Abstract
Telomerase is a ribonucleoprotein reverse transcriptase (RT) that processively synthesizes telomeric repeats onto the ends of linear chromosomes to maintain genomic stability. It has been proposed that the N terminus of the telomerase protein subunit, telomerase RT (TERT), contains an anchor site that forms stable interactions with DNA to prevent enzyme-DNA dissociation during translocation and to promote realignment events that accompany each round of telomere synthesis. However, it is not known whether human TERT (hTERT) can directly interact with DNA in the absence of the telomerase RNA subunit. Here we use a novel primer binding assay to establish that hTERT forms stable and specific contacts with telomeric DNA in the absence of the human telomerase RNA component (hTR). We show that hTERT-mediated primer binding can be functionally uncoupled from telomerase-mediated primer extension. Our results demonstrate that the first 350 amino acids of hTERT have a critical role in regulating the strength and specificity of protein-DNA interactions, providing additional evidence that the TERT N terminus contains an anchor site. Furthermore, we establish that the RT domain of hTERT mediates important protein-DNA interactions. Collectively, these data suggest that hTERT contains distinct anchor regions that cooperate to help regulate telomerase-mediated DNA recognition and elongation.
Telomeres are protective DNA-protein structures that define and “cap” the termini of eukaryotic chromosomes. Various DNA binding proteins interact with telomeric DNA in vivo and are involved in regulating telomere length and architecture, protecting against unwarranted recombination events and/or preventing chromosome termini from being recognized as damaged DNA (reviewed in reference 47). Telomeres therefore have a critical role in maintaining genetic stability (6, 16). Telomerase is the ribonucleoprotein reverse transcriptase (RT) that synthesizes telomeric repeats onto the ends of linear chromosomes (19, 20). Human telomerase minimally consists of an RNA subunit (hTR) that directs telomere synthesis and a protein subunit (human telomerase RT [hTERT]) that catalyzes telomere synthesis (11, 50).
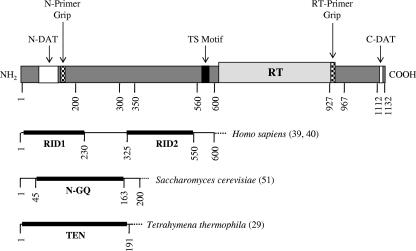
The telomerase protein subunit, TERT, shows strong sequence conservation across disparate organisms, ranging from ciliates to yeast to humans (35, 38, 44). Conceptually, the architecture of hTERT can be divided into three modular regions: a long, evolutionarily conserved N-terminal extension, a central catalytic RT domain containing seven evolutionarily conserved RT motifs, and a short C-terminal extension that shows little sequence conservation among organisms (Fig. 1) (reviewed in reference 32). One feature of the hTERT N terminus is that it contains several DAT regions that dissociate the biological and catalytic activities of telomerase (1, 7). Studies suggest that the DAT domains may be involved in recruiting telomerase to the telomere and/or regulating substrate recognition and utilization (1-3, 7, 34, 41). Furthermore, it has recently been reported that substitution mutations in the Saccharomyces cerevisiae Est2p N-DAT region (G75A, N95A, and N95D) can cause telomere overelongation in vivo without apparent defects in DNA binding or catalytic activity (31). Collectively, these studies with humans and yeast demonstrate that DAT domains are not functionally equivalent.
FIG. 1.
A schematic diagram depicting some of the known and putative functional domains of the hTERT subunit is shown at the top. The RT domain of hTERT is depicted with a light gray box, the black box depicts the conserved telomerase-specific (TS) motif, the white boxes represent previously identified N- and C-terminal DAT domains (1, 7), and the small, hatched boxes depict the N-terminal and RT primer grip regions. The schematics shown below depict functionally important regions in the TERT N terminus of other organisms, as identified in previous studies (reference numbers are indicated in parentheses to the right of the schematic).
While hTERT contains RT motifs that are conserved in other RT enzymes (35, 44), it also contains several unique regions that contribute to its distinguishing properties. Unlike other RTs, telomerase reiteratively copies an integral RNA template during catalysis (21). It is therefore of interest to investigate how variant and invariant residues influence the ability of telomerase to recognize and extend telomeric DNA substrates. Sequence alignment and mutational studies performed with Tetrahymena thermophila and S. cerevisiae have led to the following observations: (i) invariant residues between human immunodeficiency virus type 1 (HIV-1) RT and TERT are more critical for telomerase activity and processivity than are TERT-specific residues; (ii) similar to HIV-1 RT, TERT has a conserved “primer grip” region in RT motif E that is an important determinant of enzyme processivity; and (iii) the C-terminal extension of TERT is involved in DNA binding and/or enzyme activity, but its exact role is likely species specific (12, 13, 27, 37, 46). Since the primer grip sequence is conserved in hTERT (44) and single amino acid substitutions in this region decrease the processivity of S. cerevisiae telomerase (46), we were interested in investigating the functional significance of the human telomerase primer grip region.
Another unique feature of telomerase is its ability to processively synthesize telomeric repeats (repeat addition processivity or type II DNA synthesis) (17, 19, 42). There are a number of models that can explain this distinctive property (reviewed in reference 18). During RNA-dependent telomere elongation, telomerase hybridizes to the telomere through its cRNA template, and hTERT catalyzes the de novo addition of telomeric DNA via reverse transcription. Subsequently, the enzyme translocates to the newly synthesized chromosome end, where the RNA template is realigned for another round of telomere synthesis. An alternative model is one in which the newly synthesized telomeric DNA translocates away from the enzyme so that the 3′ end can be aligned with the RNA template. This model implies that some portion of the enzyme remains associated with the telomere during translocation and realignment. One plausible explanation is that an upstream region of telomeric DNA makes stable interactions with a telomerase “anchor site” domain that is independent of the catalytic domain. The theory is that these interactions would prevent complete enzyme/DNA dissociation during the translocation and realignment events that accompany each round of telomere synthesis. Similarly, by imparting stability to an otherwise energetically unfavorable enzyme-DNA complex, an anchor site-DNA interaction might help explain why telomerase can engage in RNA-independent telomere recognition and elongate nontelomeric DNA during chromosome healing (26, 43).
Collectively, studies performed with ciliates, yeast, and humans implicate at least two telomerase binding sites on telomeric DNA: one at the 3′ (template-proximal) end and another on a 5′ G-rich (template-distal) region (reviewed in reference 4). It is postulated that the 3′ end of the primer hybridizes to the telomerase RNA template to initiate telomere extension and that the 5′ region interacts with the telomerase anchor site for processive telomere elongation (reviewed in reference 4). The first direct evidence for a telomerase anchor site came from photo-cross-linking studies with Euplotes aediculatus, which show an interaction between telomeric DNA substrates and (i) a TERT-sized protein and (ii) the telomerase RNA subunit (23). In vitro studies using telomerase partially purified from transformed human 293T cells demonstrate that the enzyme-primer complex is primarily stabilized by contacts between hTERT and the template-proximal region (49). Photo-cross-linking studies with S. cerevisiae suggest that the main cross-linking site on Est2p (the yeast homologue of hTERT) is within the first 190 amino acids and that TLC1 (the yeast homologue of hTR) is required to cross-link Est2p to telomeric DNA primers (36). Most recently, Jacobs et al. (29) described the X-ray crystallography structure for an N-terminal fragment (encompassing residues 2 to 191) of T. thermophila TERT, which they termed the TERT essential N-terminal (TEN) domain. Although the most efficient cross-links between T. thermophila TERT and telomeric DNA are observed in the presence of the telomerase RNA subunit, a weak protein-DNA interaction is reported in the absence of the RNA subunit (29). Collectively, these studies with T. thermophila and S. cerevisiae telomerase indicate that the TERT N terminus contains an evolutionarily conserved telomeric DNA binding site, which may represent the telomerase anchor region. To date, it is still not clear whether hTERT can directly interact with telomeric DNA in the absence of the telomerase RNA subunit.
Much of the work that has been performed on the characterization of the human telomerase anchor site has been inferred from activity-based assays. It remains possible that in the absence of the human telomerase RNA subunit, the protein subunit physically interacts with DNA primers that are not substrates for telomerase-mediated extension, thereby uncoupling primer binding and utilization. To address this possibility, we used a direct primer binding assay to directly ascertain the protein and DNA requirements for a physical interaction between hTERT and telomeric primers in vitro. Importantly, we show that hTERT binds telomeric DNA in the absence of hTR. This strongly suggests that the anchor region(s) resides within the telomerase protein subunit. Furthermore, through extensive deletion and mutational analysis of hTERT, we identify regions within the N terminus and the RT domain that are critical for stable association with telomeric primers. We coupled the primer binding assay with the conventional telomerase activity assay (CTA) to measure total DNA synthesis and repeat addition processivity. Using this approach, we show that certain hTERT truncations bind, but do not elongate, telomeric primers. These results demonstrate that the interaction between hTERT and telomeric DNA can be functionally uncoupled from polymerization. We also find that the structurally conserved primer grip region within motif E of the RT domain of hTERT is required for telomerase activity and binding short telomeric primers. Furthermore, we identify a putative primer grip region in the hTERT N terminus. Similar to the RT domain primer grip, this region is a determinant of enzyme activity and repeat addition processivity. Finally, we provide supporting evidence that hTERT DAT regions have roles in substrate recognition and utilization. Our results suggest that defects in catalytic activity do not necessarily correlate with impaired primer binding.
MATERIALS AND METHODS
Construction of hTERT mutants.
The EcoRI-SalI DNA fragment containing FLAG-hTERT, FLAG-hTERTD712A, or FLAG-hTERTDAT (+68, +98, +122, +128, and +1127 mutations) was subcloned from the plasmid pBABE-puro-FLAG-hTERT, pBABE-hygro-FLAGhTERTD712A, or pBABE-puro-FLAG-hTERTDAT (+68, +98, +122, +128, and +1127 mutations) (generous gifts from C. M. Counter, Duke University Medical Center, Durham, NC), respectively, into the EcoRI-SalI site of plasmid pCI (Promega, Fisher Scientific Ltd., Nepean, Ontario, Canada) to create a full-length (FL) hTERT cDNA expressing an N-terminal EcoRI restriction site and FLAG epitope (DYKDDDDK) and a C-terminal SalI restriction site.
A PCR cloning approach was used to construct various hTERT mutants. These truncation mutants were selected based on previously characterized hTERT variants (9, 10). The hTERT variants used in this study were cloned from the plasmid pBABE-puro-FLAG-hTERT (a generous gift from C. M. Counter, Duke University Medical Center, Durham, NC) to express an N-terminal FLAG epitope using primers specific for the following sequences (indicated 5′ to 3′).
(i) hTERT 1-300 was made by overlap extension PCR with the primers 1-300 pr. 1, GCGTTTCTGATAGGCACCTATTG; 1-300 pr. 2, GAGCGCGCGGCGCGTCGTCCTTG; 1-300 pr. 3, CAAGGACGACGCGCCGCGCGCTC; and 1-300 pr. 4, GCTGTCGACTCAGCCCACGGATG.
First, two separate PCR products were generated from FL hTERT, one using the 1-300 pr. 1 and 2 primer set and another using the 1-300 pr. 3 and 4 primer set. Then these products were gel purified and reamplified using 1-300 pr. 1 and 4.
(ii) hTERT 1-350 was made by overlap extension PCR with the primers 1-350 pr. 1, GCGTTTCTGATAGGCACCTATTG; 1-350 pr. 2, GAGCGCGCGGCGCGTCGTCCTTG; 1-350 pr. 3, CAAGGACGACGCGCCGCGCGCTC; and 1-350 pr. 4, GCTGTCGACTCACAGAGAGCTGAG.
First, two separate PCR products were generated from FL hTERT, one using the 1-350 pr. 1 and 2 primer set and another using the 1-350 pr. 3 and 4 primer set. Then these products were gel purified and reamplified using 1-350 pr. 1 and 4.
(iii) hTERT 301-1132 was amplified from FL hTERT with the primers Δ300 forward pr., GGAATTCATGGACTACAAGGACGACGATGACAAGCGCCAGCACCACGCG, and hTERT reverse pr. 1, GCTGTCGACTCAGTCCAGGATGGTCTT.
(iv) hTERT 351-1132 was amplified from FL hTERT with the primers Δ350 forward pr., GGAATTCATGGACTACAAGGACGACGATGACAGGCCCAGCCTGACT, and hTERT reverse pr. 1 (described above).
(v) hTERT 928-1132 was amplified from FL hTERT with the primers Δ928 forward pr., GGAATTCATGGACTACAAGGACGACGATGACAAGTTCCCCTGGTGCGGC, and hTERT reverse pr. 1 (described above).
(vi) hTERT 967-1132 was amplified from FL hTERT with the primers Δ967 forward pr., GAATTCATGGACTACAAGGACGACGATGACAAGGGGAGGAACATGCGTCGC, and hTERT reverse pr. 2, GCTGTCGACTCAGTCCAGGATGG.
(vii) hTERT 505-967 was amplified from FL hTERT with the primers 505 forward pr., GGAATTCATGGACTACAAGGACGACGATGACAAGCTGCAGGAGCTGACGTG, and 967 reverse pr., GCTGTCGACTCACCCAGCCTTGAAGCCG.
(viii) hTERT 601-927 was amplified from FL hTERT with the primers 601 forward pr., GGAATTCATGGACTACAAGGACGACGATGACAAGCTGTCGGAAGCAGAGG, and 927 reverse primer, GCTGTCGACTCATAGGCCGTGGGCCGG.
(ix) FL N95A was made by overlap PCR extension with the primers FLAG forward pr., GGAATTCATGGACTACAAGGACGAC; N95A reverse pr., GGCCAGCACCCGCTTCGCGCCG; N95A forward pr., CGGCGCGAAGGCCGTGCTGGCC; and hTERT reverse pr. 2 (described above).
First, two separate PCR products were generated from FL hTERT, one using FLAG forward pr. and N95A reverse pr. and another using the N95A forward pr. and hTERT reverse pr. 2 primer set. Then these products were gel purified and reamplified using FLAG forward pr. and hTERT reverse pr. 2.
(x) FL N-GRIP was made by overlap PCR extension with the primers FLAG forward pr., GGAATTCATGGACTACAAGGACGAC; N-GRIP reverse pr., GGGCGGCGGCGGCGGCCGCCCCGCTCCCCCGCAGTG; N-GRIP forward pr., CGGCCGCCGCCGCCGCCCGCCGCGTGGGCGACGACGTG; and hTERT reverse pr. 2 (described above).
First, two separate PCR products were generated from FL hTERT, one using FLAG forward pr. and N-Grip reverse pr. and another using the N-Grip forward pri. and hTERT reverse pr. 2 primer set. Then these products were gel purified and reamplified using FLAG forward pr. and hTERT reverse pr. 2.
(xi) FL RT-GRIP was made by overlap PCR extension with the primers FLAG forward pr., GGAATTCATGGACTACAAGGACGAC; RT-GRIP reverse pr., CAGGGCGGCCGCCGCGGCGGGGAATAGGCCGTGGGCCGG; RT-GRIP forward pr., CCGCCGCGGCGGCCGCCCTGGATACCCGGACCCTGGAGG; and hTERT reverse pr. 2 (described above).
First, two separate PCR products were generated from FL hTERT, one using FLAG forward pr. and RT-GRIP reverse pr. and another using the RT-GRIP forward pr. and hTERT reverse pr. 2 primer set. Then these products were gel purified and reamplified using FLAG forward primer and hTERT reverse pr. 2.
(xii) FL N&RT-GRIP was created by EcoRI-SphI restriction digest of pCI-FL N-GRIP and pCI-FL RT-GRIP and standard cohesive-ended ligation of the 1,556- and 5,850-bp DNA fragments recovered from digested pCI-FL N-GRIP and pCI-FL RT-GRIP, respectively.
Subsequently, each hTERT mutant was cloned into the EcoRI-SalI site of plasmid pCI (Promega, Fisher Scientific Ltd., Nepean, Ontario, Canada) using standard molecular cloning techniques.
Construct identity was confirmed by EcoRI-SalI restriction enzyme digest, in vitro expression of [35S]cysteine-labeled hTERT, and DNA sequencing.
Cloning and synthesis of hTR.
The FL telomerase RNA (hTR), spanning nucleotides 1 to 451, was cloned into puc19 driven by a T7 promoter as previously described (10) and synthesized in vitro using the MEGAscript T7 transcription kit (Ambion, Inc., Austin, TX). Briefly, hTR cDNA was digested with EcoRI to obtain a linear template for the T7 transcription reaction. T7 transcription reaction mixtures (20 μl) containing 1 μg linearized template DNA, 1× reaction buffer, 7.5 mM each deoxynucleoside triphosphate, and 2 μl enzyme mix were incubated at 37°C for 2 h and then inactivated by treatment with 2 U DNase I and incubation at 37°C for 15 min. The transcription reaction products were extracted by phenol and chloroform-isoamyl alcohol (24:1), ethanol precipitated, and resuspended in nuclease-free water. hTR was purified from a 6% (wt/vol) polyacrylamide gel (19:1 [wt/wt] acrylamide-bisacrylamide) containing 11 M urea by elution in nuclease-free water (overnight at 4°C with rocking). Following elution, telomerase RNA was incubated at 60°C for 30 min (with vortexing every 10 min), centrifuged at 1,500 × g for 5 min, filtered through a Supor membrane 0.8-/0.2-μm filter (Gelman Sciences, Ann Arbor, MI), ethanol precipitated, and dissolved in nuclease-free water.
In vitro telomerase reconstitution.
Human telomerase was reconstituted with the rabbit reticulocyte lysate (RRL) in vitro transcription/translation reaction kit (Promega, Fisher Scientific Ltd., Nepean, Ontario, Canada), as per the manufacturer's instructions. Fifty-microliter total reaction volumes contained FL or truncated hTERT (0.01 μg/μl cDNA) synthesized in the presence of FL hTR (0.01 μg/μl). Reactions were incubated at 30°C for 2 h and subsequently used for the conventional telomerase extension assay (see below). Western blotting with a polyclonal anti-hTERT antibody was used to show that each RRL reaction contained equivalent amounts of hTERT protein (data not shown).
Oligonucleotide sequences and preparation.
All oligonucleotides were synthesized and gel purified by University Core DNA Services (University of Calgary). Unless otherwise noted, all oligonucleotides were designed with a 5′ biotin tag. The biotinylated oligonucleotide sequences (5′ to 3′) are listed in Table 1.
TABLE 1.
Description of the DNA primers tested for physical interaction with hTERT and telomerase-mediated elongation
| Primer | Length (nt) | Presence of 5′-biotin tag | Sequence (5′ to 3′) |
|---|---|---|---|
| bio-TELO24 | 24 | Yes | TTAGGG TTAGGG TTAGGG TTAGGG |
| bio-TELO18 | 18 | Yes | TTAGGG TTAGGG TTAGGG |
| bio-TELO12 | 12 | Yes | TTAGGG TTAGGG |
| bio-TELO6 | 6 | Yes | TTAGGG |
| bio-antiTELO18 | 18 | Yes | AATCCC AATCCC AATCCC |
| TELO24 | 24 | No | TTAGGG TTAGGG TTAGGG TTAGGG |
Oligonucleotide labeling.
Nonbiotinylated or 3′-biotinylated DNA primers (10 pmol) were 5′ end labeled with [γ-32P]ATP (3,000 Ci/mmol, 10 mCi/ml; GE Healthcare Bio-Sciences Corp., Piscataway, NJ) and T4 polynucleotide kinase (Invitrogen Biobar, University of Calgary, Calgary, Alberta, Canada) according to the manufacturers' instructions. Unincorporated nucleotides were removed using G-25 microspin columns (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Approximately 0.2 fmol of 5′ end-labeled primer was loaded for detection on a denaturing polyacrylamide gel.
Direct primer binding assay.
hTERT proteins were synthesized with the RRL in vitro transcription/translation reaction kit (Promega, Fisher Scientific Ltd., Nepean, Ontario, Canada) in the absence of the telomerase RNA. Thirty-microliter total reaction volumes contained 15 μl cell lysate, 2.4 μl reaction buffer and minus cysteine amino acid mixture, 1.2 μl RNase Out (Invitrogen Biobar, University of Calgary, Calgary, Alberta, Canada), 1.2 μl [35S]cysteine (>1,000 Ci/mmol, 10 mCi/ml; GE Healthcare Bio-Sciences Corp., Piscataway, NJ), 1.2 μl T7 polymerase, and 0.08 μg/μl hTERT cDNA. For control samples, luciferase cDNA was substituted for hTERT cDNA. Reaction mixtures were incubated at 30°C for 2 h.
A total of 100 μl of Ultralink immobilized neutravidin biotin beads (Pierce/MJS BioLynx, Inc., Brockville, Ontario, Canada) per sample was prewashed four times with 500 μl of 100-mM hypobuffer (90.7 mM HEPES [pH 8.0], 7 mM KCl, 2.3 mM MgCl2) supplemented with β-mercaptoethanol (0.05 mM) and complete EDTA-free proteinase inhibitor cocktail tablets (Roche Diagnostics Biobar, University of Calgary, Calgary, Alberta, Canada) and centrifuged at 3,300 × g for 5 min between washes. During the second wash, the beads were blocked with 5 μl RRL hTERT and incubated at 4°C for 30 min with rocking. After the final prewash, the beads were resuspended in 500 μl of 100-mM hypobuffer. The hTERT-primer complex was assembled by adding equivalent counts of [35S]cysteine-labeled, in vitro-synthesized hTERT to a mixture containing 6 μl primer (100 pmol/μl) and 10 μl (TTTTTT)3 oligonucleotide (100 pmol/μl). For samples without primer, the remaining volume was adjusted with water. Complex formation was allowed to proceed for 10 min, and then 60 μl was immobilized on neutravidin beads for 3 h at 4°C with rocking. The samples were centrifuged at 3,300 × g for 5 min and washed three times with 500 μl 100 mM hypobuffer. The second wash was incubated for 15 min at 4°C with rocking. After the final wash, the beads were resuspended in 4 μl sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye, boiled for 5 min, and centrifuged at 16,000 × g for 5 min. Reaction products were resolved by 8 or 10% wt/vol Tris-glycine SDS-PAGE, and reaction products were detected by autoradiography.
Quantification of primer binding results.
To assess the level of protein synthesis, 2 μl of each RRL reaction mixture was separated with 8 or 10% SDS-PAGE and detected using autoradiography. Bands corresponding to each hTERT mutant were quantified using Quantity One software. The volume of RRL added to each primer binding reaction was adjusted so that each experiment was performed with equivalent amounts of hTERT protein. The total protein counts in this volume were mathematically determined and considered to represent “input protein counts.” To determine the percentage of protein that was bound to a DNA oligonucleotide after the primer binding reaction (“final protein counts”), the appropriate hTERT bands were quantified using Quantity One software, adjusted for background binding by subtracting the nonbiotinylated TELO24 oligonucleotide signal, and expressed by the formula (“final protein counts”/“input protein counts”) × 100%.
Statistical analysis of primer binding results.
Primer binding results are reported as the means ± standard errors of the means (SEM) of at least three independent experiments. One-way analysis of variance with Tukey multiple comparisons posttest or unpaired t tests (Welch corrected) were performed using GraphPad InStat, version 3.00 (GraphPad Software, San Diego, CA).
CTA.
The CTA was performed as previously described (28), with some modifications. Briefly, 24.5 μl rabbit reticulocyte lysate containing reconstituted human telomerase (see above) was assayed for activity in a 40-μl reaction mixture containing 1× CTA buffer (50 mM Tris-HCl [pH 8.3], 50 mM KOAc, 1 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM spermidine), 1 mM dATP, 1 mM dTTP, 0.0125 mM dGTP, 1.5 μM telomeric primer, and 1.5 μM [α-32P]dGTP (3,000 Ci/mmol, 10mCi/ml; GE Healthcare Bio-Sciences Corp., Piscataway, NJ). The elongation reaction mixture was incubated at 30°C for 3.5 h and then terminated by adding 50 μl of stop buffer (10 mM EDTA, 2 M NaCl, 0.1 mg/ml RNase A) and incubating at 37°C for 15 min.
Seventy microliters of Streptavidin Magnasphere paramagnetic beads (Promega, Fisher Scientific Ltd., Nepean, Ontario, Canada) per sample was prewashed three times with 250 μl buffer A (10 mM Tris-HCl [pH 7.5], 1 M NaCl, and 0.5 mM EDTA) and then resuspended in buffer A. The elongation products were immobilized on resuspended magnetic beads at room temperature for 1 h, during which time the samples were mixed once every 10 to 15 min. Subsequently, the bead-elongation product complexes were washed three times with buffer A (10 mM Tris-HCl [pH 7.5], 1 M NaCl) and twice with buffer B (10 mM Tris-HCl [pH 7.5]), resuspended in 10 μl of CTA loading dye (95% deionized formamide, 10 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue), and boiled for 30 min. Telomerase elongation products were resolved by 8% polyacrylamide-urea gel electrophoresis at 1,500 V for 1.5 h and detected by autoradiography or exposure to a phosphorimaging screen.
Quantification of telomerase activity and repeat addition processivity.
Telomerase activity was measured using the conventional telomerase activity assay. The signal of each elongation product was quantified by phosphorimager analysis and Quantity One software.
Total DNA synthesis within the first telomeric repeat (Pi) was calculated using the formula Pi = (Ti + Ti+1 +… + Ti+n). T values correspond to the signal of the elongation product at position i. For the 18-nucleotide telomeric primer, the values obtained at positions Ti… Ti+6 in each independent experiment were added and expressed as a fraction of the total DNA synthesis calculated for the wild-type telomerase control. For the 6-nucleotide telomeric primer, only the most intense products within the first telomeric repeat were quantified and analyzed as described above. The mean total DNA synthesis fraction was determined from at least three independent experiments. Results are reported ± SEM (GraphPad InStat, version 3.00; GraphPad Software, San Diego, CA).
Repeat addition processivity (Pi) was calculated using methods similar to those described previously (28). The signal of the +6, +12, +18, +24, and +30 elongation products was quantified (where detectable) and applied to the formula Pi = (Ti+6)/(Ti + Ti+6), as described by Hardy et al. (24). First, we calculated the repeat addition processivity for each pair of elongation products separately. Next, the repeat addition processivity values at each position (i.e., +6, +12, +18, and +24) were multiplied by 100. Finally, the results of three independent experiments were averaged and the SEM was determined using GraphPad InStat, version 3.00 (GraphPad Software, San Diego, CA).
RESULTS
hTERT interacts with telomeric DNA oligonucleotides independently of hTR in vitro.
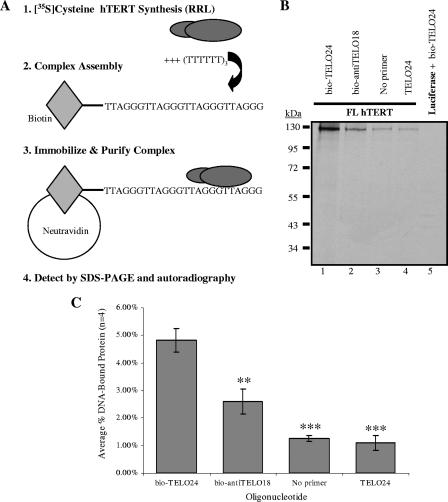
To examine whether hTERT specifically binds telomeric DNA oligonucleotides, we developed a binding assay that we have termed the direct primer binding assay. This assay utilizes biotinylated DNA oligonucleotides to test for protein-DNA interactions with methods similar to those that Hainaut et al. used to examine interactions between p53 and biotinylated DNA (22). More recently, this oligonucleotide capture assay has been used to characterize telomeric DNA binding proteins (48). Specifically, Snow et al. demonstrated that hEST1A could bind biotinylated telomeric DNA (48). Here we have established optimal reaction conditions to characterize the interactions that occur between the catalytic subunit of human telomerase, hTERT, and biotinylated telomeric DNA primers. Our direct primer binding assay uses in vitro human telomerase reconstituted in the RRL transcription and translation system. The advantage of the RRL reconstitution system is that hTERT can be synthesized in the absence of hTR, which allows us to specifically investigate hTERT-DNA interactions. We reasoned that this assay would provide us with information on anchor site interactions that occur between hTERT and telomeric (or nontelomeric) DNA, which might otherwise be masked by RNA-DNA interactions if hTR was present. We chose an in vitro approach to focus on hTERT-DNA interactions in the absence of cellular factors that may aid or inhibit this association in vivo. As illustrated in Fig. 2A, the direct primer binding assay is based on biotin-neutravidin affinity purification. We used the RRL system and 35[S]cysteine to synthesize radioactively labeled FL hTERT in the absence of hTR. FL hTERT was tested for its ability to interact with the DNA primers listed in Table 1. When combined with different 5′-biotin-labeled DNA oligonucleotides, FL hTERT showed a stable interaction with a primer containing 24 nucleotides of canonical human telomeric DNA (Fig. 2B, lane 1). This interaction was specific for telomeric DNA since we observed a statistically significant decrease (P < 0.01) in the interaction between FL hTERT and an anti-telomeric oligonucleotide containing the sequence (AATCCC)3 (Fig. 2B, lane 2, and C). We used an 18-nucleotide anti-telomeric primer instead of a 24-mer because human telomerase reconstituted with FL hTERT is more active on bio-TELO18 than on bio-TELO24 (discussed below; Fig. 3C). Furthermore, the interaction between FL hTERT and bio-TELO18 is slightly stronger than that between FL hTERT and bio-TELO24 (Fig. 3A and B; discussed below). No interaction was observed when bio-TELO24 was incubated with equivalent amounts of luciferase (Fig. 2B, lane 5), providing additional evidence that this assay can be used to specifically detect hTERT-telomeric DNA associations. Only minimal amounts of FL hTERT bound the neutravidin beads in the absence of any DNA oligonucleotide (Fig. 2B, lane 3) or in the presence of a nonbiotinylated 24-nucleotide telomeric primer (lane 4). To reduce nonspecific retention, we repeated these experiments using wash buffers containing 32 mM, 100 mM, or 200 mM salt (described in Materials and Methods) and observed optimal results after washing with 100 mM salt buffer (data not shown). To assess and correct for background binding, we measured the association of hTERT with nonbiotinylated TELO24 in each subsequent primer binding experiment (see Materials and Methods for details).
FIG. 2.
Developing and optimizing the direct primer binding assay. (A) Schematic illustration of the direct primer binding assay. (B) FL hTERT interacts specifically with a 5′-biotinylated oligonucleotide containing the telomeric DNA sequence (TTAGGG)4 (bio-TELO24; lane 1 and 1), not the corresponding nonbiotinylated oligonucleotide (TELO24; lane 4). Lane 2, FL hTERT discriminates against binding anti-telomeric DNA (AATCCC)3 (bio-antiTELO18). Lane 3, low levels of FL hTERT are nonspecifically retained on neutravidin beads in the absence of DNA; this retention is statistically similar to that observed after incubating FL hTERT with a nonbiotinylated 24-nucleotide telomeric primer, TELO24 (lane 4). As a control, luciferase does not interact with bio-TELO24 (compare lanes 1 and 5). (C) Quantification (described in Materials and Methods) and subsequent statistical analysis of the FL hTERT primer binding results shown in panel B. Error bars indicate SEM. Asterisks denote the level of statistical significance compared to FL hTERT bio-TELO24 binding: P values were <0.01 (∗∗) and <0.001 (∗∗∗).
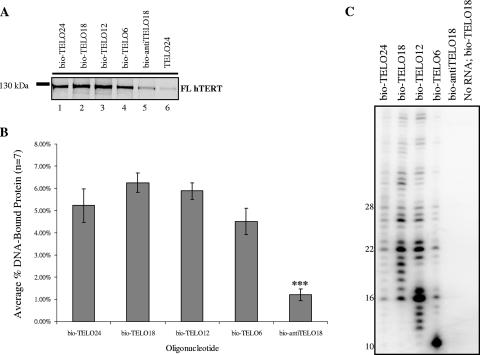
FIG. 3.
Binding and utilization of 5′-biotinylated DNA oligonucleotides containing different lengths of telomeric DNA. (A) FL hTERT physically interacts with 5′-biotinylated telomeric DNA in the absence of hTR. Each primer binding reaction contained equal amounts of RRL FL hTERT and 600 pmol of the indicated oligonucleotide (Table 1). Reaction products were immobilized on neutravidin beads, purified, resolved by 8% SDS-PAGE, and detected by autoradiography. (B) Quantification and subsequent statistical analysis of the FL hTERT primer binding results shown in panel A (see Materials and Methods for details). Error bars indicate SEM. Asterisks denote levels of statistical significance: P was <0.001 (∗∗∗). (C) Telomerase reconstituted in RRL with FL hTERT and FL hTR is able to recognize and elongate the 5′-biotinylated telomeric oligonucleotides tested in panel A (see Materials and Methods for details).
FL hTERT binds and elongates oligonucleotides containing different lengths of telomeric DNA.
We coupled the direct primer binding assay to the CTA to determine whether oligonucleotides that bound hTERT could be elongated by RRL-synthesized telomerase. This approach allowed us to directly compare the abilities of different DNA primers to physically interact with hTERT and be functionally elongated by human telomerase (see below).
It has previously been shown in vitro that human telomerase elongates DNA primers consisting of two or three TTAGGG repeats more efficiently than those consisting of four TTAGGG repeats (41, 42). Similar observations have been reported for Tetrahymena and Oxytricha telomerase (8, 14, 33). To investigate whether differences in telomere elongation were caused by differences in protein-DNA interactions, we compared the ability of FL hTERT to form stable complexes with biotinylated telomeric oligonucleotides containing one, two, three, or four consecutive TTAGGG repeats (Fig. 3A and B) with the ability of telomerase to elongate these DNA primers (Fig. 3C). We observed that FL hTERT bound each oligonucleotide with similar efficiencies (Fig. 3B) (P > 0.05); however, we cannot rule out the possibility that our assay lacks the sensitivity required to detect, with statistical confidence, subtle differences in FL hTERT binding to each DNA primer. Nonetheless, the fact that FL hTERT binds short telomeric primers significantly better than it does the longer anti-TELO18 primer (Fig. 3A and B, compare lanes 3 to 5) provides additional evidence for the specificity of the hTERT-DNA interactions measured using this binding assay. Furthermore, the ability of FL hTERT to bind various lengths of telomeric DNA with similar efficiencies shows that the 5′ biotin residue does not impede hTERT from contacting short stretches of telomeric DNA (i.e., bio-TELO6 primer).
The primer binding results shown in Fig. 3A and B suggest that differences in hTERT-telomeric DNA interactions (as per the direct primer binding assay) cannot account for the observed differences in telomerase-mediated extension of various lengths of telomeric oligonucleotides (Fig. 3C). We observe that human telomerase is more active on telomeric substrates containing two or three telomere repeats than on primers containing four or one telomere repeat. This is consistent with what has been reported previously (41, 42). Our primer binding assay, however, shows that in the absence of hTR, FL hTERT binds bio-TELO18 as efficiently as bio-TELO24 (Fig. 3C). These observations suggest that in addition to physical anchor site interactions between FL hTERT and telomeric DNA, there are also functional requirements for telomerase-mediated primer extension. Finally, our primer binding results demonstrate that FL hTERT efficiently interacts with a single telomeric repeat and that this interaction is not enhanced with longer DNA oligonucleotides that contain additional telomeric repeats. This likely suggests that only a small region of telomeric DNA is needed to form a stable hTERT-DNA complex.
Identification of hTERT regions required for stable protein-telomeric DNA interactions in vitro.
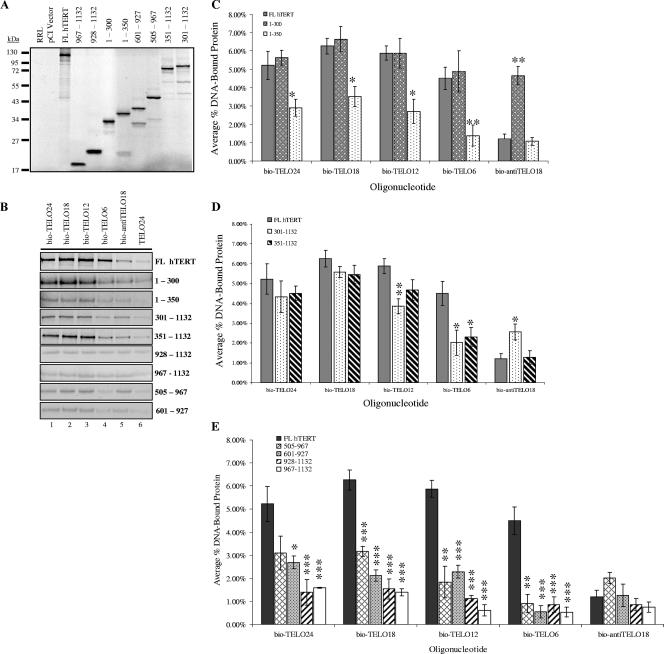
Studies with E. aediculatus, yeast, and T. thermophila show that specific regions of TERT contact telomeric DNA primers in an RNA-dependent manner (23, 29, 36). These residues map to the TERT N-terminal extension (the TEN domain) in T. thermophila (29) and the N-GQ domain in S. cerevisiae (36, 51), which align with RNA interacting domain 1 (RID-1) in hTERT (39, 40). In order to further delineate the anchor site of human TERT, we used a series of truncated hTERT constructs to identify regions of the protein that mediate stable and specific interactions with telomeric DNA primers. This panel included N-terminal fragments (spanning residues 1 to 300 and 1 to 350), mutants that lack different regions of the N terminus (spanning residues 301 to 1132 or 351 to 1132), C-terminal fragments (spanning residues 928 to 1132 and 967 to 1132), and RT variants (spanning residues 505 to 967 and 601 to 927). Many of these truncated proteins have previously been shown to bind hTR and the telomerase-associated protein TEP1. Furthermore, catalytic activity is restored by the complementary fragment in vitro and in vivo (9, 10, 39, 40). Preliminary chymotryptic proteolysis data indicate similar cleavage patterns for each fragment (unpublished data). Collectively, this indicates that each truncated protein exists in a native or nearly native conformation.
As previously reported, human telomerase reconstituted with any one of these deletion variants was inactive by the telomeric repeat amplification protocol assay and CTA, indicating that the enzyme was unable to elongate telomeric DNA in vitro (5, 10; data not shown). However, the primer binding assay showed that certain mutants physically associated with the 5′-biotinylated telomeric primers indicated in Table 1 at levels comparable to that of FL hTERT (Fig. 4B to E). Each primer binding experiment was performed with equivalent levels of input protein (assessed by SDS-PAGE and autoradiography) (Fig. 4A) to allow for accurate comparison of different hTERT variants. Additional product bands represent prematurely truncated hTERT proteins and/or hTERT proteins synthesized from noncoding start sites (Fig. 4A). These products did not interact with any of the primers listed in Table 1.
FIG. 4.
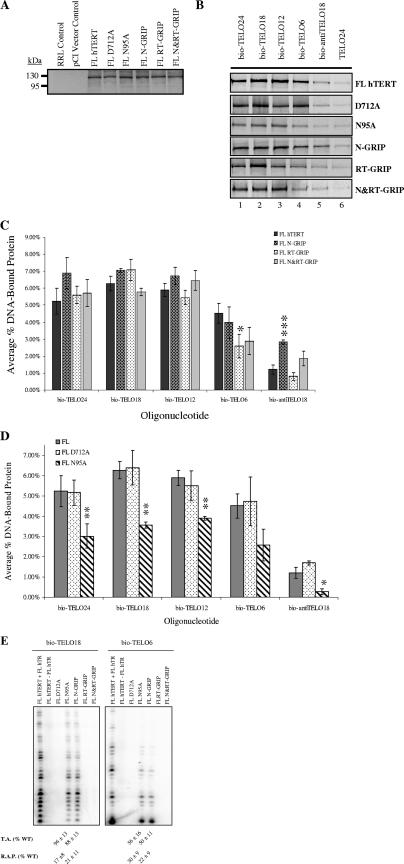
Identification of hTERT regions required to support a stable protein-DNA interaction in vitro. A series of hTERT deletion constructs were used to delineate regions of hTERT that mediate stable and specific interactions with telomeric DNA in the primer binding assay. (A) Autoradiography and 10% SDS-PAGE were used to assess the efficiency of protein production in the RRL system in the presence of [35S]cysteine, RRL without protein (RRL), or RRL with the pCI vector alone (pCI vector). Bands corresponding to the molecular mass of each hTERT mutant were quantified as described in Materials and Methods and normalized to FL hTERT. The volume of RRL added to subsequent primer binding reactions (B) was adjusted so that each reaction contained equivalent amounts of the radiolabeled hTERT deletion variant being tested. (B) The primer binding assay was used to investigate the ability of different hTERT variants to bind the oligonucleotides listed in Table 1. DNA-bound protein was immobilized on neutravidin beads and purified, and the reaction products were separated by 10% SDS-PAGE and visualized with autoradiography. (C to E) Quantification and subsequent statistical analysis of the FL hTERT primer binding results shown in panel B (see the text for a discussion). Error bars indicate SEM. Asterisks denote levels of statistical significance compared to FL hTERT binding to the corresponding 5′-biotinylated oligonucleotide: P values were <0.05 (∗), <0.01 (∗∗), and <0.001 (∗∗∗).
Interestingly, the interaction between an hTERT mutant spanning amino acids 1 to 300 and each telomeric primer was indistinguishable from that of FL hTERT, whereas the slightly larger fragment spanning amino acids 1 to 350 showed a significantly reduced telomeric DNA binding activity (Fig. 4C). Furthermore, the 1-300 mutant showed an abnormally strong interaction with anti-TELO18 relative to FL hTERT and 1-350 hTERT (Fig. 4C). This suggested that the region between residues 300 and 350 might inhibit an hTERT-DNA interaction and/or mediate protein specificity towards telomeric DNA. To expand this observation, we assessed the DNA binding properties of hTERT mutants that lacked either the first 300 amino acids (fragment 301-1132) or the first 350 amino acids (fragment 351-1132). Consistent with the above observations, an hTERT mutant spanning amino acids 301 to 1132 showed a reproducible reduction in binding short telomeric oligonucleotides (bio-TELO12 and bio-TELO6) relative to FL hTERT and the mutant spanning amino acids 351 to 1132 (Fig. 4B and D). We also observed that the 301-1132 variant showed a more stable interaction with bio-antiTELO18 than did FL hTERT and the 351-1132 fragment (Fig. 4D). This may indicate that the region spanning amino acids 300 to 350 has to function in cis with the extreme N terminus (i.e., the first 300 amino acids) to provide hTERT with specificity for telomeric DNA.
We next tested two protein fragments that lacked the N terminus and the RT domain to determine whether the hTERT C terminus was sufficient for a protein-DNA interaction in vitro. The primer binding results in Fig. 4B and E show that neither the 928-1132 hTERT nor the 967-1132 hTERT fragment formed stable interactions with telomeric DNA primers. This suggests that the hTERT C terminus does not contain a major DNA binding domain. Lastly, we considered that the RT domain might be able to support stable interactions with telomeric DNA. Two different hTERT RT mutants were tested: the 601-927 variant represents most of the RT domain, and the 505-967 mutant consists of the entire RT domain, the telomerase-specific motif, and a short stretch of residues C terminal to the RT domain. In general, both RT mutants bound the telomeric DNA primers more efficiently than did the C-terminal fragments (P < 0.05), indicating the presence of a DNA binding site in the RT domain. However, compared with FL hTERT, each RT mutant displayed significantly reduced interaction with bio-TELO18 and short telomeric primers (bio-TELO12 and bio-TELO6). Taken together, these results establish that there are regions in the RT domain that contact telomeric DNA (Fig. 4E), but N-terminal regions make a significant contribution to the overall stability of an hTERT-DNA interaction, especially with short DNA primers (Fig. 4C and D).
Distinct residues within hTERT mediate physical protein-DNA interactions and functional telomerase-DNA interactions in vitro.
We speculated that the region between residues 927 and 967 (RT motif E) was likely involved in mediating the length-dependent DNA binding demonstrated by an hTERT mutant spanning amino acids 505 to 967. This region contains a five residue “primer grip” sequence (930WCGLL934) that is highly conserved in other telomerase and RT enzymes (44). In HIV-1 RT, the primer grip sequence is required for optimal polymerization and processivity, template-primer DNA binding, and heteroduplex stability (30). Substitution of the corresponding region in S. cerevisiae Est2p results in reduced levels of total DNA synthesis and decreased enzyme processivity (46). We generated an alanine substitution mutation in the primer grip region of FL hTERT (FL RT-GRIP) and tested the ability of this mutant to recognize and utilize different lengths of telomeric DNA primers. Substitution of the RT primer grip region completely abolished telomerase-mediated elongation of all telomeric primers tested (Fig. 5E). However, this mutant showed defects only in binding a short telomeric primer (bio-TELO6) (Fig. 5B and C), suggesting that the RT primer grip region is critical for enzyme function and stabilizing the interaction between FL hTERT and short stretches of telomeric DNA. Interestingly, the N terminus of hTERT contains a putative primer grip sequence, 137WGLLL141. We hypothesized that this region might partially account for the strong DNA binding activity displayed by the hTERT mutant spanning amino acids 1 to 300. To test this hypothesis, we substituted 137WGLLL141 with 137AAAAA141 in the context of FL hTERT (mutant FL N-GRIP). In the absence of hTR, FL N-GRIP hTERT bound telomeric primers at levels similar to those of wild-type FL hTERT (Fig. 5B and C). However, telomerase reconstituted with the FL N-GRIP mutant demonstrated significantly reduced catalytic activity and repeat addition processivity on the bio-TELO18 and bio-TELO6 primers compared to wild-type telomerase (Fig. 5E). In the primer binding assay, FL N-GRIP hTERT displayed increased binding to bio-antiTELO18, indicating that the N-terminal primer grip region may mediate telomere sequence-specific substrate recognition. Lastly, we generated a FL hTERT construct with alanine substitution mutations in both the RT and the N-terminal primer grip regions (FL N&RT-GRIP). The double mutant was completely inactive and, interestingly, showed no significant alterations in its capacity to bind telomeric DNA primers (Fig. 5B, C, and E).
FIG. 5.
Distinct residues within hTERT mediate physical protein-DNA interactions and functional telomerase-DNA interactions in vitro. A series of FL hTERT substitution mutants were assessed for DNA binding defects with the direct primer binding assay (B) and telomerase activity and processivity with the conventional telomerase assay (E). (A) [35S]cysteine-labeled FL hTERT and hTERT mutants were generated with the RRL system, separated by 8% SDS-PAGE, and visualized using autoradiography. Control reactions include RRL without protein (RRL control) and RRL with the pCI vector alone (pCI vector control). Each hTERT signal was quantified (see Materials and Methods) and normalized to the intensity of FL hTERT, and the volume of RRL added to subsequent primer binding reactions was adjusted so that each reaction mixture contained equivalent amounts of protein. (B) FL hTERT and hTERT mutants were tested for a physical interaction with the indicated oligonucleotides (Table 1). DNA-bound hTERT was purified from neutravidin beads, resolved by 8% SDS-PAGE, and detected by autoradiography. (C and D) Quantification and statistical analysis of the FL hTERT primer binding results shown in panel B ± SEM (see Materials and Methods for details). Asterisks denote levels of statistical significance compared to FL hTERT binding to the corresponding DNA primer: P values were < 0.05 (*), <0.01 (**), and <0.001 (***). (E) Telomerase was assembled in RRL with mutant hTERT protein and wild-type (WT) RNA and then assayed for primer recognition and utilization with a conventional telomerase assay. Average relative telomerase activity (T.A.) and repeat addition processivity (R.A.P.) values calculated from at least three independent experiments are shown at the bottom. Error bars indicate SEM.
Consistent with its RT activity, hTERT requires a conserved triad of metal-coordinating aspartic acids (D868, D869, and D712) for telomerase catalysis (11, 25, 50). Mutational studies with hTERT indicate that these residues are directly involved in the polymerization reaction in vitro and in vivo. As expected, telomerase reconstituted with FL hTERT D712A showed no catalytic activity when tested by the CTA (Fig. 5E). However, in the absence of hTR, FL hTERT D712A bound telomeric and anti-telomeric DNA primers in a manner similar to that of wild-type FL hTERT (Fig. 5B and D). This observation provides evidence that the requirements for a physical interaction between hTERT and telomeric DNA oligonucleotides can be separated from those that mediate substrate elongation in vitro.
In S. cerevisiae Est2p, N95A substitution causes aberrant telomere lengthening without altering the enzyme's primer binding and catalytic properties in vitro (31). Since residue 95 is also an asparagine in hTERT, we were interested in looking at the DNA binding properties of the FL hTERT N95A mutant. Interestingly, we observed significantly reduced interactions between FL hTERT N95A and telomeric DNA primers (Fig. 5B and D). In parallel, we tested the activity and processivity of telomerase reconstituted with FL hTERT N95A in the CTA. We observed that the mutant telomerase showed reduced catalytic activity and defective repeat addition processivity on the bio-TELO18 and bio-TELO6 primers (Fig. 5E). This result argues that certain similar residues within the TERT N terminus, such as N95, have species-specific roles in primer binding and enzyme catalysis.
FL hTERT DAT regions are involved in primer binding and utilization in vitro.
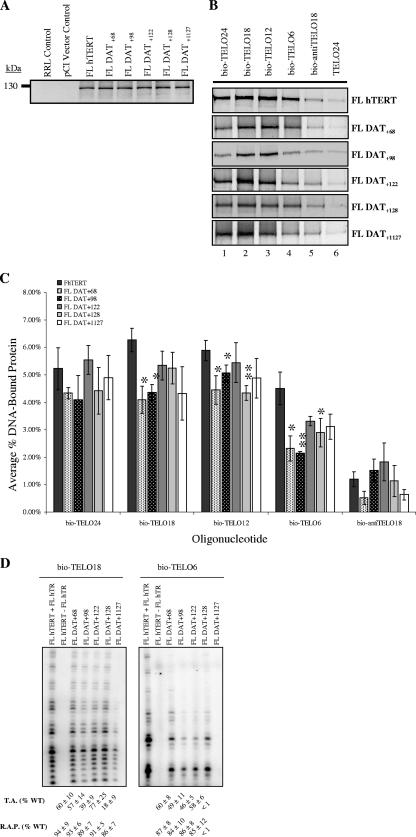
Collectively, the observations presented here and in previous studies indicate that the telomerase anchor site resides in the TERT RID1/N-GQ/TEN domain (29, 36, 41). Interestingly, using NAAIRS substitution mutations, Armbruster et al. (1) identified several DAT domains, which dissociate the biological and catalytic activities of telomerase, in the N terminus of hTERT that map to RID1. The DAT regions have been implicated in physical and functional telomerase-telomere interactions. Moriarty et al. (41) deleted nine amino acids at position 110 of hTERT and identified a RID1 DAT mutant (FL DAT+110) with defects in telomerase catalytic activity that were speculated to be caused by disruptions in the telomerase anchor site. These observations were based solely on telomerase activity assays (41). To extend these studies, we used our direct primer binding assay and the CTA to test the DNA binding and catalytic activity of five well-characterized FL hTERT DAT mutants (1, 2, 7). These include four N-terminal DAT mutants (FL DAT+68, FL DAT+98, FL DAT+122, and FL DAT+128) and one C-terminal DAT mutant (FL DAT+1127). In general, the FL hTERT DAT mutants showed a slightly less stable interaction with different lengths of telomeric primers than that of wild-type FL hTERT (Fig. 6B and C). The weakest interactions were observed between FL DAT+68 or FL DAT+98 and bio-TELO6, the shortest telomeric primer that we tested (Fig. 6B and C). Similarly, we found that FL DAT+68 and FL DAT+98 exhibited catalytic defects on both long (bio-TELO18) and short (bio-TELO6) primers, although repeat addition processivity was unaffected (Fig. 6D). FL DAT+122 and FL DAT+1127 showed minor defects in protein-telomeric DNA interactions relative to wild-type FL hTERT. However, telomerase reconstituted with these mutants had significantly reduced levels of total DNA synthesis, but not repeat addition processivity, when tested with the bio-TELO18 and bio-TELO6 primers (Fig. 6D). Strikingly, we were unable to detect any activity when we reconstituted telomerase with FL DAT+1127 and tested its ability to elongate the bio-TELO6 primer. However, in the absence of hTR, the FL DAT+1127 protein bound this short DNA primer as efficiently as did the catalytically active N-DAT mutants, arguing that the C-DAT region contributes to telomerase catalytic function independently of protein-DNA interactions (Fig. 6B to D). Finally, the N- and C-DAT hTERT mutants interacted specifically with telomeric DNA and did not show stable interactions with anti-telomeric DNA (Fig. 6B and C). Together, these results suggest that the N- and C-DAT domains function with other hTERT regions to regulate human telomerase protein-DNA interactions and catalytic function on long and short telomeric primers. Furthermore, the inability of certain telomerase mutants to utilize short telomeric primers is not a consequence of reduced or defective protein-substrate interactions.
FIG. 6.
hTERT DAT regions are involved in primer binding and utilization in vitro. (A) hTERT proteins were synthesized in the RRL system and radiolabeled with [35S]cysteine. Proteins were resolved by 8% SDS-PAGE and detected with autoradiography and analyzed so equivalent amounts of each protein were used in the primer binding experiment. Control reactions included RRL without protein (RRL control) and RRL with the pCI vector alone (pCI vector control). (B) The direct primer binding assay was used to evaluate the DNA binding properties of several previously identified FL hTERT DAT mutants using the oligonucleotides listed in Table 1. In parallel, the ability of mutant telomerase (reconstituted in RRL with FL hTERT DATx protein and wild-type [WT] RNA) to recognize and elongate these oligonucleotides was assessed with the conventional telomerase assay, as shown in panel D. (C) Quantification and statistical analysis of the data shown in panel B. Results are reported as the means (see Materials and Methods for details). Error bars indicate SEM. Asterisks denote levels of statistical significance compared to FL hTERT binding to the corresponding DNA primer: P values were <0.05 (∗) and <0.01 (∗∗). Average relative telomerase activity (T.A.) and repeat addition processivity (R.A.P.) values calculated from at least three independent experiments are shown at the bottom ± SEM.
DISCUSSION
We have developed a binding assay to directly analyze how the human telomerase protein subunit (hTERT) interacts with telomeric and nontelomeric DNA. Importantly, this is an activity-independent assay that allows us to investigate telomerase anchor site interactions in the absence of hTR, which might otherwise mask critical protein-DNA interactions. Using this assay, we show that hTERT specifically and stably interacts with telomeric DNA in the absence of hTR. Importantly, our observations are in accordance with those made by Sealey and Harrington, who have also observed that FL hTERT and various hTERT truncations (including an hTERT mutant spanning residues 1 to 350) interact specifically with biotinylated telomeric DNA in the absence of hTR (D. Sealey, and L. Harrington, personal communication). This is strong evidence for the existence of one or more DNA binding domains within hTERT. We use the direct primer binding assay in conjunction with the conventional telomerase activity assay to show that hTERT-mediated primer binding can be functionally uncoupled from telomerase-mediated primer extension. Through extensive deletion and mutational analysis, we have identified regions and residues within the protein's N terminus and RT domain that regulate the stability of hTERT-DNA interactions and those that regulate binding specificity. Finally, our results indicate that the catalytic defects of certain hTERT DAT mutants cannot be solely attributed to reduced telomerase affinity for the DNA substrate.
hTERT interacts with telomeric DNA in the absence of hTR.
Increasing evidence suggests that the N terminus of TERT contains at least one DNA binding domain. However, none of these studies investigate the nature of protein-DNA interactions in human telomerase. Here we report the development of an in vitro binding assay that allowed us to directly determine whether hTERT could bind telomeric DNA in the absence of hTR. We demonstrate that FL hTERT can form stable interactions with telomeric primers in the absence of hTR. Our primer binding results showed that there is no statistical difference in the interaction between FL hTERT and telomeric primers containing four, three, two, or one hexameric repeat. This is consistent with the hypothesis that telomerase can interact with a short stretch of single-stranded telomeric DNA in vivo (43). We did not detect an interaction between FL hTERT and anti-telomeric primers, which argues that human telomerase does not require the hTR template to discriminate between telomeric and nontelomeric DNA. Evidence to support this conclusion comes from a previous study in which Harrington and Greider show that human telomerase can elongate primers lacking 3′ telomeric DNA if G-rich or telomeric DNA is included at the 5′ end (26). The cumulative data support a model in which telomerase can recognize telomeric DNA independently of the telomerase RNA subunit by DNA-protein recognition of G-rich sequences (26, 33, 43, 49).
In contrast to the stable interaction observed with short telomeric primers, telomerase catalytic activity was optimal with an 18-nucleotide telomeric primer (consisting of three telomeric repeats) and was much reduced with primers of either 24 or 6 telomeric nucleotides. This is consistent with previously published observations (41, 42). Our results suggest that the reduced activity on a DNA substrate containing only one or four telomeric repeats is not due to a reduced hTERT-DNA interaction. The decreased activity may in part be due to the ability of telomeric primers containing four consecutive TTAGGG repeats to adopt a higher-order DNA structure known as the G-quadruplex, since our reaction conditions are similar to those in which these structures can form (15, 52). Prior to telomerase-mediated elongation, the G-quadruplex would have to be unwound to allow telomerase access to a single-stranded DNA substrate. In support of this possibility, hPOT1 has recently been shown to stimulate RecQ helicases WRN and BLM to unwind telomeric DNA (45) and increase telomerase activity on (TTAGGG)4 primers in vitro (52).
Primer binding can be functionally uncoupled from primer utilization.
Our hypothesis that the protein requirements for telomere binding are distinct from those mediating telomere elongation is supported by the finding that different catalytically inactive hTERT deletion variants (e.g., 1-300 and 351-1132) and substitution mutants (e.g., D712A and RT-GRIP) interact with telomeric primers as efficiently as does wild-type FL hTERT. This also argues that subtle alterations in physical hTERT-DNA interactions can lead to profound changes in functional telomerase-DNA interactions. In general, a complex interplay of protein-protein, protein-RNA, protein-DNA, and RNA-DNA interactions likely contributes to the ability of telomerase to efficiently recognize, bind, and elongate telomeric DNA.
Specific regions of the hTERT N terminus and the RT domain are required for primer recognition and utilization.
We studied a panel of hTERT mutants to identify regions within the protein that, when mutated or deleted, disrupt DNA binding and/or compromise hTERT's specificity for telomeric DNA by enhancing nonspecific protein-DNA interactions. Our studies show that the magnitude of a physical protein-DNA interaction is independent of the protein's size. This provides evidence that specific regions within hTERT make stable contacts with telomeric DNA. In particular, we found that the first 300 amino acids of hTERT possess strong DNA binding activity. However, this binding activity was not specific for telomeric DNA because the hTERT mutant spanning residues 1 to 300 also showed an abnormally strong interaction with anti-telomeric DNA. This contrasts with our observation that an hTERT fragment containing the first 350 amino acids showed an overall decrease in DNA binding and yet retained specificity for telomeric DNA. Interestingly, an hTERT mutant spanning residues 300 to 1132 showed an increased interaction with antitelomeric primers compared to that of an hTERT mutant spanning residues 350 to 1132. Collectively, our primer binding results suggest that the region between amino acids 300 and 350 acts in cis with the extreme N terminus of hTERT to confer specificity for telomeric DNA and restrain the protein-DNA interaction. We speculate that human telomerase may in fact benefit from a negative regulatory DNA-binding domain because an exceedingly stable hTERT-DNA interaction could “lock” the enzyme-DNA complex into an unfavorable conformation that impedes polymerization and translocation. It has previously been shown that human telomerase reconstituted in RRL with FL hTR and an hTERT mutant spanning amino acids 301 to 1132 catalyzes the synthesis of short elongation products when tested with telomeric primers in the telomeric repeat amplification protocol assay (10). In contrast, telomerase reconstituted in RRL with an hTERT mutant spanning residues 351 to 1132 is completely inactive, attesting to the functional importance of these 50 amino acids (10). This catalytic difference is not caused by defective hTERT-hTR interactions, suggesting that it can be attributed to compromised protein-DNA interactions (10). We surmise that human telomerase contains two distinct, but cooperative, anchor regions that are regulated in part by the hTERT region spanning amino acids 300 to 350. When this region is found in cis with RID2, the RT domain, and the C terminus, hTERT can bind telomeric DNA, but because the specificity of this interaction is compromised, telomerase does not engage in stable or processive DNA synthesis (this study; 10). On the other hand, when the 50-amino-acid region is found in cis with the extreme N terminus, hTERT specifically interacts with telomeric DNA (this study). Catalytic activity is restored to wild-type levels when the 1-350 hTERT fragment is combined with the 351-1132 protein, which provides the catalytic RT domain (10). Collectively, our results complement previous studies to establish the functional and structural importance of the region spanning amino acids 300 to 350.
In addition to the strong DNA binding activity associated with the first 300 amino acids of hTERT, we also provide evidence that there are additional sites within this protein that contact telomeric DNA. Specifically, we showed that hTERT mutants lacking regions of the N terminus engaged in stable interactions with telomeric primers (Fig. 3 and data not shown). Because these hTERT mutants were catalytically inactive when tested with the CTA, we propose that the N terminus is required for optimal enzyme-DNA interactions that support telomere synthesis. Furthermore, we have identified regions of hTERT that do not appear to make a significant contribution to the magnitude of a protein-DNA interaction. Specifically, the hTERT C-terminal extension was not sufficient to support a stable interaction with telomeric or antitelomeric primers, arguing that it must associate with other hTERT regions to bind DNA substrates. Our results contrast the weak nucleic acid binding activity observed in the C-terminal extension of yeast TERT (27). This difference is likely explained by the weak sequence conservation in this domain, suggesting that the TERT C terminus has evolved species-specific roles in DNA binding. Similarly, regions of the TERT RID1/N-GQ domain show low sequence conservation, suggesting that it too may engage in species-specific activities. In support, an N95A substitution in FL hTERT reduced the DNA binding and catalytic activity of human telomerase (this study) whereas the same mutation in Est2p causes telomere overelongation without altering the catalytic properties of S. cerevisiae telomerase (31).
In spite of these evolutionarily diverse regions, the TERT RT domain shows strong sequence homology to that of HIV-1 RT (44). This domain contains a conserved primer grip sequence that has been shown to regulate telomerase repeat addition processivity and telomere length in S. cerevisiae (46). We investigated the functional significance of the hTERT primer grip sequence and show that it is absolutely required for telomerase activity and a stable interaction between hTERT and short telomeric primers. A putative primer grip sequence within the hTERT N terminus is involved in regulating the specificity of hTERT for telomeric DNA and the ability of telomerase to processively elongate telomeric primers. The existence of two primer grips may be explained as follows: the evolutionarily conserved RT primer grip sequence aligns a potential DNA substrate in the catalytic site for telomerase-mediated elongation, and the N primer grip sequence regulates the strength and specificity of protein-DNA interactions. Together, optimal N and RT primer grip interactions culminate in an enzyme-DNA conformation that favors the specific and processive elongation of telomeric DNA.
Not all DATs are created equal.
In general, FL hTERT DAT mutants showed slightly reduced protein-DNA interactions when tested in the primer binding assay with different lengths of telomeric primers. Similarly, telomerase reconstituted with the DAT mutants showed significantly reduced catalytic activity but wild-type levels of repeat addition processivity. The latter finding contrasts to that reported for FL DAT+110 (41), suggesting that defects in different aspects of telomerase biology can manifest the DAT phenotype. Our results suggest that the +68, +98, +122, +128, and +1127 DAT mutations do not compromise the ability of telomerase to translocate the DNA substrate. However, we did observe some interesting differences between these DAT mutants. FL DAT+122 showed reduced catalytic activity compared to that of FL DAT+128, arguing that the former may have a more critical role in mediating telomerase activity. In contrast, FL DAT+128 exhibited reduced interaction with the bio-TELO12 and bio-TELO6 primers, indicating that this mutant requires longer telomeric primers for efficient binding and elongation. Since Lee et al. have shown that FL DAT+122 is partially active on a (GGGTTA)3 primer in vivo, it will be interesting to investigate the in vivo activity of FL DAT+128 as well as the other DAT mutants (34). Finally, our observation that FL DAT+1127 could bind, but not elongate, the bio-TELO6 primer argues that the hTERT C-terminal extension has an important role in catalytic function. These results are consistent with those obtained by Lee et al., in which the activity of FL hTERT DAT+1127 is abrogated in human cell extracts (34). Moriarty et al. have also shown that deleting hTERT residues 1123 to 1132 impairs telomerase activity in vitro (41), attesting to the importance of the hTERT C terminus. In summary, our results indicate that the DAT phenotype can manifest from alterations in different aspects of telomerase biology and that defects in DNA binding are not always sufficient to account for defects in catalytic activity and repeat addition processivity.
Summary.
We have developed a direct primer binding assay to delineate specific regions of hTERT that can bind telomeric DNA with high affinity and specificity in the absence of hTR. Our results provide evidence that the first 350 amino acids of hTERT have a critical role in regulating protein-DNA interactions, which strengthens the hypothesis that the TERT N terminus contains the telomerase anchor site. However, our results also indicate that an additional region within the RT domain of hTERT is involved in protein-DNA interactions. We surmise that the TERT N terminus, defined by residues with evolutionarily conserved and species-specific properties, contains a physical anchor region that regulates the strength and specificity of protein-DNA interactions. In addition, the RT domain contains an evolutionarily conserved functional anchor region (i.e., primer grip motif) that is critical for telomerase catalytic activity. It will be interesting to investigate how hTR affects hTERT-DNA interactions and the relationship that exists between physical and functional telomerase-DNA interactions. Furthermore, the primer binding assay can directly address how different DNA-binding proteins, such as the shelterin complex, interact with telomeric DNA in the presence or absence of hTERT and influence hTERT's DNA-binding properties. Lastly, regions of hTERT that make stable contacts with telomeric DNA may be amenable to therapeutic strategies that disrupt protein-DNA interactions and down-regulate aberrant telomerase activity in diseases like cancer.
Acknowledgments
We thank Christopher Counter for the FL hTERT, FL hTERT-DAT, and FL hTERT D712A constructs and helpful discussions; Lea Harrington and Dave Sealey for sharing unpublished data and critically reading the manuscript; Susan Lees-Miller and members of the Beattie lab for helpful discussions; and George Chaconas, Paul Bonnefin, Chris Hough, and Brant Pohorelic for technical assistance.
H.D.M.W. holds an NSERC doctoral research award, and T.L.B. is a Scholar of the AHFMR and a CIHR new investigator. This work was funded by operating grants from the National Cancer Institute of Canada and the Alberta Cancer Board to T.L.B.
Footnotes
Published ahead of print on 12 February 2007.
REFERENCES
- 1.Armbruster, B. N., S. S. Banik, C. Guo, A. C. Smith, and C. M. Counter. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21:7775-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armbruster, B. N., K. T. Etheridge, D. Broccoli, and C. M. Counter. 2003. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol. Cell. Biol. 23:3237-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armbruster, B. N., C. M. Linardic, T. Veldman, N. P. Bansal, D. L. Downie, and C. M. Counter. 2004. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 24:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Autexier, C., and N. F. Lue. 2006. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75:493-517. [DOI] [PubMed] [Google Scholar]
- 5.Bachand, F., and C. Autexier. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol. Cell. Biol. 21:1888-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey, S. M., and J. P. Murnane. 2006. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 34:2408-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banik, S. S., C. Guo, A. C. Smith, S. S. Margolis, D. A. Richardson, C. A. Tirado, and C. M. Counter. 2002. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol. 22:6234-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baran, N., Y. Haviv, B. Paul, and H. Manor. 2002. Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 30:5570-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2001. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21:6151-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11:3329-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 12.Bosoy, D., and N. F. Lue. 2001. Functional analysis of conserved residues in the putative “finger” domain of telomerase reverse transcriptase. J. Biol. Chem. 276:46305-46312. [DOI] [PubMed] [Google Scholar]
- 13.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 275:24199-24207. [DOI] [PubMed] [Google Scholar]
- 14.Collins, K., and C. W. Greider. 1993. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 7:1364-1376. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher, T. M., D. Sun, M. Salazar, and L. H. Hurley. 1998. Effect of DNA secondary structure on human telomerase activity. Biochemistry 37:5536-5541. [DOI] [PubMed] [Google Scholar]
- 16.Gilley, D., H. Tanaka, and B. S. Herbert. 2005. Telomere dysfunction in aging and cancer. Int. J. Biochem. Cell Biol. 37:1000-1013. [DOI] [PubMed] [Google Scholar]
- 17.Greider, C. W. 1991. Telomerase is processive. Mol. Cell. Biol. 11:4572-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337-365. [DOI] [PubMed] [Google Scholar]
- 19.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 20.Greider, C. W., and E. H. Blackburn. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51:887-898. [DOI] [PubMed] [Google Scholar]
- 21.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 22.Hainaut, P., A. Hall, and J. Milner. 1994. Analysis of p53 quaternary structure in relation to sequence-specific DNA binding. Oncogene 9:299-303. [PubMed] [Google Scholar]
- 23.Hammond, P. W., T. N. Lively, and T. R. Cech. 1997. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 17:296-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy, C. D., C. S. Schultz, and K. Collins. 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276:4863-4871. [DOI] [PubMed] [Google Scholar]
- 25.Harrington, L., W. Zhou, T. McPhail, R. Oulton, D. S. Yeung, V. Mar, M. B. Bass, and M. O. Robinson. 1997. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 11:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington, L. A., and C. W. Greider. 1991. Telomerase primer specificity and chromosome healing. Nature 353:451-454. [DOI] [PubMed] [Google Scholar]
- 27.Hossain, S., S. Singh, and N. F. Lue. 2002. Functional analysis of the C-terminal extension of telomerase reverse transcriptase: a putative “thumb” domain. J. Biol. Chem. 277:36174-36180. [DOI] [PubMed] [Google Scholar]
- 28.Huard, S., T. J. Moriarty, and C. Autexier. 2003. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31:4059-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs, S. A., E. R. Podell, and T. R. Cech. 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 13:218-225. [DOI] [PubMed] [Google Scholar]
- 30.Jacques, P. S., B. M. Wohrl, M. Ottmann, J. L. Darlix, and S. F. Le Grice. 1994. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem. 269:26472-26478. [PubMed] [Google Scholar]
- 31.Ji, H., M. H. Platts, L. M. Dharamsi, and K. L. Friedman. 2005. Regulation of telomere length by an N-terminal region of the yeast telomerase reverse transcriptase. Mol. Cell. Biol. 25:9103-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 33.Lee, M. S., and E. H. Blackburn. 1993. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 13:6586-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, S. R., J. M. Wong, and K. Collins. 2003. Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 278:52531-52536. [DOI] [PubMed] [Google Scholar]
- 35.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 36.Lue, N. F. 2005. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 280:26586-26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lue, N. F., Y. C. Lin, and I. S. Mian. 2003. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell. Biol. 23:8440-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 39.Moriarty, T. J., S. Huard, S. Dupuis, and C. Autexier. 2002. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriarty, T. J., D. T. Marie-Egyptienne, and C. Autexier. 2004. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 24:3720-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriarty, T. J., R. J. Ward, M. A. Taboski, and C. Autexier. 2005. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell 16:3152-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morin, G. B. 1989. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59:521-529. [DOI] [PubMed] [Google Scholar]
- 43.Morin, G. B. 1991. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature 353:454-456. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 45.Opresko, P. L., P. A. Mason, E. R. Podell, M. Lei, I. D. Hickson, T. R. Cech, and V. A. Bohr. 2005. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 280:32069-32080. [DOI] [PubMed] [Google Scholar]
- 46.Peng, Y., I. S. Mian, and N. F. Lue. 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7:1201-1211. [DOI] [PubMed] [Google Scholar]
- 47.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 48.Snow, B. E., N. Erdmann, J. Cruickshank, H. Goldman, R. M. Gill, M. O. Robinson, and L. Harrington. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13:698-704. [DOI] [PubMed] [Google Scholar]
- 49.Wallweber, G., S. Gryaznov, K. Pongracz, and R. Pruzan. 2003. Interaction of human telomerase with its primer substrate. Biochemistry 42:589-600. [DOI] [PubMed] [Google Scholar]
- 50.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 51.Xia, J., Y. Peng, I. S. Mian, and N. F. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20:5196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaug, A. J., E. R. Podell, and T. R. Cech. 2005. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 102:10864-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]