. Hughes, J. Solway, R. H. Costa, and V. V. Kalinichenko. 2005. Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J. Biol. Chem. 280:37908-37916. [DOI] [PubMed] [Google Scholar]
- 25.Lai, E., K. L. Clark, S. K. Burley, and J. E. Darnell, Jr. 1993. Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proc. Natl. Acad. Sci. USA 90:10421-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, L., V. V. Kalinichenko, J. A. Whitsett, and R. H. Costa. 2002. Fusion of right lung lobes and pulmonary vessels in mice heterozygous for the Forkhead Box f1 targeted allele. Am. J. Physiol. Lung Cell. Mol. Physiol. 282:L1012-L1022. [DOI] [PubMed] [Google Scholar]
- 27.Liu, C., E. E. Morrisey, and J. A. Whitsett. 2002. GATA-6 is required for maturation of the lung in late gestation. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L468-L475. [DOI] [PubMed] [Google Scholar]
- 28.Mahlapuu, M., S. Enerbäck, and P. Carlsson. 2001. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development 128:2397-2406. [DOI] [PubMed] [Google Scholar]
- 29.Mahlapuu, M., M. Ormestad, S. Enerback, and P. Carlsson. 2001. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128:155-166. [DOI] [PubMed] [Google Scholar]
- 30.Mahlapuu, M., M. Pelto-Huikko, M. Aitola, S. Enerback, and P. Carlsson. 1998. FREAC-1 contains a cell-type-specific transcriptional activation domain and is expressed in epithelial-mesenchymal interfaces. Dev. Biol. 202:183-195. (Erratum, 207:476, 1999.) [DOI] [PubMed] [Google Scholar]
- 31.McHugh, K. P., S. Kitazawa, S. L. Teitelbaum, and F. P. Ross. 2001. Cloning and characterization of the murine beta(3) integrin gene promoter: identification of an interleukin-4 responsive element and regulation by STAT-6. J. Cell. Biochem. 81:320-332. [DOI] [PubMed] [Google Scholar]
- 32.Milner, R., H. J. Anderson, R. F. Rippon, J. S. McKay, R. J. Franklin, M. A. Marchionni, R. Reynolds, and C. Ffrench-Constant. 1997. Contrasting effects of mitogenic growth factors on oligodendrocyte precursor cell migration. Glia 19:85-90. [DOI] [PubMed] [Google Scholar]
- 33.Myou, S., X. Zhu, E. Boetticher, S. Myo, A. Meliton, A. Lambertino, N. M. Munoz, and A. R. Leff. 2002. Blockade of focal clustering and active conformation in beta 2-integrin-mediated adhesion of eosinophils to intercellular adhesion molecule-1 caused by transduction of HIV TAT-dominant negative Ras. J. Immunol. 169:2670-2676. [DOI] [PubMed] [Google Scholar]
- 34.Nagahara, H., A. M. Vocero-Akbani, E. L. Snyder, A. Ho, D. G. Latham, N. A. Lissy, M. Becker-Hapak, S. A. Ezhevsky, and S. F. Dowdy. 1998. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 4:1449-1452. [DOI] [PubMed] [Google Scholar]
- 35.Nakae, J., W. H. Biggs III, T. Kitamura, W. K. Cavenee, C. V. Wright, K. C. Arden, and D. Accili. 2002. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 32:245-253. [DOI] [PubMed] [Google Scholar]
- 36.Overdier, D. G., A. Porcella, and R. H. Costa. 1994. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino acid residues adjacent to the recognition helix. Mol. Cell. Biol. 14:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson, R. S., L. Lim, H. Ye, H. Zhou, D. G. Overdier, and R. H. Costa. 1997. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech. Dev. 69:53-69. [DOI] [PubMed] [Google Scholar]
- 38.Rausa, F., Y. Tan, and R. H. Costa. 2003. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol. Cell. Biol. 23:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodan, S. B., and G. A. Rodan. 1997. Integrin function in osteoclasts. J. Endocrinol. 154:S47-S56. [PubMed] [Google Scholar]
- 40.Ruegg, C., O. Dormond, and A. Mariotti. 2004. Endothelial cell integrins and COX-2: mediators and therapeutic targets of tumor angiogenesis. Biochim. Biophys. Acta 1654:51-67. [DOI] [PubMed] [Google Scholar]
- 41.Ruegg, C., and A. Mariotti. 2003. Vascular integrins: pleiotropic adhesion and signaling molecules in vascular homeostasis and angiogenesis. Cell. Mol. Life Sci. 60:1135-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scatena, M., M. Almeida, M. L. Chaisson, N. Fausto, R. F. Nicosia, and C. M. Giachelli. 1998. NF-kappaB mediates alphavbeta3 integrin-induced endothelial cell survival. J. Cell Biol. 141:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shattil, S. J., and P. J. Newman. 2004. Integrins: dynamic scaffolds for adhesion and signaling in platelets. Blood 104:1606-1615. [DOI] [PubMed] [Google Scholar]
- 44.Stahl, M., P. F. Dijkers, G. J. Kops, S. M. Lens, P. J. Coffer, B. M. Burgering, and R. H. Medema. 2002. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 168:5024-5031. [DOI] [PubMed] [Google Scholar]
- 45.Tan, Y., R. H. Costa, I. Kovesdi, and R. R. Reichel. 2001. Adenovirus-mediated increase of HNF-3 levels stimulates transthyretin and sonic hedgehog expression which is associated with F9 cell differentiation toward the visceral endoderm lineage. Gene Expr. 9:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tseng, H. T., R. Shah, and M. Jamrich. 2004. Function and regulation of FoxF1 during Xenopus gut development. Development 131:3637-3647. [DOI] [PubMed] [Google Scholar]
- 47.Varani, J., W. Orr, and P. A. Ward. 1978. Comparison of cell attachment and caseinolytic activities of five tumour cell types. J. Cell Sci. 34:133-144. [DOI] [PubMed] [Google Scholar]
- 48.Varner, J. A., and D. A. Cheresh. 1996. Integrins and cancer. Curr. Opin. Cell Biol. 8:724-730. [DOI] [PubMed] [Google Scholar]
- 49.Vogt, P. K., J. Li, and B. S. Freyaldenhoven. 1997. Revelations of a captive: retroviral Qin and the oncogenicity of winged helix proteins. Virology 238:1-7. [DOI] [PubMed] [Google Scholar]
- 50.Wang, I. C., Y. J. Chen, D. Hughes, V. Petrovic, M. L. Major, H. J. Park, Y. Tan, T. Ackerson, and R. H. Costa. 2005. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25:10875-10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, X., H. Kiyokawa, M. B. Dennewitz, and R. H. Costa. 2002. The forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA 99:16881-16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warburton, D., M. Schwarz, D. Tefft, G. Flores-Delgado, K. D. Anderson, and W. V. Cardoso. 2000. The molecular basis of lung morphogenesis. Mech. Dev. 92:55-81. [DOI] [PubMed] [Google Scholar]
- 53.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26:48-56. [DOI] [PubMed] [Google Scholar]
- 54.Winnier, G. E., L. Hargett, and B. L. Hogan. 1997. The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes Dev. 11:926-940. [DOI] [PubMed] [Google Scholar]
- 55.Zaret, K. S. 2000. Liver specification and early morphogenesis. Mech. Dev. 92:83-88. [DOI] [PubMed] [Google Scholar]
- 56.Zaret, K. S. 2002. Regulatory phases of early liver development: paradigms of organogenesis. Nat. Rev. Genet. 3:499-512. [DOI] [PubMed] [Google Scholar]