Abstract
Treatment of yeast and human cells with DNA-damaging agents elicits lysine 48-linked polyubiquitylation of Rpb1, the largest subunit of RNA polymerase II (Pol II), which targets Pol II for proteasomal degradation. However, the ubiquitin ligase (E3) responsible for Pol II polyubiquitylation has not been identified in humans or the yeast Saccharomyces cerevisiae . Here we show that elongin A (Ela1) and cullin 3 (Cul3) are required for Pol II polyubiquitylation and degradation in yeast cells, and on the basis of these and other observations, we propose that an E3 comprised of elongin C (Elc1), Ela1, Cul3, and the RING finger protein Roc1 (Rbx1) mediates this process in yeast cells. This study provides, in addition to the identification of the E3 required for Pol II polyubiquitylation and degradation in yeast cells, the first evidence for a specific function in yeast for a member of the elongin C/BC-box protein/cullin family of ligases. Also, these observations raise the distinct possibility that the elongin C-containing ubiquitin ligase, the von Hippel-Lindau tumor suppressor complex, promotes Pol II polyubiquitylation and degradation in human cells.
Nucleotide excision repair (NER) is a versatile DNA repair process which operates on bulky helix-distorting lesions caused by a variety of DNA-damaging agents, including UV and chemical agents. In eukaryotes, following the initial recognition of the DNA lesion by the damage recognition factors, the damaged strand is unwound from the undamaged strand by the DNA helicases present in TFIIH to promote the dual incision of the damaged strand on the 5′ side and the 3′ side of the lesion by two separate endonucleases, which then results in the removal of a fragment of ∼30 nucleotides containing the lesion (21, 25).
In eukaryotes, the genetic controls of NER for the nontranscribed regions and the transcribed regions of the genome differ in some important respects. For example, in the yeast Saccharomyces cerevisiae, the RAD7 and RAD16 genes are specifically required for the repair of nontranscribed regions of the genome as well as for the repair of the nontranscribed DNA strand (21). The Rad7 and Rad16 proteins form a heterodimeric complex in which Rad16, a member of the Swi2/Snf2 family of ATPases, provides a DNA-dependent ATPase activity, and the Rad7-Rad16 complex binds preferentially to UV-damaged DNA in an ATP-dependent manner (8, 9). We previously suggested a role for the Rad7-Rad16 complex in the scanning of nontranscribed regions of the genome for DNA lesions and in initiating the nucleation of NER protein factors at the lesion site (8, 9). Rad16 also harbors a C3HC4 motif, a characteristic feature of ubiquitin (Ub) ligases; however, how Rad16 functions in such a role is not understood.
DNA lesions from the transcribed strand of expressed genes are removed faster than lesions from the nontranscribed strand, a phenomenon known as transcription-coupled repair (TCR) (17, 18, 29). TCR is conserved from Escherichia coli to humans, and the process in E. coli, in which a transcription repair coupling factor promotes the release of the stalled RNA polymerase along with the nascent transcript from the damage site and then recruits the NER proteins for lesion removal, is well understood (27). In humans, TCR is modulated by Cockayne syndrome A and B proteins (CSA and CSB), and in yeast, TCR is modulated by Rad26, which is the CSB counterpart (21, 25). Although it is generally accepted that the stalled RNA polymerase II (Pol II) has to be removed from the lesion site for the repair machinery to assemble there, it is not known whether Pol II is removed or translocated away from the lesion site or whether it undergoes some sort of conformational change to allow the assembly of NER proteins at the lesion site.
Another phenomenon that is observed upon treatment of yeast or human cells with DNA-damaging agents is lysine 48-linked polyubiquitylation of the Rpb1 subunit of Pol II, which targets Pol II for proteasomal degradation (6, 23, 24). It remains unclear, however, whether Pol II polyubiquitylation contributes to TCR by targeting it for degradation, thereby promoting the assembly of the NER proteins at the lesion site. Moreover, since the Ub ligase (E3) responsible for Pol II polyubiquitylation in yeast or human cells has not been identified, this phenomenon has not been amenable to genetic analysis.
Our recent observation that elongin C (Elc1) was required for Pol II polyubiquitylation and degradation in yeast cells raised the possibility that an Elc1-dependent Ub ligase might be involved in this process (24). Yeast Elc1 is a homolog of mammalian elongin C (1), which forms a heterotrimeric complex with elongins A and B (3). In mammalian cells, the elongin A, B, and C complex increases the rate of transcription elongation by suppressing Pol II pausing (4, 5). In yeast cells, however, the only elongins present are elongins A and C, and the complex of these two proteins does not stimulate transcription elongation by Pol II (14). Since Elc1 is required for Pol II polyubiquitylation and degradation in yeast cells (24), and since it exists in vivo in a complex with the Rad7 and Rad16 proteins (22), we first considered the possibility that this protein complex was involved in modulating Pol II modification. The Elc1-Rad7-Rad16 complex shares several characteristic features of the SCF (Skp1-Cullin-F box)-type Ub ligases (15). In the SCF complex, the Skp1 subunit acts as an adaptor which connects the F-box protein to the scaffold protein Cdc53/cullin, to which the RING-H2 finger protein Rbx1/Roc1 binds. The F-box protein binds the protein substrates targeted for ubiquitylation by the Ub-conjugating enzyme (E2) which assembles with the Cdc53/cullin subunit. In the Rad7-Rad16-Elc1 complex, Rad7 is an F-box protein, Rad16 is a RING-H2 protein, and Elc1 is a Skp1 homolog. However, our finding that Pol II polyubiquitylation and degradation were not affected in rad7Δ or rad16Δ yeast cells treated with UV or 4-NQO (4-nitroquinoline-1-oxide) (24) led us to consider other possibilities.
Here we show that elongin A (Ela1) and cullin 3 (Cul3) are required for Pol II polyubiquitylation and degradation in DNA-damaged yeast cells, and based upon these and other observations, we propose that an E3 comprised of Elc1, Ela1, Cul3, and Roc1 (Rbx1) mediates this process in yeast cells. Further, our genetic analyses indicate that Pol II polyubiquitylation and degradation are not a prerequisite for Rad26-mediated TCR; rather, these two processes provide separate means of lesion removal from the transcribed strand. In addition, these observations lead us to predict that in human cells, the von Hippel-Lindau (VHL) tumor suppressor complex (28, 32), comprised of elongin B, elongin C, cullin 2, and Rbx1 subunits, would function in an analogous manner in mediating Pol II polyubiquitylation and degradation.
MATERIALS AND METHODS
Yeast strains.
The strains used in this study are derivatives of the wild-type strain YRP668 (MATa his3-Δ1 leu2-3,112 trp1Δ::URA3+ ura3-52) and are as follows: EMY74-7, MATa his3-Δ1 leu2-3,112 trp1Δ ura3-52; YRP785, cul3Δ; YRP738, ela1Δ; YRP669, elc1Δ; YR7-50, rad7Δ; YR26-1, rad26Δ; YR7-73, rad7Δ elc1Δ; YR26-124, rad26Δ elc1Δ; YR7-73, rad7Δ elc1Δ; YR26-160, rad26Δ ela1Δ; YR26-124, rad26Δ elc1Δ; YR26-162, rad26Δ cul3Δ; YR7-75, rad7Δ ela1Δ; and YR7-76, rad7Δ cul3Δ. Standard genetic techniques were used for construction and growth of strains.
Deletion of the ELA1 and CUL3 genes.
Microhomology-mediated PCR targeting was used for constructing strains in which the genomic copy of the ELA1 or CUL3 gene was deleted. Briefly, DNA fragments containing the entire HIS3 gene as the selectable marker, flanked by 45 base pairs 5′ of the initiating ATG codon and 45 base pairs 3′ of the termination codons TAG and TAA for the ELA1 and CUL3 genes, respectively, were generated by PCR. These disruption cassettes were used to transform the strain EMY74.7, and His3+ colonies were screened for the correct integration pattern by PCR using appropriate primer pairs for each of the two genes.
UV and 4-NQO sensitivity.
Cells were grown in yeast extract-peptone-dextrose (YPD) and harvested when they reached exponential phase. They were then centrifuged and suspended in water at a density of 2 × 108 cells/ml. A 5-μl sample of sequential 10-fold serial dilutions was spotted onto YPD plates, and when the spots dried, the plates were UV irradiated and then incubated at 30°C in the dark. In order to check 4-NQO sensitivity, 10-fold serial dilutions of cells were spotted onto YPD plates containing the indicated amount of 4-NQO and incubated at 30°C.
UV and 4-NQO treatment of yeast cells.
For UV treatment, cells grown overnight in YPD culture were diluted in YPD to an optical density at 600 nm (OD600) of 0.2. When the culture reached an OD600 of 1.0, cells were centrifuged and resuspended to the same density in an equal volume of phosphate-buffered saline. A 50-ml cell suspension was exposed to 400 J/m2 of UV irradiation in a large petri dish (150 by 15 mm) with vigorous and continuous stirring. After irradiation, the cell suspension was mixed with 50 ml of 2× YPD medium in a flask and placed in the dark at 30°C in a shaking water bath. Aliquots of cells were removed at the indicated times for protein extraction.
For 4-NQO treatment, logarithmically growing yeast cells as described above for UV irradiation were treated with 4-NQO by adding appropriate volumes of 10 mg/ml stock solution of 4-NQO dissolved in dimethyl sulfoxide to the culture medium, followed by incubation for 30 min. After treatment, cells were centrifuged, washed, and processed for protein extraction.
Preparation of yeast cell extracts.
UV- or 4-NQO-treated yeast cells were harvested and resuspended in 1 ml Y-PER lysis buffer (Pierce, Rockford, IL) containing a protease inhibitor mixture (Mini-Complete; Roche, Mannheim, Germany), phosphatase inhibitor cocktail II (Sigma), and 10 mM N-ethylmaleimide (Sigma). One milliliter of Y-PER buffer was added per gram of cell pellet. Following breakage of cells with glass beads by using a Mini-Bead Beater (Biospec, Bartlesville, OK), which yielded greater than 90% broken cells, cellular debris and unbroken cells were removed by centrifugation at 20,000 × g for 10 min. Protein concentrations were determined by the Bio-Rad protein assay. Equivalent protein amounts (25 μg) were loaded onto a 4 to 20% gradient sodium dodecyl sulfate (SDS)-polyacrylamide gel (precast gel; Bio-Rad). Western analysis for Rpb1 was done using monoclonal antibody (MAb) 8WG16 (Promega) mouse RNA Pol II antibody. As a loading control, phosphoglycerate kinase 1 (PGK1) levels were examined in the same gel by using mouse MAb PGK1 (Molecular Probes).
In vivo ubiquitylation of Rpb1.
Yeast strains were transformed with plasmid YEp112, which encodes an epitope from hemagglutinin (HA) of influenza virus attached to the amino terminus of Ub driven by the copper-inducible CUP1 promoter. When transformed cells reached an OD600 of 1.0 in selective minimal medium, they were induced with 100 μM CuSO4 for 2 h and then treated with 4-NQO (6 μg/ml) for 30 min. For UV treatment, 50 ml of cells were placed in a 150- by 15-mm petri dish and irradiated, and yeast cell extracts were prepared as described above. The cleared cell extracts were incubated for 2 h with prewashed anti-HA affinity gel (EZview Red anti-HA affinity gel; Sigma) and washed twice with the same Y-PER lysis buffer as used for the preparation of cell extracts (see above). The affinity gel was then resuspended in the same volume of 2× SDS loading buffer and boiled for 5 min. The eluted proteins were loaded onto an SDS-6% polyacrylamide gel and electrophoresed for 3 h at 150 V. Ubiquitylated Pol II was visualized by Western analysis using mouse RNA Pol II H14 MAb (Covance).
RESULTS AND DISCUSSION
Since Ela1 exists in a complex with Elc1 in yeast cells and contains a BC-box motif characteristic of other substrate recognition subunits of the elongin C-containing Ub ligases (2, 14), it may function with Elc1 together with one of the cullins in Pol II polyubiquitylation and degradation. Yeast cells contain three cullin genes, CDC53 (CUL1), CUL3, and CUL8. Cdc53 is a component of the SCF Ub ligase which targets the ubiquitylation of G1 cyclin-dependent kinase inhibitors and of G1 cyclins and controls the transition from G1 phase to S phase in the cell cycle (15). While Cdc53 physically associates with Skp1, it shows no evidence of binding Elc1 (11). Cul3 and Cul8 are functionally distinct from Cdc53, and they do not interact with Skp1 (19). However, since cullin 8 has been suggested to be a component of an E3 required for cell cycle progression (19), we examined whether cullin 3 was involved in Pol II polyubiquitylation.
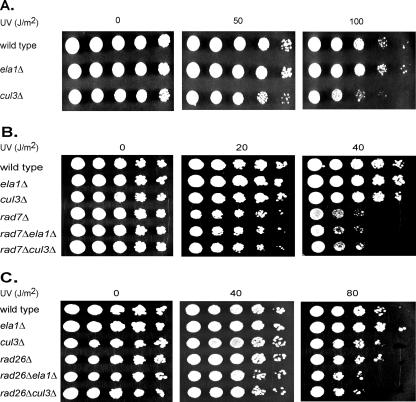
Because the UV sensitivities of rad7Δ and rad16Δ strains are not affected by the introduction of the elc1Δ mutation into them and the UV sensitivity of rad26Δ is enhanced by the elc1Δ mutation (24), we first determined whether the ela1Δ and cul3Δ mutations also sensitize yeast cells to UV damage in a way similar to the elc1Δ mutation. As we have reported previously for the elc1Δ mutation, we found that neither the ela1Δ mutation nor the cul3Δ mutation affects UV sensitivity to an appreciable degree (Fig. 1A), and the UV sensitivity of the rad7Δ strain is not affected upon the introduction of the ela1Δ or the cul3Δ mutation into it (Fig. 1B). The UV sensitivity of the rad26Δ strain, however, shows an increase upon the introduction of the ela1Δ or cul3Δ mutation (Fig. 1C).
FIG. 1.
Effects of ela1Δ and cul3Δ mutations on UV sensitivity. (A) UV sensitivity of ela1Δ and cul3Δ strains. (B) UV sensitivity conferred by the ela1Δ and cul3Δ mutations in combination with the rad7Δ mutation. (C) UV sensitivity conferred by the ela1Δ and cul3Δ mutations in combination with the rad26Δ mutation. YPD plates containing 5 μl of serial 10-fold dilutions of exponentially growing yeast cells were UV irradiated and incubated in the dark at 30°C.
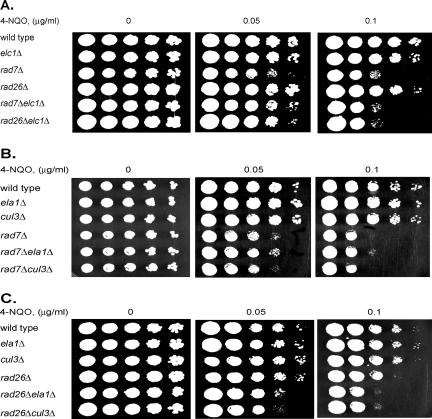
To verify that the epistatic relationships observed upon UV damage extend to other types of DNA damage, we examined the sensitivities of 4-NQO-treated yeast cells with elc1Δ, ela1Δ, and cul3Δ mutations in combination with the rad7Δ and rad26Δ mutations. Whereas all three mutations exhibited epistasis with the rad7Δ mutation, a clear enhancement of sensitivity was observed when any of these mutations was combined with the rad26Δ mutation (Fig. 2). Thus, with both UV and 4-NQO, the elc1Δ, ela1Δ, and cul3Δ mutations exhibited epistasis with the rad7Δ mutation and an enhancement in sensitivity with the rad26Δ mutation.
FIG. 2.
Effects of elc1Δ, ela1Δ, and cul3Δ mutations on sensitivity to 4-NQO. (A) 4-NQO sensitivity conferred by the elc1Δ mutation in combination with the rad7Δ and rad26Δ mutations. (B) 4-NQO sensitivity conferred by the ela1Δ and cul3Δ mutations in combination with the rad7Δ mutation. (C) 4-NQO sensitivity conferred by the ela1Δ and cul3Δ mutations in combination with the rad26Δ mutation. Five microliters of serial 10-fold dilutions of yeast cells was spotted onto YPD plates containing the indicated amounts of 4-NQO and incubated at 30°C.
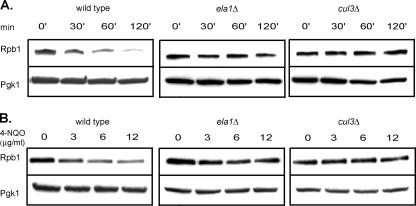
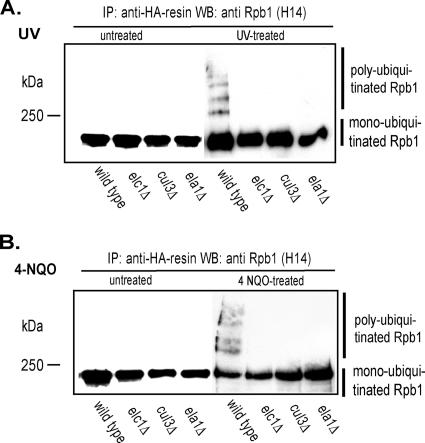
Next, we determined whether the ELA1 and CUL3 genes were required for Pol II polyubiquitylation and degradation in UV- or 4-NQO-treated yeast cells. As is shown in Fig. 3, in contrast to wild-type cells, in which the Rpb1 levels decline following treatment with UV or 4-NQO, the Rpb1 levels remained stable in ela1Δ or cul3Δ cells. Also, in contrast to the extensive polyubiquitylation of Rpb1 that occurs in UV- or 4-NQO-treated wild-type cells, no Rpb1 polyubiquitylation could be discerned in the ela1Δ or cul3Δ cells (Fig. 4). From these observations, we conclude that Ela1 and Cul3 function together with Elc1 in mediating Pol II polyubiquitylation and degradation in DNA-damaged yeast cells.
FIG. 3.
Rpb1 is not degraded in UV- or 4-NQO-treated ela1Δ or cul3Δ cells. (A) Rpb1 levels in wild-type, ela1Δ, and cul3Δ strains following UV treatment. Yeast cell extracts were prepared at the indicated times after UV irradiation. Rpb1 and the loading control PGK1 were detected by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting with MAb 8WG16 and MAb PGK1, respectively. (B) Rpb1 levels in wild-type, ela1Δ, and cul3Δ strains following treatment with 4-NQO. 4-NQO was added to liquid cultures of log-phase yeast cells at the indicated concentrations, and cells were collected at 60 min after the addition. Cell extracts were prepared, and Rpb1 and PGK1 detected by Western blot analysis as performed for the cell extracts shown in panel A.
FIG. 4.
Rpb1 polyubiquitylation does not occur in UV- or 4-NQO-treated ela1Δ and cul3Δ cells. (A) Absence of Rpb1 polyubiquitylation in UV-treated ela1Δ and cul3Δ cells. Yeast strains harboring plasmid YEp112, which expresses HA-tagged wild-type Ub, were grown to an OD600 of 1.0 in selective minimal medium, and Ub expression from the CUP1 promoter was induced with 100 μM CuSO4 for 2 h. After induction, cells in suspension were irradiated with UV (400 J/m2). Ubiquitylated proteins were purified with anti-HA resin (Sigma), separated by SDS-6% PAGE, and Rpb1 ubiquitylation analyzed with Rpb1-specific H14 antibody (Covance). (B) Absence of Rpb1 polyubiquitylation in 4-NQO-treated ela1Δ and cul3Δ cells. Methods were the same as described for panel A, except that cells were treated with 6 μg/ml of 4-NQO for 30 min. IP, immunoprecipitation; WB, Western blotting.
The yeast genome contains a single gene that encodes a RING protein, Roc1 (regulator of cullins), that binds cullins. Roc1 physically and functionally interacts with the three highly related yeast cullins, Cdc53 (Cul1), Cul3, and Cul8, and it interacts also with the distantly related cullin Apc2 (11, 13, 19, 20). Because of the requirement of Roc1 as a component of SCF and of many other Ub ligases, ROC1 is an essential gene (13). Although we have not examined the effects of ROC1 inactivation upon Pol II polyubiquitylation and degradation, the fact that Roc1 is the only RING protein in yeast that binds the various cullins, including cullin 3, we presume that Roc1 also is indispensable for Pol II polyubiquitylation and degradation.
Based upon our observations, we propose that a novel Ub ligase, comprised of the Elc1, Ela1, Cul3, and Roc1 subunits, mediates the polyubiquitylation of Rpb1 subunit in Pol II in DNA-damaged yeast cells. Further support for this proposal is provided from the observation that these four yeast proteins can in fact be assembled into a complex in vitro (Joan and Ron Conaway, personal communication). We expect Ela1, a BC-box protein (2, 14), to mediate the binding to Pol II in this Ub ligase. This study provides the first evidence for a specific function in yeast for a member of the elongin C/BC-box protein/cullin family of ligases.
Although Elc1 physically associates with the Rad7-Rad16 protein complex and a complex of these three protein can be purified, there has been no evidence that a cullin is associated with either the Rad7 (10) or Rad16 (7) protein in yeast cells, and all attempts to identify an E3 activity for this complex in vitro have been unsuccessful (22). Moreover, in coimmunoprecipitation experiments with yeast cell extracts, we found no evidence for the physical association of Ela1, Cul3, or Roc1 with the Rad7 or Rad16 protein. These various observations lead us to suggest that the Elc1-Rad7-Rad16 complex is a separate functional entity from the proposed Elc1-Ela1-Cul3-Roc1 Ub ligase, and that these two complexes affect the repair of DNA lesions from the transcribed strand in different ways.
It has been generally assumed that for repair of DNA lesions, such as those inflicted by UV irradiation, the stalled Pol II has to be removed from the lesion site so that the NER machinery can assemble there. In such a scheme, Rad26 could act by recognizing the stalled Pol II and by coordinating the assembly of the NER machinery at the lesion site. Such a model posits that Pol II polyubiquitylation, its consequent removal, and the repair of the lesion, including the involvement of Rad26, will all be interdependent processes. In that case, inactivation of any of the components of the Ub ligase required for Pol II polyubiquitylation would be expected to show epistasis with the rad26Δ mutation, since they all will affect the sequence of events in the same pathway.
Our observations, however, that the elc1Δ, ela1Δ, and cul3Δ mutations enhance the UV and 4-NQO sensitivities of rad26Δ cells run counter to the notion that Pol II removal via its polyubiquitylation is a prerequisite for TCR, and they suggest, instead, that Pol II polyubiquitylation and Rad26's role in TCR represent two separate means for effecting the repair of lesions from the transcribed strand. In fact, the elegant studies with human CSB indicating that Pol II remains stalled at the site of a cis-syn thymine dimer and that in spite of this the lesion can be removed by the NER enzyme complex suggest that removal of Pol II from the lesion site is not a prerequisite for the NER ensemble to access the lesion (26). Thus, we assume that the role of CSB in humans or of Rad26 in yeast is to modify the stalled Pol II in such a way as to increase the accessibility of the lesion to the NER ensemble, and CSB/Rad26 might achieve this by either pushing back the stalled Pol II from the lesion site or changing its conformational state (12, 30, 31). A host of proteins that mediate normal transcription elongation might be involved in such a process together with Rad26. However, in the regions of the transcribed strand where Rad26-dependent TCR may be unable to act on lesions and which we assume to occur in the promoter regions because of the absence of various transcription elongation factors or in the absence of Rad26, as in the rad26Δ strain, Pol II removal upon Elc1-Ela1-Cul3-Roc1 Ub ligase-mediated polyubiquitylation may be a necessary precondition for repair to take place. In this pathway, following Pol II removal, the repair of the transcribed strand may require the Rad7-Rad16-Elc1 complex to bind the DNA lesion and to instigate the assembly of NER proteins (8, 9).
The roles proposed above for Rad26 and for the Rad7-Rad16 protein complex in mediating the repair of the transcribed strand in two separate ways are supported from the observation that even though the fast phase of repair is diminished by the rad26Δ mutation, in rad26Δ cells the transcribed strand still continues to be repaired at the same rate as the nontranscribed strand (33). And since the repair of the transcribed strand is greatly diminished in the rad7Δ rad26Δ or rad16Δ rad26Δ double mutants compared to that in the rad26Δ mutant, the Rad7 and Rad16 proteins contribute to a very substantial portion of transcribed strand repair in the absence of Rad26 (33). Our suggestion that the removal of Pol II upon its polyubiquitylation promotes the Rad7-Rad16-dependent repair of the transcribed strand is supported by the epistasis of rad7Δ over elc1Δ, ela1Δ, and cul3Δ mutations for both UV and 4-NQO sensitivities.
In humans, the VHL Ub ligase complex, comprised of elongins B and C, cullin2, and Rbx1, could act in a manner analogous to that of yeast Elc1, Ela1, Cul3, and Roc1 (Rbx1) E3 in promoting Pol II polyubiquitylation and its subsequent degradation in DNA-damaged cells. In the VHL complex, the VHL protein, which contains a BC-box embedded within a larger SOCS motif, binds the protein substrates targeted for ubiquitylation (32). Although the evidence that the VHL complex binds specifically to hyperphosphorylated Rpb1 and targets it for ubiquitylation (16) is in line with this view, it remains to be determined whether the VHL complex in fact targets Pol II for polyubiquitylation and degradation in mammalian cells treated with DNA-damaging agents.
Acknowledgments
This work was supported by National Institutes of Health grants CA35035 and CA41261.
We are grateful to Joan and Ron Conaway for their helpful suggestions and for permission to cite their unpublished observations.
Footnotes
Published ahead of print on 12 February 2007.
REFERENCES
- 1.Aso, T., and M. N. Conrad. 1997. Molecular cloning of DNAs encoding the regulatory subunits of elongin from Saccharomyces cerevisiae and Drosophila melanogaster. Biochem. Biophys. Res. Commun. 241:334-340. [DOI] [PubMed] [Google Scholar]
- 2.Aso, T., D. Haque, R. J. Barstead, R. C. Conaway, and J. W. Conaway. 1996. The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 15:5557-5566. [PMC free article] [PubMed] [Google Scholar]
- 3.Aso, T., W. S. Lane, J. W. Conaway, and R. C. Conaway. 1995. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269:1439-1443. [DOI] [PubMed] [Google Scholar]
- 4.Bradsher, J. N., K. W. Jackson, R. C. Conaway, and J. W. Conaway. 1993. RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J. Biol. Chem. 268:25587-25593. [PubMed] [Google Scholar]
- 5.Bradsher, J. N., S. Tan, H. J. McLaury, J. W. Conaway, and R. C. Conaway. 1993. RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J. Biol. Chem. 268:25594-25603. [PubMed] [Google Scholar]
- 6.Bregman, D. B., R. Halaban, A. J. van Gool, K. A. Henning, E. C. Friedberg, and S. L. Warren. 1996. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA 93:11586-11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin, A.-C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A.-M. Michon, C.-M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M.-A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Guzder, S. N., P. Sung, L. Prakash, and S. Prakash. 1998. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J. Biol. Chem. 273:6292-6296. [DOI] [PubMed] [Google Scholar]
- 9.Guzder, S. N., P. Sung, L. Prakash, and S. Prakash. 1997. Yeast Rad7-Rad16 complex specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J. Biol. Chem. 272:21665-21668. [DOI] [PubMed] [Google Scholar]
- 10.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, D. B. Sorensen, J. Matthiesen, R. S. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, T., E. Kwon, A. M. Chachulska, and L. E. Hyman. 2000. Novel roles for elongin C in yeast. Biochim. Biophys. Acta 1491:161-176. [DOI] [PubMed] [Google Scholar]
- 12.Jung, Y., and S. J. Lippard. 2006. RNA polymerase II blockage by cisplatin-damaged DNA: stability and polyubiquitylation of stalled polymerase. J. Biol. Chem. 281:1361-1370. [DOI] [PubMed] [Google Scholar]
- 13.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. J. Kaelin, S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 14.Koth, C. M., M. V. Botuyan, R. J. Moreland, D. B. Jansma, J. W. Conaway, R. C. Conaway, W. J. Chazin, J. D. Friesen, C. H. Arrowsmith, and A. M. Edward. 2000. Elongin from Saccharomyces cerevisiae. J. Biol. Chem. 275:11174-11180. [DOI] [PubMed] [Google Scholar]
- 15.Krek, W. 1998. Proteolysis and the G1-S transition: the SCF connection. Curr. Opin. Genet. Dev. 8:36-42. [DOI] [PubMed] [Google Scholar]
- 16.Kuznetsova, A. V., J. Meller, P. O. Schnell, J. A. Nash, M. L. Ignacak, Y. Sanchez, J. W. Conaway, R. C. Conaway, and M. F. Czyzyk-Krzeska. 2003. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc. Natl. Acad. Sci. USA 100:2706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellon, I., and P. C. Hanawalt. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342:95-98. [DOI] [PubMed] [Google Scholar]
- 18.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 19.Michel, J. J., J. F. McCarville, and Y. Xiong. 2003. A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J. Biol. Chem. 278:22828-22837. [DOI] [PubMed] [Google Scholar]
- 20.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 21.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 22.Ramsey, K. L., J. J. Smith, A. Dasgupta, N. Maqani, P. Grant, and D. T. Auble. 2004. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol. Cell. Biol. 24:6362-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratner, J. N., B. Balasubramanian, J. Corden, S. L. Warren, and D. R. Bregman. 1998. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. J. Biol. Chem. 273:5184-5189. [DOI] [PubMed] [Google Scholar]
- 24.Ribar, B., L. Prakash, and S. Prakash. 2006. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:3999-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 26.Selby, C. P., R. Drapkin, D. Reinberg, and A. Sancar. 1997. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selby, C. P., and A. Sancar. 1993. Molecular mechanism of transcription-repair coupling. Science 260:53-58. [DOI] [PubMed] [Google Scholar]
- 28.Shilatifard, A., R. C. Conaway, and J. W. Conaway. 2003. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72:693-715. [DOI] [PubMed] [Google Scholar]
- 29.Sweder, K. S., and P. C. Hanawalt. 1992. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl. Acad. Sci. USA 89:10696-10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tornaletti, S., S. M. Patrick, J. J. Turchi, and P. C. Hanawalt. 2003. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J. Biol. Chem. 278:35791-35797. [DOI] [PubMed] [Google Scholar]
- 31.Tornaletti, S., D. Reines, and P. C. Hanawalt. 1999. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J. Biol. Chem. 274:24124-24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyers, M., and R. Rottapel. 1999. VHL: a very hip ligase. Proc. Natl. Acad. Sci. USA 96:12230-12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhage, R. A., A. J. van Gool, N. de Groot, J. H. J. Hoeijmakers, P. van de Putte, and J. Brouwer. 1996. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol. 16:496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]