Abstract
Bone morphogenetic protein 2 (encoded by Bmp2) has been implicated as an important signaling ligand for osteoblast differentiation and bone formation and as a genetic risk factor for osteoporosis. To initially survey a large genomic region flanking the mouse Bmp2 gene for cis-regulatory function, two bacterial artificial chromosome (BAC) clones that extend far upstream and downstream of the gene were engineered to contain a lacZ reporter cassette and tested in transgenic mice. Each BAC clone directs a distinct subset of normal Bmp2 expression patterns, suggesting a modular arrangement of distant Bmp2 regulatory elements. Strikingly, regulatory sequences required for Bmp2 expression in differentiating osteoblasts, as well as tooth buds, hair placodes, kidney, and other tissues, are located more than 53 kilobases 3′ to the promoter. By testing BACs with engineered deletions across this distant 3′ region, we parsed these regulatory elements into separate locations and more closely refined the location of the osteoblast progenitor element. Finally, a conserved osteoblast progenitor enhancer was identified within a 656-bp sequence located 156.3 kilobases 3′ from the promoter. The identification of this enhancer should permit further investigation of upstream regulatory mechanisms that control Bmp2 transcription during osteoblast differentiation and are relevant to further studies of Bmp2 as a candidate risk factor gene for osteoporosis.
The bone morphogenetic protein (BMP) family of secreted signaling molecules was originally identified based on their osteoinductive properties and their ability to promote osteoblast differentiation and bone formation (9, 74). Since their initial discovery, BMPs have also been implicated as critical determinants for numerous developmental processes, including those involved in soft tissue development (14, 28, 39, 40). Several lines of evidence show that some BMP family members employ multiple and sometimes distant cis-regulatory elements to achieve precise spatiotemporal transcription, such as the Drosophila dpp and vertebrate Bmp5 and Gdf6 genes (5, 12, 13, 53). These elements are often arranged in distinct regulatory modules (3, 17, 31) and can be dispersed over tens or hundreds of kilobases upstream, downstream, or within neighboring genes. Therefore genes with highly tissue-specific expression, such as signaling molecules or transcription factors, can require multiple and sometimes distant genomic sequences to direct normal patterns of gene expression in specific tissues (13, 36, 41, 44, 45, 49, 53, 56).
The expression of Bmp2 during development has been extensively characterized, and it plays many roles in pattern formation and morphogenesis, cell fate determination, and/or differentiation. Bmp2 is expressed dynamically in numerous embryonic locations, such as kidneys, hair follicles, tooth buds, and gut epithelium (4, 16, 43, 51, 52). The involvement of Bmp2 in limb patterning is evidenced by its expression in the apical ectodermal ridge (AER) and limb bud mesenchyme (28, 50). From 8.5 to 9.5 days postcoitus (dpc), Bmp2 is transcribed in extraembryonic tissues and heart mesoderm (51, 75), and the early embryonic lethality observed in Bmp2 knockout mice is probably due to defects in these tissues (75). There is extensive evidence for the involvement of Bmp2 in bone and cartilage formation. In developing endochondral bones, Bmp2 is expressed in hypertrophic chondrocytes of the growth plates and also in the osteogenic perichondrium (52, 60, 65). Bmp2 can also stimulate signaling cascades that are required for osteoblast differentiation (9). The recently reported genetic linkage and association of osteoporosis (OP) to the human BMP2 gene also supports its role in human bone homeostasis and disease (67). In addition, Bmp2 is of tremendous interest as a therapeutic agent for OP, fracture healing, and other ailments for which increased bone formation would be beneficial (9, 55, 69).
Despite the potential importance of Bmp2 for the proper development of bone and other tissues, little is known about the molecular mechanisms that control when and where Bmp2 is expressed in vivo. Previous reports have described the regulatory functions of Bmp2 promoter fragments in cell lines (1, 18, 23, 27, 68, 76), but these provide only limited information regarding how Bmp2 transcription is controlled in vivo. A transgene containing a 2.7-kilobase Bmp2 promoter fragment has been reported to drive reporter gene expression in skeletal perichondrium and chondrocytes in vivo (18), suggesting that some important elements for Bmp2 skeletal expression lie close to the promoter. However, the same transgene failed to confer many other sites of Bmp2 expression (S. Harris, personal communication). On the other hand, several lines of evidence suggest Bmp2 might be controlled by multiple, long-range regulatory elements outside the promoter region. First, the complex patterns of Bmp2 expression are reminiscent of those for Bmp5, Gdf6, and dpp, which are other BMP genes that each have multiple, distant cis-regulatory elements (5, 12, 13, 53). Second, Bmp2 is flanked by “stable gene deserts” (i.e., large genomic regions that are devoid of other genes yet conserved across species) (58), suggesting that evolution has preserved an arrangement of distant cis-regulatory elements flanking Bmp2.
To test the hypothesis that Bmp2 is regulated by distant cis-acting sequences in vivo, we used bacterial artificial chromosome (BAC) transgenes to survey a region of approximately 400 kilobases surrounding the mouse Bmp2 gene. To do this we engineered Bmp2 BAC clones to contain a lacZ reporter cassette and then tested the BACs in transgenic embryos for their ability to direct normal patterns of Bmp2 expression in mid-gestation mouse embryos. Here we show that individual BAC clones that extend far 5′ or 3′ to the Bmp2 gene can direct distinct subsets of Bmp2 expression patterns in vivo, strongly suggesting that different cis-acting regulatory sequences are located in separate genomic regions flanking either side of Bmp2. Surprisingly, cis-regulatory sequences that control Bmp2 expression in hypertrophic chondrocytes are very far from those controlling expression in osteoblast progenitors, which are in a distant 3′ flanking region. We also found that in the osteogenic perichondrium of the endochondral bones, Bmp2 expression is closely preceded temporally by the key osteoblast factor Runx2. By testing additional Bmp2 BACs with engineered deletions across the 3′ region, we more closely refined the location of the osteoblast progenitor regulatory element, and we could also parse distinct elements controlling Bmp2 expression in tooth buds, hair placodes, kidney, and other tissues into four separate genomic regions 3′ to Bmp2. Finally, using heterologous minigenes, we identified a conserved osteoblast progenitor Bmp2 enhancer located approximately 156 kb from the promoter. Taken together, these findings demonstrate that numerous and distant regulatory sequences direct Bmp2 expression during development and that Bmp2 transcription in differentiating osteoblasts is controlled by a distant 3′ enhancer.
MATERIALS AND METHODS
Bmp2 BACs.
Mouse BAC clones RP23-85011 and RP23-409L24 from C57BL/6J mouse BAC library 23 were obtained from the Children's Hospital Oakland Research Institute. BAC clones RP23-85011 and RP23-409L24 were renamed 5′ BAC and 3′ BAC, respectively, for this study. Both Bmp2 BACs were verified by direct end sequencing at the vector-insert junction and standard gel electrophoresis techniques to compare BamHI restriction digest patterns to those of the predicted insert sequences (35, 73).
BAC IRES-β-geo modifications.
An internal ribosome entry site (IRES)-β-geo cassette was inserted into each Bmp2 BAC, in place of Bmp2 exon 3 mature-region-coding sequences, using the homologous recombination technique of Lee et al. (47) as follows. The plasmid pIBG-FTet was generated by ligating the IRES-β-geo-SV40pA cassette from pGT1.8 (54) and an FRT-flanked tetracycline resistance cassette into a modified pBluescript II SK(+) backbone. The recombination cassette was constructed by subcloning a 50-bp 5′ recombination arm overlapping part of Bmp2 intron 2 and part of exon 3 and a 50-bp 3′ recombination arm containing part of exon 3 into pIBG-FTet such that the recombination arms flanked the IRES-β-geo-FRT-Tet-FRT cassette. The forward strand (relative to Bmp2) sequences of the 50-bp homology arms were as follows: for the 5′ arm, TTGCACCAAGATGAACACAGCTGGTCACAGATAAGGCCATTGCTAGTGACT; for the 3′ arm, GTGTCGTTAGCACAGCAAGAATAAATAAATAAATATATATATTTTAGAAAC. Both recombination arms were created by annealing polyacrylamide gel electrophoresis (PAGE)-purified oligonucleotides designed to allow overhanging ends for direct ligation to pIBG-FTet. The final cassette with recombination arms was digested from the vector, gel purified, and then recombined with Bmp2 BACs as described previously (47). Successful recombinants were selected by tetracycline resistance. The tetracycline resistance gene was then removed by l-arabinose promoter-driven FLpe recombinase excision (47, 53). Correctly modified BACs were verified by conventional and pulsed-field gel analysis of restriction digests to confirm expected banding patterns, as well as direct BAC sequencing.
Bmp2 3′ deletion lacZ-BAC modifications.
Four 3′ deletion lacZ-BACs were engineered using the mouse 3′ BAC (RP23-409L24) that contains the lacZ insertion (as described above) and the galK homologous recombination technique of Warming et al. (72) as follows. First, the galK gene was used as a selection cassette and inserted into the 3′ lacZ-BAC. Selection cassettes used for targeting the 3′ lacZ-BAC deletions were generated by PCR amplification of the galK gene with PAGE-purified primers, which also contain homology flanking the genomic regions deleted. The primer sequences (5′→3′) used are as follows (homology arms are in uppercase, and sequences that anneal within galK gene are in lowercase): for deletion 1 (D1), TGGGCACAGAGAGCAGAGCTAACAGTTGAGTGGGGAGTGTCTAAGAATTCcctgttgacaattaatcatcggca (forward) and CAATTGTATATACCCAGTGACTTTCCCTGATTCACTGGGGAGACATGGGAtcagcactgtcctgctcctt (reverse); for D2, TGTGTGTGTGTGTGTGCTTTTTTAGACTAAGGTTGGCAATGTTCCTAAGGcctgttgacaattaatcatcggca (forward) and AATGGGGAGGTGGACTTGATAGATGTGTACACTCTGCCTATTGTGTACATtcagcactgtcctgctcctt (reverse); for D3, CACAATATCAAAATGTGTCAATGGTATGAAGTAGTAGAGCTCATAATGACcctgttgacaattaatcatcggca (forward) and AGTAAGATAGGTGCACATAGAGAAAGGAGAATTGGGACAGACGGACtcagcactgtcctgctcctt (reverse); for D4, CCAAGCCCAGCTGCTTCTGACTGGGTATTCTCTGGTGACCTGTTCTTTGAcctgttgacaattaatcatcggca (forward) and ATTTGCGGCCGCTAATACGACTCACTATAGGGAGAGGATCCGCGGAATTCtcagcactgtcctgctcctt (reverse). Note that the 3′ homology arms used to generate D4 have homology to sequences present in the BAC vector. The final galK cassette with recombination arms was gel purified and then recombined with 3′ lacZ-BAC as described previously (72). Successful recombinants were selected for the ability to use galactose as a carbon source in minimal medium. Second, the galK cassettes were deleted via recombination, by using synthetic DNA fragments made by annealing PAGE-purified oligonucleotides that were homologous to sequences flanking the galK insertion site. The forward-strand (relative to Bmp2) sequences of the annealed oligonucleotides used for substitutions are as follows: for D1, TGGGCACAGAGAGCAGAGCTAACAGTTGAGTGGGGAGTGTCTAAGAATTCTCCCATGTCTCCCCAGTGAATCAGGGAAAGTCACTGGGTATATACAATTG; for D2, TGTGTGTGTGTGTGTGCTTTTTTAGACTAAGGTTGGCAATGTTCCTAAGGATGTACACAATAGGCAGAGTGTACACATCTATCAAGTCCACCTCCCCATT; for D3, CACAATATCAAAATGTGTCAATGGTATGAAGTAGTAGAGCTCATAATGACATGAGTCCGTCTGTCCCAATTCTCCTTTCTCTATGTGCACCTATCTTACT; for D4, CCAAGCCCAGCTGCTTCTGACTGGGTATTCTCTGGTGACCTGTTCTTTGAGAATTCCGCGGATCCTCTCCCTATAGTGAGTCGTATTAGCGGCCGCAAAT. Annealed oligonucleotides with recombination arms were recombined with 3′ galK+ deletion lacZ-BACs as described previously (72). Successful galK-negative recombinants were selected for resistance to 2-deoxygalactose in minimal medium with glycerol as the carbon source. Correctly modified deletion BACs were verified by conventional and pulsed-field gel analysis of restriction digests to confirm expected banding patterns, as well as direct BAC sequencing.
ECR-Hsp promoter-lacZ transgene construction.
Evolutionarily conserved region (ECR)-Hsp68 promoter-lacZ transgenes were made as follows. PCR products corresponding to genomic sequences for ECR-1 + 2 (4.5 kb), ECR-1 (656 bp), and ECR-2 (358 bp) were amplified using 3′ BAC DNA (RP23-409L24) and the following primer sequences: for ECR-1 + 2, AGAGAGAGCTAGATGGTGGAC (forward) and TGCCTTAGTCCTCTCCAGAG (reverse); for ECR-1, CGCTGCTTTTGAATGCTT (forward) and TAGACCCAAGATGGAGTATTAG (reverse); for ECR-2, TGTACTTACTCGATTGTGTGC (forward) and TGCCTTAGTCCTCTCCAGAG (reverse). Each PCR product was ligated into the pGEM-T Easy vector (Promega). A NotI restriction fragment containing each ECR fragment was ligated into pSfi-Hsp68lacZ in the forward orientation (13, 53, 63) to create ECR-Hsp68 promoter-lacZ transgenes. All ECR plasmids were verified by analysis of restriction digests and direct sequencing.
Transgenic mice.
Bmp2 BAC DNAs were purified according to established techniques and used for pronuclear injection of C57BL/6J × DBA/2J F1 hybrid embryos (12). All BACs were injected as uncut circular DNAs. Circular BACs have been shown to be as efficient as linearized BACs for generating transgenic mice (24, 53). Plasmids used for microinjection were prepared as follows. Plasmid DNA was isolated using the QIAGEN Endo-Free Maxi plasmid isolation kit. One hundred micrograms of plasmid DNA was linearized with SalI, and the enzyme was removed using Micropure EZ restriction enzyme removal columns (Millipore). The filtrate containing the DNA was then transferred to Montage PCR filter units (Millipore) and purified according to the manufacturer's protocol. Purified and linearized DNAs were then dialyzed against 3 liters of microinjection buffer (10 mM Tris-HCl [pH 7.4], 0.15 mM EDTA [pH 8.0]) using 10,000-molecular-weight-cutoff Slide-A-Lyzer dialysis cassettes (Pierce). DNAs were further dialyzed and concentrated with 30,000-molecular-weight-cutoff Centriprep centrifugal filter devices (Millipore) prior to microinjection. Injections and oviduct transfers were performed by the Vanderbilt Transgenic Core Facility using standard techniques in accordance with protocols approved by the Vanderbilt University Institutional Animal Care and Use Committee. Transgenic embryos or weanlings were verified by PCR from yolk sac or tail DNAs. All of the BAC-transgenic embryos and lines analyzed for lacZ expression in this study were positive for the β-geo insertion and BAC vector, as determined by both conventional and real-time PCRs.
Bmp2 BAC transgene copy number determination.
Genomic DNAs were harvested from mouse embryo yolk sacs or tail samples from live-born transgenic mice by using standard genomic DNA isolation methods. BAC transgene copy number was estimated using TaqMan-based (Applied Biosystems Inc.) real-time PCR on a 7900HT platform (Applied Biosystems Inc.). All reactions were performed according to the manufacturer's instructions, using 20 ng of mouse genomic DNA. Custom TaqMan primer/probe assays for the neomycin (Neo) resistance gene (β-geo insertion) and the chloramphenicol (Cam) cassette (BAC vector) were provided by Applied Biosystems (Neo and Cam primer/probe sequences are available upon request). For all mouse genomic DNA samples, the Jun gene (GenBank accession no. NM_010591.1; Applied Biosystems Inc. assay identification no. Mm00495062_s1) was used as an internal control. Copy number estimates for either Neo or Cam were derived using the triplicate ΔCT value relative to the internal Jun genomic control and compared to the ΔCT values of standard-curve samples derived from C57BL/6J × DBA/2J F1 hybrid genomic DNAs spiked with known quantities of BAC DNAs diluted to copy number equivalents (1, 2, 4, 8, and 16 BAC copies per diploid genome).
Preparation of agarose-embedded high-molecular-weight DNA from BAC-transgenic embryos.
Mouse embryonic fibroblasts (MEFs) used for the generation of agarose-embedded high-molecular-weight DNA were isolated and cultured as follows. Embryonic day 13.5 embryos were generated by crossing BAC-transgenic males with wild-type CD-1 females. Embryos were dissected at room temperature in sterile phosphate-buffered saline (PBS), and the head and viscera were removed. The head was X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) stained in parallel to identify transgenic embryos. Individual embryos were then forced through an 18-gauge needle into a 10-cm tissue culture dish containing 10 ml of MEF medium (Dulbecco's modified Eagle medium supplemented with 4.5 g/liter glucose, 2 mM sodium pyruvate, 0.5 g/liter l-glutamine, 10% fetal bovine serum, 200 units/liter of penicillin G, 200 μg/liter streptomycin sulfate, and 0.5 μg/liter of amphotericin B). Cells were incubated at 37°C in a 5% CO2 atmosphere and fed every 2 days with MEF medium until they reached confluence. Confluent cells were passaged at 1:3 dilutions with trypsin and propagated in MEF medium until sufficient cell numbers were attained. Confluent MEF cells were washed twice in sterile 1× PBS, disassociated with a rubber policeman, and resuspended in sterile cell suspension buffer (0.1 M EDTA [pH 8.0], 10 mM Tris-HCl [pH 7.6], 20 mM NaCl) at a concentration of approximately 7 × 106 cells/ml. Cells were embedded in 0.75% low-melting-point agarose (Invitrogen) using agarose plug molds (Bio-Rad). Approximately 10 plugs were then placed in 10 ml of lysis buffer (1% sodium dodecyl sulfate, 2 mg/ml proteinase K [Roche], 0.5 M EDTA [pH 8.0]) and incubated overnight in a 50°C water bath with no agitation. Plugs were sequentially washed in 50 ml of the following buffers at room temperature for 30 min each with agitation: twice in 0.1 EX buffer (0.1 M EDTA [pH 8.0], 0.01% Triton X-100), once in 10:10 buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0]), once in proteinase K inactivation buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1 mM phenylmethylsulfonyl fluoride), twice in 10:10 buffer, twice in 0.5 EX buffer (0.5 M EDTA [pH 8.0], 0.01% Triton X-100), and once in 0.5 M EDTA (pH 8.0). The plugs were then stored until use at 4°C in 0.5 M EDTA (pH 8.0).
Southern analysis of high-molecular-weight transgenic DNA.
Agarose plugs (isolated as described above) were washed twice in 50 ml of TEX buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 0.01% Triton X-100) and once in 50 ml of Tris-EDTA (pH 8.0) at room temperature with agitation. Plugs were then transferred to 2-ml screw cap tubes (one plug per tube) and equilibrated for 30 min at room temperature in 2 ml of 1× restriction enzyme buffer with agitation. For NotI enzyme, 10 mM spermidine trihydrochloride was also included in the buffer. Once equilibrated, the solution was replaced with 800 μl of 1× restriction enzyme buffer containing 160 units of NotI or FseI (NEB) and incubated for 6 to 8 h at 4°C on a Nutator rotator to allow the enzyme to infiltrate the agarose plug. After 6 to 8 h, the tubes were transferred to a 37°C dry incubator and incubated for 12 to 16 h with agitation. Plugs were then washed twice in 2 ml TEX buffer at 4°C for 30 min each with agitation and equilibrated at room temperature in 2 ml of 0.5× Tris-borate-EDTA gel electrophoresis buffer. Restriction-digested high-molecular-weight DNA was then resolved by pulsed-field gel electrophoresis in 1% agarose-0.5× Tris-borate-EDTA for 20 h at 15°C (6 V/cm; 6- to 80-s switch time). DNA fragments were depurinated with 0.25 M HCl for 20 min and transferred by downward alkaline capillary transfer onto a Zeta-Probe GT membrane (Bio-Rad) with 0.4 N NaOH for 24 h. The membrane was neutralized with 0.2 M Tris-HCl (pH 7.5)-2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 10 min and baked for 30 min at 80°C. Probe used in hybridizations was generated from a 4.7-kb XbaI fragment containing the IRES-β-geo cassette from pIBG-FTet (described above) and was labeled using Ready-to-Go labeling beads (GE/Amersham) and [α-32P]dCTP (GE/Amersham). Membranes were washed twice with sterile H2O and hybridized for 3 h with Rapid-hyb buffer (GE/Amersham) according to the manufacturer's instructions. Membranes were exposed to a Kodak phosphorimaging screen for 16 h and imaged using a Pharos FX imaging system and software (Bio-Rad).
X-Gal staining and histology.
Whole-mount X-Gal staining was performed as described previously (13, 53). Specimens that were counterstained with alizarin red were first X-Gal stained and then processed for alizarin red staining as follows. Postfixed X-Gal-stained specimens were rinsed in H2O and then cleared in 1.0% KOH for 4 to 6 h at room temperature. Ossifying regions of bone were stained with 0.05 mg/ml alizarin red S (Sigma) in 1.0% KOH for 3 h to overnight. Specimens were then rinsed several times in H2O and cleared in glycerol. Whole-mount X-Gal-stained specimens were prepared for histology by dehydrating through a graded ethanol series, clearing in Citrisolv (Fisher), and embedding in paraffin. Samples were sectioned at 10 μm and collected on Superfrost-Plus slides (Fisher). Sections for histology were counterstained with eosin or nuclear fast red.
In situ hybridization.
In situ hybridizations were performed with probes specific for mouse Bmp2 (52), Runx2, and Spp1. The Bmp2 probe template was kindly provided by B. L. M. Hogan. Mouse Runx2 and Spp1 probe templates were made by PCR amplifying fragments of the 3′ untranslated region of either gene and ligating them into the pGEM T-easy vector (Promega). The 3′ untranslated regions were inspected for nonrepetitive regions suitable for probe design by using the UCSC Genome Browser (35). Primers were as follows: for Runx2, AAAGCTTGCAGAACTCTTAGAATGA (forward) and ATCATATTAAAAAGCCAAGCACAAG (reverse); for Spp1, ACATGAAGAGCGGTGAGTCTAAG (forward) and AAATGCAGTGGCCGTTTG (reverse). Single-stranded antisense and sense RNA probes were labeled with digoxigenin by using either Sp6, T7, or T3 polymerase as described previously (29). Tissues were fixed in 4% paraformaldehyde-1× PBS at 4°C overnight, followed by dehydration through a graded methanol series. Specimens were then cleared in Citrisolv (Fisher) and embedded in paraffin. Samples were sectioned at 10 μm and collected on Superfrost-Plus slides (Fisher). Sections were deparaffinized in Citrisolv and rehydrated in 1× PBS. Tissues were permeabilized by incubation in 3 mg/ml proteinase K-1× PBS for 10 min. Probe hybridizations were performed overnight at 63°C in 40% formamide-5× SSC (pH 7.0) with a probe concentration of 100 ng/ml (Spp1), 150 ng/ml (Bmp2), or 200 ng/ml (Runx2). Posthybridization stringency washes were performed in 2× SSC and 0.2× SSC at 63°C for 1 h. Sections were then blocked with blocking reagent (Roche) and incubated in the presence of a 1:5,000 dilution of antidigoxigenin-alkaline phosphatase Fab fragments (Roche) for 1 h at room temperature. Sections were allowed to develop in an nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate solution containing 6% polyvinyl alcohol. Slides were washed and mounted using aqueous medium.
Immunohistochemistry.
Immunohistochemistry for Runx2 (also known as polyomavirus enhancer binding protein 2α [PEBP2α], Cbfa1, or AML1) was performed using a rabbit polyclonal antibody (PEBP2α A M-70; Santa Cruz Biotechnology, Inc.) (30) specific for Runx2/PEBP2α. Whole-mount X-Gal staining was performed on 10% neutral buffered formalin-fixed embryos. Whole-mount X-Gal-stained specimens were washed with PBS and postfixed in neutral buffered formalin for 3 h at 4°C. Specimens were dehydrated through a graded ethanol series. The material was then cleared in Citrisolv and embedded in paraffin. Samples were sectioned at 10 μm and collected on Superfrost-Plus slides (Fisher). Sections were deparaffinized in Citrisolv and rehydrated into H2O. Sections were placed in 10 mM sodium citrate, slowly heated until boiling, and then cooled to room temperature to aid in antigen retrieval. Endogenous peroxidase activity was quenched by placing the sections in 3% hydrogen peroxide-100% methanol for 30 min. Sections were blocked in a solution containing 3 mg/ml normal goat serum and 1% bovine sheep albumin (Jackson ImmunoResearch, Inc.) and then incubated in the presence of a 1:50 dilution of anti-PEBP2α A/Runx2 antibody for 30 min at room temperature. Secondary detection of rabbit immunoglobulin G was performed using a 1:200 dilution of biotinylated goat anti-rabbit immunoglobulin G (Vector Laboratories). Indirect enzyme-linked immunostaining was performed using a Vectastain ABC-diaminobenzidine substrate kit (Vector Laboratories). Sections were dehydrated and mounted using nonaqueous medium.
Microscopy and imaging.
Sections from X-Gal staining, in situ hybridization, and immunohistochemistry experiments were analyzed and photographed on an Olympus BX51 microscope using bright-field optics and a digital camera.
RESULTS
lacZ-BAC transgene scan to explore Bmp2 cis-regulatory function.
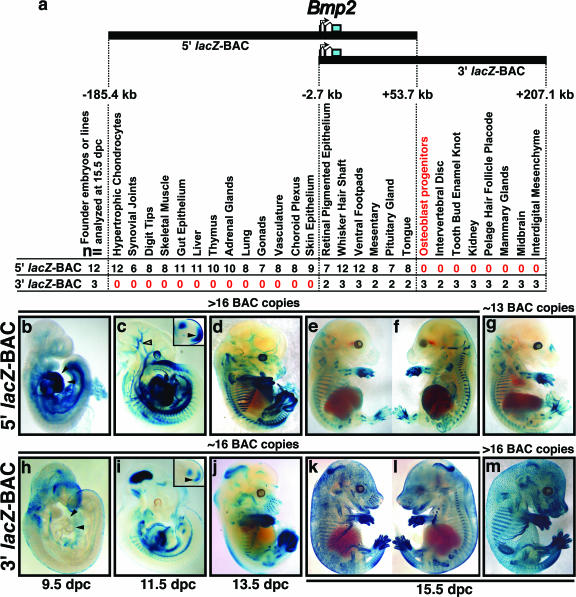
To initially determine whether sequences surrounding the mouse Bmp2 gene had important cis-regulatory functions, we utilized two overlapping clones from a C57BL/6J BAC library (Fig. 1a). The insert positions of the BACs relative to the three Bmp2 exons were determined by inspecting BAC end sequences displayed on the UCSC Genome Browser (35) and direct BAC end sequencing (data not shown). The two BACs together span an approximately 392.5-kb genomic region, including the 11-kb Bmp2 transcription unit, 188.1 kb of 5′ flanking sequence, and 193.4 kb of 3′ flanking sequence. No other genes besides Bmp2 are in either BAC, based on the UCSC Genome Browser “known genes” and Ensembl tracks (35). The BAC clone inserts overlap in a region from kb −2.7 and +53.7 that contains the entire Bmp2 transcription unit (Fig. 1a) (all distances are relative to a Bmp2 transcription start site reported previously [23]). Using homologous recombination in bacteria (47), each BAC was modified to contain a cassette containing an IRES (37) fused to the β-geo (lacZ/Neo) gene (21, 54) in place of the Bmp2 mature-peptide-coding sequence in exon 3. This design allowed translation of the β-geo protein from transgene-derived Bmp2 mRNA via the IRES and in place of functional Bmp2 protein.
FIG. 1.
lacZ-BAC transgene scan for Bmp2 cis-regulatory function. (a) UCSC Genome Browser plot of a 392.5-kb region of the mouse Bmp2 locus and BAC genomic insert positions relative to the previously reported Bmp2 transcription start site (23). The blue box in the third exon of Bmp2 denotes the relative position of the IRES-β-geo cassette in each BAC construct. The summary table shows expression data arranged in three groups to reflect the regions unique to the 5′ BAC, common to both, or unique to the 3′ BAC that contain putative regulatory elements required for expression in specific locations. The far left column shows the number of transgenic embryos or stable lines analyzed for each 5′ and 3′ BAC integration event. Twelve 5′ BAC and three 3′ BAC transgenic embryos/lines were analyzed at 15.5 dpc. Column labels indicate specific sites of lacZ expression. Numbers below each site description denote the number of transgenic embryos/lines with detectable X-Gal staining in the specific anatomical location. (b to m) Whole-mount X-Gal-stained 9.5-, 11.5-, 13.5-, and 15.5-dpc 5′ BAC (b to g) and 3′ BAC (h to m) embryos. (b) A 9.5-dpc 5′ BAC embryo showing expression in early heart mesoderm (arrowheads), ventral trunk region, and extraembryonic tissues. (h) A 9.5-dpc 3′ BAC embryo showing expression in the anterior ectoderm of the head and along the dorsal midline. No X-Gal staining is observed in the heart mesoderm (arrowheads) at this stage. (c) An 11.5-dpc 5′ BAC embryo with expression in early vasculature (open arrowhead), eye lens, trunk, and tail bud. The inset shows an 11.5-dpc 5′ BAC hindlimb bud showing expression in posterior/distal mesenchyme (arrowhead) and AER as well as an anterior/proximal domain. (d) Similar patterns in the posterior/distal mesenchyme and AER are observed in 3′ BAC hindlimb bud, but also a central domain of expression is present, unlike the anterior/proximal expression seen in panel c. (e, f, k, and l) Medial and lateral images of bisected, whole-mount 15.5-dpc embryos for the 5′ BAC (e and f) and 3′ BAC (k and l). (g and m) 5′ BAC and 3′ BAC 15.5-dpc embryos, respectively, that were derived from independent transgene injections in addition to those used to make the stable lines depicted in panels b to f and h to l. Notice the consistent patterns of lacZ expression between embryos depicted in panels e versus g and k versus m. Copy number estimates based on real-time PCR are shown for 5′ BAC (b to f) and 3′ BAC (h to l) stable lines and for the independently generated embryos (g and m).
To analyze each Bmp2 lacZ-BAC for regulatory potential during development, they were purified and injected into one-cell-stage mouse embryos. Founder embryos were either collected at 15.5 dpc for direct X-Gal whole-mount staining or allowed to develop to term for the establishment of stable transgenic lines and subsequent analysis of progeny embryos. The 15.5-dpc time point is convenient for examining multiple aspects of bone and skeletal cartilage development. In addition, previous studies have revealed Bmp2 expression in numerous skeletal and soft tissues at 15.5 dpc, such as bone, kidney, and gut (4, 16, 65). Therefore, we focused on this time point to compare Bmp2-lacZ BAC transgene expression between multiple independent founder embryos or lines. For each BAC, between 3 and 12 transgenic “founder embryos” or stable transgenic lines were analyzed for lacZ expression at 15.5 dpc by whole-mount and histological sectioning of X-Gal-stained embryos (summarized in Fig. 1). Founder embryos were sacrificed at 15.5 dpc for X-Gal staining. For stable lines, we analyzed progeny embryos at multiple time points including 15.5 dpc.
In general, for each BAC the staining patterns at 15.5 dpc were mostly identical for multiple embryos/lines generated from the same construct. However, the overall lacZ expression intensity varied such that in some embryos staining was faint or undetectable in tissues where Bmp2 expression is relatively low. This variation could in theory be due to either copy number effects, fragmentation of the BAC causing loss of cis-regulatory elements, or repression due to site-specific position effects. Since BAC transgenes are thought to be fairly resistant to position effects and it has been reported that BAC-transgenic lines that have multiple copies are very likely to contain full-length copies of the transgene (24), we therefore estimated BAC transgene copy number by using quantitative real-time PCR. We confirmed our results using assays for both the β-geo gene and the BAC vector (see Materials and Methods). This revealed that embryos or lines that had several (∼3 or more) copies of the transgene almost always had robust expression patterns that were identical to those of the other high-copy founders, while weakly stained embryos almost invariably had only one or two transgene copies (Fig. 1 and data not shown). However, real-time PCR has limited ability to confirm whether a transgenic embryo or line contains a full-length BAC transgene insertion. Therefore, we performed Southern blot analysis on high-molecular-weight DNAs isolated from both 5′ and 3′ BAC-carrying stable lines used to generate the embryos in Fig. 1b to f and h to l, using rare restriction enzyme sites present in the BACs and a transgene-specific probe (see Materials and Methods; see also Fig. S1 in the supplemental material). This analysis revealed that both 5′ and 3′ BAC-carrying stable lines most likely contained one or more intact BAC copies, as evidenced by strong bands at the expected size of the BAC transgenes (see Fig. S1 in the supplemental material). These data are consistent with previous evidence that multicopy transgenes tend to integrate as tandem concatemers (8, 19). Our results are very consistent with the idea that BACs are highly resistant to position effects and that multiple-copy BAC transgenes usually contain one or more intact BAC copies that drive highly reproducible expression (24).
Since we confirmed that our 5′ and 3′ BAC lines each had multiple-copy integrations and that embryos from these lines had identical and complete staining patterns at 15.5 dpc as listed in Fig. 1, we analyzed staining in these lines at additional time points. Progeny embryos from representative stable lines for each Bmp2 lacZ-BAC were analyzed for lacZ expression from 9.5 dpc to 17.5 dpc. Whole-mount X-Gal-stained 9.5-, 11.5-, 13.5-, and 15.5-dpc embryo specimens for each Bmp2 lacZ-BAC are shown in Fig. 1.
The overall staining patterns for each BAC were quite distinct, suggesting differences in the distribution of cis-acting regulatory sequences surrounding the Bmp2 gene in the genomic region(s) covered by the two BACs. Similar patterns of lacZ expression were consistently observed in transgenic embryos across multiple independent transgene integration events at 15.5 dpc (Fig. 1a, e, g, k, and m). These consistent lacZ patterns closely reflect normal sites of endogenous Bmp2 expression as previously reported and/or reported here (see below), suggesting that ectopic or position site effects on BAC transgene expression were very minimal or nonexistent. While many lacZ expression patterns were specific to either BAC construct, some sites of lacZ expression were obviously similar for each BAC and reflect sites of endogenous Bmp2 expression. For example, at 11.5 dpc both BACs drive lacZ expression in the AER and in limb bud mesenchyme (28) (Fig. 1c and i, insets). In addition, both 5′ BAC and 3′ BAC transgenic embryos display lacZ expression in vibrissae (whisker hair follicles) at 15.5 dpc (Fig. 1e and k) in hair shaft precursor or precortical cells adjacent to the dermal papilla (see Fig. S2 in the supplemental material) (4, 43). Likewise, both Bmp2 lacZ-BACs consistently directed lacZ expression in the retinal pigmented epithelium (16), ventral footpads (28), gut mesentery, pituitary gland (71), and dorsal tongue papillae (33) at 15.5 dpc (Fig. 1a and data not shown; see Fig. S1 in the supplemental material). Additionally, expression was also seen in the hyaline cartilage of the nonossifying costal or ventral ribs in 5′ (Fig. 1d to g) and 3′ (Fig. 1j to m) BAC embryos. These data suggest that cis-regulatory elements sufficient to drive several aspects of embryonic Bmp2 expression are located in the region shared by both BACs (i.e., between kb −2.7 and +53.7), which also contains the Bmp2 transcription unit.
However, many lacZ expression domains were associated only with the 5′ BAC or the 3′ BAC but not both, probably because distant 5′ or 3′ regions are required to upregulate Bmp2 in specific embryonic locations. For example, embryos from stable 5′ BAC lines displayed lacZ expression in heart mesoderm at 9.5 dpc (Fig. 1b), while transgenic embryos from a 3′ BAC line had no observable X-Gal staining in the heart primordia at this stage (Fig. 1h). These data suggest that the genomic region present upstream of kb −2.7 contains sequences required for Bmp2 transcription in heart mesoderm.
Other patterns consistently observed in 5′ BAC embryos but not in 3′ BAC-transgenic mice include expression in the ventral trunk and extraembryonic tissues at 9.5 dpc (Fig. 1b) and in vasculature, dorsal tail bud, and eye lens and in a segmented pattern along the trunk at 11.5 dpc (Fig. 1c). Expression in eye lens and vasculature persisted at 13.5 dpc (Fig. 1d). By 15.5 dpc, staining in thymus, lung, and liver was clearly seen, as well as expression in all endochondral bones (Fig. 1e to g; see Fig. S2 in the supplemental material). In all 12 transgenic embryos/lines carrying the 5′ BAC, expression in developing endochondral bones appeared to be concentrated to the hypertrophic chondrocyte zones (Fig. 1a and e to g). This was first noticeable in the proximal limb bones, vertebrae, and dorsal ribs at approximately 13.5 dpc (Fig. 1d). Closer inspection of 15.5-dpc whole-mount-stained 5′ BAC embryos also consistently revealed staining in digit tips, choroid plexus, skeletal muscle, adrenal glands, synovial joints, and faintly in skin epithelium (Fig. 1 and data not shown; also see Fig. 2 and 3).
FIG. 2.
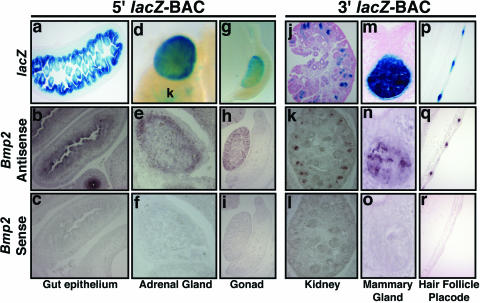
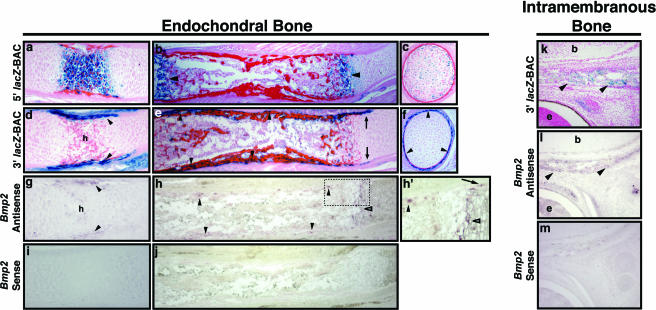
Bmp2 lacZ-BAC transgenes confer normal patterns of endogenous Bmp2 expression in soft tissues in vivo. X-Gal-stained tissues from 15.5-dpc 5′ BAC and 3′ BAC transgenic embryos are shown in comparison to sections of nontransgenic embryos hybridized with riboprobes for mouse Bmp2. Top row, transgenic embryos. Middle row, Bmp2 mRNA in nontransgenic embryos. Bottom row, Bmp2 sense probe controls. (a and b) Section through the gut of a 5′ BAC transgenic embryo, showing lacZ expression in the epithelium facing the lumen (a), similar to Bmp2 (b). (d) Whole-mount X-Gal-stained adrenal gland. k, kidney. (e) Sagittal section through adrenal gland, showing endogenous Bmp2 expression in the outer cortex layer. (g) Whole mount of X-Gal-stained gonad. (h) Sagittal section showing endogenous Bmp2, similar to lacZ in panel g. (j) Sagittal section through X-Gal-stained kidney from a 3′ BAC transgenic embryo, showing expression in comma- and s-shaped bodies. (k) Near-adjacent section to panel j, showing Bmp2 expression similar to expression in panel j. (m) Section through X-Gal-stained mammary gland, showing strong expression in the secretory epithelium, similar to that of endogenous Bmp2 (n). (p) Near-sagittal section through dorsal midline of an X-Gal-stained 3′ BAC transgenic embryo. (q) Section similar to that in panel p of a nontransgenic embryo hybridized with a Bmp2 antisense probe. lacZ and endogenous Bmp2 transcripts are present in the invaginating epithelial buds of pelage hair follicle placodes shown in panels p and q.
FIG. 3.
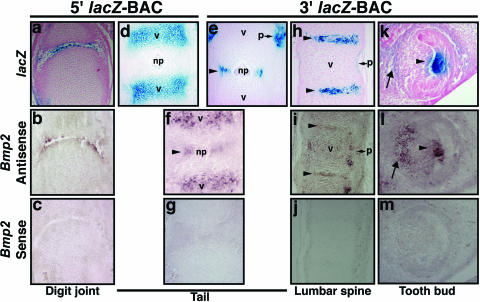
Bmp2 lacZ-BAC transgenes confer normal patterns of endogenous Bmp2 expression in skeletal joints and teeth. X-Gal-stained skeletal tissues from 5′ and 3′ BAC transgenic embryos are shown in comparison to age-matched sections of nontransgenic embryos hybridized with antisense or sense riboprobes for Bmp2. (a) Section through X-Gal-stained distal portion of hindlimb digit from a 17.5-dpc 5′ BAC transgenic embryo from a stable line. lacZ-positive cells are located in the middle or deep articular layers of the digit joint. (b) Section similar to that in panel a, showing Bmp2 mRNA in a nontransgenic embryo. (d) Sagittal section through the tail of an X-Gal-stained 15.5-dpc 5′ BAC embryo, showing expression in hypertrophic chondrocytes of vertebral bodies. (e) Sagittal section through the tail of an X-Gal-stained 15.5-dpc 3′ BAC embryo, showing expression in the annulus fibrosus layer (arrowhead) surrounding the nucleus pulposus. (f) Endogenous Bmp2 in both the annulus fibrosus layer (arrowhead) and hypertrophic chondrocytes. (h and i) Sagittal section through the lumbar spine of an X-Gal-stained 15.5-dpc 3′ BAC transgenic embryo, showing expression in the annulus fibrosus layer (h, arrowheads) similar to that of Bmp2 mRNA in nontransgenic embryos (i, arrowheads). (k and l) Near-sagittal sections through incisor tooth buds of an X-Gal-stained 15.5-dpc 3′ BAC embryo and a 15.5-dpc nontransgenic embryo. lacZ and endogenous Bmp2 are expressed in the enamel knot (arrowhead) and dental papilla mesenchyme (arrow). np, nucleus pulposus; p, perichondrium; v, vertebral body.
In contrast, several patterns were unique to the 3′ BAC line but were never observed in 5′ BAC-transgenic mice. At 9.5 dpc staining was seen in the anterior head ectoderm and along the dorsal midline, possibly in migrating neural crest (Fig. 1h). By 11.5 dpc strong staining was seen in the mesencephalon (midbrain), as well as ventrally restricted segmental expression along the trunk (Fig. 1i). The midbrain pattern continued through 15.5 dpc (Fig. 1j to m; see Fig. S2 in the supplemental material). In addition, staining was observed in the dorsal limb bud mesenchyme at 11.5 dpc (Fig. 1i, inset) and in interdigital tissue from 13.5 to 15.5 dpc (Fig. 1j to m). Also at 13.5 dpc, stripes of lacZ expression were seen demarcating the proximal limb bones (Fig. 1j). By 15.5 dpc, staining was present in the osteogenic perichondrium surrounding ossifying centers of long bones (Fig. 1k to m), as well as intramembranous bones of the head (not shown). Likewise, strong staining was observed in the kidney and intervertebral discs of the spine (Fig. 1k to m).
Additionally, 3′ BAC embryos expressed lacZ in the placodes of the forming whisker follicles at 12.5 dpc and in pelage (coat) hair follicles clustered towards the dorsal midline at 13.5 dpc (not shown). Hair placode expression continued throughout the surface ectoderm at 15.5 dpc (Fig. 1k and m). Hair placode expression is also consistent with previously reported Bmp2 expression (51). Interestingly, since both 5′ BAC and 3′ BAC clones drive expression in whisker follicle hair shaft precortical cells at 15.5 dpc, this indicates that separate regulatory sequences control Bmp2 expression early in the epithelial hair placodes versus later in hair shaft precursor cells. As indicated for regulatory elements controlling Bmp2 expression in osteoblast progenitors, midbrain, and kidney, sequences controlling expression in hair follicle placodes are likely located between kb +53.7 and +207.1 3′ to the promoter (Fig. 1a).
In summary, the analysis of 5′ and 3′ BACs indicated that many sites of embryonic Bmp2 expression require cis-regulatory elements that are more than 2.7 kb 5′ or more than 53 kb 3′ to the promoter. Regulatory elements for expression in hypertrophic chondrocytes, synovial joints, liver, lung, gut, and other tissues are in the 5′ region, whereas elements controlling expression in osteoblast progenitors, tooth buds, hair follicles, kidney, midbrain, and other locations are in a distant 3′ region; notably, only a minority of the distinct expression sites at 15.5 dpc were common to both BACs and are thus likely to be controlled by more proximal elements. Taken together, our data strongly suggest the presence of numerous enhancer elements in the 5′ and 3′ genomic regions unique to either Bmp2 lacZ-BAC, and some can affect promoter activity from large distances.
Bmp2 lacZ-BAC transgenes recapitulate endogenous expression in numerous tissues in vivo.
In order to determine if the Bmp2 lacZ-BAC transgenes were able to direct expression patterns similarly to endogenous Bmp2 mRNA, we performed in situ hybridizations on wild-type embryo sections and compared them to corresponding X-Gal-stained transgenic specimens. Figure 2 shows a comparison of lacZ directed by either the 5′ or 3′ BAC clone to endogenous Bmp2 mRNA in several developing organs. Close similarity of 5′ BAC expression to endogenous Bmp2 was seen in the gut, adrenal gland, and gonads at 15.5 dpc (Fig. 2a to i). Expression in gut epithelium was similar to previous reports of Bmp2 expression (4, 26) (Fig. 2a and b). Expression of Bmp2 in adrenal gland has not been previously reported. We found that the 5′ BAC drives robust lacZ expression similarly to Bmp2 mRNA in the adrenal gland outer cortex layer (Fig. 2d and e). Similar patterns of lacZ and endogenous Bmp2 were also observed in the liver and in lung, where lacZ-positive cells were localized to proximal airway epithelium and mesothelium at 15.5 dpc (see Fig. S2 in the supplemental material). Bmp2 is also expressed in mouse gonads at midgestation (7) (Fig. 2g and h). Previous studies indicate similarity between our 5′ BAC lacZ patterns and those for Bmp2 mRNA in digit tips, choroid plexus, and thymus (not shown) (22, 25, 28). We also detected weak 5′ BAC expression and Bmp2 mRNA in skin epithelium adjacent to hair follicle placodes (not shown).
In 3′ BAC embryos, we observed close similarities of lacZ expression to Bmp2 mRNA patterns in kidneys, mammary gland, and pelage hair follicle placodes (Fig. 2j to r). Expression in the kidney (Fig. 2j and k) was localized to comma- and s-shaped bodies (16). In the mammary gland, expression was prevalent in the developing secretory epithelium (Fig. 2m and n). Bmp2 and lacZ expression were prominent in the epithelial bud of hair follicle placodes at 15.5 dpc (Fig. 2p and q).
Sites of Bmp2 expression in numerous skeletal tissues were also recapitulated by BAC-transgenic mice (Fig. 3). The 5′ lacZ-BAC drives lacZ expression in synovial joints, in a manner very similar to that for endogenous Bmp2, within the middle or deep articular layer of 17.5-dpc digit joints (Fig. 3a and b). The 5′ BAC also directed expression in the hypertrophic cartilage of the vertebral bodies (Fig. 3d). In contrast, the 3′ lacZ-BAC drives expression in the intervertebral discs of the spine and vertebral perichondrium and adjacent connective tissue (Fig. 3e and h). Both lacZ and Bmp2 mRNA are localized to fibrous cartilaginous cells of the annulus fibrosus surrounding the nucleus pulposus (Fig. 3e, f, h, and i). Bmp2 protein expression has also been reported to occur in the annuli of adult, senescent mice (70). In 15.5-dpc developing incisor buds, 3′ BAC-directed expression in the enamel knot is very similar to that for endogenous Bmp2 mRNA (Fig. 3k and l). These patterns were also similar in molar tooth bud enamel knot (data not shown). Weak X-Gal staining and Bmp2 mRNA were also observed in the dental papilla (Fig. 3k and l). Our findings corroborate previous reports of Bmp2 mRNA expression in tooth buds (4, 51). Taken together, these results suggest that both BAC transgenes accurately reflect overlapping subsets of normal Bmp2 expression and that regulatory elements in 5′ and 3′ flanking regions within the Bmp2 locus are endogenous Bmp2 regulatory elements.
Distinct cis-regulatory regions drive Bmp2 expression in developing bone in vivo.
The endochondral skeleton, which is composed of limb bones, the chondrocranium (cartilaginous bones of the skull base), and axial components (the vertebrae and ribs), undergoes ossification via cartilage templates that prefigure the future bone elements. In contrast, the intramembranous skeleton is composed of the cranial vault (calvarial bones) and viscerocranium (facial bones) and forms via the direct conversion of mesenchyme to bone, thus bypassing a cartilage template. Although these two ossification processes are distinct, differentiating osteoblasts are present in all endochondral and intramembranous bones. Embryonic Bmp2 expression has been previously documented in both the perichondrium and hypertrophic chondrocytes of endochondral long bones and also in intramembranous bones (10, 52, 60, 65). Given the significance of Bmp2 for osteoblast differentiation, we examined the skeletal expression of both BACs in more detail.
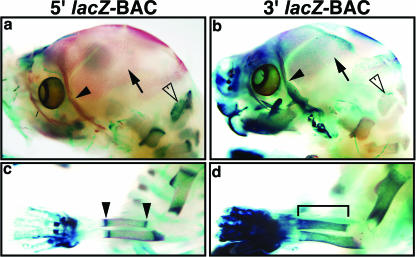
Figure 4 shows images of X-Gal-stained skeletons from 5′ lacZ-BAC and 3′ lacZ-BAC 17.5-dpc transgenic embryos generated from the stable lines described above, counterstained with alizarin red to highlight ossifying regions of bone. In 17.5-dpc 5′ BAC embryos, strong staining is present in the endochondrally derived occipital bone (Fig. 4a). No X-Gal staining is present in the intramembranous parietal bone of the calvarium or zygomatic bone of the facial skeleton in 5′ BAC embryos (Fig. 4a). In contrast, lacZ-positive cells are present in all intramembranous bones in 3′ BAC embryos at 17.5 dpc (Fig. 4b), such as the parietal and zygomatic bones of the skull, as well as the occipital bone. In the radii and ulnae of 5′ BAC embryos, stripes of X-Gal staining are localized to the ends of the ossifying bone collar (Fig. 4c). However, in 3′ BAC embryos, X-Gal staining extends throughout the entire length of the bone collar (Fig. 4d). These data suggest that the 3′ BAC can direct expression in cells in ossifying regions of both intramembranous and endochondral bones, while the 5′ BAC directs expression only in endochondral bones.
FIG. 4.
Bmp2 cis-regulation in bone. X-Gal- and alizarin red-stained 17.5-dpc whole-mount 5′ and 3′ BAC embryonic head and forelimb skeletons generated from stable lines are shown. Alizarin red staining is shown in red and labels ossified bone. (a and b) Whole-mount 17.5-dpc 5′ and 3′ head skeletons, respectively. (a) No X-Gal staining can be seen in the intramembranous parietal (arrow) or zygomatic (closed arrowhead) bones; however, staining is present in the endochondral occipital bone (open arrowhead). (b) Numerous lacZ-positive cells are present in the parietal (arrow), zygomatic (closed arrowhead), and occipital (open arrowhead) bones. (c and d) Whole-mount 17.5-dpc 5′ and 3′ forelimb skeletons, respectively. (c) Stripes of X-Gal staining can be seen at the ends of the ossified regions in the long bones, corresponding to the growth plate region (arrowheads). (d) X-Gal staining is present throughout the ossified regions (bracket) of the long bones.
We further examined histological sections of X-Gal-stained embryos. Figure 5 shows sections through stained metacarpals and radii from 5′ BAC and 3′ BAC 17.5-dpc transgenic embryos. The onset of primary ossification in the limb bones occurs in a proximal-to-distal sequence, with the primary ossification centers starting to form at about 14.5 dpc in the radius condensations in mouse (61). In the metacarpals, these centers are not obvious until approximately 16.5 dpc (61). By 17.5 dpc, lacZ expression is localized to the hypertrophic chondrocytes in the center of the metacarpal cartilage anlage (Fig. 5a). Strong eosin staining is seen in extracellular matrix surrounding the hypertrophic chondrocytes and in the flanking bone collar (Fig. 5a to f), indicating calcified cartilage and bone. In the radii of 17.5-dpc 5′ BAC embryos, lacZ expression is localized to the growth plate hypertrophic chondrocytes (Fig. 5b). However, in 5′ BAC embryos no stained cells are present in the perichondrium/bone collar surrounding the forming marrow cavity (Fig. 5a to c).
FIG. 5.
Bmp2 lacZ-BACs confer endogenous expression in developing bone. X-Gal-stained endochondral bone sections from representative 5′ and 3′ BAC transgenic embryos are shown in comparison to sections of nontransgenic embryos hybridized with Bmp2 antisense or sense probes. (a and b) Sections through X-Gal-stained metacarpal and radius condensations, respectively, from 17.5-dpc 5′ BAC transgenic embryos generated from stable lines. lacZ expression is observed in hypertrophic chondrocytes of the metacarpal (a) and growth plate regions (b) (arrowheads) of the radius. (c) Cross-section through the radius growth plate, showing lacZ-positive hypertrophic chondrocytes. No lacZ-positive cells are observed in the perichondrium/bone collar region surrounding the cartilage. (d and e) Sections through X-Gal-stained metacarpal (d) and radius (e) condensations from 17.5-dpc 3′ BAC transgenic embryos generated from stable lines. lacZ expression is observed in the perichondrium/bone collar region flanking the metacarpal hypertrophic zone in panel d. Arrowheads in panel e indicate lacZ-positive cells in the perichondrium and throughout the periosteal bone collar flanking the marrow cavity. Arrows in panel e indicate expression in perichondrium flanking the hypertrophic zone. (f) Cross-section through the radius growth plate region, showing lacZ-positive cells localized to the perichondrium/bone collar (arrowheads) surrounding the hypertrophic zone. (g) Bmp2 mRNA detected in the metacarpal of a nontransgenic 17.5-dpc embryo. Both the hypertrophic chondrocytes and the perichondrium/bone collar (arrowheads) are faintly positive for endogenous Bmp2. (h) Section through radius of a nontransgenic 17.5-dpc embryo, showing numerous Bmp2-positive cells throughout the bone collar (closed arrowheads) flanking the marrow cavity and within the hypertrophic chondrocyte zone (open arrowheads). (h′) Higher magnification of dashed box in panel h. The arrow indicates Bmp2-positive perichondrial cells flanking the hypertrophic zone. (k) Sagittal sections through orbital plate of frontal bone from an X-Gal-stained 15.5-dpc 3′ BAC embryo. (l) Age-matched section to panel k hybridized with a Bmp2 antisense probe. The arrowheads in panels k and l indicate expression in cells located in ossifying regions. b, brain; e, eye; h, hypertrophic zone.
In sharp contrast to 5′ BAC embryos, 3′ BAC embryos displayed lacZ expression in cells within the perichondrium regions flanking the hypertrophic zone of the metacarpal (Fig. 5d). Transgene expression was also observed in the perichondrium of the 17.5-dpc radius (Fig. 5e). In addition, lacZ-positive cells are dispersed throughout the bone collar (Fig. 5e), from the diaphysis to the perichondrial region flanking the hypertrophic zone (Fig. 5e). lacZ-positive cells were also on the surface and embedded within the cortical bone of the bone collar at 17.5 dpc.
To confirm whether these lacZ expression patterns matched endogenous Bmp2 expression at 17.5 dpc, we performed in situ hybridizations for Bmp2. In age-matched metacarpal sections of nontransgenic embryos, Bmp2 transcripts are present in both the hypertrophic chondrocytes and adjacent perichondrium/bone collar (Fig. 5g). These are very similar to the combined patterns of both lacZ expression domains observed in Fig. 5a and d. The apparent low levels of endogenous expression in comparison to X-Gal-stained sections may reflect difficulties in detecting low Bmp2 mRNA levels by in situ hybridization, whereas the enzymatic staining procedure used to detect lacZ is very robust. In the 17.5-dpc radius Bmp2 mRNA was detected in hypertrophic chondrocytes located in the growth plate (Fig. 5h and h′), similar to 5′ BAC lacZ expression. Numerous Bmp2-positive cells are also distributed throughout the bone collar surrounding the marrow cavity (Fig. 5h and h′) and in perichondrium flanking the hypertrophic cartilage zones (Fig. 5h′). Therefore, while both Bmp2 lacZ-BACs can confer endogenous patterns of Bmp2 expression in developing endochodral bones, they each direct complementary patterns that together reflect the complete Bmp2 regulatory domains at 17.5 dpc.
Since the 3′ BAC could also direct expression in intramembranous bones, X-Gal-stained sections through the orbital plate of frontal bones from 15.5-dpc 3′ BAC embryos (Fig. 5k) were compared to corresponding Bmp2 in situ hybridizations of wild-type embryos (Fig. 5l). This confirmed that lacZ-positive cells are distributed throughout the network of intramembranous bone in 3′ BAC embryos (Fig. 5k), similar to endogenous Bmp2 mRNA (Fig. 5l) (10). We also observed lacZ expression throughout the intramembranous calvarium and mandible at 15.5 dpc (data not shown).
Clearly, each BAC directed a different subset of normal patterns in bone; the 5′ BAC drives expression in hypertrophic chondrocytes of endochondral bones, and the 3′ BAC drives expression in differentiating osteoblasts present in the perichondrium/bone collar and intramembranous bones. Surprisingly, this implies that regulatory sequences controlling embryonic Bmp2 expression in differentiating osteoblasts are located at greater than 53 kb from the Bmp2 promoter.
Relationship between Bmp2 transcription and osteogenic markers in vivo.
The conversion of the perichondrium to an osteoblast-containing bone collar is intimately linked to osteoblast differentiation, as well as molecular and morphological changes in adjacent chondrocytes, such as proliferation and subsequent hypertrophy (34). To correlate Bmp2 and 3′ BAC transgene expression to the progression of osteoblast differentiation in vivo, we compared both to Runx2 and Spp1 expression. Runx2 (also named Cbfa1/PEBP2αA) is one of the earliest markers of, and is essential for, osteoblast differentiation (15, 42, 57). Runx2 is expressed in the mesenchyme surrounding future skeletal elements, in osteogenic cells within the perichondrium and bone collar, and in both prehypertrophic and hypertrophic chondrocytes at midgestation developmental stages (15, 38, 57). Spp1 (also named Bsp/bone sialoprotein/osteopontin) is expressed in osteoblasts and hypertrophic chondrocytes (20, 32). Its expression coincides with collagenous and noncollagenous matrix production, and in osteoblasts it is indicative of a bone matrix-producing phenotype (2). In addition, both Runx2 and Bmp2 are implicated in osteoblast-specific control of Spp1 transcription (15, 42, 46).
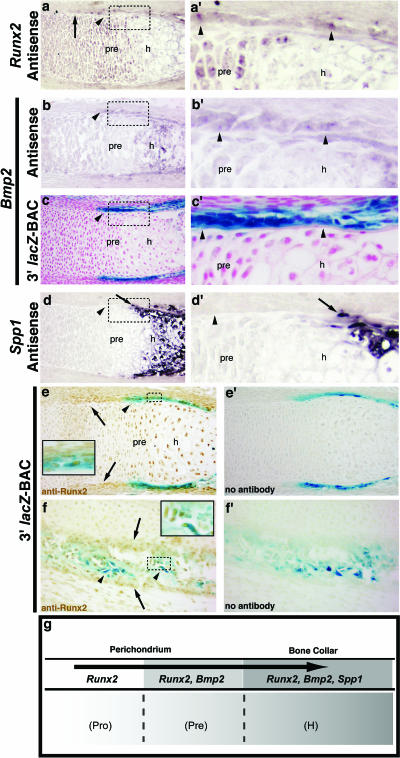
In 15.5-dpc radius condensations, Runx2 transcripts are present in cells within the perichondrium/bone collar flanking the hypertrophic and prehypertrophic zone (Fig. 6a and a ′). In the osteogenic inner perichondrial layer, domains of Runx2 expression closely resemble those for endogenous Bmp2 mRNA (Fig. 6b and b′) as well as 3′ BAC-directed (Fig. 6c and c′) lacZ expression at 15.5 dpc, as the latter both label perichondrial cells flanking the prehypertrophic and hypertrophic zones. However, the inner perichodrium region surrounding the nonhypertrophic proliferative and resting zones appears faintly positive for Runx2 (Fig. 6a, arrow) but not Bmp2-lacZ or endogenous Bmp2 transcripts. Runx2 is also transcribed in prehypertrophic chondrocytes (Fig. 6a and a′). In addition, endogenous Bmp2 mRNA is detected in the hypertrophic zone (Fig. 6b and b′). In contrast, Spp1 mRNA is undetected in perichondrial cells adjacent to the prehypertrophic chondrocytes (Fig. 6d and d′) but is upregulated in bone matrix-producing osteoblasts flanking the hypertrophic zone and lining the nascent bone collar (Fig. 6d and d′) and in late-stage hypertrophic chondrocytes. These data suggest that both Bmp2 and Runx2 are transcribed in osteoblast progenitors within the inner perichondrial layer flanking the prehypertrophic zone, prior to onset of Spp1 production by these cells. In addition, Bmp2 and Runx2 expression persists as these cells upregulate Spp1 and transition to a more mature osteoblastic phenotype capable of bone matrix production and bone collar formation (Fig. 6g).
FIG. 6.
Analysis of Bmp2 expression in comparison to osteogenic markers in vivo. (a to e and a′ to e′) Sections of 15.5-dpc radii. (f and f′) Sections of 15.5-dpc frontal bone (orbital plate). (a, b, and d) In situ hybridization of Runx2, Bmp2, and Spp1 mRNAs in nontransgenic embryos. (c) Section through X-Gal-stained 3′ BAC transgenic embryo. The boxed regions in panels a, b, c, and d are shown at a higher magnification in panels a′, b′, c′, and d′. The arrowheads in panels a, b, and c indicate expression in cells in the perichondrium/bone collar region flanking the prehypertrophic and hypertrophic zones. The arrowheads in panel d indicate lack of Spp1 expression in these regions. The arrow in panel a indicates faint Runx2 expression in the inner perichondrium flanking the zones of proliferative and resting chondrocytes. The arrows in panels d and d′ indicate Spp1-positive osteoblasts lining the outer surface of the bone collar. (e) Section from 3′ BAC embryo, showing Runx2 protein and Bmp2-lacZ colocalization in the osteogenic perichondrium/bone collar. Coexpressing cells (arrowheads in panel e and inset) are evidenced by brown nuclei (stained with anti-Runx2 antibody) and blue X-Gal stain in the cytoplasm (Bmp2-lacZ). Runx2 protein is also present in prehypertrophic and hypertrophic chondrocytes, and the arrows indicate a stronger Runx2 signal in the inner perichondium flanking the proliferative and resting chondrocytes. (e′) Section adjacent to that in panel e, showing a no-antibody control. (f) Section through frontal bone from a 3′ BAC transgenic embryo. Runx2 and Bmp2-lacZ colocalize in osteogenic cells lining newly formed bone (arrowheads and inset). The arrows indicate Runx2 in mesenchyme surrounding ossifying regions. In panels e and f, the insets show a higher magnification of the dashed-box regions. (g) Schematic representation showing the relationship of Bmp2 transcription and osteogenic markers during osteoblast differentiation. h, hypertrophic zone; pre, prehypertrophic zone; pro, proliferating zone.
In order to determine if the 3′ BAC drives lacZ expression in cells coexpressing Runx2, we performed Runx2 immunohistochemistry on X-Gal-stained radius sections. Runx2 protein localizes to both the prehypertrophic and hypertrophic chondrocytes (Fig. 6e). A strong Runx2 protein signal was also observed in perichondrial cells flanking the nonhypertrophic cartilage and extending to the perichondium/bone collar region flanking the hypertrophic cartilage (Fig. 6e, arrow), consistent with Runx2 mRNA expression (Fig. 6a) (38). Bmp2-lacZ-expressing cells in the inner perichondrium/bone collar region flanking the hypertrophic cartilage (Fig. 6e) display a flattened, rectangular morphology and have large nuclei that are also strongly positive for Runx2 protein (Fig. 6e, inset). In intramembranous bone, Bmp2-lacZ and Runx2 protein colocalization was observed in osteoblasts lining newly formed bone (Fig. 6f and inset), while the mesenchyme surrounding the ossifying regions was positive for Runx2 protein but not lacZ (Fig. 6f).
Numerous cis-regulatory sequences are dispersed throughout the distant 3′ region.
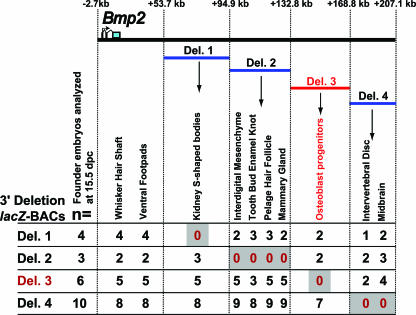
In order to further refine the location of cis-acting sequences throughout the distant 3′ region (kb +53.7 to +207.1), including that of the osteoblast progenitor element, four BACs were engineered to contain deletions in the 3′ region and these were tested for ability to drive lacZ expression at midgestation time points. Using the galK homologous recombination system in bacteria (72), serial and nonoverlapping deletions of approximately 40 kilobases in size were engineered into the full-length Bmp2-lacZ 3′ BAC (described above). We refer to these deletion BACs as D1-, D2-, D3-, and D4-BACs (see Fig. 8 for the locations of the deletions relative to the Bmp2 transcription start site [23]). This system allowed us to generate “seamless” contiguous deletions for systematically analyzing the cis-regulatory functions of the entire 153.5-kb 3′ region (from kb +53.7 to +207.1). As described above, the 3′ deletion lacZ-BAC DNAs were purified and injected into one-cell-stage mouse embryos. For each deletion BAC, between 3 and 10 transgenic founder embryos were analyzed directly for lacZ expression at 15.5 dpc. Each founder embryo represents an independent transgene integration event.
FIG. 8.
Summary of 3′ deletion lacZ-BAC mapping data. The expression data are arranged in five groups to reflect the expression patterns present in full-length 5′ and 3′ lacZ-BAC transgenes (kb −2.7 to +53.7) or lost in 3′ deletion lacZ-BAC transgenes. The far-left column shows the number of transgenic founder embryos analyzed for each deletion BAC integration event. Between 3 and 10 deletion BAC transgenic founder embryos were analyzed at 15.5 dpc. The numbers below each site of expression denote the number of transgenic founder embryos for each deletion BAC with detectable X-Gal staining within the specific anatomical location listed above.
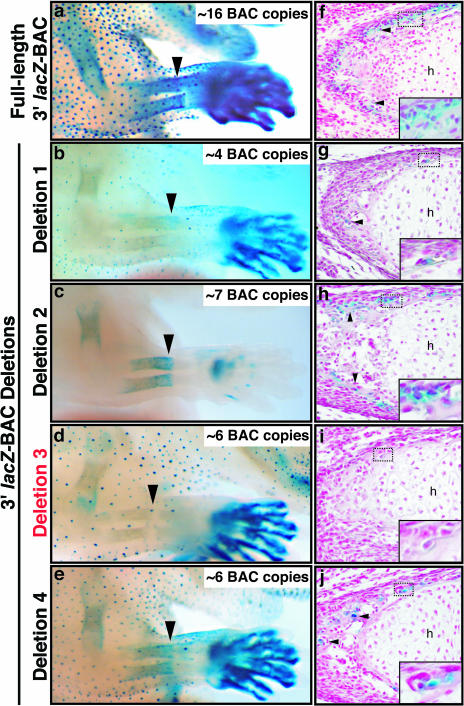
Whole-mount X-Gal-stained forelimbs as well as oblique sections through radii of selected full-length 3′ BAC and 3′ deletion BAC embryos are shown in Fig. 7. For each deletion BAC (Fig. 7b to e), some X-Gal staining patterns were quite similar to those of full-length 3′ lacZ-BAC embryos (Fig. 7a; see Fig. 1a and k to m). Each deletion BAC drove expression in ventral footpads and whisker hair shaft (Fig. 7 and 8 and data not shown), consistent with the previous data from both the full-length 5′ BAC and 3′ BAC that these regulatory elements are in the overlap region from kb −2.7 to +53.7 (Fig. 1a; see Fig. S2 in the supplemental material). However, no lacZ expression was observed in the pelage hair follicle placodes or limb interdigital mesenchyme of D2-BAC embryos (Fig. 7c and 8). Although interdigital expression was lost, expression in the ventral footpads of the autopod was still present (Fig. 7c). This strongly suggested that D2-BAC had deleted critical hair follicle and interdigital mesenchyme cis-elements.
FIG. 7.
Numerous cis-regulatory sequences are dispersed throughout the distant 3′ region. Shown are whole-mount X-Gal-stained forelimbs and oblique, near-adjacent sections through the radius from full-length 3′ lacZ-BAC and 3′ deletion lacZ-BAC embryos. The arrowheads in panels a to e indicate the distal end of the ossifying zone of the radius. (a, b, c, and e) X-Gal staining can be seen in the long bones (radius, ulna, and humerus) of the forelimbs in full-length 3′ lacZ-BAC and 3′ deletion lacZ-BAC D1, D2, and D4 embryos. (d) No staining is present the D3 embryo shown. The arrowheads in panels f, g, h, and j indicate lacZ-positive osteoblasts in ossifying regions of the radius bone sections shown. In panels f to j, the insets show a higher magnification of the dashed-box regions. (i) No staining is present in the section through the radius of the D3 embryo shown. BAC transgene copy number estimates are indicated for the embryos shown in panels a to e. h, hypertrophic zone.
Analysis of endochondral bones of D3-BAC embryos indicate that sequences present within this deletion interval are required for osteoblast progenitor-specific expression. For example, although X-Gal staining was observed in the radii of full-length 3′ BAC embryos (Fig. 7a) as well as D1-, D2-, and D4-BAC embryos (Fig. 7b, c, and e), no staining was observed in the long bones of D3-BAC embryos (Fig. 7d and 8). Sections confirmed that lacZ-positive osteoblasts are present in the ossifying regions of the radii of D1-, D2-, and D4-BAC embryos (Fig. 7g, h, and j, insets), while no lacZ-positive cells were detectable in similar sections from D3-BAC embryos (Fig. 7i, inset). Importantly, patterns of lacZ expression in the radius bones of D1-, D2-, and D4-BAC embryos (Fig. 7g, h, and j, insets) are similar to those in full-length 3′ BAC embryos (Fig. 7f, inset). We also observed lacZ-positive osteoblasts in sections through intramembranous cranial bones of D1-, D2-, and D4-BAC embryos but not D3-BAC embryos (data not shown). These data strongly suggest that during embryonic development, the genomic region covered by the D3 interval (kb +132.8 to +168.8) is required for osteoblast progenitor-specific Bmp2 expression.
The 3′ deletion BACs revealed other regions that are also essential for Bmp2-lacZ gene expression in various tissues (Fig. 8) and that were lacZ positive in full-length 3′ lacZ-BAC embryos (Fig. 1a and k to m) at 15.5 dpc. For example, lacZ expression in the kidney s-shaped bodies is absent in D1-BAC embryos (Fig. 8; see Fig. S3 in the supplemental material). In addition, X-Gal staining was lacking in the tooth bud enamel knot (Fig. 8; see Fig. S3 in the supplemental material) and mammary glands (Fig. 8; and data not shown) in all D2-BAC embryos. Expression was also observed in the intervertebral discs (Fig. 3e and h) and midbrain regions of full-length 3′ lacZ-BAC embryos (see Fig. S2 in the supplemental material) and in at least one or more each of the D1-, D2-, and D3-BAC embryos (Fig. 8 and data not shown); however, no expression was seen in the intervertebral discs in any of the 10 D4-BAC embryos analyzed (Fig. 8 and data not shown).
We analyzed lacZ transgene copy number for all of the 3′ deletion BAC founder embryos (Fig. 7, insets). Figure 7 demonstrates the general correlation between strength of X-Gal staining and copy number, (e.g., hair follicle placodes). Notably, of the embryos shown in Fig. 7, X-Gal staining is relatively weaker in the D1-BAC embryo, which also has the fewest BAC copies (∼4) yet still has detectable lacZ in bone and hair follicles. Comparison of the D3-BAC and D4-BAC embryos (both having an estimated copy number of ∼6) demonstrates that while staining in hair follicles and digits is almost identical for both, no X-Gal staining can be seen in the long bones of the D3-BAC embryo (Fig. 7d and i). Taken together, these data strongly suggest that numerous cis-regulatory sequences are dispersed throughout the distant 3′ region and that a critical osteoblast progenitor-specific Bmp2 control element is located between kb +132.8 and +168.8.
Bmp2 expression in differentiating osteoblasts is controlled by a distant 3′ enhancer.
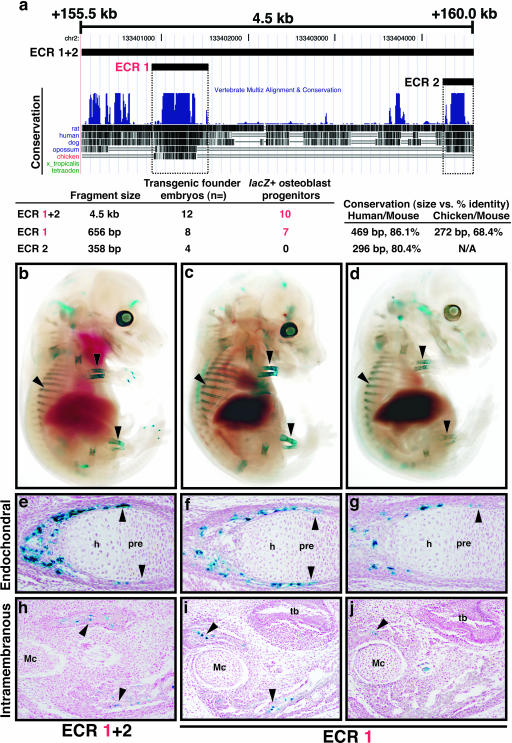
Because the deletion BAC analysis suggested that the region covered by D3 contains an osteoblast regulatory element, we next asked whether or not highly conserved sequences present within this region were capable of osteoblast progenitor-specific enhancer function. The entire D3 region covers approximately 36 kilobases of sequence (mouse February 2006 assembly; chr2, 133377031 to 133413037) (35). Based on inspection of cross-species conservation data on the UCSC genome browser (35), we focused on two regions that had the highest PhastCons conservation scores (64) in the D3 interval. These are located at approximately +156 and +160 kb from the previously reported Bmp2 transcription start site (23). Both had PhastCons scores of greater than 200, while no other regions in the D3 interval had scores over 89 (data not shown). These sequences were named ECR-1 and ECR-2 (Fig. 9a). ECR-1 is conserved across mammals and chicken, while ECR-2 conservation is detected only in mammals, based on MultiZ cross-species sequence alignments (6) (Fig. 9a); neither had alignment to frog (Xenopus tropicalis) or teleost fish (Tetraodon) (Fig. 9a). For both ECRs, the values for length of alignment versus percent identity for mouse/human and mouse/chicken comparisons were generated using the ECR browser (59) (Fig. 9a).
FIG. 9.
Bmp2 cis-regulation in osteoblast progenitors is controlled by a distant 3′ enhancer that is conserved between mammals and chicken. (a) UCSC genome browser plot of approximately 4.5 kb of mouse chromosome 2 (mouse February 2006 assembly; chr2, 133400083 to 133404598) (35), showing ECRs tested for enhancer activity using heterologous (Hsp68) promoter-lacZ minigenes. The sizes of the ECR-1 + 2, ECR-1, and ECR-2 fragments tested are drawn on the browser plot and listed below it. Also shown are vertebrate MultiZ alignment and conservation tracks (6) as well as ECR size and percent identity generated by the ECR browser (59). ECR-1 is the only region conserved between mammals and chicken. (b) Whole-mount X-Gal-stained 15.5-dpc representative ECR-1 + 2 transgenic founder embryo. (c and d) Whole-mount X-Gal-stained 15.5-dpc ECR-1 transgenic founder embryos. The 15.5-dpc embryos shown in panel d are derived from a transgene integration event independent from those depicted in panel c. The arrowheads in panels b, c, and d indicate X-Gal staining in bones of the forelimb and hindlimb and in ribs. (e to g) Near-adjacent sections through radii of the embryos shown in panels b to d. Note that lacZ-positive osteoblasts are dispersed through the ossifying regions of the radius, and this expression extends to the perichondrium region surrounding the prehypertrophic zone (arrowheads). (h to j) Near-adjacent sections through mandibles from the embryos shown in panels b to d. The arrowheads in panels h to j indicate lacZ-positive osteoblasts present throughout membranous bone. h, hypertrophic zone; pre, prehypertrophic zone; Mc, Meckel's cartilage; tb, tooth bud.
ECR-1 and ECR-2 lie within relatively close proximity (Fig. 1a); therefore, we tested three different constructs for osteoblast-specific enhancer function by cloning ECR fragments into a heterologous promoter (Hsp68)-lacZ minigene construct and analyzing for X-Gal staining at 15.5 dpc in transgenic founder embryos (13, 53, 63). The three constructs contained either each ECR separately or a 4.5-kb fragment (ECR-1 + 2) spanning both ECR-1 and ECR-2 (Fig. 9a).
Multiple transgenic embryos were analyzed for each construct at 15.5 dpc. Some embryos had localized staining in the brain, vasculature, or other soft tissues which was not consistent between any two embryos; therefore, these were likely ectopic patterns due to position effects. Most embryos had staining in the eye, which is a previously known cryptic effect of the Hsp68-lacZ vector backbone (63). However, for both the ECR-1 + 2 and ECR1 constructs, a majority of transgenic embryos (10/12 and 7/8, respectively) had obvious lacZ expression in osteoblast progenitors present in developing bone at 15.5 dpc, while no ECR-2 embryos (n = 4) had any consistent lacZ expression. One ECR-1 + 2 and two representative ECR-1 embryos are shown in Fig. 9b to d. Specific X-Gal staining can be seen in the ossifying regions of the limb long bones and ribs (Fig. 9b to d). Oblique histological sections through the radius indicated numerous lacZ-positive osteoblast progenitors in the perichondrium flanking the prehypertrophic zone and in the bone collar (Fig. 9e to g). In addition, X-Gal-stained osteoblasts were present in the intramembranous mandible (Fig. 9h to j).
We noted that in some ECR transgenic embryos, X-Gal staining in osteoblast progenitors appeared mosaic (Fig. 9d, g, and j). These data might be partly explained by somatic mosaicism of the transgene in some founder embryos or position effect variegation; alternatively, we cannot rule out that other sequences are involved in activating uniform Bmp2 expression in differentiating osteoblasts. Nevertheless, these data strongly indicate that ECR-1 contains a conserved enhancer that activates Bmp2 expression in differentiating osteoblasts of both endochondral and intramembranous bones and that this element is located 156.3 kilobases from the Bmp2 promoter.
DISCUSSION
Numerous and distant cis-regulatory elements control Bmp2 expression in vivo.
In this study, we analyzed a 392.5-kb region surrounding the mouse Bmp2 gene for cis-regulatory function in vivo and identified a critical osteoblast progenitor-specific enhancer at a great distance (156.3 kilobases) from the endogenous Bmp2 promoter. The use of BACs made it possible to identify distinct regions of cis-regulatory information that would have been missed using more traditional approaches. Furthermore, using BAC clones that extend far 5′ and 3′ while still containing the transcription unit allowed us to survey a large genomic region within the context of the endogenous Bmp2 promoter. BAC-directed reporter gene expression closely matched endogenous Bmp2 transcription in numerous key anatomical sites, including differentiating osteoblasts. Using BAC clones with engineered deletions, we were able to further map the osteoblast progenitor control element and many other cis-regulatory elements within a large interval 3′ to Bmp2.
Each deletion specifically ablated expression in certain tissues, corroborating the full-length 3′ BAC data and strongly suggesting that these endogenous Bmp2 expression patterns require modular cis-regulatory elements. By testing highly conserved sequences within the D3 region, we identified an element that can activate a heterologous promoter in osteoblast progenitors in vivo, suggesting that it normally functions as a tissue-specific enhancer. Thus, Bmp2 expression in osteoblast progenitors is controlled by a distant cis-regulatory module.
These data support the notion that Bmp2 cis-regulatory elements have evolved independently to permit new roles for Bmp2 in different structures and cell types. The large intergenic sequences flanking Bmp2 may have facilitated this process. In support of this, the large intergenic regions flanking Bmp2 show conserved synteny across vertebrates, including mammals and chicken (58). In addition, the osteoblast progenitor control sequence is highly conserved between these two clades; however, it appears to be absent in organisms that also have a bony skeleton, such as frog and teleost fish. This suggests that this sequence may have evolved in amniotes. Interestingly, endogenous Bmp2 expression in the perichondrium is similar in both mouse (data presented here) and chicken (60), suggesting that this enhancer may also function similarly in these organisms. Together, the data presented here strongly suggest that the intergenic domains surrounding Bmp2 harbor numerous, distant regulatory elements.
Bmp2 cis-regulation in differentiating osteoblasts.
It has been suggested that for endochondral bones, the perichondrium is a primary source of osteoblasts in vivo (11) and that osteoblast progenitor cells can first be identified in the inner perichondrium flanking the hypertrophic cartilage. While embryonic Bmp2 expression has been previously documented in both the perichondrium and hypertrophic chondrocytes of endochondral long bones and also in intramembranous bones (10, 52, 60, 65), our analysis of lacZ expression domains in developing endochondral and intramembranous bones of Bmp2 lacZ-BAC embryos strongly suggests that cis-regulatory elements controlling hypertrophic chondrocyte-specific Bmp2 transcription are separate from those controlling Bmp2 transcription in differentiating osteoblasts. Therefore, these data are of interest in light of previous studies of the Bmp2 proximal promoter. The activity of human and mouse Bmp2 promoter fragments have been studied in osteoblast-like cell lines (23, 27, 68, 76). In vitro binding studies suggest that the osteogenic factor Runx2 can bind to the Bmp2 promoter region, although cotransfection assays performed with rat osteosarcoma cells failed to demonstrate that Runx2 can specifically activate the Bmp2 promoter (27). However, a proximal promoter fragment can be autoregulated by Bmp2 in the 2T3 osteoblast cell line (23). Previous studies of a kb −2.7 Bmp2 promoter fragment suggested that it could drive expression in perichondrium and hypertrophic chondrocytes of midgestation mouse embryos (18) and that Gli2 binding to this sequence stimulates Bmp2 response to hedgehog signaling in osteoblast cell lines (76). Fortuitously, the 5′ end of this fragment and the upstream end of the 3′ lacZ-BAC used in our study are located at the same EcoRI restriction site at base −2712 (18) (data not shown). Therefore, both the 5′ BAC and 3′ BAC examined in this study contain this full sequence. Nevertheless, our data indicate that at least during in vivo embryonic bone development, the Bmp2 5′ lacZ-BAC containing 185.4 kb of 5′ flanking DNA, 53.7 kb of 3′ DNA, and all Bmp2 exons and introns cannot drive expression in differentiating osteoblasts, whereas the 3′ BAC cannot drive expression in hypertrophic chondrocytes. Although we cannot rule out potentially important roles for the promoter region or distant repressors in the regulation of Bmp2 in differentiating osteoblasts, our data strongly suggest that the distant 3′ enhancer element is normally required to upregulate Bmp2 expression in these cells.
Bmp2 and osteogenesis.
It is well established that Bmp2 has potent osteoinductive properties (46, 74), and BMP signaling is important for bone formation and osteoblast differentiation (9). The relationship of Bmp2 and the critical osteoblast transcription factor Runx2 is of great interest. We (Fig. 6) and others (15, 38, 57) have shown that Runx2 is expressed in the mesenchyme surrounding the future bony element, including the perichondrium of endochondral bones (in the inner perichondrium layer adjacent to the nonhypertrophic proliferative and resting chondrocyte zones, as previously reported) (38), and also in loose mesenchyme surrounding the ossifying layers of intramembranous bone. However, the Bmp2 expression domain does not extend over the nonhypertrophic zones or into the Runx2-positive mesenchyme surrounding intramembranous bone. Therefore, Runx2 expression likely precedes Bmp2 in the osteogenic mesenchyme during initial stages of osteoblast differentiation in vivo. In vitro experiments suggest that Bmp2 and other BMPs can induce Runx2 expression, while overexpression of Runx2 can upregulate Bmp2 (10, 15, 48). These data suggest a positive feedback regulatory loop between BMP signaling and Runx2. Runx2 is also genetically required for Bmp2 expression in cranial bones (10). Given these data, we speculate that Runx2 binding to the distant 3′ osteoblast enhancer is a critical step during Bmp2 upregulation during osteoblast differentiation.
Although chondrocytes that generate ossified matrix express Bmp2, Runx2, Spp1, and other markers of osteoblast differentiation, they are not thought to be the primary source of osteoblasts. Instead, the perichondrium likely contains an osteoprogenitor cell population (11, 34) throughout development. These observations suggest that similar regulatory networks controlling ossification processes are activated in chondrocytes and osteoblasts. Our Bmp2 lacZ-BACs allowed us to dissociate patterns of Bmp2 expression in developing endochondral bone and provided us with a robust means to compare Bmp2 transcription to that of other osteoblast differentiation markers. Bmp2 can activate Spp1 expression and stimulate bone matrix production in vitro (46). Expression of both Runx2 and Bmp2 precedes perichondrial cell maturation into bone matrix-associated osteogenic tissue. These findings strongly suggest that Bmp2 is upregulated in perichondrial osteoblast progenitor cells during their commitment to becoming mature, bone matrix-producing osteoblasts. Because Bmp2 is a potent inducer of osteoblast differentiation, this also suggests that a local increase of Bmp2 production by osteoblast progenitor cells likely stimulates a cascade of downstream molecular events, via autocrine or paracrine signaling, that drives their further differentiation. This upregulation may be facilitated by signals such as Ihh, which signals from the prehypertrophic chondrocytes to the perichondrium and is required for osteoblast differentiation in vivo (48, 66). Ihh misexpression can also upregulate Bmp2 in chick limb perichondrium (60). Therefore, chondrocytes may also regulate Bmp2 transcription in perichondrial cells via hedgehog signaling.
Disruption of Runx2 leads to defects in both endochondral and intramembranous osteoblast differentiation (42, 57). However, studies of Ihh mutant mice provide some evidence that intramembranous and endochondral osteoblast differentiation are distinct (66). Our observation that the osteoblast progenitor control element drives expression in cells located in both endochondral and intramembranous bones suggests that a common Bmp2 regulatory mechanism exists in bones that develop with or without a cartilage model. The identification of factors that interact with this enhancer sequence will help to better understand osteoblast differentiation during endochondral and intramembranous bone development as well as upstream pathways that regulate Bmp2 transcription during these ossification processes.
Implications of BMP2 regulation for human disease.
Although multiple factors are thought to influence the development of OP, it has become increasingly clear that the level of lifetime peak bone mass is an important measure of predisposition to OP (62). In addition, peak bone mass is attained in young adulthood, suggesting that future OP risk is partly dependent on developmental processes. To ensure proper bone formation and attainment of peak bone mass, an adequate supply of functional, bone matrix-producing osteoblasts is needed. Recently, genetic linkage of BMP2 to OP has been reported (67). Interestingly, in that study, genetic variation at BMP2 was associated with attainment of premenopausal peak bone mass as well as clinical features of OP (67), suggesting that variation in BMP2 signaling affects the production of bone early in life. In addition, BMP2 coding variants could not fully explain the OP linkage data (67). We speculate that noncoding sequence variants within the human BMP2 locus influence levels of BMP2 transcription in bone and thus could contribute to bone mass and OP by affecting osteoblast differentiation and/or activity. If this is true, such variants may influence the activity of the osteoblast enhancer identified here.
Supplementary Material
Acknowledgments
We thank Laura Selenke and Lissett Ramirez for technical assistance and Karen Deal, Maureen Gannon, Anna Means, and Laura Wilding for generously sharing equipment and advice. We thank Kyle Gurley and David Kingsley for assistance with constructing the pIBG-FTet plasmid, Scott Baldwin and Kevin Tompkins for assistance with pronuclear injections, Brigid Hogan for providing the Bmp2 in situ probe, Soren Warming and Neal Copeland for providing BAC recombination reagents, and the Vanderbilt CHGR DNA Resources Core for real-time PCR assistance and TaqMan probe/primer design. We also kindly thank Steve Harris for discussions regarding the previously reported Bmp2 promoter transgene and Nyk Reed and Michelle Southard-Smith for helpful discussions on the manuscript. We thank Michelle Southard-Smith for help with Southern blothing.
Ronald L. Chandler was supported by NIH Developmental Biology Training Grant 5T32HD07502-08. Kelly J. Chandler was supported by NIH Genetics Training Grant 1T32GM62758-03. Douglas P. Mortlock was supported by NIH Grant 1R01HD47880-01. Transgenic mice were generated by the Vanderbilt University Transgenic and ES Cell Shared Resource, which is supported by the Vanderbilt Cancer, Diabetes, Kennedy, and Vision Centers. We acknowledge use of the VUMC CHGR DNA Resources Core Facility.
Footnotes
Published ahead of print on 5 February 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abrams, K. L., J. Xu, C. Nativelle-Serpentini, S. Dabirshahsahebi, and M. B. Rogers. 2004. An evolutionary and molecular analysis of Bmp2 expression. J. Biol. Chem. 279:15916-15928. [DOI] [PubMed] [Google Scholar]
- 2.Aubin, J. E. 1998. Advances in the osteoblast lineage. Biochem. Cell Biol. 76:899-910. [PubMed] [Google Scholar]
- 3.Berman, B. P., Y. Nibu, B. D. Pfeiffer, P. Tomancak, S. E. Celniker, M. Levine, G. M. Rubin, and M. B. Eisen. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitgood, M. J., and A. P. McMahon. 1995. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 172:126-138. [DOI] [PubMed] [Google Scholar]
- 5.Blackman, R. K., M. Sanicola, L. A. Raftery, T. Gillevet, and W. M. Gelbart. 1991. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111:657-666. [DOI] [PubMed] [Google Scholar]
- 6.Blanchette, M., W. J. Kent, C. Riemer, L. Elnitski, A. F. Smit, K. M. Roskin, R. Baertsch, K. Rosenbloom, H. Clawson, E. D. Green, D. Haussler, and W. Miller. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouma, G. J., G. T. Hart, L. L. Washburn, A. K. Recknagel, and E. M. Eicher. 2004. Using real time RT-PCR analysis to determine multiple gene expression patterns during XX and XY mouse fetal gonad development. Gene Expr. Patterns 5:141-149. [DOI] [PubMed] [Google Scholar]
- 8.Brinster, R. L., H. Y. Chen, M. Trumbauer, A. W. Senear, R. Warren, and R. D. Palmiter. 1981. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 27:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., M. Zhao, and G. R. Mundy. 2004. Bone morphogenetic proteins. Growth Factors 22:233-241. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K. Y., H. J. Kim, M. H. Lee, T. G. Kwon, H. D. Nah, T. Furuichi, T. Komori, S. H. Nam, Y. J. Kim, and H. M. Ryoo. 2005. Runx2 regulates FGF2-induced Bmp2 expression during cranial bone development. Dev. Dyn. 233:115-121. [DOI] [PubMed] [Google Scholar]
- 11.Colnot, C., C. Lu, D. Hu, and J. A. Helms. 2004. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 269:55-69. [DOI] [PubMed] [Google Scholar]
- 12.DiLeone, R. J., G. A. Marcus, M. D. Johnson, and D. M. Kingsley. 2000. Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 97:1612-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiLeone, R. J., L. B. Russell, and D. M. Kingsley. 1998. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics 148:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducy, P., and G. Karsenty. 2000. The family of bone morphogenetic proteins. Kidney Int. 57:2207-2214. [DOI] [PubMed] [Google Scholar]
- 15.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 16.Dudley, A. T., and E. J. Robertson. 1997. Overlapping expression domains of bone morphogenetic protein family members potentially account for limited tissue defects in BMP7 deficient embryos. Dev. Dyn. 208:349-362. [DOI] [PubMed] [Google Scholar]
- 17.Erives, A., and M. Levine. 2004. Coordinate enhancers share common organizational features in the Drosophila genome. Proc. Natl. Acad. Sci. USA 101:3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, J. Q., L. Xing, J. H. Zhang, M. Zhao, D. Horn, J. Chan, B. F. Boyce, S. E. Harris, G. R. Mundy, and D. Chen. 2003. NF-kappaB specifically activates BMP-2 gene expression in growth plate chondrocytes in vivo and in a chondrocyte cell line in vitro. J. Biol. Chem. 278:29130-29135. [DOI] [PubMed] [Google Scholar]
- 19.Folger, K. R., E. A. Wong, G. Wahl, and M. R. Capecchi. 1982. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol. Cell. Biol. 2:1372-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzen, A., A. Oldberg, and M. Solursh. 1989. Possible recruitment of osteoblastic precursor cells from hypertrophic chondrocytes during initial osteogenesis in cartilaginous limbs of young rats. Matrix 9:261-265. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 22.Furuta, Y., D. W. Piston, and B. L. Hogan. 1997. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124:2203-2212. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh-Choudhury, N., G. G. Choudhury, M. A. Harris, J. Wozney, G. R. Mundy, S. L. Abboud, and S. E. Harris. 2001. Autoregulation of mouse BMP-2 gene transcription is directed by the proximal promoter element. Biochem. Biophys. Res. Commun. 286:101-108. [DOI] [PubMed] [Google Scholar]
- 24.Gong, S., C. Zheng, M. L. Doughty, K. Losos, N. Didkovsky, U. B. Schambra, N. J. Nowak, A. Joyner, G. Leblanc, M. E. Hatten, and N. Heintz. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425:917-925. [DOI] [PubMed] [Google Scholar]
- 25.Hager-Theodorides, A. L., S. V. Outram, D. K. Shah, R. Sacedon, R. E. Shrimpton, A. Vicente, A. Varas, and T. Crompton. 2002. Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J. Immunol. 169:5496-5504. [DOI] [PubMed] [Google Scholar]
- 26.Hardwick, J. C., G. R. Van Den Brink, S. A. Bleuming, I. Ballester, J. M. Van Den Brande, J. J. Keller, G. J. Offerhaus, S. J. Van Deventer, and M. P. Peppelenbosch. 2004. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology 126:111-121. [DOI] [PubMed] [Google Scholar]
- 27.Helvering, L. M., R. L. Sharp, X. Ou, and A. G. Geiser. 2000. Regulation of the promoters for the human bone morphogenetic protein 2 and 4 genes. Gene 256:123-138. [DOI] [PubMed] [Google Scholar]
- 28.Hogan, B. L. 1996. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10:1580-1594. [DOI] [PubMed] [Google Scholar]
- 29.Holtke, H. J., and C. Kessler. 1990. Non-radioactive labeling of RNA transcripts in vitro with the hapten digoxigenin (DIG); hybridization and ELISA-based detection. Nucleic Acids Res. 18:5843-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horner, A., L. Shum, J. A. Ayres, K. Nonaka, and G. H. Nuckolls. 2002. Fibroblast growth factor signaling regulates Dach1 expression during skeletal development. Dev. Dyn. 225:35-45. [DOI] [PubMed] [Google Scholar]
- 31.Howard, M. L., and E. H. Davidson. 2004. cis-Regulatory control circuits in development. Dev. Biol. 271:109-118. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda, T., S. Nomura, A. Yamaguchi, T. Suda, and S. Yoshiki. 1992. In situ hybridization of bone matrix proteins in undecalcified adult rat bone sections. J. Histochem. Cytochem. 40:1079-1088. [DOI] [PubMed] [Google Scholar]
- 33.Jung, H. S., V. Oropeza, and I. Thesleff. 1999. Shh, Bmp-2, Bmp-4 and Fgf-8 are associated with initiation and patterning of mouse tongue papillae. Mech. Dev. 81:179-182. [DOI] [PubMed] [Google Scholar]
- 34.Karsenty, G., and E. F. Wagner. 2002. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2:389-406. [DOI] [PubMed] [Google Scholar]
- 35.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khandekar, M., N. Suzuki, J. Lewton, M. Yamamoto, and J. D. Engel. 2004. Multiple, distant Gata2 enhancers specify temporally and tissue-specific patterning in the developing urogenital system. Mol. Cell. Biol. 24:10263-10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, D. G., H. M. Kang, S. K. Jang, and H. S. Shin. 1992. Construction of a bifunctional mRNA in the mouse by using the internal ribosomal entry site of the encephalomyocarditis virus. Mol. Cell. Biol. 12:3636-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, I. S., F. Otto, B. Zabel, and S. Mundlos. 1999. Regulation of chondrocyte differentiation by Cbfa1. Mech. Dev. 80:159-170. [DOI] [PubMed] [Google Scholar]
- 39.Kingsley, D. M. 1994. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 10:16-21. [DOI] [PubMed] [Google Scholar]
- 40.Kingsley, D. M., A. E. Bland, J. M. Grubber, P. C. Marker, L. B. Russell, N. G. Copeland, and N. A. Jenkins. 1992. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell 71:399-410. [DOI] [PubMed] [Google Scholar]
- 41.Kleinjan, D. A., and V. van Heyningen. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76:8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 43.Kulessa, H., G. Turk, and B. L. Hogan. 2000. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. EMBO J. 19:6664-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurokawa, D., H. Kiyonari, R. Nakayama, C. Kimura-Yoshida, I. Matsuo, and S. Aizawa. 2004. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development 131:3319-3331. [DOI] [PubMed] [Google Scholar]
- 45.Kurokawa, D., N. Takasaki, H. Kiyonari, R. Nakayama, C. Kimura-Yoshida, I. Matsuo, and S. Aizawa. 2004. Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development 131:3307-3317. [DOI] [PubMed] [Google Scholar]
- 46.Lecanda, F., L. V. Avioli, and S. L. Cheng. 1997. Regulation of bone matrix protein expression and induction of differentiation of human osteoblasts and human bone marrow stromal cells by bone morphogenetic protein-2. J. Cell Biochem. 67:386-396. [PubMed] [Google Scholar]
- 47.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 48.Long, F., U. I. Chung, S. Ohba, J. McMahon, H. M. Kronenberg, and A. P. McMahon. 2004. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131:1309-1318. [DOI] [PubMed] [Google Scholar]
- 49.Loots, G. G., R. M. Locksley, C. M. Blankespoor, Z. E. Wang, W. Miller, E. M. Rubin, and K. A. Frazer. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136-140. [DOI] [PubMed] [Google Scholar]
- 50.Lyons, K. M., B. L. Hogan, and E. J. Robertson. 1995. Colocalization of BMP 7 and BMP 2 RNAs suggests that these factors cooperatively mediate tissue interactions during murine development. Mech. Dev. 50:71-83. [DOI] [PubMed] [Google Scholar]
- 51.Lyons, K. M., R. W. Pelton, and B. L. Hogan. 1990. Organogenesis and pattern formation in the mouse: RNA distribution patterns suggest a role for bone morphogenetic protein-2A (BMP-2A). Development 109:833-844. [DOI] [PubMed] [Google Scholar]
- 52.Lyons, K. M., R. W. Pelton, and B. L. Hogan. 1989. Patterns of expression of murine Vgr-1 and BMP-2a RNA suggest that transforming growth factor-beta-like genes coordinately regulate aspects of embryonic development. Genes Dev. 3:1657-1668. [DOI] [PubMed] [Google Scholar]
- 53.Mortlock, D. P., C. Guenther, and D. M. Kingsley. 2003. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 13:2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mountford, P., B. Zevnik, A. Duwel, J. Nichols, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mundy, G., R. Garrett, S. Harris, J. Chan, D. Chen, G. Rossini, B. Boyce, M. Zhao, and G. Gutierrez. 1999. Stimulation of bone formation in vitro and in rodents by statins. Science 286:1946-1949. [DOI] [PubMed] [Google Scholar]
- 56.Nobrega, M. A., I. Ovcharenko, V. Afzal, and E. M. Rubin. 2003. Scanning human gene deserts for long-range enhancers. Science 302:413. [DOI] [PubMed] [Google Scholar]
- 57.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. Stamp, R. S. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765-771. [DOI] [PubMed] [Google Scholar]
- 58.Ovcharenko, I., G. G. Loots, M. A. Nobrega, R. C. Hardison, W. Miller, and L. Stubbs. 2005. Evolution and functional classification of vertebrate gene deserts. Genome Res. 15:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ovcharenko, I., M. A. Nobrega, G. G. Loots, and L. Stubbs. 2004. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32:W280-W286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pathi, S., J. B. Rutenberg, R. L. Johnson, and A. Vortkamp. 1999. Interaction of Ihh and BMP/Noggin signaling during cartilage differentiation. Dev. Biol. 209:239-253. [DOI] [PubMed] [Google Scholar]
- 61.Patton, J. T., and M. H. Kaufman. 1995. The timing of ossification of the limb bones, and growth rates of various long bones of the fore and hind limbs of the prenatal and early postnatal laboratory mouse. J. Anat. 186:175-185. [PMC free article] [PubMed] [Google Scholar]
- 62.Peacock, M., C. H. Turner, M. J. Econs, and T. Foroud. 2002. Genetics of osteoporosis. Endocrinol. Rev. 23:303-326. [DOI] [PubMed] [Google Scholar]
- 63.Portnoy, M. E., K. J. McDermott, A. Antonellis, E. H. Margulies, A. B. Prasad, D. M. Kingsley, E. D. Green, and D. P. Mortlock. 2005. Detection of potential GDF6 regulatory elements by multispecies sequence comparisons and identification of a skeletal joint enhancer. Genomics 86:295-305. [DOI] [PubMed] [Google Scholar]
- 64.Siepel, A., G. Bejerano, J. S. Pedersen, A. S. Hinrichs, M. Hou, K. Rosenbloom, H. Clawson, J. Spieth, L. W. Hillier, S. Richards, G. M. Weinstock, R. K. Wilson, R. A. Gibbs, W. J. Kent, W. Miller, and D. Haussler. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solloway, M. J., A. T. Dudley, E. K. Bikoff, K. M. Lyons, B. L. Hogan, and E. J. Robertson. 1998. Mice lacking Bmp6 function. Dev. Genet. 22:321-339. [DOI] [PubMed] [Google Scholar]
- 66.St-Jacques, B., M. Hammerschmidt, and A. P. McMahon. 1999. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13:2072-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Styrkarsdottir, U., J. B. Cazier, A. Kong, O. Rolfsson, H. Larsen, E. Bjarnadottir, V. D. Johannsdottir, M. S. Sigurdardottir, Y. Bagger, C. Christiansen, I. Reynisdottir, S. F. Grant, K. Jonasson, M. L. Frigge, J. R. Gulcher, G. Sigurdsson, and K. Stefansson. 2003. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PloS Biol. 1:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugiura, T. 1999. Cloning and functional characterization of the 5′-flanking region of the human bone morphogenetic protein-2 gene. Biochem. J. 338:433-440. [PMC free article] [PubMed] [Google Scholar]
- 69.Sykaras, N., and L. A. Opperman. 2003. Bone morphogenetic proteins (BMPs): how do they function and what can they offer the clinician? J. Oral Sci. 45:57-73. [DOI] [PubMed] [Google Scholar]
- 70.Takae, R., S. Matsunaga, N. Origuchi, T. Yamamoto, N. Morimoto, S. Suzuki, and T. Sakou. 1999. Immunolocalization of bone morphogenetic protein and its receptors in degeneration of intervertebral disc. Spine 24:1397-1401. [DOI] [PubMed] [Google Scholar]
- 71.Treier, M., A. S. Gleiberman, S. M. O'Connell, D. P. Szeto, J. A. McMahon, A. P. McMahon, and M. G. Rosenfeld. 1998. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 12:1691-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 74.Wozney, J. M., V. Rosen, A. J. Celeste, L. M. Mitsock, M. J. Whitters, R. W. Kriz, R. M. Hewick, and E. A. Wang. 1988. Novel regulators of bone formation: molecular clones and activities. Science 242:1528-1534. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, H., and A. Bradley. 1996. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 122:2977-2986. [DOI] [PubMed] [Google Scholar]
- 76.Zhao, M., M. Qiao, S. E. Harris, D. Chen, B. O. Oyajobi, and G. R. Mundy. 2006. The zinc finger transcription factor gli2 mediates bone morphogenetic protein 2 expression in osteoblasts in response to hedgehog signaling. Mol. Cell. Biol. 26:6197-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.