Abstract
The energy-dependent proteolysis of cellular proteins is mediated by conserved proteolytic AAA+ complexes. Two such machines, the m- and i-AAA proteases, are present in the mitochondrial inner membrane. They exert chaperone-like properties and specifically degrade nonnative membrane proteins. However, molecular mechanisms of substrate engagement by AAA proteases remained elusive. Here, we define initial steps of substrate recognition and identify two distinct substrate binding sites in the i-AAA protease subunit Yme1. Misfolded polypeptides are recognized by conserved helices in proteolytic and AAA domains. Structural modeling reveals a lattice-like arrangement of these helices at the surface of hexameric AAA protease ring complexes. While helices within the AAA domain apparently play a general role for substrate binding, the requirement for binding to surface-exposed helices within the proteolytic domain is determined by the folding and membrane association of substrates. Moreover, an assembly factor of cytochrome c oxidase, Cox20, serves as a substrate-specific cofactor during proteolysis and modulates the initial interaction of nonassembled Cox2 with the protease. Our findings therefore reveal the existence of alternative substrate recognition pathways within AAA proteases and shed new light on molecular mechanisms ensuring the specificity of proteolysis by energy-dependent proteases.
The cellular concentration of many regulatory proteins and the surveillance of protein quality depend on ATP-dependent proteolytic machines. These proteases form architecturally similar ring complexes which provide a sequestered environment for proteolysis (8, 33). Their conserved subunits comprise distantly related protein families, but all harbor one or two copies of an AAA+ ATPase (ATPases associated with a variety of cellular activities) module (14). This domain is generally thought to drive the unfolding of substrate proteins, allowing their entry into the proteolytic chamber.
The accuracy of substrate selection is crucial to avoid deleterious effects on cell functions. In the case of the eukaryotic 26S proteasome substrates are mostly recognized via covalently attached ubiquitin moieties (8). For other energy-dependent proteases, peptides within the native sequence of substrate proteins, often located near their termini, serve as degradation signals (4, 11). We are just beginning to understand how these signals are recognized by ATP-dependent proteolytic machines (26, 27). Conserved loop regions within the central channel formed by the ATPase subunits have been demonstrated to be involved in substrate binding (16, 34). ATP-dependent conformational changes of these loops are thought to drive the translocation, and concomitant unfolding, of associated substrates into the proteolytic chamber. Some substrates appear to interact directly with these central loops; however, others seem to bind initially to other sites before being transferred to this region. Indeed, accessory domains of ATPase subunits of bacterial Clp proteases, like N domains of ClpX and ClpA or the I domain of HslU, are critical for substrate recognition (17, 36).
AAA proteases are ubiquitously present in eubacteria as well as mitochondria and chloroplasts of eukaryotic cells (18, 30). Their subunits share a conserved domain structure: they are anchored to the membrane by one or two membrane-spanning segments at their N-terminal end which are followed by one AAA domain and a metallopeptidase domain. The inner membrane of mitochondria harbors two AAA proteases, the i-AAA protease, facing the intermembrane space, and the m-AAA protease, whose catalytic sites are exposed to the matrix space. The i-AAA protease is composed of Yme1 subunits in Saccharomyces cerevisiae (23, 41), whereas m-AAA proteases are built up of homologous Yta10 and Yta12 subunits (2).
The loss of AAA proteases causes severe phenotypes in yeast (1, 2, 5, 13, 39, 40) and leads to axonal degeneration in hereditary spastic paraplegia (6, 10). The m-AAA protease mediates the maturation of the newly imported ribosomal subunit MrpL32 and thereby controls the biogenesis of mitochondrial ribosomes and organellar translation (29). Moreover, both AAA proteases conduct the surveillance of protein quality in the inner membrane and degrade nonassembled and misfolded membrane proteins (2, 22, 28, 31, 42). Studies of the degradation of model substrate proteins have suggested that the folding state of solvent-exposed domains of mitochondrial inner membrane proteins is recognized and have assigned a crucial role in this process to the AAA domain of the i-AAA protease subunit Yme1 (24). Similar to soluble ATP-dependent proteases, a central pore loop within the AAA domains of AAA proteases appears to be crucial for proteolysis, indicating a conserved mode of action (12, 43). However, axial access to the central pore of AAA proteases is hampered by their membrane integration, suggesting that substrates might be initially bound at the outer surface of the proteolytic cylinder.
Here, we have analyzed substrate engagement by the i-AAA protease and identify surface-exposed, helical segments within the AAA and proteolytic domains of Yme1 as initial substrate binding sites. Our results reveal substrate-specific requirements of these sites for proteolysis and thereby define alternative pathways for substrate entry to the i-AAA protease.
MATERIALS AND METHODS
Cloning procedures.
IAP-1 and the hybrid proteins were fused to the mitochondrial targeting signal of Yme1 to ensure sorting to mitochondria. Chimeras of YME1 and IAP-1 were generated by gap repair in yeast cells: SNNN (Yme1 positions 1 to 248 and IAP-1 positions 228 to 739), SNNS (Yme1 positions 1 to 248, IAP-1 positions 228 to 578, and Yme1 positions 604 to 748), SSNN (Yme1 positions 1 to 495 and IAP-1 positions 470 to 739), and SSNS (Yme1 positions 1 to 495, IAP-1 positions 470 to 578, and Yme1 positions 604 to 748). DNA fragments coding for AAA protease domains to be introduced were amplified by PCR using primer pairs harboring homologous sequences (40 bp) flanking the integration site. Δyme1 cells were cotransformed with these PCR-amplified DNA fragments and linearized with pVT100U-YME1 or pVT100U-IAP-1 (20).
The following mutations were introduced into YME1: yme1E292A (A), yme1E292A,E294A (A2), yme1D287A,E288A,E292A,E294A,E295A,D298A (A6), yme1D287K,E288K,E292K,E294K,E295K,D298K (K6), and yme1D287N,E288Q,E292Q,E294Q,E295Q,D298N (Q/N6).
Yeast strains and growth conditions.
All strains used in this study are derivatives of W303. YME1 and IAP-1 genes and derivatives thereof were expressed in a Δyme1 yeast strain described previously (YCK10 [20]). Yme1 and hybrid proteins accumulated at similar levels in the different strains as revealed by immunoblotting of cellular extracts using Yme1-specific antibodies (data not shown). For the generation of Δyme1 Δcox20 cells (YMG108), the COX20 gene was disrupted in the Δyme1 strain YCK10 by PCR-based targeted homologous recombination using a kanMX4 cassette.
Assessing substrate binding to the i-AAA protease by coimmunoprecipitation.
After import of Phb1 or in organello translation, mitochondria (0.5 mg/ml) were solubilized in 2% (wt/vol) digitonin, 150 mM KAc, pH 7.4, 4 mM MgAc, 30 mM Tris-HCl, pH 7.4, and 1 mM phenylmethylsulfonyl fluoride (PMSF). After a clarifying spin for 15 min at 30,000 × g supernatants were subjected to coimmunoprecipitation and incubated for 1 h at 4°C with polyclonal antiserum directed against the N terminus of Yme1 (24). After several washing steps, bound material was eluted with sodium dodecyl sulfate (SDS)-sample buffer, analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography, and quantified by phosphorimaging. The presence of Yme1 and derivatives in the precipitate was monitored by immunoblotting to control for equal precipitation efficiencies.
Substrate binding to CH in vitro.
CH of Yme1 (amino acids 650 to 709) and IAP-1 (amino acids 625 to 684) were cloned into pKM263 and expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli BL21-CodonPlus (DE3)-RIL cells. For protein expression the cells were grown to an A600 of 0.5 to 0.8 and induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 25°C. Cells were lysed by sonication in phosphate-buffered saline, pH 7.3, containing 1 mM EDTA, 5% (vol/vol) glycerol, 5 mM dithiothreitol (DTT), and protease inhibitors. The proteins were purified using glutathione-Sepharose 4B as indicated by the manufacturer and dialyzed against 50 mM Tris-HCl (pH 7.8), 120 mM KCl, 0.5 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 5% (vol/vol) glycerol.
After Cox2 synthesis or import of Phb1, Δyme1 mitochondria (0.5 mg/ml for Cox2 and 1 mg/ml for Phb1) were lysed in 2% (wt/vol) digitonin, 5 mM DTT, 5% (vol/vol) glycerol, and 1 mM PMSF. After a clarifying centrifugation for 30 min at 16,000 × g, the soluble fraction was incubated with GST-YME1-CH, GST-IAP-1-CH, or GST (20 μM as monomer) for 10 min at 37°C. Subsequently, the sample was incubated with glutathione-Sepharose 4B (100 μl) for 1 h at 4°C and then washed extensively with lysis buffer containing 0.2% (wt/vol) digitonin and an increasing salt concentration (0.15 to 0.5 M KCl). Bound material was eluted with 20 mM reduced glutathione in 50 mM Tris-HCl, pH 8, 5% (vol/vol) glycerol, and 0.2% (wt/vol) digitonin. Eluted proteins were trichloroacetic acid precipitated and analyzed by SDS-PAGE and phosphorimaging.
RESULTS
Chimeras of orthologous i-AAA proteases Yme1 and IAP-1.
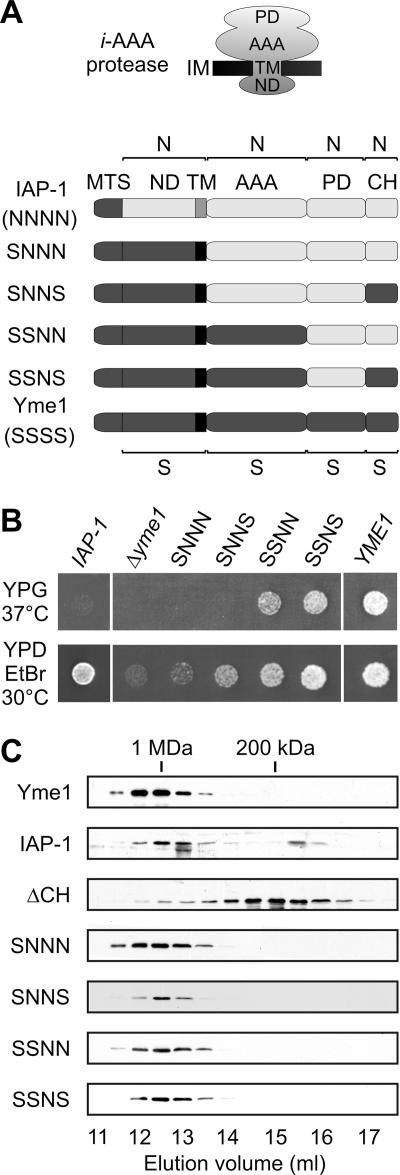
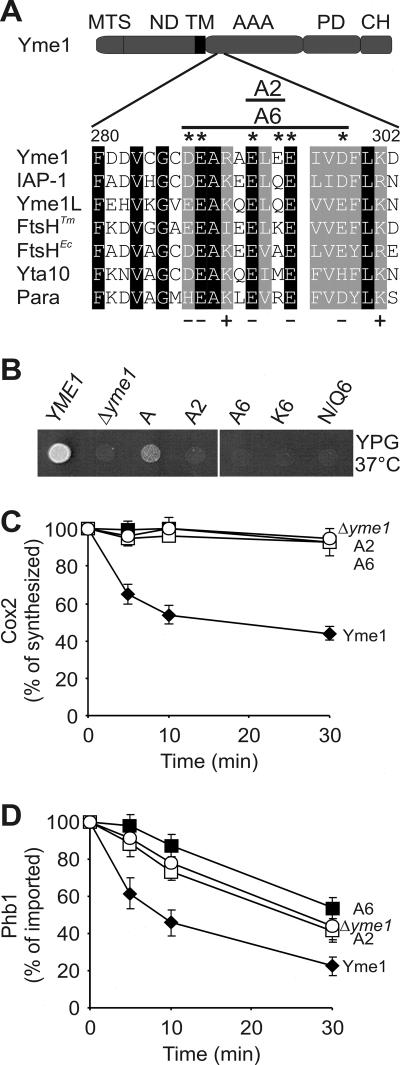
Expression of the Neurospora crassa i-AAA protease IAP-1 only partially restores growth deficiencies associated with a deletion of YME1 in yeast (20). Both proteins exhibit a conserved domain structure (Fig. 1A): they are anchored to the inner membrane by the membrane-spanning domain at the N terminus which is followed by AAA and proteolytic domains and a helical region at the C terminus. To identify domains responsible for the apparently divergent functional properties of i-AAA proteases of S. cerevisiae (S) and N. crassa (N), several chimeras of Yme1 and IAP-1 were generated and expressed in Δyme1 cells (Fig. 1A and B). Replacement of N-terminal domains of IAP-1 including the mitochondrial targeting signal and the matrix-exposed and the membrane-spanning domains, or in addition C-terminal helices (CH) (SNNS), by the corresponding domains of Yme1 (chimera SNNN) did not restore respiratory growth of Δyme1 cells at 37°C (Fig. 1B). Similarly, fermentative growth in the presence of ethidium bromide was not or only slightly promoted by these hybrids (Fig. 1B). This is in contrast to Δyme1 cells expressing IAP-1, which grow under fermenting conditions (Fig. 1B) (20), indicating negative growth effects of these hybrid proteins. However, respiratory cell growth on glycerol-containing medium was significantly improved after introduction of the AAA domain of Yme1 into the hybrid protein (chimera SSNN) (Fig. 1B). Thus, functional differences between Yme1 and IAP-1 appear to be at least partly caused by differences in their AAA domains. Notably, expression of chimeras harboring C-terminal helices of the proteolytic domain of Yme1 (chimeras SNNS and SSNS) resulted in a slightly improved cell growth in the presence of ethidium bromide, suggesting that this region also contributes to functional differences between the two proteins (Fig. 1B).
FIG. 1.
i-AAA protease chimeras. (A) Domain structure of IAP-1, various hybrid proteins, and Yme1. Individual domains were derived from N. crassa IAP-1 (N) or from S. cerevisiae Yme1 (S). MTS, mitochondrial targeting sequence; ND, N-terminal domain; TM, transmembrane segment; AAA, AAA domain; PD, proteolytic domain; CH, C-terminal helices. (B) Functional complementation of Δyme1 cells by expression of Yme1 (YME1), IAP-1, or chimeras of the two proteins. Cells were spotted onto yeast peptone medium containing 2% (wt/vol) glucose and ethidium bromide (25 μg/ml; YPD EtBr) or 3% (vol/vol) glycerol (YPG) and incubated for 3 to 4 days at 30°C or 37°C, respectively. (C) Assembly of i-AAA protease hybrids and variants. Δyme1 mitochondria (5 mg/ml) harboring Yme1, IAP-1, hybrid proteins, or Yme1(1-652) lacking C-terminal helices (ΔCH) were solubilized in 0.4% (vol/vol) Triton X-100 and fractionated by Superose 6 size chromatography. Yme1 and hybrid proteins were detected in eluate fractions by immunoblotting.
To exclude indirect effects of an impaired assembly of the hybrid proteins, complex formation of chimeric i-AAA proteases and of a truncated variant of Yme1 lacking CH (ΔCH) was assessed in Δyme1 cells. Whereas Yme1 formed a large complex of ∼1 MDa, deletion of CH impaired its assembly (Fig. 1C). However, CH regions of both Yme1 and IAP-1 allowed complex formation of all chimeras (Fig. 1C). Thus, differences in the assembly state of Yme1 and the hybrid proteins do not account for the observed functional differences.
Substrate-specific role of Yme1 CH during proteolysis.
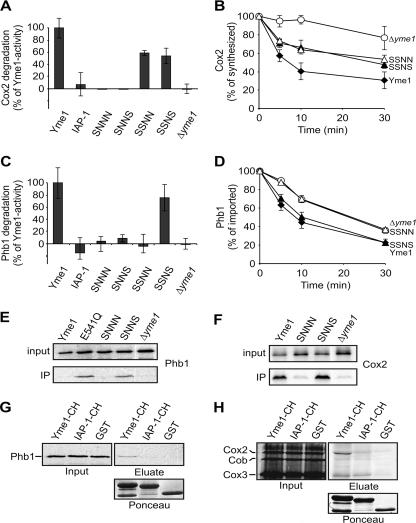
As cell growth depends on proteolysis by Yme1 (23), these complementation studies indicate proteolytic activity of the hybrid proteins SSNN and SSNS. To substantiate this conclusion, we assessed the stability of nonassembled mitochondrial proteins in the presence of various chimeras. Only two endogenous proteins have been identified as substrates of Yme1 up to now. Mitochondrially encoded subunit 2 of cytochrome c oxidase (Cox2) is rapidly degraded by Yme1 when synthesized in isolated organelles, i.e., in the absence of nucleus-encoded assembly partners (Fig. 2A and B) (28, 31, 42). IAP-1 could not substitute for Yme1 in this process (Fig. 2A). Similarly, Cox2 remained stable in the presence of the chimera SNNN or SNNS (Fig. 2A). In contrast, although at a lower rate, Cox2 was degraded upon expression of SSNN or SSNS, each containing the AAA domain of Yme1 (Fig. 2A and B). These findings are in agreement with our cell growth analysis on YPG and suggest functional differences between the AAA domains of the two i-AAA proteases.
FIG. 2.
CH dependence of proteolysis and substrate binding by the i-AAA protease. (A and B) Stability of newly synthesized Cox2. Mitochondrially encoded proteins were synthesized in the presence of [35S]methionine in Δyme1 mitochondria containing Yme1, IAP-1, or various hybrid proteins (38). The stability of Cox2 at 37°C was assessed by SDS-PAGE and phosphorimaging. The means and standard deviations of three independent experiments are indicated. (A) Stability of Cox2 after 10 min at 37°C in Δyme1 mitochondria containing Yme1, IAP-1, SNNN, SNNS, SSNN, or SSNS. Degradation of Cox2 by Yme1 was set as 100%. (B) Kinetics of Cox2 degradation in Δyme1 mitochondria (○) containing Yme1 (⧫), SSNN (▵), or SSNS (▴). (C and D) Stability of nonassembled Phb1. 35S-labeled Phb1 was imported for 20 min at 25°C into Δyme1 mitochondria containing Yme1, IAP-1, or various hybrid proteins. After trypsin digestion of nonimported Phb1, samples were incubated at 37°C. Proteolysis of Phb1 was monitored by SDS-PAGE and phosphorimaging. The means and standard deviations of three independent experiments are indicated. (C) Stability of Phb1 after 10 min at 37°C in Δyme1 mitochondria containing Yme1, IAP-1, SNNN, SNNS, SSNN, or SSNS. Yme1-dependent degradation of Phb1 was set as 100%. The means and standard deviations of three independent experiments are indicated. (D) Time course of Phb1 degradation in Δyme1 mitochondria (○) containing Yme1 (⧫), SSNN (▵), or SSNS (▴). (E) Binding of nonassembled Phb1 to Yme1 variants. 35S-labeled Phb1 was imported into Δyme1 mitochondria which harbored Yme1, Yme1E541Q (E541Q), SNNN, or SNNS. Mitochondrial membranes were solubilized in digitonin, and extracts (input, corresponding to 5% of total) were subjected to coimmunoprecipitation using antibodies directed against the N-terminal domain of Yme1. Precipitates were analyzed by SDS-PAGE and autoradiography. (F) Binding of Cox2 to Yme1 variants. Mitochondrial translation products were 35S labeled in Δyme1 mitochondria containing Yme1, SNNN, or SNNS. The association with Yme1 and variants thereof was examined by coimmunoprecipitation. (G) Phb1 binding to CH in vitro. 35S-labeled Phb1 was imported into Δyme1 mitochondria as for panel D. Digitonin extracts (input, 5%) were incubated with the GST fusion proteins Yme1-CH and IAP-1-CH or with GST and precipitated with glutathione-Sepharose beads. Bound material was analyzed by SDS-PAGE and autoradiography (eluate). The presence of equal amounts of bait proteins was monitored by Ponceau red staining (lower panel). (H) Cox2 binding to CH. After 35S labeling of mitochondrial translation products, binding to CH of Yme1 and IAP-1 was examined as in panel G.
We also examined proteolysis of prohibitin 1 (Phb1), another substrate of the i-AAA protease in mitochondria. Newly imported Phb1 fails to assemble with Phb2 subunits into prohibitin complexes in the inner membrane, resulting in its degradation by Yme1 and another, yet unidentified peptidase in the intermembrane space (Fig. 2C and D) (19). Neither expression of IAP-1, that of SNNN, nor that of SNNS accelerated proteolysis of Phb1 compared to Δyme1 cells (Fig. 2C). Moreover, in contrast to Cox2, Phb1 was not degraded by SSNN. Proteolysis occurred only in Δyme1 mitochondria in the presence of SSNS, which contains CH of Yme1 (Fig. 2C and D). Although only slightly affected, most likely due to the redundant activity of another peptidase, the kinetics of Phb1 degradation was accelerated in a statistically significant manner in the presence of SSNS and occurred as in wild-type cells (Fig. 2D). We conclude that functional differences exist between CH of i-AAA proteases and that proteolysis of some substrates is affected by CH. In agreement with these findings, mutational disruption of a C-terminal region led to impaired proteolytic activity of the related Escherichia coli AAA protease FtsH (35).
Substrate binding to Yme1 CH.
To determine a potential function of CH in substrate engagement, we analyzed the binding of substrate proteins to SNNN and SNNS by coimmunoprecipitation. Both chimeras are proteolytically inactive but differ in CH. 35S-labeled Phb1 was imported into mitochondria containing Yme1, the chimera SNNN or SNNS, or a mutant variant of Yme1 harboring a point mutation in the proteolytic center (E541Q). After solubilization of mitochondrial membranes, extracts were incubated with antibodies which recognize an epitope in the N-terminal, matrix-exposed domain of Yme1 present in both hybrid proteins. Similarly to other misfolded substrate polypeptides (23), Phb1 was precipitated efficiently only with proteolytically inactive Yme1 (E541Q) and not with wild-type Yme1 (Fig. 2E). Strikingly, Phb1 did not bind to SNNN but was found in association with SNNS harboring CH of Yme1 (Fig. 2E). These findings indicate that the CH-dependent proteolysis of Phb1 is based on CH-mediated substrate binding to the i-AAA protease.
A role of CH for substrate engagement was also revealed by analyzing Cox2 binding to Yme1 and derivatives thereof. Mitochondrially encoded proteins were synthesized in organelles isolated from wild-type cells or cells expressing different chimeras. After solubilization of mitochondrial membranes, extracts were subjected to coimmunoprecipitation. Newly synthesized, nonassembled Cox2 was efficiently bound to Yme1 and to SNNS harboring Yme1i CH, however, was not precipitated with SNNN (Fig. 2F). Thus, although not essential for proteolysis, the presence of Yme1 CH significantly increased Cox2 binding to Yme1.
Substrate specificity conferred by Yme1 CH.
These findings indicate substrate binding to CH and point to different substrate specificities of the orthologous i-AAA proteases Yme1 and IAP-1. This is remarkable considering the high sequence identities of CH in the two proteins (sequence identity, 48%). To demonstrate a direct interaction with CH of Yme1 and IAP-1, the corresponding amino acids of Yme1 and IAP-1 were fused to GST, expressed in E. coli, and purified to homogeneity. Subsequently, binding of Phb1 and Cox2 was analyzed in vitro.
35S-labeled Phb1 was imported into Δyme1 mitochondria which were subsequently solubilized. Extracts were incubated with GST-YME1-CH, GST-IAP-1-CH, or GST, and bound material was examined for the presence of radiolabeled Phb1 (Fig. 2G). A small but significant fraction of Phb1 associated with GST-Yme1-CH with low efficiency (Fig. 2G). This reflects specific binding, as Phb1 was not detected in the eluate from GST-IAP-1-CH or GST beads (Fig. 2G).
Cox2 was synthesized in Δyme1 mitochondria in further experiments, and binding to GST-CH was assessed after solubilization of mitochondria as above. Cox2 was clearly enriched in the eluate from GST-YME1-CH beads but was not associated with GST-IAP-1-CH or GST (Fig. 2H). These findings substantiate our coimmunoprecipitation experiments and demonstrate a direct and specific interaction of substrates with CH.
CH-dependent proteolysis of Cox2.
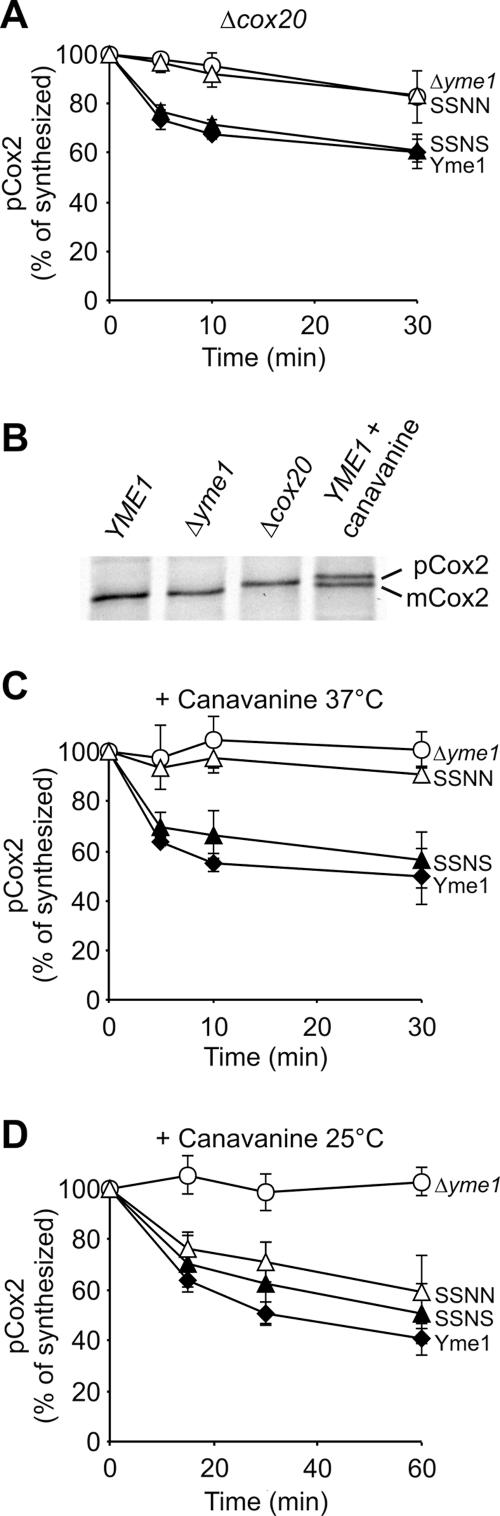
While these experiments revealed a striking substrate specificity of CH regions of Yme1 and IAP-1, they did not explain why degradation of Phb1 but not of Cox2 depended on Yme1 CH. We reasoned that substrate-specific factors might interact with Cox2, alleviating the need for an association with CH. According to this scenario, alternative pathways for substrate engagement by the i-AAA protease would exist. One candidate protein is Cox20, a membrane-bound chaperone necessary for maturation and assembly of Cox2 (15). We therefore examined the stability of Cox2 in Δyme1 Δcox20 mitochondria containing Yme1, SSNN, or SSNS. Deletion of COX20 did not impair proteolysis of Cox2 by the i-AAA protease (Fig. 3A). In contrast to wild-type mitochondria, however, Cox2 degradation depended on the presence of Yme1 CH in Δcox20 mitochondria. While Cox2 accumulated stably in the presence of SSNN, it was degraded in the presence of SSNS (Fig. 3A). Thus, Yme1 CH is required for proteolysis of Cox2 if Cox20 is absent, as it is for Phb1 degradation in wild-type mitochondria. These findings identify Cox20 as a substrate-specific factor which determines the CH dependence of Cox2 proteolysis.
FIG. 3.
CH-dependent Cox2 degradation in the absence of the substrate-specific chaperone Cox20 and upon unfolding. The stability of newly synthesized Cox2 was assessed as in Fig. 2. Similar results were obtained for the precursor (pCox2) and the mature form of Cox2. (A) CH-dependent degradation of Cox2 in Δcox20 mitochondria at 37°C. Cox2 was synthesized in Δyme1 Δcox20 mitochondria (○) harboring Yme1 (⧫), SSNN (▵), or SSNS (▴). (B) Synthesis of Cox2 in Δyme1 and Δcox20 mitochondria and in Δyme1 mitochondria complemented with Yme1 (YME1). When indicated, canavanine (12 μg/ml) was present during the translation. Four lanes of one gel are shown. The incorporation of canavanine affects electrophoretic migration of Cox2. (C) CH-dependent proteolysis of canavanine-containing Cox2 at 37°C. Mitochondrial translation products were synthesized in the presence of canavanine (12 μg/ml) in Δyme1 mitochondria (○) containing Yme1 (⧫), SSNN (▵), or SSNS (▴). (D) CH-independent proteolysis of canavanine-containing Cox2 at 25°C. After synthesis of Cox2 as in panel C mitochondria were incubated at 25°C.
How does Cox20 affect the proteolytic breakdown of Cox2? Cox20 may act as a chaperone protein and stabilize Cox2 in a conformation susceptible to proteolysis. To examine the role of Cox2 folding for CH-dependent proteolysis by Yme1, we synthesized mitochondrially encoded translation products in the presence of the arginine analogue canavanine, whose incorporation in newly synthesized Cox2 is expected to induce misfolding. Maturation of Cox2 was partially impaired in the presence of canavanine, indicating that the amino acid analogue was indeed incorporated into newly synthesized Cox2 (Fig. 3B). Both precursor form and mature Cox2 were degraded with similar kinetics in a Yme1-dependent manner (Fig. 3C; data not shown). Whereas proteolysis was impaired in Δyme1 mitochondria harboring SSNN, it was restored upon expression of SSNS containing Yme1 CH (Fig. 3C). Thus, synthesis of Cox2 in the presence of canavanine renders Cox2 proteolysis CH dependent, suggesting that this region is of particular importance for the recognition of misfolded substrate proteins.
In these experiments, proteolysis of Cox2 was analyzed at 37°C, conditions which may make folding of the intermembrane space domain of Cox2 difficult. We reasoned that decreasing the temperature to 25°C may stabilize this domain, allowing CH-independent degradation of Cox2 containing canavanine. Therefore, Cox2 was synthesized in the presence of canavanine in mitochondria harboring Yme1 or variants thereof and its stability was examined at 25°C (Fig. 3D). Strikingly, precursor and mature forms of Cox2 were degraded by SSNN, i.e., independently of Yme1 CH (Fig. 3D). Thus, whereas degradation of Cox2 harboring canavanine depends on CH, this requirement is alleviated by lowering the temperature, resulting most likely in a stabilization of the Cox2 fold. These results strongly suggest that the folding state of a substrate protein is one factor which determines the CH requirement of proteolysis.
CH-independent degradation of membrane-embedded Phb1.
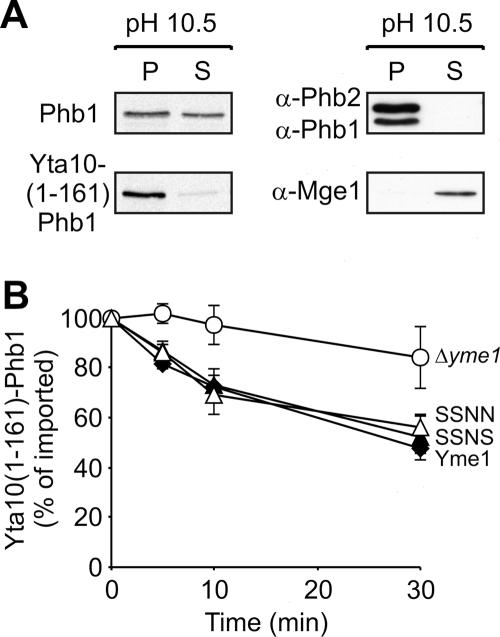
In contrast to Cox2, degradation of newly imported Phb1 was found to depend on Yme1 CH. We noted that Phb1 does not efficiently insert into the inner membrane upon import into mitochondria (Fig. 4A). Whereas endogenous Phb1 and Phb2 were exclusively recovered from the membrane pellet upon alkaline extraction of mitochondrial membranes, ∼50% of newly imported Phb1 was released to the supernatant fraction under these conditions, thus behaving like soluble mitochondrial proteins (Fig. 4A). We reasoned that impaired membrane insertion of Phb1 upon import into mitochondria not only prevents its assembly with Phb2 but also renders its degradation CH dependent. To examine this possibility, we attached Phb1 to the transmembrane domain of Yta10 [Yta10(1-161)-Phb1] to ensure membrane anchoring. The hybrid protein was indeed inserted into the inner membrane after import into mitochondria, as demonstrated by alkaline extraction of mitochondrial membranes, and exposed the Phb1 domain to the intermembrane space (Fig. 4A; data not shown). Both Phb1 and Yta10(1-161)-Phb1 were degraded by Yme1 (Fig. 4B). In contrast to Phb1 (Fig. 2C), however, proteolysis of Yta10(1-161)-Phb1 occurred in a CH-independent manner and was mediated both by SSNS and by SSNN (Fig. 4B). This is not due to recognition of the membrane-embedded moiety derived from Yta10, as newly imported Yta10(1-161) was not recognized by Yme1 and accumulated stably in mitochondria (data not shown). Thus, membrane anchoring of newly imported Phb1 alleviates the CH dependence of its degradation, identifying membrane insertion as yet another property of a substrate protein which affects the requirement of Yme1 CH for proteolysis.
FIG. 4.
CH-independent degradation of membrane-embedded Phb1. (A) Impaired membrane insertion of newly imported Phb1. 35S-labeled Phb1 or Yta10(1-161)-Phb1 was imported into isolated mitochondria, and membrane association of the imported proteins was examined by alkaline extraction (pH 10.5). Membrane and soluble fractions were split by centrifugation and analyzed by SDS-PAGE and autoradiography (left panel) or by immunoblotting (right panel) with antibodies directed against endogenous Phb1, Phb2, and matrix-localized Mge1. (B) CH-independent proteolysis of membrane-inserted Phb1. 35S-labeled Yta10(1-161)-Phb1 was imported into Δyme1 mitochondria (○) containing Yme1 (⧫), SSNN (▵), or SSNS (▴). The stability of Yta10(1-161)-Phb1 at 37°C was analyzed by SDS-PAGE and phosphorimaging.
Crucial role of N-terminal helices of the AAA domain for Yme1 activity.
These experiments assign an important function for substrate engagement to Yme1 CH. However, additional substrate recognition sites appear to exist, as Cox2 or membrane-embedded Phb1 is degraded in a CH-independent manner. We have previously demonstrated that helices at the N-terminal end of the AAA domain of Yme1 (NH; amino acids 268 to 313) are sufficient to stabilize a misfolded model substrate in vitro (24). Notably, this region is enriched in negatively charged amino acid residues (Fig. 5A). We systematically replaced glutamate and aspartate residues in Yme1 NH with alanine by site-directed mutagenesis. The mutant proteins were expressed in Δyme1 cells, and their respiratory growth was assessed (Fig. 5B). Alteration of E292 to alanine partially impaired respiratory growth at 37°C, which was further inhibited if additionally E294 (A2) or up to five negatively charged amino acid residues were mutated (A6) (Fig. 5B). The presence of negatively charged amino acids in NH appears to be essential for Yme1 activity, as respiratory competence was not maintained upon introduction of lysine residues (K6) or glutamine and asparagine residues (N/Q6) at these positions (Fig. 5B).
FIG. 5.
Essential role of Yme1 NH for proteolysis. (A) Sequence alignment of a negatively charged amino acid region within NH of various AAA protease subunits. Corresponding sequences of S. cerevisiae Yme1 (amino acids 280 to 302), N. crassa IAP-1 (amino acids 254 to 276), Homo sapiens Yme1L (amino acids 281 to 303), T. maritima FtsH (amino acids 160 to 182), E. coli FtsH (amino acids 151 to 173), S. cerevisiae Yta10 (amino acids 287 to 309), and H. sapiens paraplegin (amino acids 308 to 330) were aligned using the ClustalW program. Identical amino acid residues are boxed in black; homologous residues are boxed in gray. Positions with predominantly charged amino acid residues are indicated. Amino acids mutated in S. cerevisiae Yme1 mutants A2 and A6 are marked with asterisks. The domain structure of Yme1 is as described for Fig. 1A. (B) Mutational analysis of Yme1 NH. Amino acid residues E292 (A), E292-E294 (A2), and D287-E288-E292-E294-E295-D298 within the NH region of Yme1 were replaced by alanine (A6), lysine (K6), or asparagine and glutamine (N/Q6) residues. Yme1 and the mutant variants were expressed in Δyme1 cells, and the respiratory competence of the cells at 37°C was assessed as for Fig. 1B. (C and D) Stability of Cox2 and Phb1 in the presence of Yme1 harboring mutant NH. (C) The stability of newly synthesized Cox2 at 37°C was analyzed in Δyme1 mitochondria (○) and in Δyme1 mitochondria harboring Yme1 (⧫), Yme1A2 (A2, □), or Yme1A6 (A6, ▪) as for Fig. 2B. (D) 35S-labeled Phb1 was imported into Δyme1 mitochondria (○) and in Δyme1 mitochondria harboring Yme1 (⧫), Yme1A2 (A2, □), or Yme1A6 (A6, ▪), and its stability at 37°C was analyzed as for Fig. 2D.
Consistently, neither the presence of A2 nor that of A6 allowed degradation of Cox2 to occur in Δyme1 mitochondria (Fig. 5C). Similarly, the variants did not promote proteolysis of Phb1 (Fig. 5D). None of the NH mutations affected oligomerization of Yme1 subunits, excluding indirect effects of an impaired protease assembly (data not shown). These results therefore point to a crucial role for NH in Yme1-mediated proteolysis in vivo.
Substrate binding to NH and CH of Yme1.
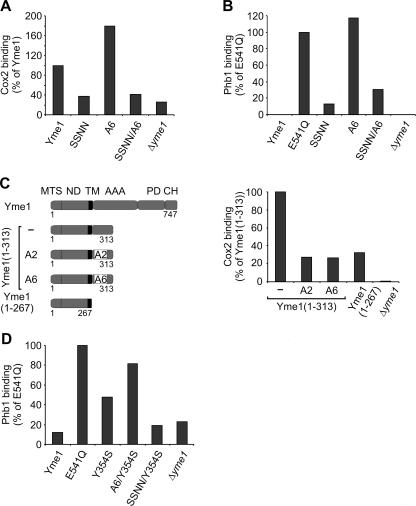
Binding of substrate proteins to NH variants of Yme1 was analyzed by coimmunoprecipitation. Mutations in NH (A6) did not impair Cox2 binding and stabilized Phb1 in association with the i-AAA protease as observed for proteolytically inactive Yme1E541Q (Fig. 6A and B). Notably, Cox2 was found in association with mutant Yme1 regardless of the presence of Cox20, which renders its proteolysis CH dependent (data not shown).
FIG. 6.
Substrate binding to NH and CH of Yme1. (A and B) Substrate binding to Yme1 variants with mutant NH and CH. The association of 35S-labeled Cox2 (A) and Phb1 (B) with Yme1 and derivatives thereof was analyzed by coimmunoprecipitation as in Fig. 2. Mitochondria contained Yme1 variants with mutations in NH (A6), CH (SSNN), or both (SSNN/A6). For control, Phb1 binding to Yme1E541Q was examined in parallel. (C) Impaired substrate binding to mutant Yme1 NH of Yme1(1-313). The domain structure of Yme1 and C-terminally truncated Yme1(1-313) and Yme1(1-267) is shown. Mutations present in A2 and A6 were introduced in NH of Yme1(1-313). MTS, mitochondrial targeting sequence; ND, N-terminal domain; TM, transmembrane segment; AAA, AAA domain; PD, proteolytic domain; CH, C-terminal helices. Binding of Cox2 was examined by coimmunoprecipitation as in Fig. 2. For control, Δyme1 and Δyme1 mitochondria containing Yme1(1-267) were analyzed in parallel. (D) Accumulation of Phb1 at CH upon mutating Y354 within the central pore loop. Y354S was introduced into A6 and SSNN, and binding of Phb1 was examined by coimmunoprecipitation.
These findings may indicate that CH or other recognition sites in Yme1 subunits mask the effect of NH mutations on substrate binding. We therefore analyzed the role of NH for the interaction with substrate proteins using truncated variants of Yme1 (Fig. 6C). Whereas Yme1(1-313) was found to interact with a misfolded model substrate with efficiencies similar to those of Yme1E541Q, an association with the substrate was no longer observed for Yme1(1-267) (24). Consistently, Cox2 binding to Yme1(1-267) was strongly reduced while it interacted with similar efficiencies with Yme1(1-313) and Yme1 (Fig. 6C; data not shown). Replacement of E292 and E294 with alanine or the removal of six negatively charged amino acids within NH abolished Cox2 binding to Yme1(1-313) (Fig. 6C). Similar results were obtained for Phb1 (data not shown). These findings demonstrate a crucial role of negatively charged amino acids for substrate binding and strongly suggest a direct interaction of substrate proteins with Yme1 NH.
As substrates associate with Yme1 after mutating NH in the context of full-length Yme1, we reasoned that they interact with CH under these conditions. To examine this possibility, we replaced the C-terminal domain of A6 with the corresponding domains of IAP-1, generating SSNN/A6. This hybrid protein contains IAP-1 CH, which binds neither Cox2 nor Phb1 (Fig. 2), and harbors a binding-incompetent NH region. In contrast to A6, Cox2 was not precipitated with SSNN/A6 (Fig. 6A). Cox2 binding was similarly impaired in Δcox20 mitochondria in these experiments, i.e., under conditions of CH-dependent proteolysis of Cox2 (data not shown). Like Cox2, Phb1 was found in association with A6 but not with SSNN/A6 lacking Yme1 CH (Fig. 6B). These results demonstrate that substrate polypeptides are bound to CH if NH is mutated and, therefore, confirm the crucial role of CH for substrate binding.
Of note, mutations in CH but not NH abolished Phb1 binding to the i-AAA protease. Cox2 binding to Yme1 was similarly affected in the absence of the assembly factor Cox20 (data not shown). These findings therefore suggest that substrates degraded in a CH-dependent manner can initially interact with Yme1 CH before they are bound to NH.
After initial interaction with the i-AAA protease substrates are thought to enter the proteolytic chamber via the central pore formed by hexameric AAA ring complexes. Consistently, replacement of Y354 in the central pore loop with serine impaired proteolysis but not binding of a misfolded model substrate to the i-AAA protease (12). We reasoned that Phb1 remains associated with CH if transfer through the central pore is abolished. To examine this possibility, we introduced the mutation Y354S both into A6 lacking binding-competent NH and into SSNN lacking binding-competent CH. Phb1 was imported into isolated mitochondria containing the variant A6/Y354S or SSNN/Y354S, and binding to the AAA protease variants was assessed by coimmunoprecipitation (Fig. 6D). In agreement with previous findings (12), mutating Y354 in Yme1 did not inhibit Phb1 binding but rather resulted in the accumulation of Phb1 at Yme1 (Fig. 6D). Notably, Phb1 was also found in association with A6/Y354S but not with SSNN/Y354S, demonstrating that Phb1 interacts with CH under these conditions (Fig. 6D). These findings strongly suggest that Phb1 interacts with binding sites at the surface of the proteolytic cylinder prior to the insertion into the central pore.
DISCUSSION
AAA proteases conduct the surveillance of protein quality control in the inner membrane of mitochondria and degrade specifically misfolded polypeptides. However, molecular structures involved in substrate engagement remained poorly described. Here, we have identified two substrate binding sites in the i-AAA protease subunit Yme1 and provide evidence for substrate-specific pathways for the entry of polypeptides into the proteolytic chamber of AAA proteases.
Substrates can encounter the i-AAA protease by interaction with conserved helices at the N-terminal end of the AAA domain (NH) and at the C-terminal end of the proteolytic domain (CH) of Yme1 subunits. Substrate binding to these subdomains is demonstrated first by biochemical experiments in vitro, which revealed a substrate-specific association of misfolded proteins with these helices (Fig. 2) (24), and second by a mutational analysis of these regions in vivo. In agreement with strong intersubunit contacts between proteolytic domains in the crystal structure of Thermotoga maritima FtsH (3), deletion of the CH region of Yme1 abolished assembly of the i-AAA protease. However, assembled protease complexes were formed upon replacement of CH with the corresponding helices of N. crassa IAP-1 and thereby allowed us to analyze the role of CH for substrate binding. These experiments revealed differences in substrate recognition between CH of S. cerevisiae Yme1 and N. crassa IAP-1, despite a sequence identity of 48% in this region. Similarly, differences appear to exist between the AAA domains of the two orthologues, as only chimeras harboring the AAA domain of Yme1 restored Cox2 degradation and respiratory growth of Δyme1 cells.
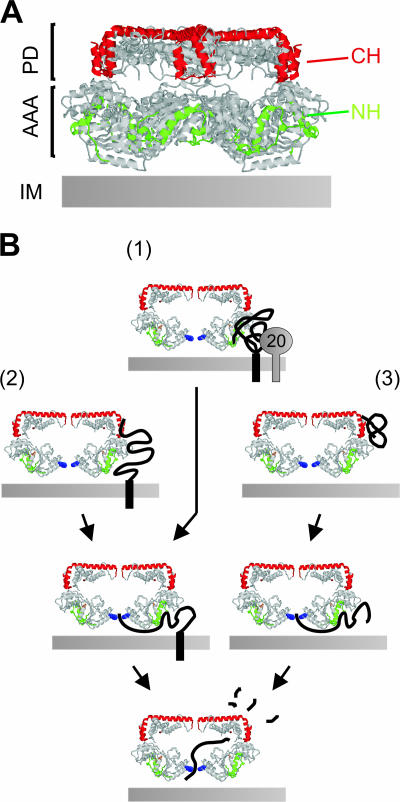
The recent elucidation of the crystal structure of the bacterial AAA protease FtsH (3, 37) allows NH and CH to be located within the hexameric ring structure of AAA proteases (Fig. 7A). NH and CH form helix-loop-helix structures at lateral surfaces of AAA and proteolytic domains, respectively, which are vertically aligned with each other (Fig. 7A). These structures are followed by extended helices at the apical surfaces of the protease cylinder which protrude from the periphery of the complex towards the central pore. NH and CH thus form a cage-like, surface-exposed lattice, ideally suited for an initial encounter with solvent-exposed domains of substrate proteins. Our results therefore suggest that substrates are recognized at the outer surface of the proteolytic cylinder of AAA proteases before they enter the proteolytic chamber of AAA proteases.
FIG. 7.
Substrate engagement by the i-AAA protease. (A) CH and NH regions form a lattice-like surface on AAA proteases. NH (amino acids 148 to 193, green) and CH (amino acids 544 to 603, red) are marked in the crystal structure of T. maritima FtsH (3). AAA, AAA domains; PD, proteolytic domains; IM, inner membrane. (B) Alternative substrate entry pathways to the proteolytic chamber of the i-AAA protease. (1) Cox20 (“20”) allows association of Cox2 with NH; (2) misfolded and (3) peripheral membrane proteins associate with CH before pore insertion. Only one pair of AAA protease subunits is shown. Y354 within the central pore is indicated in blue.
We observed a differential requirement of NH and CH for proteolysis by Yme1, indicating that substrates can enter the proteolytic chamber along different pathways (Fig. 7B). Whereas NH mutations impaired proteolysis of nonassembled Phb1 and Cox2, only Phb1 degradation depended on the presence of Yme1 CH. Several characteristics of substrate proteins appear to determine the requirement for CH. Anchoring of newly imported Phb1 to the inner membrane alleviated the CH dependence of proteolysis, identifying membrane insertion of substrates, i.e., their distance from NH at the membrane surface, as one crucial feature. Enhanced binding to CH, which is located more distantly from the membrane surface (Fig. 7A), seems to be especially important for peripheral membrane proteins. However, membrane insertion by itself does not override the CH requirement in all cases. Although membrane anchored, degradation of misfolded Cox2 containing canavanine still depended on CH. These findings substantiate our analysis of the turnover of nonassembled Phb1, which is made difficult by the fact that an unknown peptidase can degrade Phb1 in the absence of Yme1. Similarly to misfolded Cox2, degradation of nonassembled Phb1 requires CH, indicating that the observed CH dependence does not reflect an altered functional interaction of Yme1 with the unknown peptidase. We therefore conclude from our studies on both the turnover of Cox2 and that of Phb1 that CH plays a crucial role for the recognition of peripheral and misfolded membrane proteins by the i-AAA protease (Fig. 7B).
The CH requirement of proteolysis is also determined by the presence of a substrate-specific factor within mitochondria (Fig. 7B). In contrast to Phb1, degradation of Cox2 occurred in a CH-independent manner due to the action of Cox20. Understanding the role of Cox20 should therefore provide insight into the substrate-specific function of CH in Yme1-mediated proteolysis. Cox20 is required for the maturation of newly synthesized Cox2 proteins by the IMP processing peptidase and interacts directly with both the precursor and mature form of Cox2 (15). It is therefore conceivable that Cox20 triggers the folding of newly synthesized Cox2, allowing its subsequent assembly into cytochrome c oxidase complexes. Consequently, the absence of Cox20 would impair folding, leading to CH-dependent degradation by Yme1. Alternatively, Cox20 may not affect Cox2 folding but exert a holdase function which overlaps with the activity of Yme1 CH during proteolysis. According to this scenario, CH is required for the proteolysis of substrates which are only weakly bound by NH. Regardless, Cox20 is not generally essential for proteolysis of Cox2, due to its functional overlap with CH. It therefore appears to be distinct from known cofactors of bacterial Clp proteases, which determine their substrate specificity by promoting substrate binding (25, 27, 44), as well as from Mgr1, which has recently been isolated as a cofactor of Yme1-mediated proteolysis (9).
After initial interaction with surface-exposed helices, substrates are transported into the proteolytic chamber through the central pore of hexameric AAA proteases. Mutations in the central pore loop of Yme1 have substrate-specific effects (12), which can be rationalized by the differential requirement of CH for substrate binding. Mutating tyrosine 354 in the pore loop of Yme1 does not affect the binding of misfolded proteins like Phb1, as they still interact with CH. Cox2, on the other hand, was found not to interact with Yme1 harboring a mutant pore loop (12). In light of our present findings, Cox2 likely remains associated with Cox20 and does not associate with Yme1, presumably as ATP- and pore loop-dependent substrate translocation into the proteolytic chamber of Yme1 is impaired.
AAA+ proteases in general exert chaperone-like properties and ensure protein quality control. Therefore, the question arises whether the proteases achieve substrate binding merely by hydrophobic contacts, similarly to many molecular chaperone proteins, or by sequence-specific recognition. Our results define initial steps of substrate engagement by the i-AAA protease within mitochondria and identify two helical and surface-exposed substrate binding domains within Yme1. Helical regions have been implicated in substrate recognition by class I chaperonins or by the bacterial AAA+ proteins HslU and ClpB (7, 21, 36), pointing to a conserved mode of interaction. Whether substrates are stabilized by hydrophobic contacts to helical segments in HslU or ClpB, however, remains open, as evidence for a direct interaction of substrates with these regions is still missing. Notably, the NH region of AAA protease subunits is generally characterized by the presence of a large number of negatively charged amino acid residues, which is difficult to reconcile with the sole recognition of hydrophobic surfaces. This is reminiscent of various molecular chaperones which recognize both hydrophobic and positively charged residues (32). Therefore, sequence-specific elements in substrate polypeptides may contribute to their recognition by AAA proteases. This is also suggested by the apparently divergent substrate specificity of closely related CH regions in orthologous i-AAA proteases. The identification of two substrate binding sites in Yme1 allows us now to further characterize molecular mechanisms ensuring the specificity of proteolysis by these conserved proteolytic machines.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB635), the European Union (6th Framework Programme), and the German Israeli Project Cooperation (DIP grant F.5.1) to T.L.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Arlt, H., G. Steglich, R. Perryman, B. Guiard, W. Neupert, and T. Langer. 1998. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 17:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlt, H., R. Tauer, H. Feldmann, W. Neupert, and T. Langer. 1996. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85:875-885. [DOI] [PubMed] [Google Scholar]
- 3.Bieniossek, C., T. Schalch, M. Bumann, M. Meister, R. Meier, and U. Baumann. 2006. The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, R. E., T. A. Baker, and R. T. Sauer. 2005. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 12:245-251. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, C. L., N. Tanaka, K. H. White, and P. E. Thorsness. 1994. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol. Biol. Cell 5:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casari, G., M. De-Fusco, S. Ciarmatori, M. Zeviani, M. Mora, P. Fernandez, G. DeMichele, A. Filla, S. Cocozza, R. Marconi, A. Durr, B. Fontaine, and A. Ballabio. 1998. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93:973-983. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., and P. B. Sigler. 1999. The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell 99:757-768. [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover, A. 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6:79-87. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, C. D., M. S. Lee, F. A. Spencer, and R. E. Jensen. 2006. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell 17:213-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreirinha, F., A. Quattrini, M. Priozzi, V. Valsecchi, G. Dina, V. Broccoli, A. Auricchio, F. Piemonte, G. Tozzi, L. Gaeta, G. Casari, A. Ballabio, and E. I. Rugarli. 2004. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Investig. 113:231-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 12.Graef, M., and T. Langer. 2006. Substrate specific consequences of central pore mutations in the i-AAA protease Yme1 on substrate engagement. J. Struct. Biol. 156:101-108. [DOI] [PubMed] [Google Scholar]
- 13.Guélin, E., M. Rep, and L. A. Grivell. 1996. Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in the degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 381:42-46. [DOI] [PubMed] [Google Scholar]
- 14.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 15.Hell, K., A. Tzagoloff, W. Neupert, and R. A. Stuart. 2000. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 275:4571-4578. [DOI] [PubMed] [Google Scholar]
- 16.Hinnerwisch, J., W. A. Fenton, K. J. Furtak, G. W. Farr, and A. L. Horwich. 2005. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121:1029-1041. [DOI] [PubMed] [Google Scholar]
- 17.Hinnerwisch, J., B. G. Reid, W. A. Fenton, and A. L. Horwich. 2005. Roles of the N-domains of the ClpA unfoldase in binding substrate proteins and in stable complex formation with the ClpP protease. J. Biol. Chem. 280:40838-40844. [DOI] [PubMed] [Google Scholar]
- 18.Ito, K., and Y. Akiyama. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59:211-231. [DOI] [PubMed] [Google Scholar]
- 19.Kambacheld, M., S. Augustin, T. Tatsuta, S. Müller, and T. Langer. 2005. Role of the novel metallopeptidase MOP112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J. Biol. Chem. 280:20132-20139. [DOI] [PubMed] [Google Scholar]
- 20.Klanner, C., H. Prokisch, and T. Langer. 2001. MAP-1 and IAP-1, two novel AAA proteases with catalytic sites on opposite membrane surfaces in the mitochondrial inner membrane of Neurospora crassa. Mol. Biol. Cell 12:2858-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S., M. E. Sowa, Y. H. Watanabe, P. B. Sigler, W. Chiu, M. Yoshida, and F. T. Tsai. 2003. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115:229-240. [DOI] [PubMed] [Google Scholar]
- 22.Leonhard, K., B. Guiard, G. Pellechia, A. Tzagoloff, W. Neupert, and T. Langer. 2000. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 23.Leonhard, K., J. M. Herrmann, R. A. Stuart, G. Mannhaupt, W. Neupert, and T. Langer. 1996. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15:4218-4229. [PMC free article] [PubMed] [Google Scholar]
- 24.Leonhard, K., A. Stiegler, W. Neupert, and T. Langer. 1999. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 398:348-351. [DOI] [PubMed] [Google Scholar]
- 25.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354-2356. [DOI] [PubMed] [Google Scholar]
- 26.Maurizi, M. R., and D. Xia. 2004. Protein binding and disruption by Clp/Hsp100 chaperones. Structure 12:175-183. [DOI] [PubMed] [Google Scholar]
- 27.Mogk, A., D. Dougan, J. Weibezahn, C. Schlieker, K. Turgay, and B. Bukau. 2004. Broad yet high substrate specificity: the challenge of AAA+ proteins. J. Struct. Biol. 146:90-98. [DOI] [PubMed] [Google Scholar]
- 28.Nakai, T., T. Yasuhara, Y. Fujiki, and A. Ohashi. 1995. Multiple genes, including a member of the AAA family, are essential for the degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol. Cell. Biol. 15:4441-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolden, M., S. Ehses, M. Koppen, A. Bernacchia, E. I. Rugarli, and T. Langer. 2005. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123:277-289. [DOI] [PubMed] [Google Scholar]
- 30.Nolden, M., B. Kisters-Woike, T. Langer, and M. Graef. 2006. Quality control of proteins in the mitochondrion. Top. Curr. Genet. 16:119-147. [Google Scholar]
- 31.Pearce, D. A., and F. Sherman. 1995. Diminished degradation of yeast cytochrome c by interactions with its physiological partners. Proc. Natl. Acad. Sci. USA 92:3735-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rousseau, F., L. Serrano, and J. W. Schymkowitz. 2006. How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 355:1037-1047. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, R. T., D. N. Bolon, B. M. Burton, R. E. Burton, J. M. Flynn, R. A. Grant, G. L. Hersch, S. A. Joshi, J. A. Kenniston, I. Levchenko, S. B. Neher, E. S. Oakes, S. M. Siddiqui, D. A. Wah, and T. A. Baker. 2004. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlieker, C., J. Weibezahn, H. Patzelt, P. Tessarz, C. Strub, K. Zeth, A. Erbse, J. Schneider-Mergener, J. W. Chin, P. G. Schultz, B. Bukau, and A. Mogk. 2004. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11:607-615. [DOI] [PubMed] [Google Scholar]
- 35.Shotland, Y., D. Teff, S. Koby, O. Kobiler, and A. B. Oppenheim. 2000. Characterization of a conserved alpha-helical, coiled-coil motif at the C-terminal domain of the ATP-dependent FtsH (HflB) protease of Escherichia coli. J. Mol. Biol. 299:953-964. [DOI] [PubMed] [Google Scholar]
- 36.Song, H. K., C. Hartmann, R. Ramachandran, M. Bochtler, R. Behrendt, L. Moroder, and R. Huber. 2000. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA 97:14103-14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suno, R., H. Niwa, D. Tsuchiya, X. Zhang, M. Yoshida, and K. Morikawa. 2006. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol. Cell 22:575-585. [DOI] [PubMed] [Google Scholar]
- 38.Tatsuta, T., and T. Langer. 2007. Studying proteolysis within mitochondria. Methods Mol. Biol. 372:343-360. [DOI] [PubMed]
- 39.Thorsness, P. E., K. H. White, and T. D. Fox. 1993. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzagoloff, A., J. Yue, J. Jang, and M. F. Paul. 1994. A new member of a family of ATPases is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269:26144-26151. [PubMed] [Google Scholar]
- 41.Weber, E. R., T. Hanekamp, and P. E. Thorsness. 1996. Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Mol. Biol. Cell 7:307-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, E. R., R. S. Rooks, K. S. Shafer, J. W. Chase, and P. E. Thorsness. 1995. Mutations in the mitochondrial ATP synthase gamma subunit suppress a slow-growth phenotype of yme1 yeast lacking mitochondrial DNA. Genetics 140:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada-Inagawa, T., T. Okuno, K. Karata, K. Yamanaka, and T. Ogura. 2003. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J. Biol. Chem. 278:50182-50187. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]