Abstract
Recent evidence suggests that fibrocystin/polyductin (FPC), polycystin-1 (PC1), and polycystin-2 (PC2) are all localized at the plasma membrane and the primary cilium, where PC1 and PC2 contribute to fluid flow sensation and may function in the same mechanotransduction pathways. To further define the exact subcellular localization of FPC, the protein product encoded by the PKHD1 gene responsible for autosomal recessive polycystic kidney disease (PKD) in humans, and whether FPC has direct and/or indirect cross talk with PC2, which, in turn, is pivotal for the pathogenesis of autosomal dominant PKD, we performed double immunostaining and coimmunoprecipitation as well as a microfluorimetry study of kidney tubular epithelial cells. FPC and PC2 are found to completely or partially colocalize at the plasma membrane and the primary cilium and can be reciprocally coimmunoprecipitated. Although incomplete removal of FPC by small interfering RNA and antibody 803 to intracellular epitopes of FPC did not abolish flow-induced intracellular calcium responses, antibody 804 to extracellular epitopes of FPC blocked cellular calcium responses to flow stimulation. These findings suggest that FPC and polycystins share, at least in part, a common mechanotransduction pathway.
Inherited polycystic kidney diseases (PKD) are a large group of diseases characterized by the development of multiple fluid-filled cysts in the kidney, starting from infancy or adulthood, which gradually lead to end-stage renal disease (ESRD). About 10 percent of ESRD cases are caused by PKD. In humans, there are two major forms of PKD, i.e., autosomal dominant PKD (ADPKD) and autosomal recessive PKD (ARPKD).
ADPKD is the most common type, with an incidence of 1 in 500 to 1 in 1,000 adults (4). The causative genes, PKD1 and PKD2, account for ∼85% and ∼15% of all cases, respectively (28a). The genes are similar in size, but the encoded proteins polycystin-1 and -2 (PC1 and PC2) are vastly different. PC1 is a 4,303-amino-acid (aa) protein with a large extracellular amino (N) terminus, 7 to 11 transmembrane domains, and a 200-aa intracellular carboxyl (C) terminus (12, 15), while PC2 is a 968-aa and 6-transmembrane protein, with 30% identity and 50% homology to the C-terminal transmembrane region of PC1 and 25% homology to transient receptor potential channels (23). Both PC1 and PC2 are widely distributed in different tissues. PC1 has been considered to reside on the plasma membrane and be involved in cell-cell and cell-matrix interactions (9, 13, 28). Subcellular localization of PC2 remains controversial (3, 7), but it is likely to serve as a channel at both the endoplasmic reticulum and the plasma membrane (18). PC1 and PC2 may interact with each other through coiled-coil domains and produce nonselective cation conductance at the plasma membrane (10). PC1 and PC2 were reported to be colocalized at the primary cilium of the kidney tubular epithelial cells (26, 38) and contribute to fluid flow sensation in the same mechanotransduction pathway (26). The loss or dysfunction of PC1 or PC2 may therefore lead to PKD, owing to the inability of cells to sense mechanical cues that normally regulate tissue morphogenesis (26, 26a).
ARPKD is a monogenic genetic disorder found mainly in infancy, with a prevalence of 1 in 20,000 newborns (30, 40). Apart from the extrarenal phenotype, such as biliary dysgenesis, hepatic fibrosis, portal and systemic hypertension, oligohydramnios, and pulmonary hypoplasia, bilateral enlarged polycystic kidneys are the major finding. The mortality rate thus far still reaches 30% of infants (29). However, half of the children who survived the neonatal period finally developed ESRD (8). Cysts in the kidney arise mainly from the collecting ducts. More than 250 mutations in the causative gene PKHD1 have been identified thus far (1). The longest transcript in humans is 16,235 bp with an open reading frame of 12,222 bp (27, 34, 37). The encoded protein, designated fibrocystin/polyductin (FPC), is a large 4,074-aa protein with a calculated molecular mass of 447 kDa. FPC was proposed to be a novel single transmembrane protein with a 192-aa intracellular C terminus and a very large extracellular N terminus containing such domains as TIG, TIG-like, TMEM2 homolog, and DKFZ homolog (34). The longest open reading frame of the mouse ortholog of PKHD1 encodes a protein of 4,059 aa; the mouse and human protein sequences are 73% identical overall and 55% identical in the C-terminal tail (27). A study of the expression pattern in mouse metanephros by in situ hybridization showed that Pkhd1 transcripts are not expressed in metanephric mesenchyme but are strongly expressed in the branching ureteric bud (25). In postnatal kidney tissue, strong Pkhd1 expression was found in collecting ducts, with lower levels in proximal and distal tubules (25). Recently, FPC was localized to the primary cilium/basal body and plasma membrane in the renal epithelium (33, 35, 41) and is thought to be a transcriptional target of hepatocyte nuclear factor 1β (11). The inhibition of Pkhd1 impaired the tubulogenesis in cultured inner medullary collecting duct 3 (IMCD-3) cells (19).
The clinical features between ADPKD and ARPKD and the subcellular localization of FPC, PC1, and PC2 prompted us to hypothesize that these proteins function in a coordinated fashion. Here, we report, through morphological, biochemical, and physiological studies, that the proteins for ADPKD and ARPKD are in the same protein complex and share, at least in part, the same signaling pathways.
MATERIALS AND METHODS
Antibodies.
Anti-E-cadherin monoclonal antibody (MAb) was obtained from Pharmingen. Anti-acetylated α-tubulin and anti-β-actin MAb were purchased from Sigma. Anti-myc MAb was purchased from Invitrogen. Normal rabbit immunoglobulin G (IgG) and IgY were obtained from Santa Cruz Biotechnology and Aves Labs. Texas Red-fluorescein isothiocyanate-labeled secondary antibodies were obtained from Molecular Probes. Donkey anti-rabbit immunoglobulin and rabbit anti-chicken IgY conjugated with horseradish peroxidase were obtained from Amersham Biosciences and Aves Labs, respectively. FPC (4883, 803, and 804), PC2 (96525), and PC1 (96521) polyclonal antibodies have been characterized previously (18, 26, 33; J. Zhang and J. Zhou, unpublished data). Antibody 5249 to an intracellular epitope of mouse FPC (RPDLRQERKQGQEPSQLDK) was raised in hens and affinity purified (Aves Labs). Kif3a and Kif3b antibodies were purchased from BD Biosciences and Santa Cruz Biotechnology, respectively. Biotinylated Dolichos biflorus agglutinin (DBA), Lotus tetragonolobus lectin (LTL), and Texas Red streptavidin were purchased from Vector Laboratories.
All of these studies were completely in compliance with human and animal research guidelines, and Institutional Review Board approval of the Brigham and Women's Hospital and Harvard Medical School was obtained.
Cell culture.
IMCD-3 cells, MDCK cells, and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium-F12-10% fetal bovine serum and Dulbecco's modified Eagle's medium-10% fetal bovine serum (Gibco). DBA-positive mouse embryonic kidney (MEK) cells, developed in our laboratory, were derived from embryonic day 15.5 kidneys from wild-type and Pkd1null/null mice bearing temperature-sensitive simian virus large T antigen (17). MEK cells were usually cultured at 33°C in simian virus 40 epithelium medium for 1 to 5 days and, when needed, moved to 37°C for differentiation without gamma interferon.
Immunostaining and immunofluorescence microscopy.
The methods for immunostaining in kidney cells and tissues have been described in detail elsewhere (33). In brief, cultured cells were fixed with 3% paraformaldehyde (Merck)-2% sucrose (Sigma) and permeabilized with 0.2% Triton X-100 (Sigma). Permeabilized cells were incubated with the primary antibody for 1 h, washed completely, and incubated with a labeled secondary antibody for 1 h. For double labeling, antibodies against two different antigens were concomitantly incubated. After thorough washing, one or two labeled secondary antibodies were added for 1 h. Before being mounted with ProLong antifade medium (Molecular Probes), the cells were incubated with DAPI (4′,6′-diamidino-2-phenylindole; Sigma) for 5 min. For human kidney tissue staining, 3% paraformaldehyde-prefixed tissues were embedded in Tissue-Tek O.C.T. compound (Ted Pella), sectioned at 3 to 6 μm, and further fixed in acetone at −20°C for 5 min. The other procedures were the same as for the cells, except without Triton X-100. A Zeiss Axioskop 2 Plus fluorescence microscope (Carl Zeiss) and the SPOT camera system (Diagnostic Instruments) were used for image analysis. All the experiments were done at room temperature.
RNA isolation, reverse transcription (RT), and PCR.
Total RNA was isolated according to Chomczynski's and Sacchi's one-step method (5) by using TRIzol reagent (Invitrogen). For PCR, Taq DNA polymerase (Clontech) was used after first-strand cDNA synthesis with Superscript II RNase H reverse transcriptase (Clontech) in the presence of oligo(dT)12-18 (Clontech). PKHD1/Pkhd1 primers were designed according to the human and mouse sequences (Tables 1 and 2). Primers for mouse β-actin, Pkd1, and Pkd2 were designed according to their respective DNA sequences (Table 2). The PCR amplification began with denaturation at 94°C for 1 min and continued with annealing at 56 to 60°C for 1 min and elongation at 72°C for 1 to 3 min, followed by 15 to 40 PCR cycles. For abundance comparison of the gene transcripts, different numbers of PCR cycles (15, 20, 25, 30, and 35) were used.
TABLE 1.
PCR primer sequences for DNA constructs
| DNA construct | Primer sequence (5′-3′)a | Enzyme |
|---|---|---|
| pGADT7/ hPKHD1CT | GGCCGAATTCAAAAGAAGCAAAAGCAGAAAA | EcoRI |
| AGTCCTCGAGTCACAGTTGCTCCTGAATAGTTTCCGGGTG | XhoI | |
| pGADT7/hPKD2TM2-3 | GGCCGAATTCTTCTTCCTGGCAGCCTGTGAG | EcoRI |
| AGTCCTCGAGTATGTTAATTCCTATAGCTACCACTGACAGCAC | XhoI | |
| pGADT7/hPKD2TM4-5 | GGCCGAATTCTTCAACAATATAGCTGCTGTCACAGTATTTTTT | EcoRI |
| AGTCCTCGAGAAGGTATGCCAACTGAGCATACGC | XhoI | |
| pGBKT7/ hPKHD1CT | CAAGGAATTCAAAAGAAGCAAAAGCAGAAAAACAAAACCTGAAGAGATTC | EcoRI |
| AGTCCTGCAGTCACAGTTGCTCCTGAATAGTTTCCGGGTG | PstI | |
| pcDNA3.1/ mPkhd1NT2 | TACGAAGCTTCCACCATGGCAAACACCCCATTGCTGGTTCAT | HindIII |
| AGTCTCTAGAGCGGCATTAACAGCAAGGCCTGACG | XbaI |
The upper and lower sequences of each construct represent forward and reverse primers, respectively.
TABLE 2.
PCR primers used for real-time PCR and RT-PCR
| Gene name | Primer name | Primer sequence (5′-3′) | GenBank accession no. |
|---|---|---|---|
| Pkhd1 | F1 | TGTACCCTGGCAGAAACACGGGAT | NM_153179 |
| R1 | GCAACGTAACCAGTTTGAACCCCA | ||
| F2 | AAGAAAAGCAAAACCAGAAAAATAAAACCAGAA | ||
| R2 | CTGGATGGTTTCTGGTGGAGTAGTGTGGAG | ||
| PST-031C-Fa | ACTGAATTTCAGTAAGCAGGAACTG | ||
| PST-031C-Ra | CACTGCAGATACAACATCAGCTAAC | ||
| Pkd1 | F1 | CTTGGTGTGGCCTATGCACA | NM_013630 |
| R1 | TGAAGCTTCTGAGCCTGAGC | ||
| Pkd2 | F1 | TCCCCAGAAGCCTGGATGAC | NM_008861 |
| R1 | TTTGCGAAGCTGCATCATCC | ||
| β-Actin | F | CATTGTTACCAACTGGGACGACAT | X03672 |
| R | ATCTCCTGCTCGAAGTCTAGAGCA | ||
| Gapdh | PST-052-Fa | GATGCCCCCATGTTTGTGAT | M32599 |
| PST-052-Ra | TTGTCATGGATGACCTTGGC |
These primers were used for real-time PCR, while the others were used for RT-PCR.
In order to determine the copy number of Pkhd1 relative to that of Gapdh (glyceraldehyde-3-phosphate dehydrogenase) cDNA molecules, we performed real-time PCR analysis by use of the LightCycler system (Roche) with the following conditions and primers. Each cycle included denaturation for 15 s at 94°C, annealing for 8 s at 53°C, and extension for 8 s at 72°C. Melting curve analysis was performed after each run, and data were analyzed using Roche LightCycler data analysis software (v3.5.28). Results were expressed as a percentage compared to the control. The primer sequences for Pkhd1 and Gapdh are listed in Table 2.
DNA constructs.
All other DNA constructs except pGBKT7/hPKD2NT and pBD-c-PKD2 were made by the RT-PCR method (Table 1). The RT-PCR products were digested with restriction enzymes (Table 1) and ligated with T4 DNA ligase into the respective pGBKT7, pGADT7, and pcDNA3.1 vectors according to the protocol supplied by New England Biolabs. For the construction of pGBKT7/hPKD2NT, NcoI was used to cut out 274 nucleotides at the 5′ end of human PKD2 and this fragment was subcloned into the pGBKT7 vector. pBD-c-PKD2 was made by subcloning the C terminus of PC2 containing aa 679 to 969 into the pGBKT7 vector (16). All DNA constructs were confirmed to be correct by DNA sequencing.
siRNA constructs and establishment of FPC knockdown stable cell lines.
For FPC knockdown, small interfering RNA (siRNA) constructs were made according to the protocol provided by Ambion. Briefly, two selected top and bottom oligonucleotides (Table 3) were synthesized and dissolved at a concentration of 1 μg/μl and then incubated at 37°C for 1 h with annealing buffer (100 mM K-acetate, 30 mM HEPES-KOH, pH 7.4, and 2 mM Mg-acetate). Before incubation, the mixture was denatured at 90°C for 3 min. Finally, double strands of oligonucleotides were cloned into the BamHI/HindIII or ApaI/EcoRI site of the pSilencer2.1/U6 or BS/U6/GFP vector, respectively. Two pairs (F939 and F1417) of oligonucleotides were inserted into the BS/U6/GFP vector (32), and another four pairs (F2141, F2850, F11689, and F11813) were inserted into pSilencer2.1/U6. The constructs were transformed into competent Escherichia coli DH5α by electroporation (Gene Pulser II/E. coli Pulser system) according to the protocol provided by Bio-Rad.
TABLE 3.
Sequences for the generation of FPC siRNA constructs
| siRNA construct | Strand | Sequence (5′-3′) |
|---|---|---|
| BS/U6/GFP/F939 | Top | ATGAAGCAAGGCTCACTGCTTCAAGAGAGCAGTGAGCCTTGCTTCATTTTTTT |
| Bottom | AATTAAAAAAATGAAGCAAGGCTCACTGCTCTCTTGAAGCAGTGAGCCTTGCTTCATGGCC | |
| BS/U6/GFP/F1417 | Top | CACCTGGTTGAATCCAGATTTCAAGAGAATCTGGATTCAACCAGGTGTTTTTT |
| Bottom | AATTAAAAAACACCTGGTTGAATCCAGATTCTCTTGAAATCTGGATTCAACCAGGTGGGCC | |
| pSilencer2.1/U6/F2141 | Top | GATCCGCTGATTCTGGAAGAGCCTTTCAAGAGAAGGCTCTTCCAGAATCAGCTTTTTTGGAAA |
| Bottom | AGCTTTTCCAAAAAAGCTGATTCTGGAAGAGCCTTCTCTTGAAAGGCTCTTCCAGAATCAGCG | |
| pSilencer2.1/U6/F2850 | Top | GATCCCTGGTTTCCCTAGAGACACTTCAAGAGAGTGTCTCTAGGGAAACCAGTTTTTTGGAAA |
| Bottom | AGCTTTTCCAAAAAACTGGTTTCCCTAGAGACACTCTCTTGAAGTGTCTCTAGGGAAACCAGG | |
| pSilencer2.1/U6/F11689 | Top | GATCCGAAGAATACCCATAATTCCTTCAAGAGAGGAATTATGGGTATTCTTCTTTTTTGGAAA |
| Bottom | AGCTTTTCCAAAAAAGAAGAATACCCATAATTCCTCTCTTGAAGGAATTATGGGTATTCTTCG | |
| pSilencer2.1/U6/F11813 | Top | GATCCGCACTCAATGGATGGAGTGTTTCAAGAGAACACTCCATCCATTGAGTGTTTTTTGGAAA |
| Bottom | AGCTTTTCCAAAAAACACTCAATGGATGGAGTGTTCTCTTGAAACACTCCATCCATTGAGTGCG |
To test the effects of siRNA on FPC transcripts, IMCD-3 cells were transfected with FuGENE 6 reagent. Two days after transfection, the cells were used for real-time and RT-PCR analyses. In addition, HEK 293T cells were cotransfected with pcDNA3.1/mPkhd1NT2 and pSilencer2.1/U6/F2141 or pSilencer2.1/U6/F2850, and after 2 days, proteins were extracted for further analysis.
For the establishment of FPC knockdown stable cell lines, after a 1-day transfection with pSilencer2.1/U6/F2141 or pSilencer2.1/U6/F2850, cells were split into five 10-cm plates and followed by selection with G418 (0.75 μg/ml). Twenty-five and 30 clones were isolated, respectively, for pSilencer2.1/U6/F2850 and pSilencer2.1/U6/F2141 knockdown after a 21-day culture.
Protein extraction and immunoprecipitation (IP).
HEK 293T cells were harvested and solubilized with M-PER mammalian protein extraction reagent (Pierce) containing the appropriate protease inhibitors (Roche) and 1 mM phenylmethylsulfonyl fluoride (Sigma). IMCD-3, MEK, and MDCK cells were lysed with radioimmunoprecipitation assay lysis buffer (Upstate) or M-PER mammalian protein extraction reagent and centrifuged at 10,000 × g for 15 min at +4°C. For immunoprecipitation, the supernatants were precleared with protein A/G agarose beads (Invitrogen) for rabbit/mouse antibody or agarose-coupled goat anti-chicken IgY (Aves Labs) for chicken antibody for 1 h at +4°C before the addition of the primary antibody. Subsequently, fresh agarose beads and the antibody were added and incubated at +4°C overnight, followed by centrifugation at 10,000 × g for 1 min at +4°C. The beads were washed twice with the lysis buffer. Finally, 30 μl of sample buffer was added, and the samples were analyzed with immunoblotting (IB).
Laemmli gel and IB.
The Laemmli gel and IB method has been described in detail previously (33). Briefly, the protein samples were electrophoresed in a 5% or 12% acrylamide Laemmli resolving gel (Bio-Rad) and transferred to Hybond ECL nitrocellulose membranes (Amersham Pharmacia Biotech). After being blocked with 5% nonfat dry milk (Bio-Rad) in phosphate-buffered saline, the filter was incubated with the primary antibody and washed three times with phosphate-buffered saline-0.1% Tween 20 (Bio-Rad). The filters were finally incubated with donkey anti-rabbit, -mouse, or -chicken immunoglobulin conjugated with horseradish peroxidase-linked secondary antibody. After being washed thoroughly, the bound antibodies were detected with the ECL Western blotting analysis system (Amersham Pharmacia Biotech). If needed, the same filter was stripped with Restore Western blot stripping buffer and reblotted according to the protocol provided by Pierce. Quantification of the protein band intensity was accomplished with NIH Image software (version 1.63).
Yeast two-hybrid analysis.
For FPC-PC2 interaction studies, a pGBKT7 bait construct and a pGADT7 prey construct were simultaneously cotransformed into fresh-made competent Saccharomyces cerevisiae strain AH109 according to the protocol provided by Clontech. The clones were screened for ADE2, HIS3, and MEL1 expression by high-stringency strategy on minimal standard dropout agar base lacking adenine (Ade), histidine (His), leucine (Leu), and tryptophan (Trp). In addition, pGBKT7-53 and pGADT7-T were used as positive controls. All the plates were incubated at 30°C for at least 1 week.
Calcium microfluorimetry.
Wild-type MEK cells were cultured for at least 3 days in the absence of gamma interferon to induce optimal differentiation. Parental IMCD-3 cells (or vector-transfected stable cells) and FPC knockdown cells were cultured as described above. In transient FPC knockdown cells with either BS/U6/GFP/F939 or BS/U6/GFP/F1417, intracellular calcium responses were measured in green fluorescent protein (GFP)-positive cells after a 3-day transfection. In antibody-blocking experiments, MEK or IMCD-3 cells were incubated with antibodies against FPC (803 and 804) at dilutions of 1:50, 1:100, 1:500, and 1:1,000 for at least 30 min and then washed at least three times with HEPES-Na. For control experiments, normal rabbit IgG and antibody 804 preincubated with its peptide were used. A nonfluorescent, CO2-independent medium was used for Fura-2 Ca2+ imaging as described previously (26). For Ca2+ imaging, cells were incubated for 30 min with the Ca2+-sensitive probe Fura2-AM (5 μM) (Molecular Probes) at 37°C, then washed three times to remove excess Fura2-AM, and placed in a perfusion chamber with a thickness of 0.0254 cm and a width of 1.0 cm (GlycoTech). The chamber was positioned under a Nikon Diaphot microscope equipped with a charge-coupled-device camera using IPLab software for Macintosh. Paired Fura images were captured every 5 s at excitation wavelengths of 340 nm and 380 nm. The emitted fluorescence was filtered at 500 nm, and the ratio of the emitted light at an excitation wavelength of 340 nm to that at 380 nm was calculated automatically as a measure of intracellular Ca2+. After equilibration in the microscopy media for at least 10 min, the primary cilia of these cells were stimulated at a fluid shear stress of 0.75 dynes cm−2. The Ca2+ level was radiometrically calculated relative to the baseline value, using Rmin and Rmax values of 0.3 and 6.0, respectively.
RESULTS
FPC localization in the plasma membrane and cytosol of collecting duct/tubule cells in normal human kidney tissues.
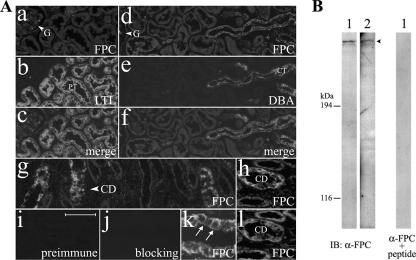
As reported previously, FPC is localized to primary cilia, with concentration in the basal body area (33). Here, we further examined the expression of FPC in normal human kidney tissue so as to define whether FPC is also localized at the plasma membrane with our antibodies. In the human kidney cortex, double labeling with FPC, LTL (a marker for the proximal tubule), or DBA (a marker for the collecting tubule/duct) localized FPC to collecting tubules/ducts (Fig. 1A, panels a to f), whereas no staining was detectable along proximal tubules and glomeruli (Fig. 1A, panels a to c). In the human kidney medulla, FPC was expressed at collecting ducts with FPC antibodies (both 803 and 804), and both the apical and basolateral membrane domains were immunoreactive (Fig. 1A, panels g, h, and k). The staining patterns of both FPC antibodies (803 and 804) were similar in normal human kidney tissue (Fig. 1A, panels h and l). No obvious membrane and cytoplasmic signals were observed with the preimmune serum (Fig. 1A, panel i). Preincubation of FPC antibody with its immunogen completely blocked the membrane and cytoplasmic staining (Fig. 1A, panel j). FPC antibody detected a predominant band at ∼450 kDa in kidney medulla tissue extracts from two normal human individuals, and this band was not detectable if FPC antibody was preincubated with its immunogen (Fig. 1B).
FIG. 1.
FPC is detectable at the apical and basolateral domains of the plasma membrane in human adult kidneys. (A) Double labeling with FPC (804) (a and d), LTL (b), or DBA (e) showed no immunoreactivity in glomeruli (G) and proximal tubules (PT) but did show immunoreactivity in the apical and basolateral areas of collecting tubules (CT) in the human kidney cortex (c and f). (g, h, and k) Immunoreactivity of FPC (804) was found along collecting ducts (CD) in the medulla of the human kidney (arrows). (i) No signals were observed with the preimmune serum (804). (j) Preincubation of FPC antibody (α-FPC) (804) with its immunogen completely blocked the membrane and cytosol labeling. 804 (h) and 803 (l) FPC antibodies gave similar staining patterns in the medulla of the human kidney. (B) 804 recognized a band at ∼450 kDa by IB in tissue extracts of human kidney medullas from two normal individuals (1 and 2), and this band was not detectable if FPC antibody was preincubated with its immunogen. Bars, 100 μm (a to g, i, and j), 50 μm (h and l), and 25 μm (k).
FPC colocalization with PC2 at the plasma membrane and the cilium in MDCK cells.
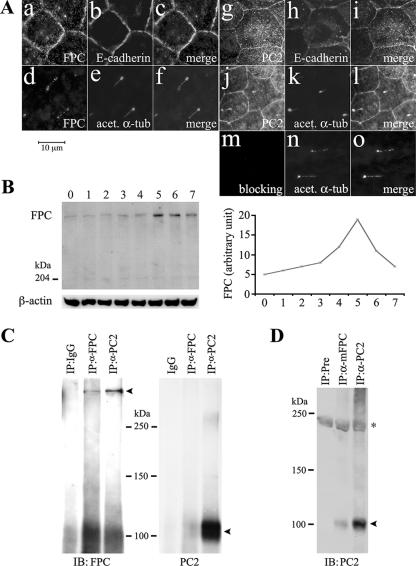
In order to examine whether FPC and PC2 are colocalized, we double immunostained MDCK cells. Because our antibodies against FPC and PC2 were all generated in rabbits, we adopted an indirect colocalization approach to study whether both FPC and PC2 are colocalized at the plasma membrane and the primary cilium. We used E-cadherin as a plasma membrane marker and acetylated α-tubulin as a cilium marker. Either FPC or PC2 was found to colocalize largely with E-cadherin at the plasma membrane (Fig. 2A). Interestingly, FPC became detectable in the ciliary shaft when cells cultured under certain conditions (Fig. 2A, panel d), coinciding with its change at protein levels (Fig. 2B). PC2, however, was regularly situated at the shaft of the primary cilium (Fig. 2A, panel j).
FIG. 2.
Results of studies on FPC and PC2 interaction in MDCK cells. (A) Double labeling of FPC (803) (a and d) or PC2 (96525) (g and j) with E-cadherin (b and h) or acetylated antitubulin (acet. α-tub) (e and k) showed that both FPC and PC2 were localized at the plasma membrane (c and i) and on the primary cilium (f and l). Preincubation of 803 with its immunogen completely abolished the staining on the cilium and at the plasma membrane (m to o). (B) Western blot analysis showed that FPC expression increased gradually from day 0 to day 4 postconfluence, peaked at day 5, and then decreased rapidly (left). The line graph (right) showed the expression level of FPC after normalization with β-actin by using NIH Image software (version 1.63). (C) FPC and PC2 were reciprocally immunoprecipitated in MDCK cells. One band at ∼450 kDa and one at ∼110 kDa (arrowheads) were detected with FPC (804) and PC2 antibody after IP with the respective FPC (4883) and PC2 antibodies; IgG was used as a control. (D) PC2 can be coimmunoprecipitated by anti-mouse FPC antibody (α-mFPC) (5249) in mouse kidney tissues. Preimmune (Pre) and PC2 antibodies were used as negative and positive controls, respectively. The asterisk indicates a nonspecific band.
Coimmunoprecipitation of FPC and PC2 in MDCK cells and mouse kidney tissues.
The colocalization of FPC and PC2 prompted us to further explore whether these two PKD molecules are in the same protein complex. Therefore, we performed coimmunoprecipitation experiments with well-differentiated MDCK cells. FPC antibody precipitated a band recognized by PC2 antibody at ∼110 kDa, and a band precipitated with PC2 antibody was recognized by FPC antibody at ∼450 kDa, indicating that FPC and PC2 can be reciprocally coimmunoprecipitated. The specificity of PC2 and FPC interaction was confirmed using IgG as a control (Fig. 2C). This interaction was further confirmed in vivo with mouse kidney tissues. PC2 was detected in FPC antibody (5249)-precipitated tissue lysate (Fig. 2D).
Interactions between FPC and PC2.
The coimmunoprecipitation of endogenous FPC and PC2 encouraged us to test whether FPC and PC2 interact physically with each other. First of all, we took advantage of Matchmaker GAL4 two-hybrid system 3 and generated an FPC C-terminal tail construct (aa 3883 to 4074) and four PC2 yeast constructs, including the N terminus (aa 1 to 274), the C terminus (aa 679 to 969), and two intracellular loops (aa 471 to 526 and 558 to 617). Yeast (AH109) cotransformation of the FPC bait/prey construct with each prey/bait construct of all four PC2 intracellular domains was performed. No colony grew on high-stringency (lacking Ade-His-Leu-Trp) selection plates.
Characterization of FPC knockdown stable cell lines.
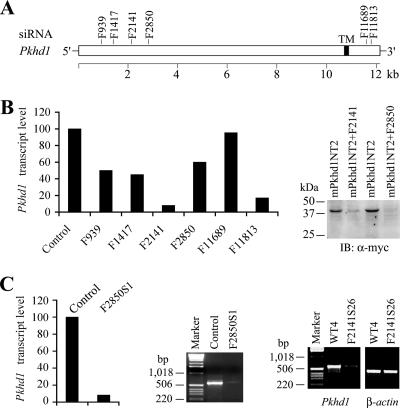
Six siRNA constructs (BS/U6/GFP/F939, BS/U6/GFP/F1417, pSilencer2.1/U6/F2141, pSilencer2.1/U6/F2850, pSilencer/U6/F11689, and pSilencer/U6/F11813) (Fig. 3A) were transiently transfected into IMCD-3 cells. All constructs showed knockdown effects by real-time PCR, but their efficiencies varied greatly (Fig. 3B, left). Considering the possibility of the existence of multiple splicing variants and the splicing variants likely lacking various 3′ fragments, we further tested the effects of two constructs (pSilencer2.1/U6/F2141 and pSilencer2.1/U6/F2850) by cotransfection with a mini-construct containing the target sequence adjacent to the 5′ end of the FPC transcript (Fig. 3B, right). Both constructs had knockdown effects on the exogenous FPC fusion protein, and pSilencer2.1/U6/F2850 completely eliminated the expression of exogenous FPC. Thus, we used pSilencer2.1/U6/F2141 and pSilencer2.1/U6/F2850 to make FPC knockdown stable cells. For pSilencer2.1/U6/F2850, 25 clones were selected and 4 were proven to effectively knock down endogenous FPC by RT-PCR (data not shown). By RT-PCR, IP, and IB, one clone, designated F2850S1, showed ∼90% reduction of FPC at the transcript and protein levels (Fig. 3C and 4B). However, IB of FPC immunoprecipitates did not show a significant reduction of FPC-associated PC2 in FPC knockdown cells. IP with PC2 antibody provided similar patterns for PC2-associated FPC in parental and knockdown IMCD-3 cells (Fig. 4B). These data suggested that either PC2 is a preferred binding partner of FPC or FPC knockdown did not affect the isoforms of FPC that bind to PC2.
FIG. 3.
Characterization of FPC stable knockdown in IMCD-3 cells. (A and B) Six siRNA constructs (BS/U6/GFP/F939, BS/U6/GFP/F1417, pSilencer2.1/U6/F2141, pSilencer2.1/U6/F2850, pSilencer2.1/U6/F11689, and pSilencer2.1/U6/F11813) were made and tested for transiently transfected FPC knockdown in IMCD-3 cells (B, left). (B) F2141 and F2850 were also tested by transient cotransfection with the FPC mini-construct pcDNA3.1/mPkhd1NT2 containing the target sequences (right). Compared with the control, five of the six constructs with the exception of F11689 had significant knockdown effects on FPC transcripts, especially F2141 and F11813, by real-time PCR (left). Expression of the FPC recombinant protein was also significantly reduced by the tested constructs (F2141 and F2850), particularly the latter one (right). (C) In stable FPC knockdown cells (F2850S1), FPC transcripts were significantly reduced (90%) compared to those in the parental cells (control) by real-time and standard RT-PCR. In F2141S26 knockdown cells, FPC transcripts were dramatically decreased compared to those in the control cells (WT4) stably transfected with empty vector. TM, nucleotides for the transmembrane domain; α-myc, anti-myc.
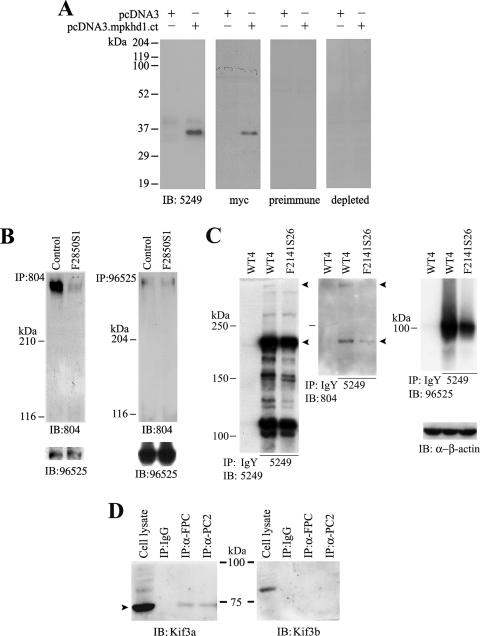
FIG. 4.
Characterization of a new anti-mouse FPC antibody and FPC and PC2 interaction in FPC knockdown IMCD-3 cells. (A) Affinity-purified antipeptide antibody (5249) was raised against the intracellular domain of mouse FPC. Cell lysates of HEK 293T cells, transfected with the cytosolic C-terminal tail of mouse FPC (pcDNA3.mpkhd1.ct) with a myc tag or empty vector, were blotted with the indicated antibodies. Both 5249 and anti-myc tag (α-myc) antibodies, but not preimmune and antibody-depleted IgY, recognized the recombinant FPC fragment. (B and C) PC2-FPC interaction in FPC knockdown cells. (B) IB following IP with antibody to the N terminus of FPC (804) revealed a significant reduction of FPC. However, the amounts of FPC-associated PC2 were similar in control and F2850S1 knockdown cells (left). With PC2 IP, no obvious difference of either PC2 or FPC was noticed between control and knockdown cells (right). (C) PC2-FPC interaction in another FPC knockdown clone targeted at a different site of Pkhd1 (F2141S26). Two bands (arrowheads) immunoprecipitated by the FPC C-terminal tail antibody 5249 were apparently absent or reduced in the knockdown cells when blotted with both N-terminal (804) and C-terminal (5249) antibodies. Blotting with PC2 antibody (96525) showed a slight reduction of FPC-associated PC2 in this knockdown clone, compared to that in cells expressing the siRNA vector alone (WT4). IgY was used as a control for IP, and β-actin was used as a loading control for cell lysates used for IP. (D) Detection of Kif3a (arrowhead) and Kif3b in immunoprecipitates of FPC and PC2 in MDCK cells. Cell lysate and IgG were used as controls.
In order to further test this hypothesis, we made another FPC knockdown cell line, F2141S26. A new antibody raised against the C-terminal tail of mouse FPC (5249), which recognized a recombinant protein of the mouse FPC C-terminal tail (Fig. 4A), precipitated a number of bands, and the band with the highest molecular mass, likely the endogenous full-length FPC, was apparently absent in the knockdown cells, along with a reduction of the 230-kDa band which may be an alternatively spliced form of FPC. Both bands were detected by Western blotting with both N-terminal (804) and C-terminal (5249) FPC antibodies. Blotting with PC2 antibody (96525) showed a mild reduction of FPC-associated PC2 in this clone compared to that in cells expressing the siRNA vector alone (WT4) (Fig. 4C), possibly due to the presence of multiple splicing variants as suggested previously (27). We believe that some, if not all, bands precipitated by 5249 are posttranscriptional or posttranslational forms of FPC, although some cross-reactivity to proteins with similar epitopes cannot be completely excluded.
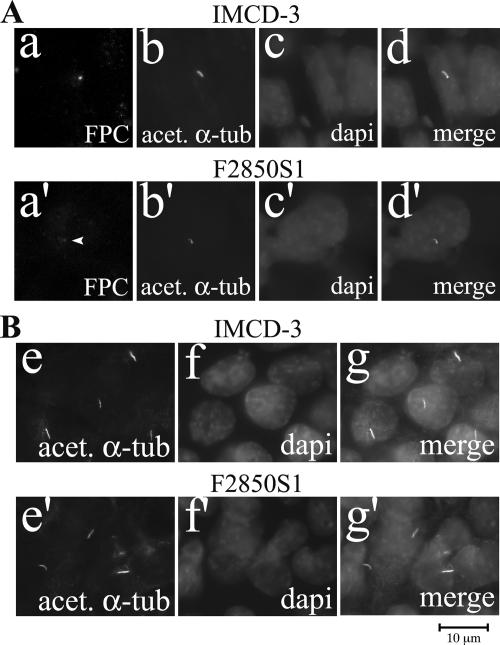
To evaluate the effect of FPC knockdown in F2850S1 and F2141S26 cells, we examined the localization of FPC in parental and FPC knockdown cells. A reduction of FPC signal in the basal body area was observed in F2850S1 (Fig. 5A) and F2141S26 (data not shown) cells compared to that in parental cells.
FIG. 5.
Effect of FPC knockdown on ciliogenesis. (A) In IMCD-3 cells, FPC was expressed mainly in the basal body area (a and a′) and partially colocalized with acetylated α-tubulin (acet. α-tub) (b and b′). FPC staining in the basal body area was moderately decreased in FPC knockdown cells (d and d′). (B) With acetylated α-tubulin staining (e and e′), the number and length of the primary cilia were not affected by FPC knockdown. Nuclei were stained with DAPI (c, c′, f, and f′). Panels d, d′, g, and g′ are merged images.
Effects of FPC knockdown on ciliogenesis.
We calculated the cilium number by counting 100 cells in both parental cells and F2850S1 cells. No significant difference of cilium number between these two groups was found. Comparison of cilium lengths in cells with and without knockdown of FPC revealed no significant changes (Fig. 5B). In F2141S26 cells, no obvious changes of cilium length and number were found (data not shown).
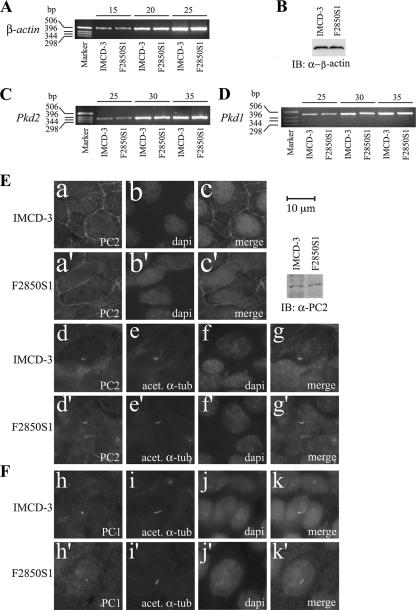
Effects of FPC knockdown on PC2 and PC1.
The effects of FPC knockdown on β-actin were studied at both the transcript and protein levels. Neither transcripts nor proteins of β-actin were affected by knockdown of FPC (Fig. 6A and B). There was no significant impact of FPC knockdown on PC1 and PC2 at the transcription level (Fig. 6C and D). By immunostaining, we found no changes in PC2 and PC1 expression and subcellular localization (Fig. 6E and F). No differences were found in PC2 expression levels between parental and FPC knockdown IMCD-3 cells by Western analyses (Fig. 6E).
FIG. 6.
Effects of FPC knockdown on PC2 and PC1. (A and B) Transcripts and protein expression of β-actin were evaluated by semiquantitative RT-PCR and IB as a control. No difference was found between parental IMCD-3 cells and FPC knockdown cells (F2850S1). PC1 and PC2 were also examined by semiquantitative RT-PCR (C and D), IB, and immunostaining (E and F). (C and D) Transcript levels of neither PC2 nor PC1 were changed after FPC knockdown. The upper numbers (15, 20, 25, 30, and 35) indicate the number of PCR cycles. (E) By immunocytochemistry, plasma membrane localization patterns of PC2 appeared to be similar between IMCD-3 and F2850S1 cells (a and a′). The cilium staining pattern and intensity of PC2 (E, d and d′) and PC1 (F, h and h′) were not changed with knockdown of FPC. (E) No changes in PC2 expression were confirmed with IB of parental IMCD-3 and F2850S1 cells. The cilium was stained with acetylated antitubulin (acet. α-tub) (E, e and e′, and F, i and i′). Nuclei (b, b′, f, f′, j, and j′) were stained with DAPI. Panels c, c′, g, g′, k, and k′ are merged images.
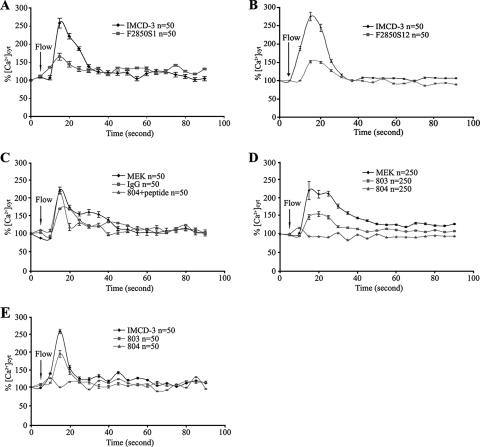
Effects of FPC knockdown and antibody blockage on intracellular calcium responses to fluid flow shear stress.
We first examined the effect of FPC reduction on intracellular calcium responses in F2850S1 and F2850S12 cells. Interestingly, these cells remained responsive to fluid flow shear stress compared with the control group (Fig. 7A and B). Two GFP-positive siRNA constructs (BS/U6/GFP/F939 and BS/U6/GFP/F1417) gave results similar to those for F2850S1 and F2850S12 (data not shown). Because both FPC and PC2 are expressed at the ciliary shaft and in the basal body area of MEK cells that are well characterized for a flow assay and are derived from the embryonic kidney when ARPKD cysts initiate, and IMCD-3 cells are of adult origin, we chose to test the effects of FPC antibody on intracellular calcium responses in MEK cells. With the increasing concentration of 804, intracellular calcium responses decreased in a concentration-dependent manner, and at a 1:50 dilution of the antibody (26 μg/ml), the response was blocked completely (Fig. 7D). Importantly, antibody 803, raised to intracellular epitopes of FPC that were not accessible, and normal rabbit IgG, at the same concentration as 804, did not block the calcium responses to fluid flow shear stress under all tested dosages. Furthermore, preincubation of antibody 804 with its peptide immunogen did not block the calcium responses in MEK cells (Fig. 7C). These data provided further evidence on the specificity of the effects of antibody 804. Similar effects of antibodies 803 and 804 were also obtained with IMCD-3 cells (Fig. 7E). All the experiments were repeated at least two times.
FIG. 7.
Effects of FPC knockdown and FPC antibodies on intracellular calcium responses to fluid flow-induced sheer stress. (A and B) In two IMCD-3 clones, F2850S1 and F2850S12, intracellular calcium responses were reduced compared with those in the parental IMCD-3 cells. A mean of 50 cells in one representative experiment out of five is shown. (C) The application of normal rabbit IgG or 804 antibody preincubated with its peptide immunogen had no effects on the calcium response in MEK cells. (D) Flow-induced calcium response was abolished in MEK cells incubated with antibody 804 (to extracellular epitopes of FPC) but not 803 (to intracellular epitopes of FPC). The results of the antibody experiments with MEK cells show the averages for the five independent experiments performed on different days (50 cells were randomly chosen for each experiment). (E) Antibodies 804 and 803 had similar effects on the calcium response in IMCD-3 cells, as in MEK cells. The results of one representative experiment out of three are shown. %[Ca2+]cyt, percent increase in cytosolic calcium.
DISCUSSION
We previously reported that FPC was localized to the shaft of the primary cilium, with concentration in the basal body area (33). We herein report that a pool of FPC is localized at the plasma membrane. In normal human kidney tissues, we identified FPC expression in the distal parts of tubules, i.e., the collecting tubules and ducts, a distribution pattern in agreement with pathological features of ARPKD and previous reports (22, 35, 41). Prolonged differentiation in MDCK cells resulted in ciliary shaft expression of FPC and an elevation of its expression, indicating that the subcellular localization of FPC is dynamic and likely associated with cellular differentiation. In IMCD-3 cells, we found FPC in the basal body area but not at the plasma membrane, and this finding is in agreement with a recent report (41). In MEK cells, FPC was detected at the plasma membrane and in the basal body as in MDCK cells (33). The differences of FPC subcellular localization in these two mouse cell lines may reflect the existence of a highly regulated specific domain between the ciliary shaft and the basal body area, restricting the entrance of proteins to the ciliary shaft compartment. The regulations may be affected by the developmental stage origin, the effect of the transformation, culture conditions, and fluid stimulations. Expression of FPC at the basolateral membrane both in the kidney tubules and in cultured cells suggests a role for FPC in cell-cell and cell-matrix interactions, where it may function as an adhesive molecule in concert with other junction molecules, such as the cadherin family members. Very recently, it was reported that disruption of laminin 5, an extracellular matrix protein, led to cystic kidney disease (31). Thus, the potential roles of FPC in tubulogenesis and cystogenesis deserve to be studied in detail.
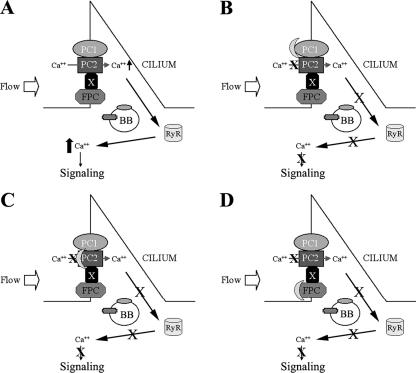
Colocalization of FPC and PC2 at the plasma membrane and on the cilium led us to hypothesize FPC-PC2 interaction. Reciprocal coimmunoprecipitation experiments verified this hypothesis. A remaining question is whether this interaction is direct or mediated by a third molecule. Our in vivo studies of FPC-PC2 in yeast did not support a direct interaction between their intracellular domains; however, the possibility for a direct interaction remains to be excluded, as physical interactions between the extracellular domains of FPC and PC2 may occur. Our finding that Kif3a but not Kif3b was coimmunoprecipitated by both FPC and PC2 antibodies indicates that Kif3a is likely to be one of the molecular candidates mediating the interaction between FPC and PC2. Because Kif3a and Kif3b usually function as a heterodimer, these results are in agreement with a recent report (36). Studies of interactions between the extracellular domains of these molecules and the identification of molecules binding to both FPC and PC2 will further clarify the nature of this interaction (Fig. 8).
FIG. 8.
A model of the polycystin-FPC complex at the primary cilium. (A) Upon fluid flow stimulation, mechanical shear stress is transmitted into the cell via membrane proteins on the cilium, which consequently activates a calcium channel such as PC2 and allows calcium entry and subsequently activates the ryanodine receptor (RyR) through a calcium-induced calcium release intracellular signaling mechanism. (B and C) Disruption of the PC1/PC2 protein complex abolishes the Ca2+ entry signal. (D) Blocking of FPC may diminish the Ca2+ signal through the modulation of the polycystin complex and downstream effectors such as RyR. BB, basal body; X, unknown molecule(s).
In order to explore the effects of FPC knockdown on ciliogenesis, expression of PC1 and PC2, and their functional cross talk, we stably knocked down FPC in IMCD-3 cells via RNA interference (6). Unlike the biliary ductal epithelial cells (20, 21), however, successful reduction of FPC by ∼90% at the RNA and protein levels did not result in obvious defects in ciliogenesis, suggesting that either ∼10% of the residual FPC is sufficient for normal ciliogenesis or there might be other specific splicing events of Pkd1 as suggested previously (27). The latter is certainly true for Pkhd1-like molecules (39). Indeed, knockout of Pkhd1 resulted in liver but not kidney cysts (24). We do not exclude a subtle change in cilium length that is beyond the resolution of our detection method. Because ADPKD and ARPKD share common clinical and pathological features, we were interested in determining the effect of FPC knockdown on PC1/PC2 expression and trafficking. RT-PCR, IB, and immunostaining did not yield any hints that PC1/PC2 was altered, suggesting that the integrity of FPC does not affect the expression levels of polycystins.
Because no morphological differences of the primary cilium were found between FPC knockdown cells and the parental cells, we went on to determine whether cilium dysfunction existed as in Pkd1 knockout cells. In FPC knockdown cells, intracellular calcium responses to flow were not abolished. This observation was unexpected but not surprising because ARPKD is a recessive disease that results from the functional loss of FPC from both alleles, and the knockdown of FPC did not completely remove FPC in IMCD-3 cells. To complement this approach, we used FPC antibodies to study the cellular calcium responses to fluid flow, which we successfully used in studying PC2 function in flow-induced calcium responses (26). Because the density of FPC molecules on a primary cilium is unknown, we tested the effects of two antibodies (803 and 804), raised, respectively, to intra- and extracellular epitopes of FPC, at different dosages. In contrast to the absence of effects on flow-induced calcium responses with 803, a dose-dependent blockage effect of 804 was evident. Complete blocking of flow-induced calcium signal was achieved when 804 was used at a dilution of 1:50. These data were obtained with both MEK and IMCD-3 cells. Therefore, disruption of either FPC or the PC1/PC2 complex at the primary cilium results in defects in mechanosensation of fluid flow. Future studies on the molecular details of FPC interaction with polycystins and the ARPKD and ADPKD signaling pathways should shed light on the molecular mechanism of cilium-mediated mechanochemical signaling.
Acknowledgments
We thank L. Contrino, Q. Xi, and P. Finnerty in the laboratory of J. Zhou for technical assistance.
This work was supported by grants from the National Institutes of Health to J. Zhou (DK40703, DK53357, and P50DK074030). S. Wang was a recipient of a postdoctoral fellowship from the American Heart Association.
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Bergmann, C., F. Kupper, C. Dornia, F. Schneider, J. Senderek, and K. Zerres. 2005. Algorithm for efficient PKHD1 mutation screening in autosomal recessive polycystic kidney disease (ARPKD). Hum. Mutat. 25:225-231. [DOI] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Cai, Y., Y. Maeda, A. Cedzich, V. E. Torres, G. Wu, T. Hayashi, T. Mochizuki, J. H. Park, R. Witzgall, and S. Somlo. 1999. Identification and characterization of polycystin-2, the PKD2 gene product. J. Biol. Chem. 274:28557-28565. [DOI] [PubMed] [Google Scholar]
- 4.Calvet, J. P., and J. J. Grantham. 2001. The genetics and physiology of polycystic kidney disease. Semin. Nephrol. 21:107-123. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 6.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 7.Foggensteiner, L., A. P. Bevan, R. Thomas, N. Coleman, C. Boulter, J. Bradley, O. Ibraghimov-Beskrovnaya, K. Klinger, and R. Sandford. 2000. Cellular and subcellular distribution of polycystin-2, the protein product of the PKD2 gene. J. Am. Soc. Nephrol. 11:814-827. [DOI] [PubMed] [Google Scholar]
- 8.Fonck, C., D. Chauveau, M. F. Gagnadoux, Y. Pirson, and J. P. Grunfeld. 2001. Autosomal recessive polycystic kidney disease in adulthood. Nephrol. Dial Transplant 16:1648-1652. [DOI] [PubMed] [Google Scholar]
- 9.Geng, L., Y. Segal, B. Peissel, N. Deng, Y. Pei, F. Carone, H. G. Rennke, A. M. Glucksmann-Kuis, M. C. Schneider, M. Ericsson, S. T. Reeders, and J. Zhou. 1996. Identification and localization of polycystin, the PKD1 gene product. J. Clin. Investig. 98:2674-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanaoka, K., F. Qian, A. Boletta, A. K. Bhunia, K. Piontek, L. Tsiokas, V. P. Sukhatme, W. B. Guggino, and G. G. Germino. 2000. Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature 408:990-994. [DOI] [PubMed] [Google Scholar]
- 11.Hiesberger, T., Y. Bai, X. Shao, B. T. McNally, A. M. Sinclair, X. Tian, S. Somlo, and P. Igarashi. 2004. Mutation of hepatocyte nuclear factor-1beta inhibits Pkhd1 gene expression and produces renal cysts in mice. J. Clin. Investig. 113:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, J., C. J. Ward, B. Peral, R. Aspinwall, K. Clark, J. L. San Millan, V. Gamble, and P. C. Harris. 1995. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 10:151-160. [DOI] [PubMed] [Google Scholar]
- 13.Ibraghimov-Beskrovnaya, O., W. R. Dackowski, L. Foggensteiner, N. Coleman, S. Thiru, L. R. Petry, T. C. Burn, T. D. Connors, T. Van Raay, J. Bradley, F. Qian, L. F. Onuchic, T. J. Watnick, K. Piontek, R. M. Hakim, G. M. Landes, G. G. Germino, R. Sandford, and K. W. Klinger. 1997. Polycystin: in vitro synthesis, in vivo tissue expression, and subcellular localization identifies a large membrane-associated protein. Proc. Natl. Acad. Sci. USA 94:6397-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.International Polycystic Kidney Disease Consortium. 1995. Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell 81:289-298. [DOI] [PubMed] [Google Scholar]
- 16.Li, X., Y. Luo, P. G. Starremans, C. A. McNamara, Y. Pei, and J. Zhou. 2005. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat. Cell Biol. 7:1102-1112. [DOI] [PubMed] [Google Scholar]
- 17.Loghman-Adham, M., S. M. Nauli, C. E. Soto, B. Kariuki, and J. Zhou. 2003. Immortalized epithelial cells from human autosomal dominant polycystic kidney cysts. Am. J. Physiol. Renal Physiol. 285:F397-F412. [DOI] [PubMed] [Google Scholar]
- 18.Luo, Y., P. M. Vassilev, X. Li, Y. Kawanabe, and J. Zhou. 2003. Native polycystin 2 functions as a plasma membrane Ca2+-permeable cation channel in renal epithelia. Mol. Cell. Biol. 23:2600-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mai, W., D. Chen, T. Ding, I. Kim, S. Park, S. Y. Cho, J. S. Chu, D. Liang, N. Wang, D. Wu, S. Li, P. Zhao, R. Zent, and G. Wu. 2005. Inhibition of Pkhd1 impairs tubulomorphogenesis of cultured IMCD cells. Mol. Biol. Cell 16:4398-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masyuk, T. V., B. Q. Huang, A. I. Masyuk, E. L. Ritman, V. E. Torres, X. Wang, P. C. Harris, and N. F. LaRusso. 2004. Biliary dysgenesis in the PCK rat, an orthologous model of autosomal recessive polycystic kidney disease. Am. J. Pathol. 165:1719-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masyuk, T. V., B. Q. Huang, C. J. Ward, A. I. Masyuk, D. Yuan, P. L. Splinter, R. Punyashthiti, E. L. Ritman, V. E. Torres, P. C. Harris, and N. F. LaRusso. 2003. Defects in cholangiocyte fibrocystin expression and ciliary structure in the PCK rat. Gastroenterology 125:1303-1310. [DOI] [PubMed] [Google Scholar]
- 22.Menezes, L. F., Y. Cai, Y. Nagasawa, A. M. Silva, M. L. Watkins, A. M. Da Silva, S. Somlo, L. M. Guay-Woodford, G. G. Germino, and L. F. Onuchic. 2004. Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int. 66:1345-1355. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki, T., G. Wu, T. Hayashi, S. L. Xenophontos, B. Veldhuisen, J. J. Saris, D. M. Reynolds, Y. Cai, P. A. Gabow, A. Pierides, W. J. Kimberling, M. H. Breuning, C. C. Deltas, D. J. Peters, and S. Somlo. 1996. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272:1339-1342. [DOI] [PubMed] [Google Scholar]
- 24.Moser, M., S. Matthiesen, J. Kirfel, H. Schorle, C. Bergmann, J. Senderek, S. Rudnik-Schoneborn, K. Zerres, and R. Buettner. 2005. A mouse model for cystic biliary dysgenesis in autosomal recessive polycystic kidney disease (ARPKD). Hepatology 41:1113-1121. [DOI] [PubMed] [Google Scholar]
- 25.Nagasawa, Y., S. Matthiesen, L. F. Onuchic, X. Hou, C. Bergmann, E. Esquivel, J. Senderek, Z. Ren, R. Zeltner, L. Furu, E. Avner, M. Moser, S. Somlo, L. Guay-Woodford, R. Buttner, K. Zerres, and G. G. Germino. 2002. Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J. Am. Soc. Nephrol. 13:2246-2258. [DOI] [PubMed] [Google Scholar]
- 26.Nauli, S. M., F. J. Alenghat, Y. Luo, E. Williams, P. Vassilev, X. Li, A. E. Elia, W. Lu, E. M. Brown, S. J. Quinn, D. E. Ingber, and J. Zhou. 2003. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33:129-137. [DOI] [PubMed] [Google Scholar]
- 26a.Nauli, S. M., and J. Zhou. 2004. Polycystins and mechanosensation in renal and nodal cilia. Bioessays 26:844-856. [DOI] [PubMed] [Google Scholar]
- 27.Onuchic, L. F., L. Furu, Y. Nagasawa, X. Hou, T. Eggermann, Z. Ren, C. Bergmann, J. Senderek, E. Esquivel, R. Zeltner, S. Rudnik-Schoneborn, M. Mrug, W. Sweeney, E. D. Avner, K. Zerres, L. M. Guay-Woodford, S. Somlo, and G. G. Germino. 2002. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am. J. Hum. Genet. 70:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters, D. J., A. van de Wal, L. Spruit, J. J. Saris, M. H. Breuning, J. A. Bruijn, and E. de Heer. 1999. Cellular localization and tissue distribution of polycystin-1. J. Pathol. 188:439-446. [DOI] [PubMed] [Google Scholar]
- 28a.Reeders, S. T., M. H. Breuning, K. E. Davies, R. D. Nicholls, A. P. Jarman, D. R. Higgs, P. L. Pearson, and D. J. Weatherall. 1985. A highly polymorphic DNA marker linked to adult polycystic kidney disease on chromosome 16. Nature 317:542-544. [DOI] [PubMed] [Google Scholar]
- 29.Roy, S., M. J. Dillon, R. S. Trompeter, and T. M. Barratt. 1997. Autosomal recessive polycystic kidney disease: long-term outcome of neonatal survivors. Pediatr. Nephrol. 11:302-306. [DOI] [PubMed] [Google Scholar]
- 30.Shaikewitz, S. T., and A. Chapman. 1993. Autosomal recessive polycystic kidney disease: issues regarding the variability of clinical presentation. J. Am. Soc. Nephrol. 3:1858-1862. [DOI] [PubMed] [Google Scholar]
- 31.Shannon, M. B., B. L. Patton, S. J. Harvey, and J. H. Miner. 2006. A hypomorphic mutation in the mouse laminin alpha5 gene causes polycystic kidney disease. J. Am. Soc. Nephrol. 17:1913-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, S., Y. Luo, P. D. Wilson, G. B. Witman, and J. Zhou. 2004. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J. Am. Soc. Nephrol. 15:592-602. [DOI] [PubMed] [Google Scholar]
- 34.Ward, C. J., M. C. Hogan, S. Rossetti, D. Walker, T. Sneddon, X. Wang, V. Kubly, J. M. Cunningham, R. Bacallao, M. Ishibashi, D. S. Milliner, V. E. Torres, and P. C. Harris. 2002. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 30:259-269. [DOI] [PubMed] [Google Scholar]
- 35.Ward, C. J., D. Yuan, T. V. Masyuk, X. Wang, R. Punyashthiti, S. Whelan, R. Bacallao, R. Torra, N. F. LaRusso, V. E. Torres, and P. C. Harris. 2003. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 12:2703-2710. [DOI] [PubMed] [Google Scholar]
- 36.Wu, Y., X. Q. Dai, Q. Li, C. X. Chen, W. Mai, Z. Hussain, W. Long, N. Montalbetti, G. Li, R. Glynne, S. Wang, H. F. Cantiello, G. Wu, and X. Z. Chen. 2006. Kinesin-2 mediates physical and functional interactions between polycystin-2 and fibrocystin. Hum. Mol. Genet. 15:3280-3292. [DOI] [PubMed] [Google Scholar]
- 37.Xiong, H., Y. Chen, Y. Yi, K. Tsuchiya, G. Moeckel, J. Cheung, D. Liang, K. Tham, X. Xu, X. Z. Chen, Y. Pei, Z. J. Zhao, and G. Wu. 2002. A novel gene encoding a TIG multiple domain protein is a positional candidate for autosomal recessive polycystic kidney disease. Genomics 80:96-104. [DOI] [PubMed] [Google Scholar]
- 38.Yoder, B. K., X. Hou, and L. M. Guay-Woodford. 2002. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 13:2508-2516. [DOI] [PubMed] [Google Scholar]
- 39.Yuasa, T., A. Takakura, B. M. Denker, B. Venugopal, and J. Zhou. 2004. Polycystin-1L2 is a novel G-protein-binding protein. Genomics 84:126-138. [DOI] [PubMed] [Google Scholar]
- 40.Zerres, K., S. Rudnik-Schoneborn, C. Steinkamm, J. Becker, and G. Mucher. 1998. Autosomal recessive polycystic kidney disease. J. Mol. Med. 76:303-309. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, M. Z., W. Mai, C. Li, S. Y. Cho, C. Hao, G. Moeckel, R. Zhao, I. Kim, J. Wang, H. Xiong, H. Wang, Y. Sato, Y. Wu, Y. Nakanuma, M. Lilova, Y. Pei, R. C. Harris, S. Li, R. J. Coffey, L. Sun, D. Wu, X. Z. Chen, M. D. Breyer, Z. J. Zhao, J. A. McKanna, and G. Wu. 2004. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc. Natl. Acad. Sci. USA 101:2311-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]