Abstract
In amphibian oocytes, the maternal nuclear factor NF7 associates with the elongating pre-mRNAs present on the numerous lateral loops of the lampbrush chromosomes. Here, we have purified NF7 from an oocyte extract by using a combination of ion-exchange chromatography and gel filtration chromatography and demonstrated for the first time that nucleoplasmic NF7 exists primarily as free homotrimers. We confirmed the in vivo homotrimerization of NF7 by using a glutaraldehyde cross-linking assay, and we further showed that it only requires the coiled-coil domain of the NF7 tripartite motif/RBCC motif. Interestingly, we also obtained evidence that NF7 is recruited to the nucleus as a homotrimer, and expression of several mutated forms of NF7 in oocytes demonstrated that both the coiled coil and B box of NF7 are required for its chromosomal association. Together, these data strongly suggest that the interaction of NF7 with the active transcriptional units of RNA polymerase II is mediated by a trimeric B box. Finally, and in agreement with a role for NF7 in pre-mRNA maturation, we obtained evidence supporting the idea that NF7 associates with Cajal bodies.
The tripartite motif, or TRIM, defines a large family of proteins (more than 75 members) exhibiting a highly conserved modular organization that includes a TRIM associated with one or several variable domains positioned in their carboxyl-terminal half. While the TRIM was described as the RBCC (RING finger or A-box, B-box, coiled-coil) motif more than a decade ago, its function(s) remains essentially unknown. Yet, the interest in the TRIM protein family is growing for its apparent implication in various human disease states, including cancers and developmental disorders (36), and more recently in the human immunodeficiency virus cycle (43). Several discernible functional clues, however, are given by the three different motifs that make up a TRIM. The RING finger (really interesting new gene), also described as the C3HC4 motif in reference to the several conserved cysteine and histidine residues, is a zinc binding domain for which the ratio of two zinc cations per RING finger was previously established for several protein family members (2, 6, 8, 18, 21). Initially proposed to be a DNA binding motif (23), the RING finger is now considered a protein domain implicated in ubiquitin ligase activities, and it was recently proposed that the TRIM family represents a subgroup of the RING-type E3 ligase family (28). In addition to the RING finger, TRIMs contain one or two B boxes and a coiled-coil region. There are two types of B-box domain (B1 and B2) containing several highly conserved cysteine and histidine residues involved in the chelation of zinc cations. While all TRIM proteins have a B2 domain, a subgroup of TRIM family members also has a B1 domain that is invariably positioned between the RING finger and the B2 domain. The solution structures of only two B boxes have been elucidated, which surprisingly revealed two distinct protein folds. The B2 domain of amphibian nuclear factor 7 (NF7) was characterized a decade ago (9). It presents a unique compact structure and was found to chelate one zinc cation. The B1 domain of the human protein MID1 (midline 1) was obtained very recently (26) and displays a fold similar to that of the RING finger with, in particular, the chelation of two zinc cations. Overall, the function of both B1 and B2 remains largely uncharacterized. The coiled-coil region, a domain involved in protein multimerization, is always positioned after the B boxes and was shown for several TRIM family members to be necessary for the formation of homomultimers and in some cases for subcellular distribution (reviewed in reference 28). The linear arrangement, in the following order, of the RING finger, B box(es), and coiled-coil region (hence the name RBCC) is conserved among all of the TRIM family members and strongly suggests that the TRIM functions as an integrated unit. However, how the three distinct domains of a TRIM influence each other's structures and the overall function of the TRIM itself remain unclear.
NF7 is one of the very first TRIM proteins described and was characterized concurrently in two amphibian species, Xenopus laevis (35) and Pleurodeles waltl (5) (the recently corrected newt sequence is available through GenBank accession number L04190). In addition to the TRIM, NF7 has a chromodomain (CHD) and an RFP (Ret finger protein)-like domain in its N-terminal and C-terminal regions, respectively. The RFP domain (also referred to as the B-30.2 or PRY-SPRY domain) was defined on the basis of a remarkable conservation of its primary sequence among a small group of TRIM proteins, including the oncogene RFP. This modular structure, common to all TRIM proteins, suggested early on that NF7 (PwA33 in newts) may have multiple cellular functions. Supporting this idea is its apparent implication in pre-mRNA maturation in the oocyte (5), as well as its regulatory role in dorsal-ventral patterning during early development (15). The multifunctional aspect of NF7 was recently expanded even further with two novel associated activities. First, it was demonstrated that NF7 can affect, presumably through its newly described E3 ligase activity and interaction with the anaphase-promoting complex, the speed with which a Xenopus egg extract progresses though mitosis during the cell cycle (13). Second, NF7 was described as a protein that contributes to mitotic-spindle integrity through its microtubule-binding property (25). Clearly, NF7 is integral to multiple cellular processes.
Studies investigating a role for NF7 in gene regulation found that during oogenesis, NF7 is exclusively nuclear and associates with the active transcription units of RNA polymerase II (RNAPII) (3, 5, 6). In amphibian oocytes, these active transcriptional sites are readily observable by light microscopy as they correspond to the numerous lateral loops of the lampbrush chromosomes (LBCs), which are extended diplotene bivalents (reviewed in reference 31). The LBC loops are composed of a decondensed euchromatin axis surrounded by a thick ribonucleoprotein (RNP) matrix, which results from the association of an ensemble of proteins with the nascent RNAPII transcripts. Because of its specific association with the RNP matrix of the LBC loops, and despite the lack of a demonstrated RNA-binding activity, NF7 is likely to have a role in pre-mRNA maturation. Interestingly, a large fraction of nuclear NF7 is also found in the nucleoplasm either free or in association with small nucleoplasmic granules (6, 34). These free granules presumably correspond to RNP granules formed on the LBC loops and released into the nucleoplasm upon transcription termination (37). We report here that most of the free nucleoplasmic NF7 is in a homotrimeric form. We further show that NF7 homotrimers form in the cytoplasm and remain stable once in the nucleus. Importantly, NF7 trimerization is required but not sufficient for its targeting to chromosomal loops. Finally, deletion analysis revealed a possible transient association of NF7 with Cajal bodies (CBs), discrete nuclear organelles that were previously implicated in the maturation of all nuclear RNAs (16).
MATERIALS AND METHODS
Oocytes and nuclear-spread preparations.
Small fragments of ovary were obtained by surgery from female adult frogs (X. laevis) previously anesthetized in 0.15% tricaine methane sulfonate (MS222; Sigma Chemical Co., St. Louis, MO). The ovarian fragments were incubated for 2 h at room temperature in the saline buffer OR2 (40) containing 0.2% collagenase (type II; Sigma Chemical Co., St. Louis MO). Newly defolliculated stage IV to V oocytes were selected and maintained in OR2 at 18°C. Nuclear spreads were prepared as described previously by Bellini and Gall (4) and were fixed for 1 h at room temperature in phosphate-buffered saline (PBS) containing 2% paraformaldehyde and 1 mM MgCl2.
Antibodies and immunofluorescence.
Nuclear spreads were blocked in 1× PBS containing 0.5% bovine serum albumin (Sigma, St. Louis, MO) and 0.5% gelatin (from cold-water fish) for 10 min. Spreads were then incubated with primary and secondary antibodies diluted in the blocking buffer for 1 h, respectively, washed in 1× PBS, and counterstained with picogreen (Invitrogen, Carlsbad, CA) at a dilution of 1:10,000 in 1× PBS for 10 min before they were mounted in 50% glycerol containing 1 mg/ml of phenylenediamine (Sigma, St. Louis, MO). Fluorescence microscopy was carried out with a Leica DMR (Leica, Heidelberg, Germany) and a monochrome Spot RT charge-coupled device camera for image capture (Diagnostic Instruments, Sterling Heights, MI). Primary antibodies were the anti-NF7 mouse monoclonal antibody (MAb) 37-1A9, used at a concentration of 370 ng/μl; the anti-NF7 mouse polyclonal antibody M2, raised against the newt NF7 and used at a 1/2,000 dilution; the antihemagglutinin (anti-HA) antibody MAb 3F10 (Roche, Mannheim, Germany), used at a concentration of 20 ng/μl; and anti-XCAP-D2 rabbit polyclonal serum G (14), used at a dilution of 1/2,000. All secondary Alexa Fluor-conjugated antibodies (Invitrogen, Carlsbad, CA) were used at a concentration of 2.5 μg/ml, and they were Alexa 488-labeled-goat anti-mouse IgG, Alexa 594-labeled goat anti-rabbit IgG, and Alexa 594-labeled goat anti-rat IgG.
Cloning and oocyte microinjection.
The newt NF7 cDNA (accession no. L04190) was used as the template for PCR-mediated mutagenesis to generate all of the deletions used in the present study. The extent of each deletion is indicated at the amino acid level in Table 1. All constructs were N terminally tagged with the HA epitope (YPYDVPDYA) and cloned into pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA). A simian virus 40 nuclear localization sequence (NLS) (PKKKRKV) was also added immediately upstream of the HA tag to constructs in which the endogenous NLS was deleted. Capped sense strand RNAs were obtained in vitro with either the T7 or SP6 RNAP as described by Bellini et al. (6). Between 10 and 30 ng of RNA was microinjected into the cytoplasm of stage IV to V oocytes. Glass needles were prepared with a P-97 horizontal pipette puller (Sutter Instrument, Novato, CA). All injections were performed under a dissecting microscope (Leica S6) with a nanojet II (Drummond, Broomall, PA). Oocytes were maintained in OR2 for 18 h postinjection before the preparation of nuclear spreads and protein extracts.
TABLE 1.
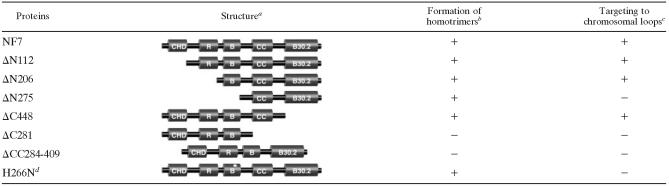
A trimeric B-box is required for the association of NF7 with chromosomal loops
R. RING finger; B. B-box; CC, coiled coil; B30.2. RFP-like domain. ΔN and ΔC indicate deletions from the N-terminal and C-terminal regions, respectively. The numbers indicate the amino acid residues where deletions stop.
Glutaraldehyde cross-linking assay.
Indirect immunofluorescence on nuclear spreads.
Described in reference 6.
In vitro translation and glutaraldehyde cross-linking.
Radiolabeled HA-tagged NF7 proteins were produced with the TNT coupled transcription-translation system from Promega (Madison, WI) in the presence of [35S]methionine (ReadiVue; GE Healthcare, Fairfield, CT) under the conditions suggested by the manufacturer. Translation products were analyzed by Western blotting to ensure that full-length proteins were produced and to estimate their concentrations. No further purification of the newly translated proteins was performed before cross-linking. A 0.02% glutaraldehyde solution was prepared fresh from a 70% glutaraldehyde stock solution (Sigma, St. Louis, MO) and used immediately. For each newly made protein, five identical cross-linking reaction mixtures were prepared with 1× PBS, 0.01% glutaraldehyde, and 2 μl of TNT product in a final volume of 40 μl. Reactions were stopped at various time points (see Fig. 3) by adding 10 μl of 5× Laemmli protein loading buffer. Samples were heated at 95°C for 5 min and then run on a 5 to 15% polyacrylamide continuous-gradient gel. Proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA), and radiolabeled proteins were detected with an FLA-3000 phosphorimager (Fuji Medical Systems, Stamford, CT). In the case of the S-200 peak fraction, 20 μl of protein sample was used instead of the TNT product. For endogenous NF7, each cross-linking reaction was performed on the nucleoplasmic protein fraction of 20 nuclei prepared as follows. Nuclei of 20 stage IV to V oocytes were manually isolated in 5:1 buffer (83 mM KCl, 17 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1 mM MgCl2, 1 mM dithiothreitol [DTT]). Nuclei were then mechanically disrupted; all of the nuclear organelles, including the chromosomes, were discarded by centrifugation at 22,000 × g at 4°C for 10 min; and the nucleoplasmic supernatant was recovered.
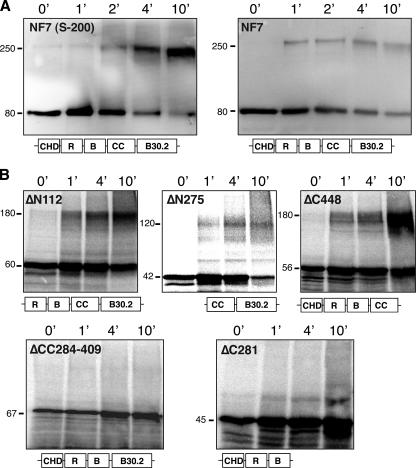
FIG. 3.
The coiled coil is essential for NF7 trimerization. (A) The cross-linking of endogenous NF7 trimers by 0.01% glutaraldehyde is demonstrated in the S-200 peak fractions and in a crude nucleoplasmic extract by Western blot analysis with MAb 37-1A9. Over time, a new band with an apparent molecular mass of ∼250 kDa is detected in addition to the monomeric NF7 form of 80 kDa. (B) Several modified forms of NF7 were produced in vitro in the presence of [35S]methionine, and their ability to trimerize was tested by glutaraldehyde cross-linking. A schematic representation of the expressed proteins is given under each autoradiograph (R, RING finger; B, B box; CC, coiled coil; B30.2, RFP-like domain). Note that the only domain involved in multimerization is the coiled-coil region. The values on the left are molecular sizes in kilodaltons.
Oocyte extract and biochemical purification of nucleoplasmic NF7.
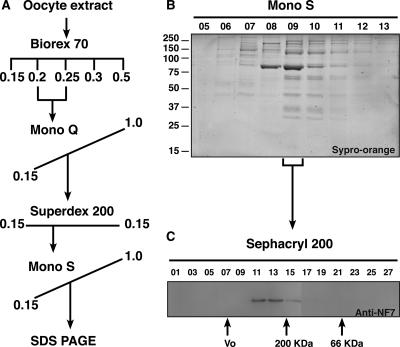
An oocyte extract was prepared, by high-speed ultracentrifugation essentially as previously described (20), from ovaries obtained from six female adult frogs (X. laevis). The soluble fraction was loaded onto a BioRex70 (Na+) column (Bio-Rad, Hercules, CA) equilibrated with 0.1 M buffer A (100 mM NaCl, 20 mM HEPES [pH 7.5], 10 mM β-glycerophosphate, 1.5 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 10% glycerol,1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 2 μg/ml pepstatin) at 10 mg of protein per ml of packed column volume. Unbound protein was removed by washing with 3 column volumes of buffer A (0.1 M), and bound protein was eluted in steps with buffer A containing increasing concentrations of NaCl (0.15 M, 0.2 M, 0.25 M, 0.3 M, and 0.5 M). All of the detectable NF7was found in the 0.2 and 0.25 step elutions. The fractions were pooled and dialyzed against 100 volumes of buffer A (0.1 M) for 2 h at 4°C. The protein pool was then fractionated by fast protein liquid chromatography by binding to a MonoQ HR10/10 column (GE Biotech) and elution in a linear gradient of 0.1 to 1 M buffer A in 20 column volumes. All of the detectable NF7 eluted in a single peak; the fractions were pooled and further fractionated by fast protein liquid chromatography on a Superdex 200 HR16/60 column in 0.15 M buffer A-0.1% Triton X-100. The single peak of NF7-containing fractions was pooled and again fractionated by ion-exchange chromatography over a MonoS HR5/5column with bound proteins eluted in 0.5-ml fractions from 10 column volumes of a linear gradient of 0.15 to 1.0 M buffer A. The MonoS fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 5 to 15% polyacrylamide gradient), stained with Sypro Orange (Invitrogen, Carlsbad, CA), and scanned with an FLA3000 fluoroimager (Fuji Medical Systems, Stamford, CT) with a 473-nm laser. During purification, fractions were tested by Western blotting as described below for the presence of NF7 with MAb 37-1A9.
Mass spectrometry.
In-gel digestion of the putative NF7 protein band was performed according to Hellman et al. (19), with the exception that ProGest at 0.6 mg/ml (Waters, Milford, MA) was used in conjunction with trypsin (sequencing grade; Promega, Madison, WI) during digestion. Mass spectrometry was performed with a Waters Q-ToF Global Ultima with a CapLC on a Symmetry C18 (3.5-μm particles; 75 m by 100 mm) NanoEase column (Waters, Milford, MA) at a flow rate of ∼400 nl/min with a gradient of acetonitrile containing 0.1% formic acid. The spectrometer was set to perform tandem mass spectrometry on the three most prominent ions (Big-3) during acquisition. The spectrum was processed with Protein Lynx Global Server 2.1 (Waters, Milford, MA) and PEAKS (Bioinformatics Solutions Inc., Waterloo, Ontario, Canada).
Protein samples and Western blot assays.
Whole nuclei and cytoplasms were hand isolated in ice-cold 5:1 buffer (83 mM KCl, 17 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1 mM MgCl2, 1 mM DTT) with fine tweezers under a dissecting microscope. Whole nuclear protein extracts were prepared by directly adding Laemmli protein sample buffer. Nucleoplasmic and organelle protein extracts were prepared by centrifuging the isolated nuclei at 22,000 × g for 15 min at 4°C. The resulting nuclear organelle pellets were directly resuspended in 1× Laemmli protein sample buffer. Supernatants corresponded to nucleoplasmic fractions. Cytoplasms were centrifuged at 22,000 × g for 30 min at 4°C to discard yolk and pigment granules. The recovered supernatants were used as cytoplasmic fractions. Proteins were separated by SDS-PAGE (5 to 15% polyacrylamide gradient gels) under denaturing conditions and transferred onto 0.2-μm-pore-size polyvinylidene difluoride membranes (Immun-Blot; Bio-Rad, Hercules, CA) for 2 h at 200 mA in Tris-glycine buffer containing 20% methanol with a wet-cell apparatus (Bio-Rad, Hercules, CA). Membranes were blocked in 1× PBS containing 5% fat-free dry milk (Carnation), rinsed briefly in 1× PBS, and then incubated with the primary antibody in 1× PBS containing 0.025% Tween 20 for 1 h at room temperature. Membranes were washed three times with 1× PBS containing 0.05% Tween 20 and incubated with the secondary antibody in 1× PBS containing 0.025% Tween 20 for 1 h at room temperature. After three washes with 1× PBS containing 0.05% Tween 20, detection was performed with an ECF kit (GE Healthcare, Fairfield, CT) and an FLA3000 fluoroimager (Fuji Medical Systems, Stamford, CT).
RESULTS
Nucleoplasmic NF7 exists primarily as a homotrimer.
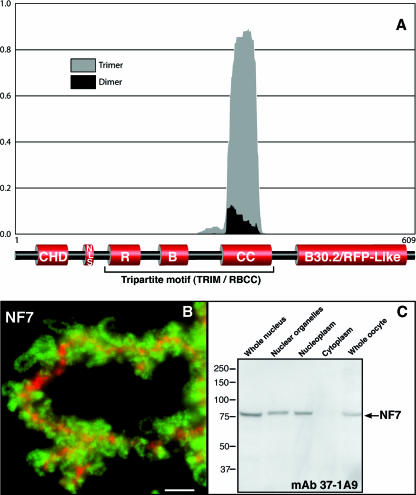
The primary sequence of NF7 displays a coiled coil, as predicted by the COILS algorithm of Lupas et al. (24), which encompasses residues 285 to 408. With the Multicoil program of Wolf et al. (41), we were able to further classify the predicted NF7 coiled coil as trimeric with a high degree of confidence (Fig. 1A). Because the coiled coil in TRIM proteins was previously shown for several family members to promote the assembly of high-molecular-weight complexes (36), we took advantage of the large nucleoplasmic pool of NF7 in the oocyte (Fig. 1C) and biochemically purified it under conditions that would maintain the native interactions with any NF7-interacting factors. A soluble oocyte extract containing the soluble nucleoplasmic NF7 protein was obtained by differential centrifugation as described by Jones et al. (20). During that procedure, the pool of NF7 associated with the chromosomal loops and RNP granules (Fig. 1B) is discarded. Fractionation by a combination of ion-exchange chromatography and gel filtration chromatography indicated a single distinct pool of NF7 protein, as detected by immunoblotting with a MAb to NF7 (37-1A9) (Fig. 2A). Size exclusion chromatography showed that all of the detectable NF7 protein fractionated at a size significantly larger (>200 kDa) than expected for NF7 alone (80 kDa), suggesting that NF7 was a component of a multiprotein complex. However, SDS-PAGE analysis of the composition of the NF7-containing fractions from the final step of purification surprisingly did not identify any potential NF7-interacting polypeptides precisely cofractionating with NF7 (Fig. 2B). A single major polypeptide with an apparent molecular mass of 80 kDa was readily detected. This polypeptide was excised from the gel, trypsin digested, and positively identified by liquid chromatography-tandem mass spectrometry as NF7. Together, these results suggested that most nucleoplasmic NF7 exists as homomultimers. To gain a more accurate estimate of the size of the suspected multimer, the purified NF7-containing fractions were fractionated over a Sephacryl 200 size exclusion column. Elution of NF7 at a peak size of ∼240 kDa was observed (Fig. 2C) and, in agreement with the presence of a predicted trimeric coiled coil in the NF7 primary sequence, indicated that endogenous NF7 assembles into homotrimers. To further confirm this trimeric stochiometry, the NF7 peak fractions from the Sephacryl 200 column were incubated at 20°C in the presence of 0.01% glutaraldehyde, a chemical cross-linker. At this very low concentration of glutaraldehyde, only direct protein interactions are expected to be cross-linked. Proteins were then analyzed in Western blot assays with MAb 37-1A9. Figure 3A shows the appearance over time of a new band with an apparent molecular mass of ∼250 kDa, in addition to the usual 80-kDa band corresponding to monomeric NF7, which is consistent with the cross-linking of an NF7 trimer.
FIG. 1.
NF7 is predicted to form trimers and associates with RNAPII active transcriptional units. (A) The Multicoil algorithm was used on Xenopus NF7 with a scanning window of 28 amino acid residues. A trimeric conformation was strongly predicted, with more than 80% probability for most of the coiled-coil region. This prediction is consistent with the data presented in Fig. 2 and 3, in which NF7 is demonstrated to form homotrimers. (B) Fluorescence micrograph showing a narrow region of an LBC labeled with anti-NF7 MAb 37-1A9 (green) and anti-XCAP-D2 (red). XCAP-D2, which is a subunit of condensin I, is restricted to the transcriptionally inactive heterochromatin domains of the LBCs (3) and was used here to define the chromosomal axes. In contrast, NF7 associates specifically with the RNP matrix of the chromosomal loops, which correspond to RNAPII active transcriptional units. In addition, NF7 is present on nucleoplasmic granules, which are out of focus here because of their small size (0.2 to 0.3 μm). Scale bar, 5 μm. (C) Western blot analysis with MAb 37-1A9 indicates that NF7 is primarily nuclear, and while it is found associated with chromosomes, it is also found soluble in the nucleoplasm. Proteins from 10 whole nuclei and organelles and nucleoplasm from 10 nuclei, five cytoplasms, or five whole oocytes were used. The values on the left are molecular sizes in kilodaltons.
FIG. 2.
Nucleoplasmic NF7 exists as a homotrimer. (A) Flow chart indicating the fractionation of nucleoplasmic NF7, which was followed through each step of purification by Western blot analysis with MAb 37-1A9. (B) Sypro orange-stained gel with each lane containing an equal volume of elution fractions 5 to 13 from the MonoS column. The major band with an apparent molecular mass of 80 kDa of fraction 09 was identified as NF7 by tandem mass spectrometry. The values on the left are molecular sizes in kilodaltons. (C) Fraction 09 from the MonoS column was further fractionated on a Sephacryl 200 column, and NF7 eluted at 240 kDa.
The extract used to purify NF7 was produced by gentle homogenization of oocytes in low-salt buffer lacking detergents that was designed to preserve native molecular interactions. However, one could not exclude the possibility that an initial large complex containing an NF7 trimer was disrupted during chromatography. To test for the presence of an eventual large nucleoplasmic NF7-containing complex, nuclei were manually isolated into a physiological buffer, the nuclear organelles were discarded by centrifugation, and the resulting crude nucleoplasmic extract was incubated in 0.01% glutaraldehyde. Western blot analysis revealed that NF7 is cross-linked over time to yield a complex of ∼250 kDa (Fig. 3A). Taken together, these data indicate that nucleoplasmic NF7 exists as a stable homotrimer.
The glutaraldehyde cross-linking assay was further used on several mutated forms of NF7, generated by in vitro translation, to confirm the role of the coiled coil in trimer formation (Table 1). The results obtained with five different deletion-containing forms of NF7, ΔN112, ΔN275, ΔC281, ΔC448, and ΔCC284-409, are presented in Fig. 3. The ΔN112, ΔN275, and ΔC448 proteins, which were expressed with apparent molecular masses of 60, 42, and 56 kDa, respectively, were all cross-linked effectively by glutaraldehyde over time to produce trimeric forms of ∼180, ∼120, and ∼180 kDa, respectively (Fig. 3B). In contrast, the two forms of NF7 that lack a functional coiled coil, ΔCC284-409 and ΔC281, failed to be cross-linked (Fig. 3B). These data, together with the data summarized in Table 1, demonstrate that the coiled coil appears to be the only domain critical for the formation of NF7 homotrimers.
NF7 trimer formation is required for its association with transcriptional units.
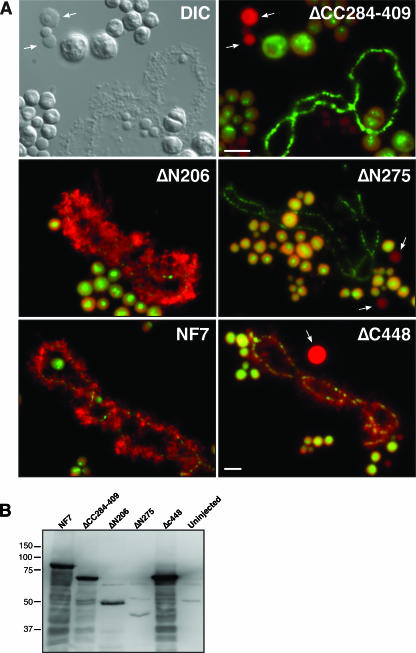
Many of the TRIM proteins display specific subcellular distributions, which appear to depend directly on the presence of the coiled-coil region (36). As previously mentioned, NF7 associates with the active transcriptional units of RNAPII and small nucleoplasmic granules, which are most likely RNP granules (34, 37) (Fig. 1B). To determine the importance of the trimerization of NF7 in its chromosomal localization, several deletion-containing forms of HA-tagged NF7 were expressed in Xenopus oocytes and their distributions were determined by indirect immunofluorescence microscopy on nuclear spreads (Fig. 4 and Table 1). NF7 displays a unique NLS that resides between its CHD and RING motifs. It is a bipartite NLS that was shown to be both necessary and sufficient for the nuclear import of NF7 in amphibian oocytes and embryos (22). As a result, several of the deletion-containing forms of NF7 that lack the NLS were unable to enter the nucleus upon their expression in the oocyte. A simian virus 40 NLS was thus fused to the amino-terminal end of these NF7 mutants to reestablish their nuclear import. In vitro-synthesized transcripts coding for HA-NF7 and the mutated forms presented in Table 1 were injected into the cytoplasm of stage V oocytes, and nuclei were isolated 18 h later. Western blot analysis indicated that in each case a major band of the expected molecular weight was detected by MAb 3F10 in the nucleus (Fig. 4B). Once in the nucleus, only the proteins that contained an intact B box and a coiled-coil region were able to associate with the nascent RNAPII transcripts of the chromosomal loops and the nucleoplasmic RNP granules. Yet, as we have shown, the B box is dispensable for NF7 trimerization. Thus, these data strongly suggest that while NF7 must form a homotrimer to interact with the RNAPII active transcriptional units, it is, however, not sufficient. All of the newly expressed NF7 mutants and, to a lesser extent, NF7 itself were found to interact with the numerous nucleoli of the oocyte nucleus, most likely in a nonspecific way. More surprisingly, several deletion-containing forms of NF7 (ΔC448, ΔCC284-409, and ΔN275) were also found to accumulate within CBs. This aberrant association with CBs was particularly obvious for ΔC448; while the chromosomal loops were labeled with MAb 3F10, as well as in the case of full-length HA-NF7, their correct exposure in Fig. 4A resulted in complete image saturation of the CBs (in panel ΔC448). We therefore revisited the distribution of endogenous NF7 with four distinct antibodies. As previously documented with another anti-NF7 antibody (L24) (17), the only nuclear domains detected by three of them (two mouse polyclonal sera and one MAb, B6) corresponded to the chromosomal loops and the free RNP granules. Interestingly, the fourth one (MAb 37-1A9), which also appears to be monospecific in immunoblot assays (Fig. 1), was found to label CBs well in addition to the RNP matrix of the chromosomal loops and the nucleoplasmic RNP granules (Fig. 5). Together, these data suggest an unexpected but physiologically relevant association of NF7 with CBs, which are discrete organelles previously implicated in the transcription and processing of all nuclear RNAs (16).
FIG. 4.
Trimerization and an intact B box are required for the association of NF7 with RNAPII active transcriptional units. (A) Transcripts of several modified forms of HA-tagged NF7 were injected into the cytoplasm of stage V oocytes, and nuclear spreads were prepared 18 h later. Newly made proteins (in red) were detected with anti-HA MAb 3F10 and an Alexa 594-conjugated secondary antibody. In all preparations, DNA was counterstained with picogreen (nucleoli and chromosomal axes are the only structures labeled) and the merged images are presented. A differential interference contrast image and its corresponding fluorescence micrograph are presented for ΔCC284-409 to emphasize the fact that while chromosomal loops are present, they are not labeled by MAb 3F10. Note that the two domains required for chromosomal association are the B box (ΔN275) and the coiled coil (ΔCC284-409). CBs, which were found to accumulate several of the newly expressed proteins, are indicated by arrows. Scale bars are10 μm. (B) Western blot analysis of the newly expressed protein with MAb 3F10. Each lane corresponds to 10 nuclei of stage V oocytes. The values on the left are molecular sizes in kilodaltons.
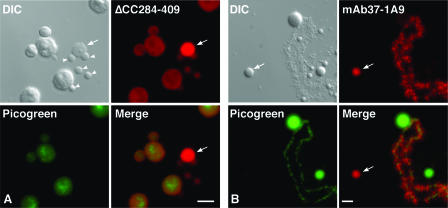
FIG. 5.
Association of NF7 with CBs. Differential interference contrast (DIC) and corresponding fluorescence micrographs of nuclear spreads. The DNA in both preparations was counterstained with picogreen. (A) The transcript coding for ΔCC284-409 was injected into the cytoplasm of stage V oocytes, and nuclear spreads were prepared 18 h later. The newly made protein was detected with MAb 3F10 (in red). Nucleoli, which are well labeled with picogreen, are weakly stained by MAb 3F10. In contrast, ΔCC284-409 is detected at a high concentration within CBs, as exemplified by the one present in the field (arrow). Note that the B snurposomes (arrowheads), which are the analogous structures of the somatic interchromatin granule clusters, are weakly labeled. (B) Immunostaining of endogenous NF7 with MAb 37-1A9 (in red). The lateral chromosomal loops and CBs (arrow) are well labeled, while nucleoli (green) and B snurposomes are negative. Scale bars are 5 μm.
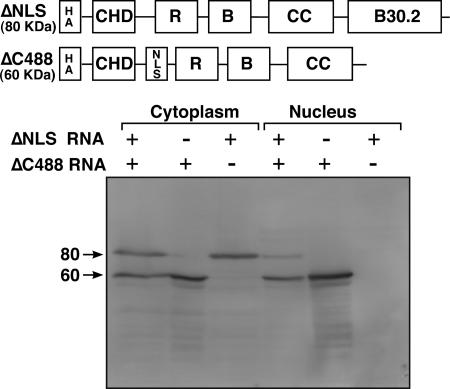
NF7 trimerization occurs in the cytoplasm prior to nuclear import.
The fact that nucleoplasmic NF7 exists as a homotrimer, together with the finding that the coiled coil is essential for the association of NF7 with the chromosomal loops suggested a model in which homotrimerization of NF7 precedes its association with the RNAPII active transcriptional units. To test this hypothesis further, we examined when trimer formation occurs by following the cellular fate of two modified forms of NF7 coexpressed in oocytes. The first one was named ΔNLS as it corresponds to full-length NF7 with the 15 residues forming the bipartite NLS deleted. As shown in Fig. 6, newly made ΔNLS, which has an apparent molecular mass of ∼80 kDa, is unable to enter the cell nucleus when expressed alone. The second one corresponds to ΔC448, which lacks the entire RFP/B30.2 domain. Because it still has the endogenous bipartite NLS, however, newly made ΔC448 is recruited well to the nucleus (Fig. 6). Both the ΔNLS and ΔC448 proteins have an intact coiled coil and can therefore be involved in the formation of trimers. Importantly, because the amount of newly synthesized endogenous NF7 is very low in the cytoplasm, it is unlikely to participate in heterotrimer formation with either ΔC448 or ΔNLS polypeptides. Interestingly, when ΔC448 and ΔNLS synthetic transcripts were coinjected into the cytoplasm of oocytes, both newly made proteins were recruited to the nucleus. We concluded that the nuclear targeting of ΔNLS was the result of heterotrimer formation with ΔC448. Together, these data demonstrate that NF7 trimerization can occur in the cytoplasm immediately after translation and strongly support a paradigm in which NF7 is recruited to the nucleus as a homotrimer that subsequently associates with active transcriptional units (Fig. 7).
FIG. 6.
NF7 trimerization occurs in the cytoplasm. Western blot assay showing the cellular distribution of two modified forms of NF7 upon expression in stage V oocytes. Both proteins were tagged with the HA epitope, and MAb 3F10 was used for their detection. Whether the transcripts coding for ΔNLS (80 kDa) and ΔC448 (60 kDa) were injected individually or coinjected is indicated by the plus and minus signs above the lanes. Cytoplasmic and nuclear protein samples were prepared 18 h after injection, and each lane corresponds to either five cytoplasms or 10 nuclei. The values on the left are molecular sizes in kilodaltons.
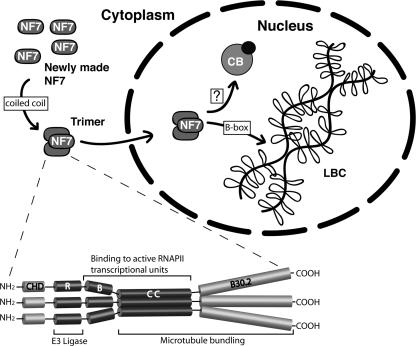
FIG. 7.
Paradigm of NF7 trimerization and association with chromosomal loops. In this model, NF7 trimerization mediated by the coiled-coil region occurs in the cytoplasm. NF7 homotrimers are then recruited to the nucleus, where they associate with the chromosomal loops through their trimeric B box. Whether NF7 transits through CBs remains uncertain.
DISCUSSION
A large pool of nucleoplasmic NF7.
In the present study, biochemical purification of NF7 revealed that it is an abundant protein of the amphibian oocyte (∼100 μg of the nucleoplasmic NF7 was purified from 400 mg of oocyte extract). Such a large pool of NF7 was expected, as it can be readily detected in immunoblot assays from fewer than 10 nuclei. It is also consistent with the fact that NF7 is a maternal nuclear factor, which is transmitted to the egg cytoplasm at the time of the nucleus breakdown and must reenter all ∼4,000 embryonic nuclei at the mid-blastula transition (1, 29). Such a situation is not unusual in the amphibian oocyte, which was previously shown to maintain in its nucleus high concentrations of many regulatory proteins, for example, RNAPII. Interestingly, many factors that are stored during oogenesis, such as transcription factors TFIIIA and FRGY2, are also functionally active (38). In the case of NF7, its association with the elongating RNAPII transcripts on the loops of the LBCs provides direct in vivo evidence for a role in pre-mRNA transcription and/or processing. In agreement with such a role, which is often ignored for NF7, its reentry into embryonic nuclei corresponds to the time when zygotic gene expression starts.
Homotrimerization of NF7 depends primarily on its coiled-coil domain.
Gel filtration and glutaraldehyde cross-linking analyses further demonstrated that all detectable nucleoplasmic NF7 exists as a homotrimer. Several TRIM family members were previously characterized as forming homo- and/or heteromultimers, and while the coiled coil is essential for their multimerization, the RING finger, the B box, and the B30.2 domain were also shown to be implicated. In particular, the multimerization of two proteins closely related to NF7 in their primary sequences, RFP and TRIM5α, depends on the B box and the B30.2 domain, respectively. In the case of TRIM5α, the B30.2 domain contributes greatly to the formation and/or the stability of TRIM5α trimers and its deletion results in the accumulation of TRIM5α dimers instead (30). In contrast, the homomultimerization of RFP and its heteroassociation with the PML (promyelocytic leukemia) oncogene directly involve its B box (11, 12). In addition, while homo- and heterodimeric RING complexes have been described in non-TRIM proteins such as BRCA1 and BARD1 (10), the transcriptional corepressor KAP-1 (KRAB-associated protein 1) is an example of a TRIM protein for which oligomerization involves all three subdomains of the TRIM motif, including the RING finger (33). Importantly, then, NF7 distinguishes itself as the coiled coil was found in our deletion analysis to be the primary motif involved in its trimerization.
Association of NF7 trimers with RNAPII transcriptional units requires the B box.
The coiled coil was also shown here for the first time to be implicated in the association of NF7 with the chromosomal loops. Trimerization is not sufficient, however, as deletion of the B box resulted in the same loss of NF7 chromosomal targeting. This is consistent with a previous study where the point mutation H266N, which prevents the B box from chelating zinc and presumably from folding correctly, inhibited NF7 chromosomal targeting (Table 1 and reference 6). We further demonstrated that the B box and the coiled coil are the only two domains essential for the association of NF7 with chromosomal loops. In addition, the fact that ΔN275 is still efficiently cross-linked by glutaraldehyde into a trimeric form but fails to target the RNAPII transcriptional units supports a model in which the interaction of NF7 with the RNP matrix of the loops is mediated by the B box (Fig. 7). In this model, the coiled coil is required to form a trimeric B box, which then can associate with components of the RNP matrix on chromosomal loops. A similar paradigm has been recently presented for the recruitment of the transcriptional corepressor KAP-1 to gene promoters (33). In this case, however, an intact TRIM motif is required to mediate the interaction between KAP-1 homotrimers and KRAB domain-containing transcriptional repressors. The lack of involvement of the RING finger in NF7 chromosomal targeting, however, does not antagonize the emerging concept of a functionally integrated TRIM motif (28). Indeed, an interesting possibility is that both the coiled coil and B box are necessary to bring NF7 to transcriptional units where the RING finger plays its newly demonstrated role of a ubiquitin E3 ligase. Yet, in the light of the recent demonstration that the coiled coil and the B30.2 domain are also implicated in the recruitment of NF7 to the mitotic spindle (25), one could argue that a key to the apparent multifunctions of NF7 resides in its oligomerization, in addition to its modular organization (Fig. 7). In such a model, the coiled coil would also be used independently of the TRIM motif to produce trimeric versions of the other domains defined in NF7. In the case of the B30.2 domain, its multimerization would offer NF7 the opportunity to contact several microtubules simultaneously, resulting in the microtubule-bundling activity presented in reference 25.
NF7 trimerization occurs in the cytoplasm.
Deletions and/or mutations of the B box have an effect on NF7 subnuclear distribution, but they do not influence its oligomerization. While newly expressed B-box mutants are likely to enter the nucleus as homotrimers, one cannot exclude the possibility of a subsequent heterotrimerization with the endogenous nuclear NF7. The fact that B-box mutants do not associate with the RNP matrix of the loops could therefore suggest a dominant negative effect of such mutants on NF7 chromosomal targeting. Yet another interesting possibility is that little heterotrimerization between endogenous NF7 and an expressed B-box mutant occurs. In support of that idea is the demonstration that the interaction between full-length KAP-1 and the KAP-1 TRIM domain requires both polypeptides to be coexpressed in Sf9 cell cultures, as the mixing of both polypeptides expressed individually could not promote the formation of heterooligomers (33). Similarly, it was reported that wild-type PML could oligomerize with various PML mutants only if these polypeptides were coexpressed in reticulocyte lysates (7). Together, these data suggest that the multimerization of these TRIM proteins might be coupled to translation or occur immediately after. Importantly, they also underscore a high stability of the TRIM homomultimers. Consistent with such a model, the demonstration that the nuclear targeting of newly expressed NF7-ΔNLS can be rescued by coexpression of ΔC448, presumably by heterotrimer formation, strongly suggests that NF7 trimers form in the cytoplasm immediately after translation and are subsequently recruited to the nucleus, where they remain stable.
Transient interaction of NF7 with CBs?
In addition to a loss of chromosomal targeting of the various B-box and coiled-coil mutants tested, a striking relocalization of these newly made proteins to nucleoli and CBs was observed. While an aberrant association with nucleoli of a large variety of expressed proteins (both wild-type and mutant proteins) is commonly seen in the oocyte and attributed to nonspecific interactions, it is not the case for CBs. The nucleolar factor NO38 is another example in amphibian oocytes of a protein, normally absent from CBs, for which some mutants were observed to accumulate within CBs (32). The reason for such a redistribution was not investigated and remains unclear, however. For NF7, this effect is not correlated with the lack of interaction between the mutated proteins and the chromosomal loops since Δ448 targets the RNP matrix of RNAPII transcriptional units well and displays the highest level of association with CBs. Interestingly, another component of the RNP matrix, the U1 snRNP, is normally detected in oocyte CBs by in situ hybridization at a level slightly higher than background levels obtained with control probes (42). Yet, upon truncation of its first 20 nucleotides, endogenous U1 accumulates in CBs (39). Because the splicing snRNPs traffic through CBs (27), one likely interpretation is that the exchange kinetics of U1 between CBs and the nucleoplasm are changed, resulting in an accumulation of U1 in CBs. Similarly, and consistent with a role in pre-mRNA processing, our data suggest that NF7 associates specifically but transiently with CBs.
Acknowledgments
We thank Natalya Novikova for expert technical assistance and Snehal Patel for helpful discussions of the present work. We are very grateful to Peter Yau and the on-campus proteomic facility for the sequencing of protein NF7 by tandem mass spectrometry.
MAb 37-1A9, developed by Christine Dreyer, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa Department of Biological Sciences, Iowa City. This work was supported by a Career Award from the National Science Foundation.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Abbadie, C., D. Boucher, J. Charlemagne, and J.-C. Lacroix. 1987. Immunolocalization of three oocyte nuclear proteins during oogenesis and embryogenesis in Pleurodeles. Development 101:715-728. [Google Scholar]
- 2.Barlow, P. N., B. Luisi, A. Milner, M. Elliott, and R. Everett. 1994. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237:201-211. [DOI] [PubMed] [Google Scholar]
- 3.Beenders, B., E. Watrin, V. Legagneux, I. Kireev, and M. Bellini. 2003. Distribution of XCAP-E and XCAP-D2 in the Xenopus oocyte nucleus. Chromosome Res. 11:549-564. [DOI] [PubMed] [Google Scholar]
- 4.Bellini, M., and J. G. Gall. 1998. Coilin can form a complex with the U7 small ribonucleoprotein. Mol. Biol. Cell 9:2987-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellini, M., J.-C. Lacroix, and J. G. Gall. 1993. A putative zinc-binding protein on lampbrush chromosome loops. EMBO J. 12:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellini, M., J.-C. Lacroix, and J. G. Gall. 1995. A zinc-binding domain is required for targeting the maternal nuclear protein PwA33 to lampbrush chromosome loops. J. Cell Biol. 131:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 8.Borden, K. L., M. N. Boddy, J. Lally, N. J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P. S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 14:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borden, K. L., J. M. Lally, S. R. Martin, N. J. O'Reilly, L. D. Etkin, and P. S. Freemont. 1995. Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J. 14:5947-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brzovic, P. S., P. Rajagopal, D. W. Hoyt, M. C. King, and R. E. Klevit. 2001. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8:833-837. [DOI] [PubMed] [Google Scholar]
- 11.Cao, T., K. L. Borden, P. S. Freemont, and L. D. Etkin. 1997. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J. Cell Sci. 110:1563-1571. [DOI] [PubMed] [Google Scholar]
- 12.Cao, T., E. Duprez, K. L. Borden, P. S. Freemont, and L. D. Etkin. 1998. Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J. Cell Sci. 111:1319-1329. [DOI] [PubMed] [Google Scholar]
- 13.Casaletto, J. B., L. K. Nutt, Q. Wu, J. D. Moore, L. D. Etkin, P. K. Jackson, T. Hunt, and S. Kornbluth. 2005. Inhibition of the anaphase-promoting complex by the Xnf7 ubiquitin ligase. J. Cell Biol. 169:61-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cubizolles, F., V. Legagneux, R. Le Guellec, I. Chartrain, R. Uzbekov, C. Ford, and K. Le Guellec. 1998. pEg7, a new Xenopus protein required for mitotic chromosome condensation in egg extracts. J. Cell Biol. 143:1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Hodiri, H. M., W. Shou, and L. D. Etkin. 1997. xnf7 functions in dorsal-ventral patterning of the Xenopus embryo. Dev. Biol. 190:1-17. [DOI] [PubMed] [Google Scholar]
- 16.Gall, J. G. 2003. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 4:975-980. [DOI] [PubMed] [Google Scholar]
- 17.Gall, J. G., and C. Murphy. 1998. Assembly of lampbrush chromosomes from sperm chromatin. Mol. Biol. Cell 9:733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanzawa, H., M. J. de Ruwe, T. K. Albert, P. C. van Der Vliet, H. T. Timmers, and R. Boelens. 2001. The structure of the C4C4 RING finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J. Biol. Chem. 276:10185-10190. [DOI] [PubMed] [Google Scholar]
- 19.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 20.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 21.Katoh, S., C. Hong, Y. Tsunoda, K. Murata, R. Takai, E. Minami, T. Yamazaki, and E. Katoh. 2003. High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides. J. Biol. Chem. 278:15341-15348. [DOI] [PubMed] [Google Scholar]
- 22.Li, X., and L. D. Etkin. 1993. Xenopus nuclear factor 7 (xnf7) possesses an NLS that functions efficiently in both oocytes and embryos. J. Cell Sci. 105:389-395. [DOI] [PubMed] [Google Scholar]
- 23.Lovering, R., I. M. Hanson, K. L. Borden, S. Martin, N. J. O'Reilly, G. I. Evan, D. Rahman, D. J. Pappin, J. Trowsdale, and P. S. Freemont. 1993. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 90:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 25.Maresca, T. J., H. Niederstrasser, K. Weis, and R. Heald. 2005. Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr. Biol. 15:1755-1761. [DOI] [PubMed] [Google Scholar]
- 26.Massiah, M. A., B. N. Simmons, K. M. Short, and T. C. Cox. 2006. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J. Mol. Biol. 358:532-545. [DOI] [PubMed] [Google Scholar]
- 27.Matera, A. G., and K. B. Shpargel. 2006. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 18:317-324. [DOI] [PubMed] [Google Scholar]
- 28.Meroni, G., and G. Diez-Roux. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27:1147-1157. [DOI] [PubMed] [Google Scholar]
- 29.Miller, M., B. A. Reddy, M. Kloc, X. X. Li, C. Dreyer, and L. D. Etkin. 1991. The nuclear-cytoplasmic distribution of the Xenopus nuclear factor, xnf7, coincides with its state of phosphorylation during early development. Development 113:569-575. [DOI] [PubMed] [Google Scholar]
- 30.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan, G. T. 2002. Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res. 10:177-200. [DOI] [PubMed] [Google Scholar]
- 32.Peculis, B. A., and J. G. Gall. 1992. Localization of the nucleolar protein NO38 in amphibian oocytes. J. Cell Biol. 116:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, H., G. E. Begg, D. C. Schultz, J. R. Friedman, D. E. Jensen, D. W. Speicher, and F. J. Rauscher III. 2000. Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 295:1139-1162. [DOI] [PubMed] [Google Scholar]
- 34.Pyne, C. K., F. Simon, M. T. Loones, G. Geraud, M. Bachmann, and J. C. Lacroix. 1994. Localization of antigens PwA33 and La on lampbrush chromosomes and on nucleoplasmic structures in the oocyte of the urodele Pleurodeles waltl: light and electron microscopic immunocytochemical studies. Chromosoma 103:475-485. [DOI] [PubMed] [Google Scholar]
- 35.Reddy, B. A., M. Kloc, and L. Etkin. 1991. The cloning and characterization of a maternally expressed novel zinc finger nuclear phosphoprotein (xnf7) in Xenopus laevis. Dev. Biol. 148:107-116. [DOI] [PubMed] [Google Scholar]
- 36.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommerville, J. 1981. Immunolocalization and structural organization of nascent RNP. Cell Nucleus 8:1-57. [Google Scholar]
- 38.Tafuri, S. R., and A. P. Wolffe. 1993. Dual roles for transcription and translation factors in the RNA storage particles of Xenopus oocytes. Trends Cell Biol. 3:94-98. [DOI] [PubMed] [Google Scholar]
- 39.Tsvetkov, A., M. Jantsch, Z. Wu, C. Murphy, and J. G. Gall. 1992. Transcription on lampbrush chromosome loops in the absence of U2 snRNA. Mol. Biol. Cell 3:249-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace, R. A., D. W. Jared, J. N. Dumont, and M. W. Sega. 1973. Protein incorporation by isolated amphibian oocytes. III. Optimum incubation conditions. J. Exp. Zool. 184:321-333. [DOI] [PubMed] [Google Scholar]
- 41.Wolf, E., P. S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, Z., C. Murphy, H. G. Callan, and J. G. Gall. 1991. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J. Cell Biol. 113:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]