Abstract
Cellular senescence is an irreversible proliferation arrest triggered by short chromosome telomeres, activated oncogenes, and cell stress and mediated by the pRB and p53 tumor suppressor pathways. One of the earliest steps in the senescence program is translocation of a histone chaperone, HIRA, into promyelocytic leukemia (PML) nuclear bodies. This relocalization precedes other markers of senescence, including the appearance of specialized domains of facultative heterochromatin called senescence-associated heterochromatin foci (SAHF) and cell cycle exit. SAHF represses expression of proliferation-promoting genes, thereby driving exit from the cell cycle. HIRA bound to another histone chaperone, ASF1a, drives formation of SAHF. Here, we show that HIRA's translocation to PML bodies occurs in response to all senescence triggers tested. Dominant negative HIRA mutants that block HIRA's localization to PML bodies prevent formation of SAHF, as does a PML-RARα fusion protein which disrupts PML bodies, directly supporting the idea that localization of HIRA to PML bodies is required for formation of SAHF. Significantly, translocation of HIRA to PML bodies occurs in the absence of functional pRB and p53 tumor suppressor pathways. However, our evidence indicates that downstream of HIRA's localization to PML bodies, the HIRA/ASF1a pathway cooperates with pRB and p53 to make SAHF, with the HIRA/ASF1a and pRB pathways acting in parallel. We present evidence that convergence of the HIRA/ASF1a and pRB pathways occurs through a DNAJ-domain protein, DNAJA2.
Cellular senescence is characterized by an irreversible arrest of cell proliferation. Senescence has been most widely studied in fibroblasts in vitro, where it is characterized by several molecular and cytological markers, such as a large flat morphology, expression of a senescence-associated β-galactosidase activity (SA β-Gal), and formation of domains of heterochromatin, called senescence-associated heterochromatin foci (SAHF) (11, 32, 66). In addition, senescence is well defined in epithelial cells grown in culture. Other cell types suggested to undergo senescence include hematopoietic and neural progenitor cells (22, 48). Senescence in vivo is an important tumor suppression process. Activated oncogenes trigger cell senescence, thereby blocking progression to a transformed cell phenotype, most notably in human melanocytes, human prostatic epithelium, mouse lymphocytes, and premalignant mouse lung adenomas (10, 12, 13, 17, 41, 58). As well as activated oncogenes, senescence is caused by shortened telomeres that result from repeated rounds of cell division, cellular stresses, and inadequate growth conditions (11, 32, 52, 66). Senescence in vivo is also thought to contribute to tissue aging through exhaustion of renewable tissue stem cell populations (11, 32, 34, 38, 42, 66).
A large body of evidence indicates that the pRB and p53 tumor suppressor pathways are master regulators of senescence. Inactivation of these two pathways abolishes senescence in mouse and human cells, regardless of the initial senescence trigger (11, 32, 66). Although human cells lacking pRB and p53 circumvent senescence, most such cells ultimately still cease proliferation through “crisis” due to erosion of telomeres to a critically short length (14). The pRB pathway is generally considered to be comprised of at least four proteins whose activity is frequently perturbed by genetic mutations or altered levels of expression in human cancers—p16INK4a, cyclin D1, cdk4, and pRB (47). By inhibiting cyclinD/cdk4 kinases, p16INK4a activates pRB. The pRB pathway inhibits cell proliferation through numerous downstream effectors. For example, pRB inhibits the E2F family of transcription factors, whose target genes are necessary for progression through S phase (47). The p53 pathway is comprised of at least three proteins whose activity is altered in human cancer: p53, p19ARF, and hdm2 (61). The p53 pathway exerts its effects through activation of downstream target genes, including the cell cycle inhibitor p21cip1, whose expression is increased in senescent cells.
SAHF are specialized domains of facultative heterochromatin that form in many senescent human cells (45, 69). SAHF contain several molecular markers of transcriptionally silent heterochromatin, including a compact structure visible by DNA staining, hypoacetylated histones, methylation of histone H3 on lysine 9, heterochromatin-associated protein 1 (HP1), and the histone variant macroH2A. Like the pRB pathway, SAHF repress expression of proliferation-linked genes, such as cyclin A, thereby contributing to senescence-associated cell cycle arrest. Indeed, Narita and coworkers (45) showed that a functional pRB pathway is required for efficient formation of SAHF, and pRB showed partial colocalization with SAHF in senescent cells.
Recently, we showed that two chromatin regulators, HIRA and ASF1a, drive formation of SAHF in human cells (69). HIRA and ASF1a are the human orthologs of proteins known to create transcriptionally silent heterochromatin in yeast, flies, and plants (35, 43, 50, 60, 62). Saccharomyces cerevisiae Asf1p is required for heterochromatin-mediated silencing of telomeres and mating loci and has histone deposition activity in vitro (37, 59, 62, 64). Yeast Asf1p is also required for histone eviction and subsequent replacement at transcribed genes (2, 36, 57) and also plays a role in DNA replication-coupled chromatin assembly (21, 26, 44, 56, 64). Yeast Hir1p and Hir2p share several biological and biochemical properties with Asf1p. Like Asf1p, they are required for heterochromatin-mediated silencing of telomeres and mating loci and are also required for formation of proper pericentromeric chromatin (but in an Asf1p-independent manner) (35, 37, 59, 60). In vitro, HIRA has been shown to act as a histone chaperone, specifically depositing the histone variant, histone H3.3, in nucleosomes (24, 39, 51, 53, 63). Consistent with their partially overlapping functions, yeast Asf1p and Hir proteins physically interact, and this interaction is necessary for telomeric silencing (15, 24, 59).
Indicative of a role for HIRA in regulation of senescence, one of the earliest cytological markers of an impending senescence is translocation of HIRA into a subnuclear organelle, called the promyelocytic leukemia (PML) nuclear body (69). Most human cells contain 20 to 30 PML nuclear bodies, which are typically 0.1 to 1 μm in diameter and enriched in the protein PML, as well as many other nuclear regulatory proteins (9, 55). PML bodies have been previously implicated in various cellular processes, including tumor suppression and cellular senescence (16, 18, 49). At a molecular level, they have been proposed to serve as sites of assembly of macromolecular regulatory complexes and protein modification (19, 27, 49). HIRA enters PML bodies prior to the appearance of other markers of senescence, such as cell cycle exit, SAHF, a large flat morphology, and SA β-Gal activity (69). Moreover, HIRA translocates into PML bodies at the same time as another protein, HP1, which ultimately exits PML bodies and is stably incorporated into SAHF (69). Ectopic expression of HIRA accelerates formation of SAHF (69). This activity requires binding of HIRA to a human ortholog of Asf1p, ASF1a, and short hairpin RNA (shRNA)-mediated knock-down of ASF1a blocks formation of SAHF triggered by an activated Ras oncogene (69). Thus, it seems likely that PML bodies serve as a molecular “staging ground” where HIRA-containing complexes are assembled or modified prior to their translocation to chromatin and formation of SAHF.
Since translocation of HIRA to PML bodies is one of the earliest events in the senescence program, we set out to identify the senescence triggers that initiate HIRA's localization to PML bodies. In addition, we have tested the requirement for the pRB and p53 tumor suppressor pathways at different steps of the HIRA/ASF1a-mediated SAHF-assembly pathway. We report that all senescence triggers tested cause HIRA's relocalization to PML nuclear bodies. We also show, by disruption of PML bodies and using a dominant negative HIRA mutant, that HIRA's localization to PML bodies is necessary for formation of SAHF. Remarkably, our data indicate that HIRA's translocation to PML bodies and initial activation of the HIRA/ASF1a SAHF-forming pathway occurs in the absence of functional pRB and p53 tumor suppressor pathways. However, formation of SAHF itself does depend on active pRB and p53 pathways. We propose that the HIRA and ASF1a histone chaperones cooperate with the pRB and/or p53 tumor suppressor proteins to drive formation of SAHF, acting in parallel, interdependent pathways. We present evidence that the HIRA/ASF1a pathway and the pRB pathway converge on a DNAJ domain protein, DNAJA2.
MATERIALS AND METHODS
Cell culture and plasmids.
WI38 cells and BJ cells were cultured according to protocols provided by the ATCC. pBabe-puro, pBabe-puro-hTERT, pBabe-puro-H-RasV12, pBabe-neo-rasV12, pBabe-zeocin simian virus 40 (SV40) (early region) ER, and pBabe-zeocin were gifts of Robert A. Weinberg and William Hahn. pBabe-neo-SV40 T antigen, pBabe-neo-SV40 T-antigen K1, and pBabe-neo-T-antigen Δ434-444 were a gift of William Hahn. A p16INK4a expression plasmid was a gift of Greg Enders. p16-1/MSCVpuro-HpaI (shRNA to p16) was a gift of Scott W. Lowe. pcDNA3-DNAJA2 (HIRIP4) has been described previously (40). pBABEpuroPML-RARα has been described previously (5). pQCXIP-HA-HIRA and mutants, pQCXIP-HA-ASF1a, pQCXIP-p16, pQCXIH-GFP, pQCXIH-GFP-shp16 (shRNAs to p16), pQCXIH-GFP-shASF1a (shRNA to ASF1a), pQCXIH-GFP-shLuc (shRNA to luciferase), have been published previously (69) or were generated by standard molecular biology techniques. Lentivirus-encoded shRNAs to HIRA, in vector pLKO1, were purchased from Open Biosystems. Details are available on request.
Immunological and immunofluorescence techniques.
Anti-Ras antibodies were purchased from BD Transduction Laboratories. Anti-p16 antibodies were purchased from BD Pharmingen. Anti-PML and anti-HP1β antibodies were purchased from Chemicon. Anti-p53 antibodies were purchase from Oncogene Research Products. Antihemagglutinin (HA; Y11) and anti-SV40 T antigen (pab108) were purchased from Santa Cruz. Anti-5′ bromodeoxyuridine (BrdU)-fluorescein isothiocyanate was purchased from Becton-Dickinson.
Anti-HIRA, anti-ASF1a, and anti-ASF1b antibodies have been described previously (30, 69). Anti-macroH2A antibodies were raised to the macrodomain of macroH2A1.2 fused to glutathione S-transferase, following a protocol described previously (31).
Immunoprecipitations, Western blotting, immunofluorescence, and measurement of 5′ BrdU incorporation were performed as described previously (30, 67, 69), using the antibodies described above. EBC buffer contains the following: 50 mM Tris (pH 8.0), 0.5% NP-40, 100 mM NaF, 0.2 mM sodium orthovanadate, 50 μg/ml phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 5 μg/ml leupeptin 500 mM NaCl. The radioimmunoprecipitation assay buffer consists of 50 mM Tris (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, and the same protease and phosphatase inhibitors as in EBC.
RT-PCR.
Total RNA was prepared using Trizol (Invitrogen) reagent, according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) to detect human telomerase reverse transcriptase (hTERT) was performed using a QIAGEN one-step RT-PCR kit, according to the manufacturer's instructions. Primer sequences have been described previously (28).
Retrovirus infections and transfections.
Retrovirus-mediated gene transfer was performed as described previously (33), using Phoenix packaging cells (Gary Nolan, Stanford University) to package the infectious viruses. Cells were infected with viruses encoding resistance to puromycin, neomycin, and zeocin and then selected in 2 μg/ml, 500 μg/ml, and 1,200 μg/ml of the respective drug. Retrovirus infections were performed on WI38 cells between population doubling 30 (PD30) to PD40. Transient transfection of U2OS cells was performed by the calcium phosphate method, as described previously (30, 46).
FISH.
Telomeric fluorescent in situ hybridization (FISH) was performed as described previously (68), using a Cy3-labeled telomeric repeat probe (DAKO).
RESULTS
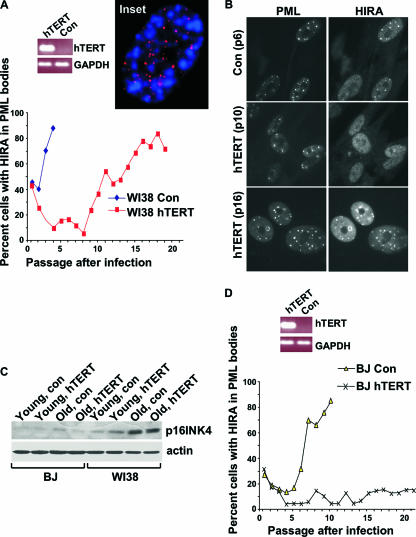
Previously, we showed that, in primary human WI38 cells, HIRA localizes to PML nuclear bodies prior to senescence induced by either activated oncogenes or extended growth in culture (69). In WI38 cells grown under standard culture conditions and ambient (20%) oxygen, senescence after extended growth in culture is thought to occur due to a cell stress response rather than to critically short telomeres (20). Previous studies showed that ectopic expression of hTERT in WI38 fibroblasts maintains telomere length, but this is insufficient for immortalization (20, 66). Therefore, to test whether relocalization of HIRA in WI38 cells under these conditions is linked to cell stress or shortening of telomeres, we suppressed the latter by infecting the cells with a virus encoding the catalytic subunit of human telomerase, hTERT. In young mock-infected WI38 cells, HIRA was predominantly diffused throughout the nucleoplasm, although a small proportion of cells did contain HIRA in PML bodies, as usual. As the cells aged in culture, the proportion of cells with HIRA in PML bodies steadily increased, until 90% of the cells contained HIRA in PML bodies (Fig. 1A and B). Expression of hTERT did not abolish relocalization of HIRA to PML bodies although it did delay it. The observation that HIRA relocalizes to PML bodies even in WI38 cells expressing hTERT supports the idea that relocalization of HIRA to PML bodies occurs even in cells whose telomeres are not short enough to trigger senescence. Consistent with the idea that there is not an obligatory link between short telomeres and the SAHF-assembly pathway, we confirmed the previous observations of Narita and coworkers that SAHF are spatially distinct from telomeres (45), this time using a telomeric DNA FISH probe (Fig. 1A, inset). In WI38 cells ectopically expressing hTERT, senescence is thought to be due to upregulation of p16INK4a by the stress of extended cell culture (4, 20, 33). Indeed, expression of p16INK4a was upregulated during prolonged cell culture of WI38 cells, regardless of ectopic expression of hTERT (Fig. 1C).
FIG. 1.
Recruitment of HIRA to PML bodies is linked to senescence induced by short telomeres and cell stress. (A) WI38 cells were infected with a retrovirus encoding hTERT or a control virus and selected for infected cells with puromycin, and the number of cells with HIRA localized to PML bodies was determined by immunofluorescence at subsequent culture passages. Expression of hTERT and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were assayed by RT-PCR. (Inset) A confocal image of a WI38 cell stained by FISH to detect telomeres (red) and with DAPI to detect DNA (blue). (B) Immunofluorescence images of control or hTERT-infected WI38 cells of indicated passage number (pN) from panel A stained with antibodies to HIRA and PML. (C) Lysates were prepared from WI38 and BJ fibroblasts infected with a retrovirus encoding hTERT or a control virus and subjected to Western blotting to detect expression of p16INK4a. “Young” lysates were all prepared at PD2 after virus infection. “Old” lysates were prepared when the cells became senescent (20PD after infection in case of WI38-hTERT) or 20PD after infection in the case of immortal BJ-hTERT. (D) Results of the same experiment as in panel A but performed in BJ fibroblasts. Con, control.
However, the fact that hTERT expression delayed localization of HIRA to PML bodies suggests that relocalization is also linked to telomere shortening to some extent. In contrast to WI38 cells, BJ fibroblasts grown in ambient oxygen are thought to undergo senescence solely due to shortened telomeres. Indeed, in contrast to WI38 cells, these cells can be immortalized by hTERT expression and do not upregulate p16INK4a after extended cell culture (Fig. 1C) (4, 8). Therefore, to directly test whether HIRA also relocalizes to PML bodies in response to short telomeres, we examined HIRA localization as BJ cells were passaged toward senescence. In noninfected and mock-infected BJ cells, HIRA also relocalized to PML bodies as the cells approached senescence (Fig. 1D and data not shown). However, in striking contrast to WI38 cells, in BJ cells relocalization of HIRA to PML bodies was abolished by ectopic expression of hTERT. Together, these data indicate that relocalization of HIRA to PML bodies is linked to senescence triggered by short telomeres in WI38 and BJ cells and by stress-induced expression of p16INK4a in WI38 cells. Previously, we also showed that relocalization of HIRA to PML bodies is induced by an activated Ras oncogene (69). We conclude that a wide range of senescence triggers causes HIRA's localization to PML bodies.
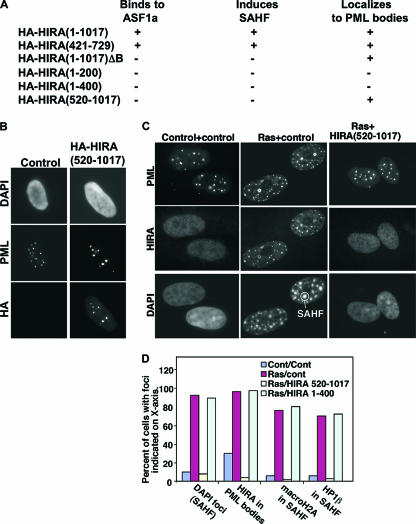
We previously reported that in WI38 cells, localization of HIRA to PML bodies is a precursor to formation of SAHF (69). However, as originally reported by Narita and coworkers, BJ cells do not form SAHF during replicative senescence (45). This indicates that localization of HIRA to PML bodies is not a sufficient trigger for formation of SAHF. Therefore, we probed the relationship between HIRA's localization to PML bodies and formation of SAHF in WI38 cells in more detail. To do this, we sought a way to block HIRA's localization to PML bodies in WI38 cells. Assuming that HIRA is specifically targeted to PML bodies, we reasoned that we ought to be able to identify a mutant of HIRA that is unable to form SAHF but competes out localization of the endogenous protein to PML bodies. As reported previously, wild-type HA-tagged HIRA efficiently localized to PML bodies when ectopically expressed in cells by retrovirus infection (69) (Fig. 2A). Next, we tested a panel of HIRA deletion mutants in this assay. The results are summarized in Fig. 2A. Significantly, we identified two HIRA mutants which we previously showed are unable to bind to ASF1a and induce SAHF (69) but which are still recruited normally to PML bodies (Fig. 2B and data not shown). One HIRA mutant consists of residues 1 to 1017 with a 37-amino-acid deletion of the ASF1a-binding B domain (consisting of residues 439 to 475) [HIRA(1-1017)ΔB], and the other, HIRA(520-1017), carries a large N-terminal truncation and also lacks the B domain. We reasoned that these mutants might compete out the localization of endogenous HIRA to PML bodies and, because the mutant itself cannot induce SAHF, would block the induction of SAHF by endogenous HIRA. Indeed, both mutants blocked localization of endogenous HIRA to PML bodies and formation of SAHF, induced by an activated Ras oncogene (Fig. 2C and D and data not shown). In contrast, another mutant HIRA(1-400), which does not bind to ASF1a or induce SAHF and did not block localization of endogenous HIRA to PML bodies, did not affect formation of SAHF (Fig. 2D).
FIG. 2.
HIRA mutants that block localization of endogenous HIRA to PML bodies also prevent formation of SAHF. (A) WI38 cells were infected with retroviruses encoding the indicated HA-tagged wild-type and mutant HIRA proteins and then selected in puromycin for 8 days. Localization of the HA-tagged protein to PML bodies and formation of SAHF was scored by immunofluorescence and DAPI staining, respectively, and positive (+) or negative (−) results are summarized. ASF1a binding data are taken from Zhang et al. (69). (B) WI38 cells were infected with a retrovirus encoding HA-HIRA(520-1017) or a control virus and stained with antibodies to PML and HA and with DAPI to visualize DNA (cells were preextracted with EBC buffer prior to fixation [1]). (C) WI38 cells were infected with the indicated retroviruses; the infected cells were selected by growth in puromycin (HIRA) and neomycin (Ras) and then stained with antibodies to HIRA and PML and with DAPI to visualize DNA. The antibody WC119 used to detect endogenous HIRA does not detect HIRA(520-1017) (data not shown). (D) Results from panel C were quantitated in terms of cells with DAPI foci (SAHF), HIRA localized to PML bodies, and macroH2A and HP1β localized to SAHF. Results are representative of more than three independent experiments. Cont, control.
Concordant with these results, we showed previously by shRNA knock-down that HIRA's binding partner, ASF1a, is required for formation of SAHF (69). We next attempted to confirm a role for HIRA in formation of SAHF using lentivirus-expressed shRNAs to knock down its expression. However, we were unable to identify shRNAs that stably knocked down HIRA for up to 8 days but did not kill the cells (see Fig. S1 in the supplemental material). Regardless of this result, the data obtained with the HIRA dominant negative mutant, HIRA(520-1017), support the idea that, in WI38 cells, localization of HIRA to PML bodies is necessary for formation of SAHF.
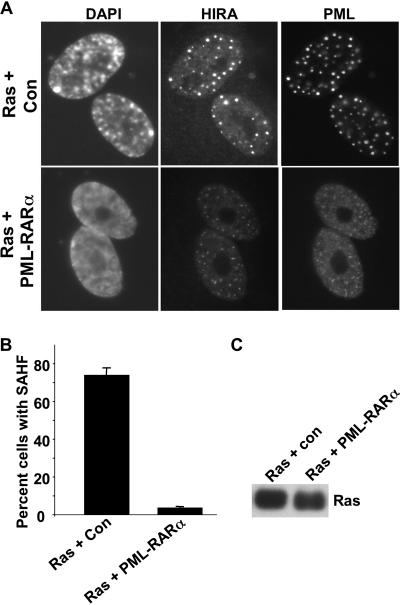
To directly test a role for PML bodies in formation of SAHF, we used a PML-RARα fusion protein to inactivate PML bodies. The PML-RARα fusion protein is expressed in many acute PML cells due to a chromosomal translocation that fuses the two proteins (55). Expression of PML-RARα causes dispersal of the normally 20 to 30 PML nuclear bodies into a large number of much smaller PML-containing foci or speckles, and this is thought to inactivate PML bodies (55). As expected, ectopic retrovirus-mediated expression of PML-RARα in WI38 cells disrupted PML bodies (Fig. 3A). Interestingly, the HIRA foci characteristic of senescent cells were also disrupted, and HIRA was distributed in speckles throughout the nucleus that colocalized with PML. Strikingly, PML-RARα virtually eliminated formation of SAHF in WI38 cells expressing an activated Ras oncogene (Fig. 3A and B). These results suggest that the normal undisrupted integrity of PML bodies is essential for formation of SAHF.
FIG. 3.
PML-RARα disrupts PML bodies and formation of SAHF. (A) WI38 cells were infected with a retrovirus expressing activated Ras, together with a control virus or a virus encoding PML-RARα. Eight days later the cells were stained with antibodies to HIRA and PML and with DAPI to visualize DNA. (B) A total of 100 cells from panel A were scored for the absence or presence of DAPI foci (SAHF). (C) Lysates were prepared from the cells in panel A and subjected to Western blotting to detect expression of activated Ras. Con, control.
Taken together, these results and results published previously indicate a key role for HIRA's translocation to intact PML bodies in induction of SAHF in senescent WI38 cells (69). On the other hand, results presented here and published previously also indicate that in BJ fibroblasts translocation of HIRA to PML bodies is not a sufficient trigger for SAHF (45). Beausejour and coworkers have suggested that at least one difference between WI38 cells and BJ cells is in the activity of the pRB tumor suppressor pathway. Specifically, BJ cells express low levels of the pRB activator, p16INK4a, and so have a less active pRB pathway (4) (Fig. 1C). Based on this idea, we hypothesized that the pRB pathway might cooperate with HIRA and ASF1a in induction of SAHF at a point downstream of HIRA's translocation to PML nuclear bodies.
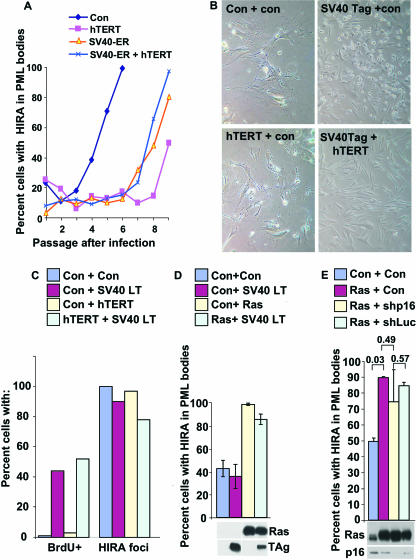
To test this, we analyzed HIRA's localization to PML bodies and formation of SAHF in cells lacking functional p53 and/or pRB. WI38 cells were infected with retroviruses encoding hTERT alone, the ER of the SV40 virus, hTERT plus SV40 ER, or a mock virus. SV40 ER encodes two oncoproteins, small t antigen and large T antigen. SV40 large T antigen binds to and inactivates both the pRB and p53 tumor suppressor proteins (3). In additional experiments, cells were infected with a virus expressing SV40 large T antigen alone instead of SV40 ER. After infection, the cells were drug selected to enrich for the infected cells and cultured over subsequent passages, and the percentage of cells with HIRA localized to PML bodies was determined by immunofluorescence at each passage. As before (Fig. 1A), expression of hTERT delayed, but did not abolish, localization of HIRA to PML bodies (Fig. 4A). These hTERT-expressing cells exhibited an extended life span in culture but ultimately became senescent, as expected, and assumed a large flat morphology similar to the control infected cells (Fig. 4B). Senescent control and hTERT-infected cells did not incorporate 5′ BrdU, an indicator of DNA synthesis in proliferating cells (Fig. 4C). Strikingly, infection with SV40 ER or SV40 large T antigen did not abrogate recruitment of HIRA to PML bodies (Fig. 4A and C) in either the absence or presence of hTERT. As shown previously, human WI38 fibroblasts expressing both hTERT and SV40 ER or large T antigen were immortal; they never assumed a senescent morphology and could be maintained indefinitely as a culture that readily labels with 5′ BrdU (29) (Fig. 4B and C). In contrast, WI38 cells expressing either SV40 ER or SV40 large T-antigen alone entered crisis, characterized by a large number of rounded dead cells and many cells that incorporate 5′ BrdU (54) (Fig. 4B and C). The ability of SV40 ER and large T antigen to drive cells into crisis or immortalization, in the absence and presence of hTERT, respectively, confirms the biological activity of large T antigen in these cells. To further verify the activity of ectopically expressed SV40 large T antigen in these cells, we tested it for binding to the p53 tumor suppressor protein. Immunoprecipitation of SV40 large T antigen coprecipitated p53 and vice versa, showing that large T antigen is functional in these cells and physically bound to p53 and, presumably, pRB (see Fig. S2 in the supplemental material). Together, these results show that HIRA is still localized to PML bodies in cells in which pRB and p53 are inactivated by SV40 large T antigen. In the experiments shown in Fig. 4A to C, extended growth in culture acted as the trigger for HIRA's relocalization to PML bodies. Previously, we showed that HIRA also relocalizes to PML bodies during senescence triggered by an activated Ras oncogene (45, 69). To test whether relocalization of HIRA under these conditions is also independent of pRB and p53, cells were coinfected with retroviruses encoding activated Ras and SV40 large T antigen, and the cells were then drug selected for the infected cells. Immunofluorescence was performed to determine HIRA's localization. Large T antigen did not affect localization of HIRA to PML bodies, even though Western blotting confirmed that both T antigen and activated Ras were expressed as expected (Fig. 4D). Finally, to confirm by another method that the pRB pathway is not required for HIRA's recruitment to PML bodies, we used a retrovirus to deliver an shRNA that knocks down p16INK4a. Like ectopic expression of large T antigen, this had no significant effect on HIRA's localization to PML bodies compared to activated Ras alone or activated Ras plus a control shRNA to luciferase (Fig. 4E). In sum, these experiments show that recruitment of HIRA to PML bodies does not require functional pRB or p53 tumor suppressor pathways.
FIG. 4.
pRB and p53 tumor suppressor proteins are not required for recruitment of HIRA to PML bodies. (A) WI38 cells were infected with retroviruses encoding SV40 ER, hTERT, both, or a control virus, as indicated. The infected cells were selected in puromycin (for hTERT and control) and zeocin (SV40 ER), and the number of cells with HIRA localized to PML bodies was determined by immunofluorescence over the subsequent passages. (B) WI38 cells were infected with a control retrovirus or viruses expressing SV40 large T antigen, hTERT, or SV40 large T antigen and hTERT. The infected cells were selected in puromycin (for control and hTERT) and neomycin (for control and SV40 large T antigen) and then passaged until senescence (control and hTERT) or crisis (SV40 large T antigen) or more than PD150 in case of immortal SV40 plus hTERT. (C) Cells from panel B were pulse-labeled with 5′ BrdU for 2 h and then immunofluorescence stained to detect 5′ BrdU and HIRA. A total of 100 cells were scored for the absence or presence of 5′ BrdU incorporation or HIRA foci (in PML bodies). (D) WI38 cells were infected with retroviruses encoding activated Ras, SV40 large T antigen, or control viruses as indicated, and the infected cells were selected in puromycin (for Ras and control) and neomycin (for SV40 large T antigen and control) for 8 days. The cells were stained with antibodies to HIRA and PML, and the percentage of cells with HIRA localized to PML bodies was determined by scoring 100 cells for colocalization of HIRA and PML. Results are the mean of three independent experiments. Whole-cell lysates were prepared and subjected to Western blotting to detect Ras and SV40 large T-antigen (TAg). (E) WI38 cells were infected with control, activated Ras, and shp16 and shLuc retroviruses; cells were then selected in puromycin (for Ras) for 8 days and stained with antibodies to HIRA and PML (shp16 and shLuc are retroviruses encoding shRNAs to p16INK4a and firefly luciferase, respectively, as well as green fluorescent protein and resistance to hygromycin). The percentage of cells with HIRA localized to PML bodies was determined by scoring 100 green fluorescent protein-positive cells for colocalization of HIRA and PML. Results are the mean of three independent experiments. P values obtained with Student's t test are shown. Another aliquot of the infected cells was selected in puromycin and hygromycin for 8 days; whole-cell extracts were prepared and subjected to Western blotting with antibodies to Ras and p16INK4a. Con, control.
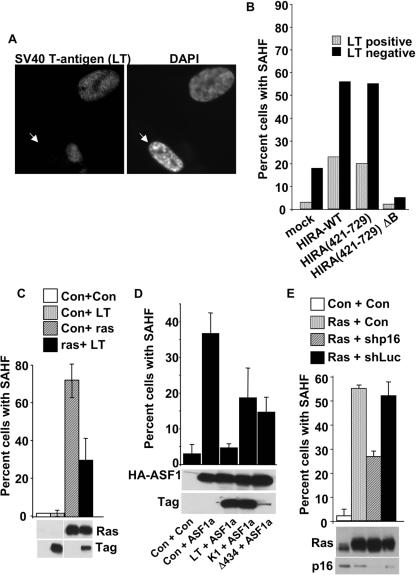
Next, we asked whether pRB and p53 are required for formation of SAHF. Previously, we showed that ectopic expression of wild-type HIRA and some mutants, e.g., HIRA(421-729), accelerates formation of SAHF (69). WI38 cells were infected with retroviruses encoding wild-type HIRA or HIRA(421-729), together with a virus encoding SV40 large T antigen. The cells were drug selected for infection by the HIRA virus and scored for expression of SV40 large T antigen and formation of SAHF by immunofluorescence. Ectopic expression of wild-type HIRA or HIRA(421-729) increased the number of cells with SAHF (Fig. 5A and B). Expression of a mutant lacking the ASF1a-binding B domain, HIRA(421-729)ΔB, did not induce SAHF. Coexpression of SV40 large T antigen largely abolished formation of SAHF induced by HIRA (Fig. 5A and B). Likewise, large T antigen largely abolished formation of SAHF induced by an activated Ras oncogene (Fig. 5C). Since SV40 large T antigen binds to and inactivates at least two key tumor suppressor proteins, pRB and p53, we tested a pair of T-antigen mutants to determine which pathway, if either, is responsible for formation of SAHF. As before, wild-type large T antigen efficiently blocked formation of SAHF, this time induced by ectopic expression of ASF1a (Fig. 5D). By comparison, the SV40 large T-antigen K1 mutant that is specifically impaired in binding to pRB (and related proteins p107 and p130) was impaired in its ability to block SAHF. Likewise, another mutant, T-antigen Δ434, which is impaired in its ability to bind to p53, was also impaired (Fig. 5D). This indicates that both the pRB and p53 pathways contribute to formation of SAHF. Because SV40 T-antigen Δ434 was underexpressed compared to wild-type T antigen, to confirm a role for the pRB pathway we used a retrovirus-encoded shRNA to knock down p16INK4a. Comparable to the large T-antigen Δ434 mutant that also inactivates the pRB pathway but not the p53 pathway, knock-down of p16INK4a reduced SAHF by about 50% (Fig. 5E). We conclude that efficient formation of SAHF depends on active pRB and p53 pathways in WI38 cells.
FIG. 5.
pRB and p53 proteins are required for formation of SAHF. (A) WI38 cells were infected with retroviruses encoding HIRA(421-729) and SV40 large T antigen, selected by growth in puromycin for infection with the HIRA virus, and then stained with antibodies to SV40 large T antigen and with DAPI to visualize DNA. The white arrow indicates a cell with DAPI foci that does not express SV40 large T antigen. (B) WI38 cells were infected with retroviruses encoding SV40 large T antigen in the absence or presence of the indicated HA-tagged HIRA mutants. Cells were selected in puromycin for infection by the HIRA or corresponding control virus, and then 100 individual cells were scored as positive or negative for expression of SV40 T antigen and formation of SAHF, judged by DAPI foci. Data are the percentages of SAHF-positive cells in both the T-antigen-positive and -negative populations of cells. Results are representative of at least three independent experiments. (C) WI38 cells were infected with retroviruses encoding activated Ras, SV40 large T antigen, or control as indicated, and then selected for 8 days in puromycin (for Ras) and neomycin (for SV40 large T antigen). Cells were stained with DAPI to visualize formation of SAHF, and 100 individual cells were scored for SAHF. Results are the mean of three independent experiments. Whole-cell extracts were subjected to Western blotting with antibodies to Ras and SV40 T antigen. This experiment was done in parallel with the experiment described in the legend of Fig. 4D, so the Western blot is the same. (D) WI38 cells were infected with retroviruses encoding SV40 large T antigen or the indicated mutant (K1 and Δ434) and/or ASF1a and/or control vectors. The cells were double-drug selected in puromycin (for ASF1a) and neomycin (for SV40 large T antigen), and 100 cells were scored for the absence or presence of SAHF. Whole-cell extracts were prepared and subjected to Western blotting to detect ASF1a and SV40 large T antigen. (E) WI38 cells were infected with control, activated Ras, shp16, and shLuc retroviruses; cells were selected in puromycin (for Ras) for 8 days and then stained with DAPI to visualize DNA. Shp16 and shLuc are retroviruses encoding shRNAs to p16INK4a and firefly luciferase, respectively, as well as green fluorescent protein and resistance to hygromycin. The percentage of cells with SAHF was determined by scoring 100 green fluorescent protein-positive cells for condensed chromatin by DAPI stain. Results are the mean of three independent experiments. Another aliquot of the infected cells was selected in puromycin and hygromycin for 8 days; whole-cell extracts were prepared and subjected to Western blotting with antibodies to Ras and p16INK4a. This experiment was done in parallel with the experiment described in the legend of Fig. 4E, so the Western blot is the same. LT, large T antigen; Con, control.
In light of the possibility that differential formation of SAHF in WI38 cells and BJ cells might depend on the relative activity of the pRB pathway in the two cell types, we were particularly interested in the role of this pathway. Based on our results so far, the pRB pathway might be required for formation of SAHF because the HIRA/ASF1a pathway activates the pRB pathway and this, in turn, makes SAHF. Consistent with this idea, pRB is known to recruit heterochromatin-forming enzymes to chromatin (70). Alternatively, the HIRA/ASF1a pathway and the pRB pathway might converge and cooperate to make SAHF at a point downstream of HIRA's localization to PML bodies. To differentiate between these two models, we asked whether SAHF induced by forced activation of the pRB pathway requires the histone chaperone, ASF1a. Lowe and coworkers previously showed that ectopic expression of the pRB activator, 16INK4a, induces SAHF. This is consistent with either of the two models outlined above (45). However, only the second “convergent/cooperating pathways” model predicts that inactivation of ASF1a will prevent induction of SAHF in cells in which the pRB pathway is activated. According to this second model, formation of SAHF requires simultaneous activity of both pathways.
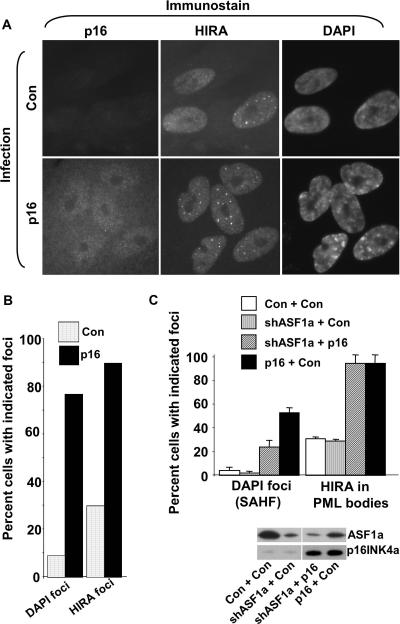
As shown previously, forced expression of p16INK4a in WI38 cells induced formation of SAHF, judged by 4′,6′-diamidino-2-phenylindole (DAPI) staining and visualization of condensed chromatin (Fig. 6A and B). However, coinfection of the p16INK4a-encoding virus with a virus encoding an shRNA targeted to ASF1a reduced formation of SAHF (Fig. 6C). Interestingly, ectopic expression of p16INK4a also induced localization of HIRA to PML bodies (Fig. 6A to C). The mechanism behind this is not clear. However, we observed that shRNA-mediated knock-down of ASF1a did not affect HIRA localization (Fig. 6C). This indicates that in cells ectopically expressing p16INK4a but lacking ASF1a due to its shRNA-mediated knock-down, p16INK4a activity is still elevated. In sum, these data show that even in the presence of elevated p16INK4a/pRB activity, formation of SAHF requires ASF1a activity. This supports the “convergent/cooperating pathways” model (see Fig. 9).
FIG. 6.
Formation of SAHF induced by p16INK4a requires ASF1a. (A) WI38 cells were infected with a retrovirus encoding p16INK4a or a control virus, the cells were selected for infected cells in puromycin for 8 days, and then cells were stained with antibodies to p16INK4a and HIRA and with DAPI to visualize DNA. (B) A total of 100 cells from panel A were scored for localization of HIRA to nuclear foci (PML bodies) and formation of SAHF. (C) WI38 cells were infected with retroviruses encoding p16INK4a and/or an shRNA targeted to ASF1a and/or appropriate control viruses. The virus encoding shRNA to ASF1a also encodes green fluorescent protein and resistance to hygromycin. The cells were then selected in puromycin (for p16INK4a) for 8 days. The cells were stained with antibodies to HIRA and with DAPI to visualize DNA. The percentage of cells with HIRA foci (in PML bodies) and DAPI foci was determined by scoring 100 green fluorescent protein-positive cells. Another aliquot of the infected cells was selected in puromycin and hygromycin for 8 days, and whole-cell extracts were prepared and subjected to Western blotting with antibodies to ASF1a and p16INK4a.
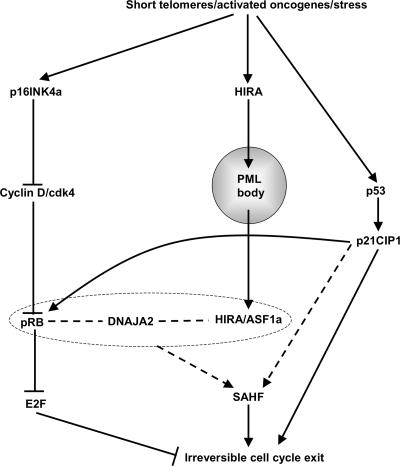
FIG. 9.
pRB- and p53-dependent and -independent steps in formation of SAHF. We propose that DNAJA2 facilitates the cooperative formation of SAHF by the pRB and HIRA/ASF1a pathways. pRB, DNAJA2, and HIRA/ASF1a are enclosed within a dashed box to indicate the activity of these proteins in a protein complex and/or a mutually interdependent network. Dashed lines indicate that the molecular details of SAHF formation remain to be determined. Data in the text also indicate that p16INK4a, pRB, and p53 also contribute to HIRA's localization to PML bodies. However, because these proteins are not essential for this relocalization, these functional interactions have been omitted from the figure for simplicity. See text for details.
Based on these data, we hypothesized that the HIRA/ASF1a and pRB pathways converge downstream of HIRA's localization to PML bodies. Remarkably, a single protein, DNAJA2, has been independently cloned as a HIRA and pRB binding protein in two-hybrid screens performed by two different laboratories. Specifically, Lorain and coworkers identified DNAJA2 as an HIRA-binding protein (40). Subsequently, Benevolenskaya and coworkers identified DNAJA2 in a differential two-hybrid screen intended to identify proteins that contribute to pRB-mediated cell differentiation (6). Importantly, the BIND database (www.bind.ca/Action) lists no other protein-protein interactions for DNAJA2, suggesting that the reported interactions with HIRA and pRB are relatively specific.
To be able to ask whether DNAJA2 physically interacts with pRB and HIRA in vivo, we raised a rabbit polyclonal antibody to DNAJA2. The purified antibody specifically detected DNAJA2 in Western blot analysis, as judged by reaction with a single polypeptide close to the predicted molecular mass of 46 kDa, whose expression was knocked down by specific small interfering RNA toward DNAJA2 (see Fig. S3 in the supplemental material). Unfortunately, we were unable to immunoprecipitate DNAJA2 from cell extracts, unless the extracts were first boiled in denaturing buffer containing 1% sodium dodecyl sulfate and then diluted (see Fig. S3 in the supplemental material). This indicates that DNAJA2 is probably contained in protein complexes and not accessible to antibodies, therefore preventing us from asking whether endogenous DNAJA2 is bound to HIRA and pRB in cells.
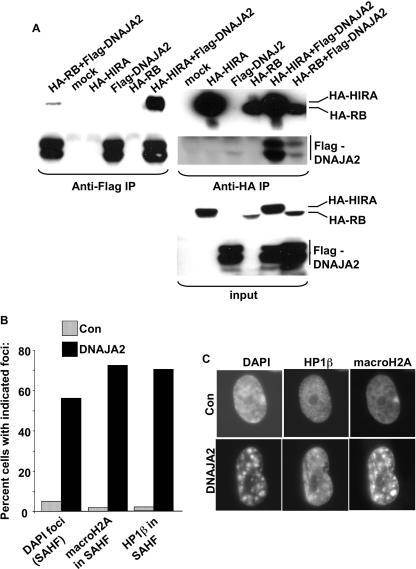
As an alternative approach to ask whether DNAJA2 physically interacts with HIRA and pRB in vivo, we ectopically expressed Flag-tagged DNAJA2 with either HA-tagged HIRA or HA-pRB in U2OS cells and tested for interactions by coimmunoprecipitation analysis. When coexpressed with Flag-DNAJA2, both HA-HIRA and HA-pRB efficiently coprecipitated in an anti-Flag immunoprecipitation but not when ectopically expressed Flag-DNAJA2 was omitted from the reaction (Fig. 7A). Similarly, Flag-DNAJA2 coimmunoprecipitated in an anti-HA immunoprecipitation when coexpressed with either HA-pRB or HA-HIRA (Fig. 7A). As an additional specificity control, Flag-DNAJA2 did not bind to the largest subunit of the chromatin assembly factor-I complex, p150CAF-I, when it was overexpressed as an HA-tagged protein (see Fig. S3 in the supplemental material). We conclude that DNAJA2 specifically binds to pRB and HIRA when the proteins are coexpressed in U2OS cells.
FIG. 7.
DNAJA2 is a candidate effector of pRB- and HIRA-mediated SAHF formation. (A) U2OS cells were transfected with plasmids encoding the indicated epitope-tagged proteins. Anti-Flag and anti-HA immunoprecipitations were performed and subjected to Western blotting with anti-HA or anti-Flag to detect the coimmunoprecipitating proteins. (B) WI38 fibroblasts were infected with a retrovirus encoding DNAJA2 or a control virus, and the infected cells were selected in puromycin. Eight days later, the cells were stained with antibodies to HP1β and macroH2A and with DAPI to visualize DNA. A total of 100 cells were scored for presence or absence of SAHF (DAPI foci), macroH2A in SAHF, and HP1β in SAHF, and the results are expressed as a histogram. (C) Representative cells from panel B, infected with control and DNAJA2-expressing retroviruses. The single large domain of intense macroH2A staining in control cells is the inactive X chromosome. IP, immunoprecipitation; Con, control.
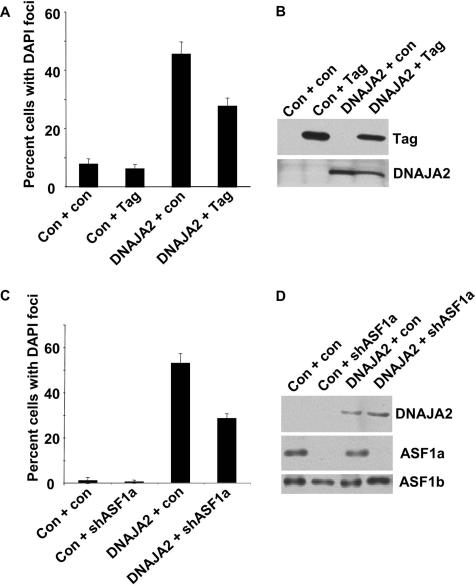
Next, we wanted to ask whether DNAJA2 plays a role in formation of SAHF. First, we set out to ask whether DNAJA2 is necessary for formation of SAHF. Since formation of SAHF takes several days or weeks, depending on whether it is induced by activated oncogenes or extended growth in culture, we attempted to generate retroviruses encoding shRNAs to stably knock down DNAJA2. Unfortunately, we were unable to generate a virus-encoded shRNA that stably knocks down DNAJA2 without killing the infected cells (data not shown). Thus, DNAJA2 appears to have one or more essential functions that preclude us from specifically testing a requirement for SAHF formation. Therefore, to address a role for DNAJA2 in formation of SAHF, we asked whether ectopic expression of DNAJA2 is sufficient to trigger formation of SAHF. As shown in Fig. 7B and C, ectopic expression of DNAJA2 efficiently promoted formation of SAHF, as indicated by formation of DAPI foci enriched in HP1 proteins and macroH2A. DNAJA2-induced SAHF was only partially abolished by inactivation of pRB and p53 by SV40 large T antigen (Fig. 8A and B) or by shRNA-mediated knock-down of ASF1a (Fig. 8C and D), suggesting that DNAJA2 does not act exclusively up- or downstream of HIRA and pRB. In sum, DNAJA2 physically interacts with pRB and HIRA, and its ectopic expression accelerates formation of SAHF. These data strongly suggest that DNAJA2 plays a role in formation of SAHF, perhaps by integrating signals from the pRB and HIRA/ASF1a pathways (Fig. 9).
FIG. 8.
DNAJA2-induced SAHF is partially abolished by SV40 large T-antigen or knock-down of ASF1a. (A) WI38 cells were infected with retroviruses encoding DNAJA2, SV40 large T antigen, or control viruses as indicated. The infected cells were selected in puromycin (for DNAJA2 and control) and neomycin (for SV40 large T antigen and control). Eight days later the cells were stained with DAPI, and 100 cells were scored for formation SAHF (DAPI foci). (B) Lysates were prepared from the cells in panel A and subjected to Western blotting to detect expression of SV40 large T antigen and DNAJA2. (C) WI38 cells were infected with retroviruses encoding DNAJA2, an shRNA to ASF1a, or control viruses as indicated. The cells were selected in puromycin (for DNAJA2 or the control). The virus encoding shRNA to ASF1a (or the control) also encodes green fluorescent protein and resistance to hygromycin. The cells were stained with DAPI to visualize DNA. A total of 100 green fluorescent protein-expressing cells were scored for formation of SAHF. (D) Another aliquot of the infected cells was selected in puromycin and hygromycin for 8 days, and whole-cell extracts were prepared and subjected to Western blotting with antibodies to ASF1a, ASF1b, and DNAJA2. Con, control; Tag, large T antigen.
DISCUSSION
Here we have dissected the steps in the SAHF assembly process and tested the causal relationship of these steps to each other and the pRB and p53 tumor suppressor pathways. We find that relocalization of HIRA to PML bodies, a presumed upstream step in the SAHF assembly process (69), occurs in response to a variety of senescence triggers, namely, activated oncogenes, short telomeres, and cell stress. Through use of dominant negative mutants that block HIRA's localization to PML bodies and a PML-RARα fusion protein, we provide additional evidence that translocation of HIRA to PML bodies is necessary for formation of SAHF. Surprisingly, we show that translocation of HIRA to PML bodies occurs in the absence of functional pRB and p53. In contrast, a downstream step, formation of SAHF by the HIRA/ASF1a histone chaperone complex, requires activity of both pRB and p53. However, formation of SAHF induced by activation of p16INK4a also requires ASF1a, implying that the HIRA/ASF1a pathway and the pRB pathway cooperate as independent pathways to make SAHF at a point downstream of HIRA's localization to PML bodies. Consistent with this model, we have identified a protein, DNAJA2, which physically interacts with HIRA and pRB and induces SAHF when ectopically expressed in cells. DNAJA2 is a good candidate to contribute to at least some of the convergent activities of pRB and HIRA.
Previously, we reported that relocalization of HIRA to PML bodies is an early step in the senescence program in WI38 cells induced to senesce by an activated Ras oncogene or extended growth in culture (69). Relocalization of HIRA to PML bodies occurs before other markers of senescence, such as expression of SA β-Gal, formation of SAHF, and cell cycle exit. Consequently, it is important to understand the molecular triggers that initiate relocalization. Senescence of WI38 cells cultured in ambient oxygen is a complex process involving at least two triggers, namely, short telomeres and upregulation of p16INK4a due to the stress of in vitro culture (4, 20, 33, 66). In contrast, BJ fibroblasts senesce solely due to short telomeres (4, 8). Here, we report that this early step in the senescence program—localization of HIRA to PML bodies—is linked to both short telomeres and upregulation of p16INK4a. In sum, relocalization of HIRA appears to accompany senescence, regardless of whether the trigger is an activated oncogene, short telomeres, or cell stress.
Previously, we proposed that relocalization of HIRA to PML bodies is an early step in formation of SAHF (69). This proposal was based on the following observations: (i) HIRA relocalized to PML bodies prior to formation of SAHF; (ii) HIRA relocalized to PML bodies coincident with another family of proteins, HP1 proteins, that are ultimately deposited in SAHF; and (iii) ectopic expression of HIRA accelerates formation of SAHF. Here, we have further strengthened the link between HIRA's localization to PML bodies and formation of SAHF in two ways. First, we showed that HIRA mutants that block localization of endogenous HIRA to PML bodies also block formation of SAHF. Second, disruption of PML bodies themselves by PML-RARα also blocks formation of SAHF. The role played by HIRA's localization to PML bodies in formation of SAHF is not yet clear. However, PML bodies have been previously implicated in induction of senescence (16, 18, 49) and proposed to serve as sites of protein modification and sites of assembly of macromolecular protein complexes (19, 27, 49). Therefore, in PML bodies HIRA is likely to be posttranslationally modified or recruited into a macromolecular complex before being exported to sites of nascent SAHF, where it contributes to formation of mature SAHF.
Our data show that multiple senescence triggers, including activated oncogenes, short telomeres, and cell stress, induce relocalization of HIRA to PML bodies. Each of these triggers is typically thought to exert its effects through pRB and/or p53 (11, 32, 66). Indeed, localization of HIRA to PML bodies is delayed in cells expressing SV40 ER, consistent with the idea that in normal cells pRB and p53 directly or indirectly contribute to HIRA's relocalization. However, in the absence of functional pRB and p53, HIRA is ultimately recruited to PML bodies. Remarkably, even in immortalized cells expressing SV40 ER and hTERT, HIRA was still recruited to PML nuclear bodies. This indicates that there are pRB- and p53-independent mechanisms to target HIRA to PML bodies and that these still function in immortalized cells. This is highly significant because most, if not all, other senescence signaling processes are thought to be abolished in cells lacking pRB and p53 and in immortalized cells (11, 32, 66). Clearly, it is important to define the cell signaling process that instigates HIRA's relocalization in the absence of pRB and p53, since this is likely to be a novel signaling process in senescent cells that may also regulate other events during senescence of human cells. We are actively working to this end.
In contrast to HIRA's relocalization to PML bodies, formation of SAHF was strongly dependent upon the integrity of the pRB and p53 pathways. Likewise, formation of SAHF by constitutive activation of pRB via ectopic expression of p16INK4a also required the HIRA/ASF1a pathway. This suggests that the pRB and HIRA/ASF1a pathways are mutually dependent upon each other and cooperate to make SAHF. Consistent with this idea, the pRB tumor suppressor protein and the HIRA and ASF1a histone chaperones are all well-established regulators of chromatin structure and function. The pRB protein facilitates formation of heterochromatin at its target genes and other sites throughout the genome (23, 70). For example, pRB recruits the histone methylase SUVAR39H to chromatin, and this enzyme is required for oncogene-induced formation of heterochromatin in murine T cells (10). HIRA and ASF1a are two histone chaperones whose physical interaction is required to form transcriptionally silent heterochromatin from yeast to humans (7, 25, 35, 37, 50, 59, 69). One simple model to describe the cooperation of the pRB and HIRA/ASF1a pathways is that pRB initiates heterochromatin formation at the promoters of its target genes, and then HIRA and ASF1a generate the more extensive domains of heterochromatin characteristic of SAHF. Consistent with these models, E2F target genes, such as cyclin A, are incorporated into SAHF, and pRB has been reported to partially colocalize with SAHF (45).
Alternatively, pRB and HIRA might both act in concert with a single protein that plays a key role in SAHF formation. One such candidate is the J domain protein, DNAJA2. J domain-containing proteins regulate the activity of 70-kDa heat-shock proteins (Hsp70s) (65). Hsp70s bind to short unfolded hydrophobic regions of substrate polypeptides, promoting assembly or disassembly of macromolecular protein-containing complexes. J domain proteins are known to regulate a wide variety of cellular processes, including protein translation, endocytosis, mRNA splicing, and mitochondrial biogenesis. We showed that DNAJA2 interacts with HIRA and pRB when ectopically expressed in cells. Moreover, DNAJA2 accelerates formation of HP1 and macroH2A-containing SAHF when ectopically expressed in primary human fibroblasts. The ability of DNAJA2 to drive formation of SAHF appears to be partially independent of pRB and p53 and ASF1a. In sum, although technical hurdles prevented us from unambiguously assigning a physiological role in SAHF formation to DNAJA2, these data clearly point to a role for DNAJA2 in formation of SAHF that is worthy of further investigation. Conceivably, the chaperone activity of DNAJA2 facilitates chromatin remodeling and SAHF formation by HIRA and pRB.
Supplementary Material
Acknowledgments
We thank Scott Lowe, Bill Hahn, Greg Enders, and Bob Weinberg for reagents and members of the Adams laboratory for critical discussions.
This work was supported by grants from the NIH (CA104429 and GM062281) and DOD (DAMD17-02-1-0726) to P.D.A. P.D.A. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Published ahead of print on 22 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. Kaelin, Jr. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 3.Ahuja, D., M. T. Saenz-Robles, and J. M. Pipas. 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24:7729-7745. [DOI] [PubMed] [Google Scholar]
- 4.Beausejour, C. M., A. Krtolica, F. Galimi, M. Narita, S. W. Lowe, P. Yaswen, and J. Campisi. 2003. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 22:4212-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellodi, C., K. Kindle, F. Bernassola, D. Dinsdale, A. Cossarizza, G. Melino, D. Heery, and P. Salomoni. 2006. Cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha. J. Biol. Chem. 281:14465-14473. [DOI] [PubMed] [Google Scholar]
- 6.Benevolenskaya, E. V., H. L. Murray, P. Branton, R. A. Young, and W. G. Kaelin, Jr. 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18:623-635. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell, C., K. A. Martin, A. Greenall, A. Pidoux, R. C. Allshire, and S. K. Whitehall. 2004. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 24:4309-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 9.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braig, M., S. Lee, C. Loddenkemper, C. Rudolph, A. H. Peters, B. Schlegelberger, H. Stein, B. Dorken, T. Jenuwein, and C. A. Schmitt. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660-665. [DOI] [PubMed] [Google Scholar]
- 11.Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120:513-522. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z., L. C. Trotman, D. Shaffer, H. K. Lin, Z. A. Dotan, M. Niki, J. A. Koutcher, H. I. Scher, T. Ludwig, W. Gerald, C. Cordon-Cardo, and P. P. Pandolfi. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436:725-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collado, M., J. Gil, A. Efeyan, C. Guerra, A. J. Schuhmacher, M. Barradas, A. Benguria, A. Zaballos, J. M. Flores, M. Barbacid, D. Beach, and M. Serrano. 2005. Tumour biology: senescence in premalignant tumours. Nature 436:642. [DOI] [PubMed] [Google Scholar]
- 14.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daganzo, S. M., J. P. Erzberger, W. M. Lam, E. Skordalakes, R. Zhang, A. A. Franco, S. J. Brill, P. D. Adams, J. M. Berger, and P. D. Kaufman. 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13:2148-2158. [DOI] [PubMed] [Google Scholar]
- 16.de Stanchina, E., E. Querido, M. Narita, R. V. Davuluri, P. P. Pandolfi, G. Ferbeyre, and S. W. Lowe. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13:523-535. [DOI] [PubMed] [Google Scholar]
- 17.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic Ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 19.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsyth, N. R., A. P. Evans, J. W. Shay, and W. E. Wright. 2003. Developmental differences in the immortalization of lung fibroblasts by telomerase. Aging Cell 2:235-243. [DOI] [PubMed] [Google Scholar]
- 21.Franco, A. A., W. M. Lam, P. M. Burgers, and P. D. Kaufman. 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19:1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiger, H., and G. Van Zant. 2002. The aging of lympho-hematopoietic stem cells. Nat. Immunol. 3:329-333. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalo, S., M. Garcia-Cao, M. F. Fraga, G. Schotta, A. H. Peters, S. E. Cotter, R. Eguia, D. C. Dean, M. Esteller, T. Jenuwein, and M. A. Blasco. 2005. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 7:420-428. [DOI] [PubMed] [Google Scholar]
- 24.Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu, J. R. Yates III, and P. D. Kaufman. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol 15:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenall, A., E. S. Williams, K. A. Martin, J. M. Palmer, J. Gray, C. Liu, and S. K. Whitehall. 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J. Biol. Chem. 281:8732-8739. [DOI] [PubMed] [Google Scholar]
- 26.Groth, A., D. Ray-Gallet, J. P. Quivy, J. Lukas, J. Bartek, and G. Almouzni. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17:301-311. [DOI] [PubMed] [Google Scholar]
- 27.Guo, A., P. Salomoni, J. Luo, A. Shih, S. Zhong, W. Gu, and P. P. Pandolfi. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730-736. [DOI] [PubMed] [Google Scholar]
- 28.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 29.Hahn, W. C., S. K. Dessain, M. W. Brooks, J. E. King, B. Elenbaas, D. M. Sabatini, J. A. DeCaprio, and R. A. Weinberg. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Herbig, U., and J. M. Sedivy. 2006. Regulation of growth arrest in senescence: telomere damage is not the end of the story. Mech. Ageing Dev. 127:16-24. [DOI] [PubMed] [Google Scholar]
- 33.Itahana, K., Y. Zou, Y. Itahana, J. L. Martinez, C. Beausejour, J. J. Jacobs, M. Van Lohuizen, V. Band, J. Campisi, and G. P. Dimri. 2003. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol. Cell. Biol. 23:389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janzen, V., R. Forkert, H. E. Fleming, Y. Saito, M. T. Waring, D. M. Dombkowski, T. Cheng, R. A. Depinho, N. E. Sharpless, and D. T. Scadden. 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16(INK4a). Nature 443:421-426. [DOI] [PubMed] [Google Scholar]
- 35.Kaufman, P. D., J. L. Cohen, and M. A. Osley. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korber, P., S. Barbaric, T. Luckenbach, A. Schmid, U. J. Schermer, D. Blaschke, and W. Horz. 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 281:5539-5545. [DOI] [PubMed] [Google Scholar]
- 37.Krawitz, D. C., T. Kama, and P. D. Kaufman. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnamurthy, J., M. R. Ramsey, K. L. Ligon, C. Torrice, A. Koh, S. Bonner-Weir, and N. E. Sharpless. 2006. p16(INK4a) induces an age-dependent decline in islet regenerative potential. Nature 443:453-457. [DOI] [PubMed] [Google Scholar]
- 39.Loppin, B., E. Bonnefoy, C. Anselme, A. Laurencon, T. L. Karr, and P. Couble. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437:1386-1390. [DOI] [PubMed] [Google Scholar]
- 40.Lorain, S., J. P. Quivy, F. Monier-Gavelle, C. Scamps, Y. Lecluse, G. Almouzni, and M. Lipinski. 1998. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell. Biol. 18:5546-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michaloglou, C., L. C. Vredeveld, M. S. Soengas, C. Denoyelle, T. Kuilman, C. M. van der Horst, D. M. Majoor, J. W. Shay, W. J. Mooi, and D. S. Peeper. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436:720-724. [DOI] [PubMed] [Google Scholar]
- 42.Molofsky, A. V., S. G. Slutsky, N. M. Joseph, S. He, R. Pardal, J. Krishnamurthy, N. E. Sharpless, and S. J. Morrison. 2006. Increasing p16(INK4a) expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443:448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myung, K., V. Pennaneach, E. S. Kats, and R. D. Kolodner. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 46.Nelson, D. M., C. Hall, H. Santos, T. Ma, G. D. Kao, T. J. Yen, J. W. Harper, and P. D. Adams. 2002. Coupling of DNA synthesis and histone synthesis in S-phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22:7459-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 48.Palmer, T. D., P. H. Schwartz, P. Taupin, B. Kaspar, S. A. Stein, and F. H. Gage. 2001. Cell culture. Progenitor cells from human brain after death. Nature 411:42-43. [DOI] [PubMed] [Google Scholar]
- 49.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 50.Phelps-Durr, T. L., J. Thomas, P. Vahab, and M. C. Timmermans. 2005. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17:2886-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prochasson, P., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramirez, R. D., C. P. Morales, B. S. Herbert, J. M. Rohde, C. Passons, J. W. Shay, and W. E. Wright. 2001. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 15:398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 54.Rubelj, I., and O. M. Pereira-Smith. 1994. SV40-transformed human cells in crisis exhibit changes that occur in normal cellular senescence. Exp. Cell Res. 211:82-89. [DOI] [PubMed] [Google Scholar]
- 55.Salomoni, P., and P. P. Pandolfi. 2002. The role of PML in tumor suppression. Cell 108:165-170. [DOI] [PubMed] [Google Scholar]
- 56.Schulz, L. L., and J. K. Tyler. 2006. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J. 20:488-490. [DOI] [PubMed] [Google Scholar]
- 57.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22:415-422. [DOI] [PubMed] [Google Scholar]
- 58.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 59.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 60.Sharp, J. A., A. A. Franco, M. A. Osley, P. D. Kaufman, D. C. Krawitz, T. Kama, E. T. Fouts, and J. L. Cohen. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherr, C. J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 62.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 64.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 65.Walsh, P., D. Bursac, Y. C. Law, D. Cyr, and T. Lithgow. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright, W. E., and J. W. Shay. 2002. Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 20:682-688. [DOI] [PubMed] [Google Scholar]
- 67.Ye, X., A. A. Franco, H. Santos, D. M. Nelson, P. D. Kaufman, and P. D. Adams. 2003. Defective S-phase chromatin assembly causes DNA damage, activation of the S-phase checkpoint and S-phase arrest. Mol. Cell 11:341-351. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, R., S.-T. Liu, W. Chen, B. Bonner, J. Pehrson, T. J. Yen, and P. D. Adams. 2007. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol. Cell. Biol. 27:949-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 8:19-30. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, L. 2005. Tumour suppressor retinoblastoma protein Rb: a transcriptional regulator. Eur. J. Cancer 41:2415-2427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.