Abstract
The eleven Fanconi anemia (FA) proteins cooperate in a novel pathway required for the repair of DNA cross-links. Eight of the FA proteins (A, B, C, E, F, G, L, and M) form a core enzyme complex, required for the monoubiquitination of FANCD2 and the assembly of FANCD2 nuclear foci. Here, we show that, in response to DNA damage, Chk1 directly phosphorylates the FANCE subunit of the FA core complex on two conserved sites (threonine 346 and serine 374). Phosphorylated FANCE assembles in nuclear foci and colocalizes with FANCD2. A nonphosphorylated mutant form of FANCE (FANCE-T346A/S374A), when expressed in a FANCE-deficient cell line, allows FANCD2 monoubiquitination, FANCD2 foci assembly, and normal S-phase progression. However, the mutant FANCE protein fails to complement the mitomycin C hypersensitivity of the transfected cells. Taken together, these results elucidate a novel role of Chk1 in the regulation of the FA/BRCA pathway and in DNA cross-link repair. Chk1-mediated phosphorylation of FANCE is required for a function independent of FANCD2 monoubiquitination.
Fanconi anemia (FA) is an inherited cancer susceptibility disorder resulting from germ line disruption of 1 of 11 FA genes (10). The FA proteins cooperate in a common cellular pathway, the FA/BRCA pathway, and disruption of this pathway results in chromosome instability and mitomycin C (MMC) hypersensitivity. Eight of the FA proteins (A, B, C, E, F, G, L, and M) are assembled in a multifunctional FA core complex (complex 1) (22). The complex contains a translocase activity, encoded by the FANCM gene, which is required for its interaction with chromatin (20, 21). The complex also has a monoubiquitin E3 ligase activity, encoded by the FANCL gene, (19) and interacts with a novel E2 ubiquitin (Ub) conjugating enzyme (14). The putative substrate of the Ub ligase is the FANCD2 protein. Monoubiquitinated FANCD2 functions in a downstream complex (complex 2) containing FANCD1/BRCA2 (11, 29). The role of FANCD2-Ub in cross-link repair remains unknown.
FANCD2 monoubiquitination occurs during normal S-phase progression (28) or in response to DNA damage (5). ATR-defective (Seckel) cells (1) are defective in the DNA damage-inducible monoubiquitination of FANCD2 and FANCD2 nuclear foci assembly. ATR activates the checkpoint kinase, Chk1 (13, 31, 32), suggesting that Chk1 may play a role in the FA/BRCA pathway. Consistent with this model, disruption of Chk1 by small interfering RNA (siRNA) or by small-molecule inhibitors (12, 15) results in DNA cross-linker hypersensitivity and chromosome instability, a phenotype reminiscent of FA cells (2). In this paper, we address the role of Chk1 in the regulation of the FA/BRCA pathway.
MATERIALS AND METHODS
Cell culture.
HeLa cells, U2OS cells, GM0637 cells, and HEK293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 15% heat-inactivated fetal calf serum in a humidified 5% CO2 incubator at 37°C. DF1179 (FA-E) fibroblasts derived from an FA-E patient were cultured in Chang medium (Irvine Scientific) (generously provided by Akiko Shimamura, Children's Hospital, Harvard Medical School, Boston, MA). Epstein-Barr virus-transformed lymphoblasts EUFA130 (FA-E) were maintained in RPMI 1640 medium with 15% fetal calf serum.
Generation of DNA damage.
Cells were UV irradiated with a Stratalinker (Stratagene) at 50% to 70% confluence without any medium after being washed with phosphate-buffered saline (PBS) once in a 100-mm dish without the lid. After UV irradiation, fresh medium was added, and cells were continuously cultured for the indicated time before lysis. Gamma irradiation was delivered using a Gammacell 40 apparatus (MDS Nordion). For MMC (Sigma) treatment, cells were continuously exposed to the drug for the indicated time before lysis. MMC sensitivity assays of human fibroblasts and lymphoblasts were performed essentially as described previously with the modifications given below (5, 7). Human fibroblasts and lymphoblasts were seeded in duplicate in 96-well microplates at a density of 1,000 cells/well in appropriate medium. MMC was added at a final concentration of 0 to 200 μM. Cells were then incubated at 37°C in a 5% CO2 incubator for 5 days, and cell survival was then determined by staining nucleic acids with a proprietary dye (CyQUANT; Molecular Probes) and subsequently analyzed by a fluorescence microplate reader according to the manufacturer's protocol.
Plasmids and purification of recombinant proteins.
Human FANCE cDNA (3) (generously provided by J. deWinter and H. Joenje, Free University Medical Center, Amsterdam, The Netherlands) was subcloned into the retroviral vector pMMP-puro (24) by adding the FLAG tag at the amino terminus of FANCE to generate pMMP-puro-FLAG-FANCE. Specific single and double mutations (pMMP-FLAG-T346A, pMMP-FLAG-S374A, and pMMP-FLAG-T346A/S374A [TS/AA]) were introduced by using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. For construction of pGEX-FANCE consisting of residues 332 to 365 [pGEX-FANCE(332-365)], pGEX-FANCE(349-382), and pGEX-FANCE(332-382), PCR products of the corresponding fragments were ligated to the EcoRI/NotI sites of the plasmid pGEX4T-1 (Pharmacia). The cDNAs of FANCE(T346A), FANCE(S374A), and the double mutant FANCE(TS/AA) were produced with a QuikChange site-directed mutagenesis kit (Stratagene). A recombinant FANCE (consisting of residues 149 to 536) wild-type (rFANCEwt) construct was cloned to the EcoRI/HindIII sites of pET32a-PPS vector (Novagen). For the recombinant double mutant protein FANCE(rTS/AA), the fragment consisting of residues 149 to 536 with the changes T346A and S374A was produced by PCR using pMMP-FLAG-TS/AA as a template, and the product was cloned to the EcoRI/HindIII sites of pET32a-PPS vector. All constructs and mutants were confirmed by DNA sequencing.
Glutathione S-transferase (GST)-FANCE constructs spanning the FANCE sequence of residues 332 to 382 were expressed in E. coli BL21 cells. A GST-Cdc25C(200-256) construct (containing residues 200 to 256 of Cdc25C) (generously provided by Michael Yaffe, Massachusetts Institute of Technology, Boston, MA) was used as a positive control for in vitro kinase assays. The GST fusion proteins were then purified on glutathione S-Sepharose beads and used as substrates in the in vitro kinase reaction. The rFANCEwt and double mutant FANCE(rTS/AA) proteins were expressed and induced in E. coli BL21(DE3)RP cells and then were purified by metal affinity chromatography using polyhistidine (His) binding HiTrap chelating HP columns (Pharmacia). After recombinant proteins were eluted from the column by incubation with precision protease to cleave the N-terminal His tag sequence, further purification by HiTrapQFF anion exchange columns (Pharmacia) and S-200 gel filtration columns was performed.
Retroviral infection.
Production of pMMP retroviral supernatants and infection of fibroblasts or lymphoblasts were performed as previously described (5, 27).
Generation of anti-FANCE, anti-pT346-FANCE, and anti-pS374-FANCE antibodies.
A rabbit polyclonal antibody against FANCE was generated by Invitrogen (Zymed) using a C-terminal peptide of residues 521 to 536 of FANCE as an antigen source. For generation of phosphospecific antibodies (anti-pT346-FANCE and anti-pS374-FANCE), rabbits were immunized with a keyhole limpet hemocyanin-conjugated FANCE phosphopeptide, SDLGLLRLC(pT)WL or phosphopeptide LFLGRIL(pS)LTSS, derived from amino acids 337 to 348 or 367 to 378 of FANCE, respectively. Antibodies were affinity purified using the corresponding phosphorylated and nonphosphorylated peptide-conjugated gels.
Immunoblotting.
Cells were lysed, and whole-cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and subjected to Western blot analysis (5). The following antibodies were used: anti-FANCD2 (FI-17) (Santa Cruz Biotechnology), antihemagglutinin (anti-HA) (HA.11; Babcock), anti-FLAG (M2) (Sigma), anti-β-tubulin (Santa Cruz Biotechnology), anti-phopsho-317-Chk1 (Cell Signaling Technology), and anti-Chk1(G-4) (Santa Cruz Biotechnology).
In vitro kinase assay.
The GST fusion proteins of FANCE (2 μg) were incubated with purified recombinant Chk1 (100 ng) (Upstate Technology) in 30 μl of kinase buffer (20 mM Tris HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mm dithiothreitol, 10 μM ATP) containing 10 μCi of [γ32P]ATP. Kinase reactions were incubated for 30 min at 30°C, stopped by the addition of SDS sample buffer, boiled for 5 min, and then analyzed by SDS-PAGE and X-ray film autoradiography. In vitro kinase assays were performed using GST-Cdc25C(200-256) and GST for positive and negative controls. Assay conditions were the same as described above. The rFANCEwt and FANCE(rTS/AA) (3 μg each) proteins were incubated without or with 100 ng of purified recombinant Chk1, Chk2, mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2; also known as MK2) (Upstate Technology) or GST in 30 μl of kinase buffer (20 mM Tris HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mm dithiothreitol, 1 mM ATP) for 30 min at 30°C; the kinase reaction was stopped by the addition of SDS sample buffer, boiled for 5 min, and then analyzed by SDS-PAGE, followed by Western blotting with anti-pT346-FANCE, anti-pS374-FANCE antibodies.
Immunoprecipitation.
Immunoprecipitation was performed as previously described (29).
SiRNA and transfection.
Expression of targeted genes was knocked down by transient expression of siRNA directed against green fluorescent protein (GFP) (5′-AACACTTGTCACTACTTTCTC-3′), Chk1 (5′-AAGAAGCAGTCGCAGTGAAGA-3′), and ATR (5′-CAGGCACTAATTGTTCTTCAA-3′). Transfection of siRNAs was performed using Hiperfect (QIAGEN) according to the manufacturer's protocol. At 72 h of transfection, cells were exposed to DNA damage.
Immunofluorescence microscopy.
Preparation of cells for immunofluorescence microscopy was performed as described previously (29). Lymphoblast cell lines were grown on culture slides coated with poly-d-lysine (BD Bioscience) to promote adhesion for 36 h before treatment. Images were acquired using an Axioplan 2 imaging microscope (Carl Zeiss) equipped with a digital camera and processed using Openlab software.
Fluorescence-activated cell sorter analysis.
G2/M checkpoint analysis and DNA replication were performed as described previously (30). To detect a sub-G1 population, cells were harvested at 0, 24, 48, and 72 h after MMC (160 ng/ml) treatment, washed with PBS, fixed in 70% ethanol (106 cells per ml) for at least 1 h at 4°C, and permeabilized in 0.25% Triton X-100-PBS at 4°C for 15 min. Following washing with PBS, cells were resuspended in PBS containing 25 ml/μg of propidium iodide (Sigma) and 0.1 mg/ml of RNase A (Sigma) prior to fluorescence-activated cell sorter analysis using a Becton Dickinson FACSCalibur flow cytometer. Cell death was measured as the sub-G1 (less than 2N DNA content) population.
In vivo ubiquitination assay.
U2OS cells stably expressing empty vector, FLAG-FANCEwt, and FLAG-TS/AA were transfected with pMT123 (HA-Ub) using FuGENE6 (Roche) as described previously (29). After 48 h of transfection, cells were treated with UV (60 J/m2) and incubated for 2 h; then cells were lysed in 100 μl of denaturation buffer (20 mM Tris HCl [pH 7.5], 50 mM NaCl, 1% SDS) and heated to 95°C for 10 min. Debris was removed by centrifugation. The remaining portions of denatured lysates were diluted to 1.1 ml in TBS buffer (20 mM Tris HCl [pH 7.5], 50 mM NaCl, 1% Triton X-100) supplemented with protease and phosphatase inhibitors (10 μg/ml pepstatin A, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 25 mM NaF) and incubated for 30 min on ice to allow renaturation. Lysates were precleared with protein G-agarose beads (Sigma) for 30 min at 4°C. Cleared extracts were then incubated with anti-FLAG antibody overnight on the shaker at 4°C before protein G-agarose beads were added. The immunoprecipitated materials were washed four times with Tris-buffered saline buffer and separated by SDS-PAGE, followed by Western blotting with the indicated antibodies.
RESULTS
Phosphorylation of FANCE on two conserved sites is required for MMC resistance.
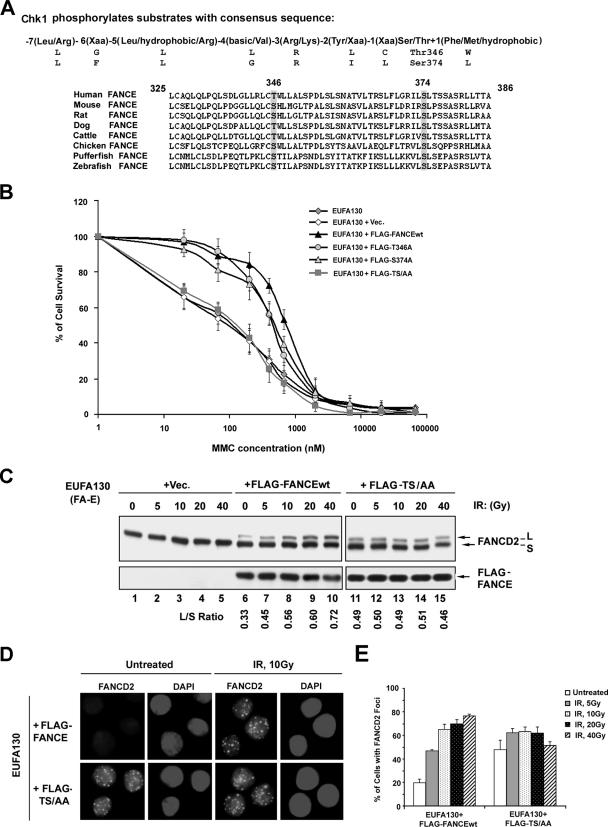
To examine the role of Chk1 in the FA/BRCA pathway, we scanned the primary amino acid sequence of eleven FA proteins (A, B, C, D1, D2, E, F, G, L, M, and J) for the Chk1 phosphorylation consensus sequence [−7(Leu/Arg) −6(Xaa) −5(Leu/hydrophobic/Arg) −4(basic/Val) −3(Arg/Lys) −2(Tyr/Xaa) −1(Xaa) Ser/Thr +1(Phe/Met/hydrophobic), where Xaa indicates no strong amino acid preference] (23). Two highly conserved phosphorylation sites were identified in the carboxy terminal region of the FANCE protein, Thr346 and Ser374 (Fig. 1A). To determine the functional relevance of these putative phosphorylation sites, we mutated each site, either individually or in combination, within the full-length FANCE protein. Patient-derived FA-E cell lines, EUFA130 lymphoblasts and DF1179 fibroblasts, were retrovirally transduced with the cDNA encoding either wt FANCE (FLAG-FANCEwt), mutant FANCE (carrying FLAG-T346A or FLAG-S374A), or the double point mutant FANCE carrying the FLAG tag [FLAG-FANCE(TS/AA)] (Fig. 1B) (see Fig. S1 in the supplemental material). While cells expressing FLAG-FANCEwt were MMC resistant, cells expressing the double mutant FLAG-FANCE(TS/AA) remained hypersensitive to MMC.
FIG. 1.
Two highly conserved Chk1 phosphorylation sites on FANCE. (A) Alignment of sequences surrounding the phosphorylation motif for Chk1 [R-X-X-(S/T)] in human FANCE with FANCE sequences in other organisms. (B) Complementation of MMC sensitivity of an FA-E lymphoblast cell line, EUFA130, with empty vector (pMMP), pMMP-FLAG-FANCE, pMMP-FLAG-T346A, pMMP-FLAG-S374A, and pMMP- FLAG-TS/AA. The indicated retroviral supernatants were generated and used to transduce EUFA130 cells. Puromycin-resistant cells were selected, and MMC sensitivity was determined as described in Materials and Methods. The values shown are the means ± standard deviations from four separate experiments. (C) Restoration of monoubiquitination of FANCD2. The indicated stably transduced FA-E lymphoblast cell lines were either untreated or exposed to IR at different doses, as indicated, and harvested after 6 h. Western blotting was performed with anti-FANCD2 or anti-FLAG antibodies. (D) Restoration of FANCD2 nuclear foci formation. EUFA130 lymphoblasts stably expressing FLAG-FANCEwt (EUFA130+FLAG-FANCE) and the double mutant FLAG-FANCE(TS/AA) (EUFA130+FLAG-TS/AA) were either untreated or treated with IR (10 Gy) and fixed 6 h later; immunofluorescence was determined using anti-FANCD2 (FI-17) antibody. Magnification, ×630. (E) Quantification of FANCD2 foci. Cells with more than four distinct foci were counted as positive; 200 cells/sample were analyzed. The values shown are the means ± standard deviations from three separate experiments. Vec, empty vector.
We examined the ability of the FANCE mutant proteins to restore FANCD2 monoubiquitination (Fig. 1C). As previously described (3, 27), FLAG-FANCEwt restored FANCD2 monoubiquitination (Fig. 1C, lanes 6 to 10), and DNA damage generated by ionizing radiation (IR) further activated FANCD2 monoubiquitination. The double mutant FLAG-FANCE(TS/AA) also restored monoubiquitination of FANCD2. Cells expressing FLAG-FANCE(TS/AA) had elevated basal levels of FANCD2 monoubiquitination (Fig. 1C, compare lanes 6 and 11) and failed to exhibit further FANCD2 monoubiquitination following DNA damage (Fig. 1C, lanes 11 to 15).
We next determined the assembly of FANCD2 nuclear foci in the FA-E cells expressing the double mutant FLAG- FANCE(TS/AA) (Fig. 1D and E). Both FLAG-FANCEwt and FLAG-FANCE(TS/AA) restored FANCD2 foci formation (Fig. 1D). Cells expressing FLAG-FANCE(TS/AA) had increased basal levels of FANCD2 foci and failed to upregulate FANCD2 foci after DNA damage (Fig. 1E), thus correlating with FANCD2 monoubiquitination levels (determined by Western blotting) (Fig. 1C). Taken together, these results suggest that phosphorylation of Thr346 and Ser374 on FANCE is not required for FANCD2 monoubiquitination and foci formation but is required for MMC resistance.
Phosphorylation of FANCE by Chk1 in vitro and in vivo.
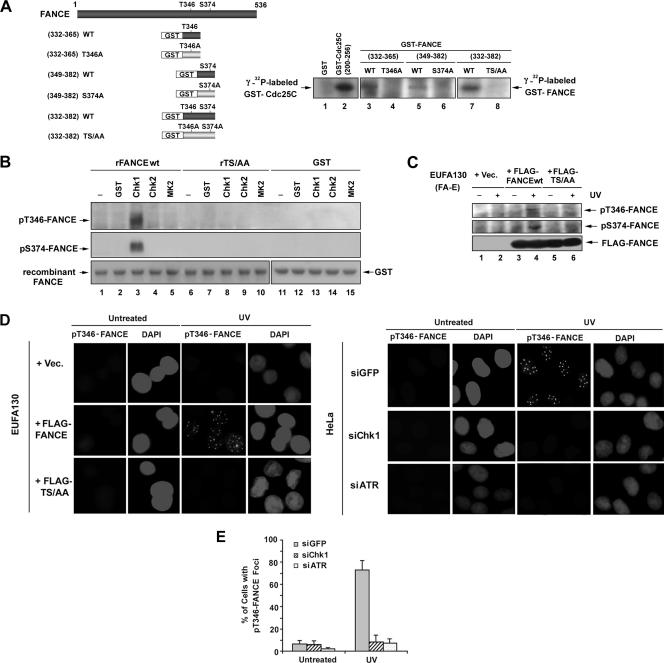
To demonstrate whether Chk1 directly phosphorylates FANCE, we examined phosphorylation of the two highly conserved threonine and serine residues by Chk1 in vitro. We generated GST peptide fusion proteins containing different regions of FANCE (Fig. 2A, left panel). Three GST peptide fusion proteins containing either Thr346 or Ser374 or both residues and consisting of the wt sequence of residues 332 to 365, 349 to 382, or 332 to 382 (but not the GST fusion proteins with single or double mutations of Thr to Ala or Ser to Ala) were phosphorylated by Chk1 in vitro (Fig. 2A, right panel).
FIG. 2.
Phosphorylation of FANCE by Chk1 in vitro and in vivo. (A) GST-FANCE peptide fusion proteins, containing the indicated regions of FANCE, were generated and used as substrates for in vitro kinase assays; the threonine (346) and/or serine (374) residues that were mutated to alanine are shown (left panel). GST-FANCE peptide fusion proteins were incubated with [γ-32P]ATP and purified recombinant Chk1 as described in Materials and Methods. The reaction was stopped by the addition of SDS sample buffer before analysis by SDS-PAGE and autoradiography (right panel). The GST-Cdc25C(200-256) peptide fusion protein and GST were used as positive and negative control substrates, respectively, for Chk1 to demonstrate efficient in vitro kinase assays. (B) In vitro kinase assays using GST, purified recombinant Chk1, Chk2, or MAPKAPK2 (MK2) to phosphorylate the recombinant proteins (containing residues 149 to 536) rFANCEwt and the double mutant FANCE(rTS/AA) were performed and analyzed by SDS-PAGE, followed by immunoblotting with the pT346-FANCE and pS374-FANCE antibodies. The GST-Cdc25C(200-256) peptide fusion protein and GST were used as positive and negative control substrates, respectively, for Chk1, Chk2, and MK2 to demonstrate efficient in vitro kinase assays (data not shown). (C) Chk1 phosphorylates FANCE in vivo. EUFA130 lymphoblasts were stably expressed with empty vector (Vec), FLAG-FANCEwt, and FLAG-FANCE(TS/AA) as indicated. Cells were either untreated or treated with UV (60 J/m2); after 3 h, immunoprecipitation was performed using anti-FLAG antibody, followed by SDS-PAGE and then Western blotting with anti-pT346-FANCE and anti-pS374-FANCE phosphospecific antibodies and anti-FLAG antibody. (D) EUFA130 lymphoblasts stably expressing empty vector (EUFA130+Vec) and FLAG-FANCEwt (EUFA130+FLAG-FANCE) were either untreated or treated with UV (60 J/m2) and fixed 2 h later; immunofluorescence was performed using anti-pT346-FANCE antibody (left panel). Magnification, ×630. HeLa cells were transiently transfected with siRNA targeted against GFP (control), Chk1, or ATR. After 72 h of transfection, cells were either untreated or treated with UV (60 J/m2) and incubated for 2 h before fixation; immunofluorescence was performed using anti-pT346-FANCE antibody (right panel). Magnifi- cation, ×400. (E) Quantification of pT346-FANCE foci. Cells with more than four distinct foci were counted as positive; 100 cells/sample were analyzed. The values shown are the means ± standard deviations from three separate experiments. The formation of pT346 FANCE foci with the treatment of UV (60 J/m2) was strongly decreased in HeLa cells in which ATR or Chk1 had been suppressed with siRNA.
To further study the phosphorylation of FANCE on T346 and S374 by Chk1, we raised rabbit polyclonal antisera against FANCE peptides containing the putative phosphorylated residues, SDLGLLRLCpT(346)WL (anti-pT346) or against LFLGRILpS(374)LTSS (anti-pS374). Purified rFANCEwt or the double mutant FANCE(rTS/AA) protein was incubated with Chk1 or with the two related checkpoint kinases, Chk2 and MAPKAP2 (17) in vitro (Fig. 2B). The anti-phosphospecific antibodies for FANCE (anti-pT346 and anti-pS374) specifically recognized the rFANCEwt protein phosphorylated by Chk1 in vitro (Fig. 2B, lane 3). rFANCEwt was not phosphorylated by Chk2 or MAPKAP2 in vitro (Fig. 2B, lanes 4 and 5). The specificity of the antibodies was demonstrated by the lack of reactivity with the double mutant protein FANCE(rTS/AA) and GST (Fig. 2B, lanes 6 to 15).
We then determined whether FANCE is phosphorylated in vivo following DNA damage (Fig. 2C). Using anti-pT346 and anti-pS374, we immunoblotted FLAG-FANCE immune complexes from FA-E cells stably expressing FLAG-FANCEwt or the double mutant FLAG-FANCE(TS/AA). Following cellular exposure to UV light, a potent activator of the FA/BRCA pathway (4), the anti-pT346 and anti-pS374 antisera detected the FLAG-FANCEwt but not FLAG-FANCE(TS/AA). Taken together, these results confirm the specificity of the antibodies and the DNA damage-inducible phosphorylation of these two residues.
We next used immunofluorescence to demonstrate the in vivo phosphorylation of FANCE by Chk1. The anti-pT346 antiserum detected activated FANCE protein in corrected FA-E lymphoblasts (Fig. 2D, EUFA130+FLAG-FANCE) after cellular exposure to DNA damage but not in FA-E cells stably expressing vector and the double mutant FLAG- FANCE(TS/AA) (Fig. 2D, left panel). Interestingly, the phosphorylated FANCE protein, detected by anti-pT346 antibody, assembled in DNA damage-inducible nuclear foci (Fig. 2D; see also Fig. S2A and B in the supplemental material). UV also activated pT346-FANCE foci in HeLa cells, and siRNA directed against ATR or Chk1 decreased the UV-inducible pT346-FANCE foci (Fig. 2D and E). Pretreatment with Chk1 inhibitors (Gö6976 or SB218078) decreased the pT346-FANCE foci formation after UV exposure (see Fig. S2C and D in the supplemental material).
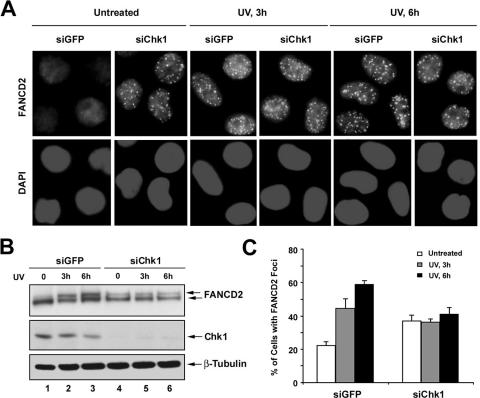
siRNA knockdown of Chk1 results in an elevated basal level of FANCD2 monoubiquitination and FANCD2 foci.
We next examined the effect of siRNA knockdown of Chk1 on FANCD2 monoubiquitination and FANCD2 focus formation after DNA damage (Fig. 3). siRNA knockdown of Chk1 resulted in an elevated basal level of FANCD2 focus formation (Fig. 3A) and FANCD2 monoubiquitination (Fig. 3B, lane 4). Monoubiquitination and nuclear focus formation of FANCD2 were not further increased following DNA damage (Fig. 3B, lanes 4 to 6, and C). These results indicate that the disruption of Chk1 activity mimics the cellular phenotype of the double mutant FANCE(TS/AA).
FIG. 3.
Knockdown of Chk1 results in an increased basal level of FANCD2 foci assembly. (A and B) HeLa cells were transfected with siRNAs targeted against GFP (control) or Chk1. After 72 h of transfection, cells were treated with UV (60 J/m2) and incubated for 3 h or 6 h before fixation and lysis; immunofluorescence was performed using anti-FANCD2 antibody. Magnification, ×400 (A) Whole-cell extracts were analyzed by Western blotting with the indicated antibodies. An anti-β-tubulin blot was used as a loading control (B). (C) Quantification of FANCD2 foci. Cells with more than four distinct foci were counted as positive; 200 cells/sample were analyzed. The values shown are the means ± standard deviations from three separate experiments.
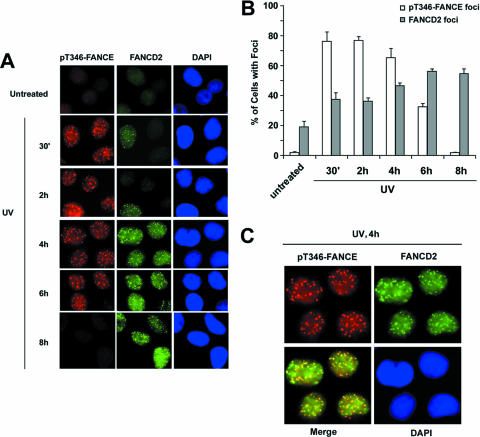
Colocalization of FANCD2 foci and pT346-FANCE foci.
We next examined the assembly of pT346-FANCE foci and FANCD2 foci following DNA damage (Fig. 4). Untreated HeLa cells exhibited no pT346-FANCE foci or FANCD2 foci (Fig. 4A). DNA damage from UV light activated the phosphorylation of FANCE on T346 and foci formation by 30 min, and these foci were no longer observed after 8 h. FANCD2 foci began to accumulate at 30 min after UV and peaked at 8 h. The kinetics of pT346-FANCE focus formation and FANCD2 focus formation are shown graphically in Fig. 4B. Double staining revealed that pT346-FANCE foci and FANCD2 foci colocalize (Fig. 4C) but exhibit distinct kinetics (Fig. 4B).
FIG. 4.
Colocalization of pT346-FANCE foci and FANCD2-Ub foci after DNA damage. (A) The kinetics of pT346-FANCE foci and FANCD2 foci were followed after DNA damage. HeLa cells were either untreated or treated with UV (60 J/m2) and incubated for different periods of time (30 min, 2 h, 4 h, 6 h, or 8 h) as indicated before fixation; immunofluorescence was performed using anti-pT346-FANCE and anti-FANCD2 (FI-17) antibodies. Magnification, ×400. (B) Cells with more than four distinct foci were counted as positive; 200 cells/sample were analyzed. The values shown are the means ± standard deviations from three separate experiments. (C) After 4 h of UV irradiation, colocalization of pT346-FANCE foci and FANCD2 foci in HeLa cells is shown. Magnification, ×630.
Chk1-mediated phosphorylation of FANCE is required for MMC resistance but not required for DNA replication or normal cell cycle progression.
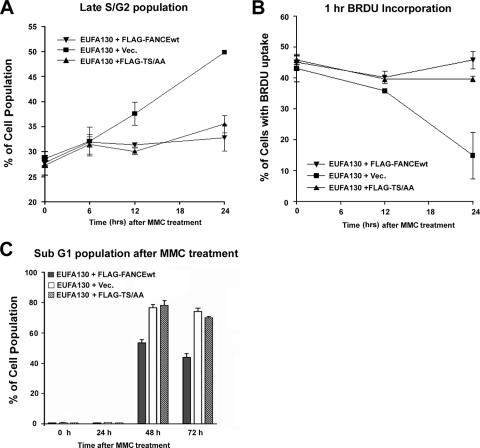
Recent studies indicate that the FA core complex has additional replication and checkpoint activities which are discrete from FANCD2 monoubiquitination (18). To determine whether phosphorylation of FANCE by Chk1 is required for normal S-phase progression, we compared the ability of FLAG-FANCEwt or the double mutant FLAG-FANCE(TS/AA) to restore DNA replication and S-phase progression (Fig. 5 A and B). After 24 h of MMC treatment, FA-E cells (EUFA130) expressing either FLAG-FANCEwt) or FLAG-FANCE(TS/AA) were equally competent for DNA replication and did not accumulate in the late S and G2 cell cycle phase. As expected, FA-E cells containing the empty vector arrested and accumulated in late S and G2 phase and demonstrated decreased DNA synthesis (Fig. 5A and B). Moreover, the double mutant FLAG-FANCE(TS/AA) stabilized the FA core complex and restored FANCD2 monoubiquitination (Fig. 1C; see also Fig. S3A in the supplemental material).
FIG. 5.
FANCE phosphorylation by Chk1 does not correct MMC-mediated cell death but corrects cell cycle progression and promotes DNA synthesis following MMC treatment. (A) FA-E cells stably expressing wt FANCE (EUFA130+FLAG-FANCEwt) or the double mutant of FANCE (EUFA130+FLAG-TS/AA) do not accumulate in the late S/G2 phase of the cell cycle after 24 h of MMC treatment compared to cells stably expressing empty vector (EUFA130+Vec). The values shown are the means ± standard deviations from three separate experiments. (B) FA-E cells stably expressing empty vector (EUFA130+Vec) have decreased DNA synthesis after 24 h of MMC 160 ng/ml treatment compared to FA-E cells stably expressing wt FANCE (EUFA130+FLAG-FANCEwt) or the double mutant of FANCE (EUFA130+FLAG-TS/AA). The values shown are the means ± standard deviations from three separate experiments. (C) FA-E cells stably expressing the double mutant of FANCE (EUFA130+FLAG-TS/AA) or empty vector (EUFA130+Vec) demonstrate a higher percentage of sub-G1 cells after 48 and 72 h following MMC (160 ng/ml) treatment than cells expressing wt FANCE. Values shown are the means ± standard deviations from three separate experiments.
We next tested whether the double mutant of FANCE could prevent MMC-mediated cell death when expressed in the FA-E cell line. Cell death results in nuclear fragmentation (the sub-G1 population), and this was assessed by flow cytometry following MMC treatment. As shown in Fig. 5C, equivalent levels of sub-G1 cells were present in FA-E cells stably expressing empty vector and the double mutant FLAG-FANCE(TS/AA) following MMC treatment. In contrast, FA-E cells corrected with FLAG-FANCEwt demonstrated a significantly lower percentage of cells with fragmented nuclei. These results demonstrate that the intact FA core complex containing FLAG-FANCE(TS/AA) can correct the cell cycle abnormalities of FA-E cells but cannot confer resistance to MMC, confirming that Chk1-mediated phosphorylation of FANCE is required for cross-linker resistance (Table 1).
TABLE 1.
Characterization of FA-E cells stably expressing empty vector, FANCEwt, or the double mutant FANCE(TS/AA)
| Feature | Characterization of EUFA130 cells expressing:
|
||
|---|---|---|---|
| Empty vector | FLAG-FANCEwt | FLAG-FANCE(TS/AA) | |
| MMC sensitivity | Sensitive | Resistant | Sensitive |
| FA core complex assembly | − | + | + |
| FANCD2 monoubiquitination | − | + | + |
| DNA damage-inducible FANCD2 monoubiquitination | − | + | − |
| Cell cycle progression after MMC treatment for 24 h | Late S/G2 accumulation | Normal | Normal |
| Relative percentage of sub-G1 population after MMC treatment | High | Low | High |
Chk1-mediated phosphorylation of FANCE promotes FANCE degradation.
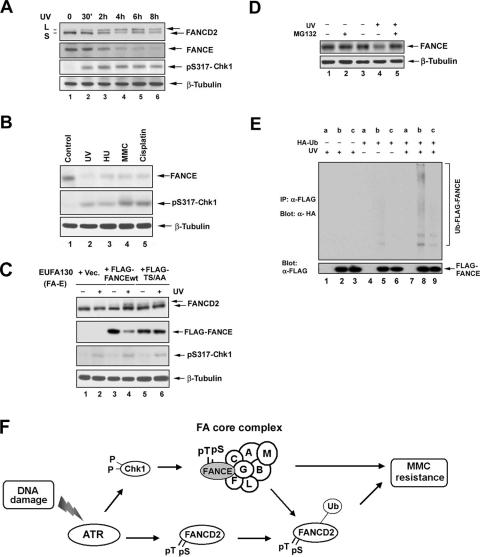
The disappearance of pT346-FANCE foci after DNA damage (Fig. 4A) suggested that FANCE may undergo regulated proteolysis. To test this hypothesis, we examined the cellular levels of FANCE following UV damage (Fig. 6A); after only 30 min, Chk1 activation and FANCD2 monoubiquitination were observed (Fig. 6A, lane 2). By 4 to 6 h, FANCD2 monoubiquitination was maximal, and FANCE levels had decreased (Fig. 6A, lanes 4 to 6); FANCE degradation was also observed in a UV dose-dependent manner (data not shown).
FIG. 6.
FANCE phosphorylation by Chk1 promotes its degradation. (A) HeLa cells were either untreated or treated with UV irradiation at 60 J/m2 and incubated for different periods of time as indicated before lysis. Whole-cell extracts were immunoblotted with the indicated antibodies. An anti-β-tubulin blot was used as a loading control. (B) HeLa cells were synchronized by a double-thymidine block and then released into S phase. One hour after release, the cells were either untreated (Control) or treated with UV (60 J/m2, 15 h), HU (2 mM, 24 h), MMC (160 ng/ml, 24 h), or cisplatin, (10 μM, 24 h). Whole-cell extracts were analyzed by Western blotting with the indicated antibodies. An anti-β-tubulin blot was used as a loading control. (C) EUFA130 lymphoblasts were stably expressed with pMMP (empty vector [Vec]), FLAG-FANCEwt, or FLAG-FANCE(TS/AA) (the double mutant) as indicated. Cells were either untreated or treated with UV (60 J/m2); after 8 h, whole-cell extracts were analyzed by Western blotting with the indicated antibodies. An anti-β-tubulin blot was used as a loading control. (D) U2OS cells were either untreated or treated with UV (60 J/m2) and incubated for 3 h with or without the addition of 25 μM MG132 to the indicated samples during the final 2 h in cell culture. Whole-cell extracts were analyzed by Western blotting with the indicated antibodies. An anti-β-tubulin blot was used as a loading control. (E) FANCE ubiquitination in vivo. U2OS stably expressing empty vector (a), FLAG-FANCEwt (b), or and FLAG-FANCE(TS/AA) (c) were transiently transfected without or with a cDNA encoding HA-Ub; after 48 h of transfection, cells were untreated or treated with UV (60 J/m2) and incubated for 2 h before cells were lysed in SDS denaturation buffer. FLAG-FANCEwt and the double mutant protein were isolated by anti-FLAG antibody immunoprecipitation. Immune complexes were run on SDS-PAGE gels and immunoblotted with anti-HA or anti-FLAG antibodies. (F) Schematic model showing the activation of the FA/BRCA pathway by the ATR-Chk1 pathway. DNA damage or replication arrest (MMC, UV, IR, or HU) activates the ATR-dependent phosphorylation of FANCD2 (6) and the Chk1-dependent phosphorylation of FANCE. Both monoubiquitinated FANCD2 and phosphorylated FANCE are required for MMC resistance. A nonubiquitinated mutant of FANCD2 (K561R) or the nonphosphorylated mutant FANCE(TS/AA) fails to correct MMC hypersensitivity.
We synchronized HeLa cells at the G1/S boundary with a double-thymidine block and released the cells into S phase for 1 h before DNA damage treatment. The level of FANCE protein was significantly decreased when a genotoxic stress (UV, hydroxyurea [HU], MMC, or cisplatin) was delivered to cells undergoing DNA replication (Fig. 6B, lanes 2 to 5). UV-inducible FANCE degradation was also observed in other cell lines, including U2OS, GM0637, and HEK293T cells (see Fig. S3B in the supplemental material).
We then tested whether Chk1-dependent phosphorylation regulates FANCE stability (Fig. 6C). We examined FA-E (EUFA130) cells stably expressing FLAG-FANCEwt or the double mutant FLAG-FANCE(TS/AA). After UV treatment, FLAG-FANCEwt was degraded (Fig. 6C, compare lanes 3 and 4), but FLAG-FANCE(TS/AA) was stable (Fig. 6C, compare lanes 5 and 6).
We next examined the role of the Ub-proteasome pathway in the degradation of FANCE (Fig. 6D). Cells were treated with UV in the absence or presence of the proteasome inhibitor, MG132. MG132 treatment blocked the UV-inducible degradation of FANCE (Fig. 6D, lane 3). We transiently transfected a cDNA encoding an HA-Ub construct into U2OS cells stably expressing FLAG-FANCEwt or the double mutant FLAG-FANCE(TS/AA). FLAG-tagged FANCE was immunoprecipitated and immunoblotted with anti-HA and anti-FLAG antibodies. Following UV exposure, the high-molecular-weight ladder of polyubiquitinated products of the FLAG-FANCEwt was enhanced by UV exposure (Fig. 6E, lane 8), which does not present in cells expressing empty vector or FLAG-FANCE(TS/AA) (Fig. 6E, lanes 7 and 9, respectively), indicating that, following DNA damage, FANCE phosphorylation precedes its polyubiquitination. Taken together, these results indicate that the Chk1-mediated phosphorylation of FANCE promotes the Ub-mediated degradation of FANCE. The specific E3 Ub ligase complex required for FANCE degradation remains unknown.
DISCUSSION
Our results elucidate several properties of FANCE in the FA/BRCA pathway. FANCE is assembled in the FA core complex and promotes the monoubiquitination of FANCD2 (Fig. 6F). FANCE is phosphorylated by Chk1 at an early stage, within 30 min following DNA damage; however, Chk1-dependent phosphorylation of FANCE is not required for FANCD2 monoubiquitination and nuclear focus formation since the nonphosphorylated, double mutant FANCE(TS/AA) is competent for restoring FANCD2 monoubiquitination and FANCD2 foci in the FA-E-deficient cells. Therefore, phosphorylation of FANCE by Chk1 is required for an unknown function independent of FANCD2 monoubiquitination.
Failure to phosphorylate FANCE, as in Chk1-depleted cells or in cells expressing the FANCE(TS/AA) mutant protein, results in an elevated basal level of FANCD2 monoubiquitination and nuclear foci. This failure also results in persistent cellular hypersensitivity to MMC. The elevated basal level of FANCD2 foci may result from increased spontaneous endogenous DNA damage or from failure of the mutant FANCE protein to function independently of FANCD2 monoubiquitination. Paradoxically, our previous studies have shown that ATR inactivation has little or no effect on the basal level of FANCD2 monoubiquitination (1). Our findings are consistent with recent studies (S. J. Boulton, personal communication) which indicate that the depletion of ATR and Chk1 may have opposite effects on the FA/BRCA pathway function. Taken together, our results from this study and our previous work (1) suggest that ATR and Chk1 have both overlapping and distinct functions in the regulation of the FA/BRCA pathway.
The function of FANCE phosphorylation by Chk1 in the FA/BRCA pathway remains unknown, and several possible mechanisms exist. First, pT346-FANCE may participate with FANCD2-Ub in recruiting other DNA repair proteins to sites of DNA cross-link repair. Second, pT346-FANCE may be required to stabilize FANCD2-Ub or to carry it to a specific nuclear compartment where cross-link repair occurs. Third, phosphorylation of FANCE may be required for its timed degradation, and this degradation may be essential for the proper function of the FA/BRCA pathway. Consistent with this notion, pT346-FANCE foci disappear 8 h after DNA damage, while FANCD2 foci persist. Also consistent with this hypothesis, phosphorylation of FANCE by Chk1 promotes its degradation. The timed degradation of FANCE may be a negative regulatory mechanism for switching off the FA/BRCA pathway after the completion of DNA repair. The cellular role of phosphorylation of FANCE and FANCD2-Ub in cross-link repair remains an important unanswered question for the FA research field.
Based on these observations, FANCE may have a dual regulatory role in the FA/BRCA pathway. On the one hand, FANCE positively regulates the assembly of the FA core complex (A/B/C/E/F/G/L/M complex) and tethers the FANCD2 protein to the FA core complex. FA complex assembly promotes the basal levels of monoubiquitination of FANCD2 observed during S-phase progression. On the other hand, FANCE may also negatively regulate the FA/BRCA pathway. In response to DNA damage or replication arrests, Chk1-mediated phosphorylation of FANCE may trigger its degradation. The timed degradation of FANCE may dissociate FANCD2 from the FA core complex and thus prevent further FANCD2 monoubiquitination.
The double mutant FANCE(TS/AA) protein functions normally in other aspects of the FA/BRCA pathway. The mutant protein stabilizes the FA core complex and corrects the DNA replication and cell cycle abnormalities observed in FA-E cells after 24 h of MMC treatment. These results suggest that the basal levels of FANCD2 monoubiquitination, observed in FA-E cells stably expressing FANCE(TS/AA), are sufficient to correct FA-associated cell cycle abnormalities after DNA damage but that the downstream function of FANCE is required to prevent cell death.
The failure of the FA core complex containing the FANCE(TS/AA) mutant protein to correct FA-E cells is consistent with other recent studies. Matsushita et al. (18) demonstrated that a FANCD2-monoubiquitin fusion protein corrects the DNA cross-linker hypersensitivity of FANCD2-deficient cells but not of core complex-deficient cells. These authors concluded that the FA core complex has additional functions beyond the monoubiquitination of FANCD2. The Chk1-mediated phosphorylation of FANCE may be required for one of these additional functions.
FANCE joins a growing list of proteins that are phosphorylated by Chk1 following DNA damage. Chk1 phosphorylates the phosphatase Cdc25A (25), thereby promoting its immediate degradation by a Cul1/β-TCRP-mediated mechanism (8, 16). Degradation of Cdc25A prevents dephosphorylation of Cdc2 and arrests cell cycle progression at G1/S (8, 16). Recent studies have identified additional substrates of the Chk1 kinase, including RAD51 (26) and MDMX1 (9). These substrates demonstrate that Chk1 can regulate checkpoint activity (Cdc25A), DNA repair activity (FANCE and RAD51), or transcriptional events (MDMX1). Finally, the anti-pT346 FANCE antibody described in this study may be a useful reagent in evaluating the activity of Chk1 inhibitors, a new class of anticancer therapeutics.
Supplementary Material
Acknowledgments
We thank Wade Harper and members of the D'Andrea laboratory for helpful discussions.
This work was supported by NIH grants RO1-HL52725, RO1-DK43889, and PO1-HL54785 (A.D.D.); the NIH Training Grant Program (T43CA09361) (X.W.); and a Susan G. Komen Postdoctoral Grant (R.D.K.).
Footnotes
Published ahead of print on 12 February 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andreassen, P. R., A. D. D'Andrea, and T. Taniguchi. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18:1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach, A. D. 1993. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp. Hematol. 21:731-733. [PubMed] [Google Scholar]
- 3.de Winter, J. P., F. Leveille, C. G. van Berkel, M. A. Rooimans, L. van Der Weel, J. Steltenpool, I. Demuth, N. V. Morgan, N. Alon, L. Bosnoyan-Collins, J. Lightfoot, P. A. Leegwater, Q. Waisfisz, K. Komatsu, F. Arwert, J. C. Pronk, C. G. Mathew, M. Digweed, M. Buchwald, and H. Joenje. 2000. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am. J. Hum. Genet. 67:1306-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn, J., M. Potter, A. Rees, and T. M. Runger. 2006. Activation of the Fanconi anemia/BRCA pathway and recombination repair in the cellular response to solar ultraviolet light. Cancer Res. 66:11140-11147. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 6.Ho, G., S. Margossian, T. Taniguchi, and A. D. D'Andrea. 2006. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol. Cell. Biol. 26:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houghtaling, S., C. Timmers, M. Noll, M. Finegold, S. Jones, M. S. Meyn, and M. Grompe. 2003. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17:2021-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin, J., T. Shirogane, L. Xu, G. Nalepa, J. Qin, S. J. Elledge, and J. W. Harper. 2003. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17:3062-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin, Y., M. S. Dai, S. Z. Lu, Y. Xu, Z. Luo, Y. Zhao, and H. Lu. 2006. 14-3-3γ Binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 25:1207-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy, R. D., and A. D. D'Andrea. 2005. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19:2925-2940. [DOI] [PubMed] [Google Scholar]
- 12.Kohn, E. A., C. J. Yoo, and A. Eastman. 2003. The protein kinase C inhibitor Go6976 is a potent inhibitor of DNA damage-induced S and G2 cell cycle checkpoints. Cancer Res. 63:31-35. [PubMed] [Google Scholar]
- 13.Lin, S. Y., K. Li, G. S. Stewart, and S. J. Elledge. 2004. Human claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. USA 101:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida, Y. J., Y. Machida, Y. Chen, A. M. Gurtan, G. M. Kupfer, A. D. D'Andrea, and A. Dutta. 2006. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23:589-596. [DOI] [PubMed] [Google Scholar]
- 15.Mack, P. C., D. R. Gandara, A. H. Lau, P. N. Lara, Jr., M. J. Edelman, and P. H. Gumerlock. 2003. Cell cycle-dependent potentiation of cisplatin by UCN-01 in non-small-cell lung carcinoma. Cancer Chemother. Pharmacol. 51:337-348. [DOI] [PubMed] [Google Scholar]
- 16.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 17.Manke, I. A., A. Nguyen, D. Lim, M. Q. Stewart, A. E. Elia, and M. B. Yaffe. 2005. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell 17:37-48. [DOI] [PubMed] [Google Scholar]
- 18.Matsushita, N., H. Kitao, M. Ishiai, N. Nagashima, S. Hirano, K. Okawa, T. Ohta, D. S. Yu, P. J. McHugh, I. D. Hickson, A. R. Venkitaraman, H. Kurumizaka, and M. Takata. 2005. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell 19:841-847. [DOI] [PubMed] [Google Scholar]
- 19.Meetei, A. R., J. P. de Winter, A. L. Medhurst, M. Wallisch, Q. Waisfisz, H. J. van de Vrugt, A. B. Oostra, Z. Yan, C. Ling, C. E. Bishop, M. E. Hoatlin, H. Joenje, and W. Wang. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35:165-170. [DOI] [PubMed] [Google Scholar]
- 20.Meetei, A. R., A. L. Medhurst, C. Ling, Y. Xue, T. R. Singh, P. Bier, J. Steltenpool, S. Stone, I. Dokal, C. G. Mathew, M. Hoatlin, H. Joenje, J. P. de Winter, and W. Wang. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosedale, G., W. Niedzwiedz, A. Alpi, F. Perrina, J. B. Pereira-Leal, M. Johnson, F. Langevin, P. Pace, and K. J. Patel. 2005. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 12:763-771. [DOI] [PubMed] [Google Scholar]
- 22.Niedernhofer, L. J., A. S. Lalai, and J. H. Hoeijmakers. 2005. Fanconi anemia (cross)linked to DNA repair. Cell 123:1191-1198. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill, T., L. Giarratani, P. Chen, L. Iyer, C. H. Lee, M. Bobiak, F. Kanai, B. B. Zhou, J. H. Chung, and G. A. Rathbun. 2002. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 277:16102-16115. [DOI] [PubMed] [Google Scholar]
- 24.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez, Y., C. Wong, R. S. Thoma, R. Richman, Z. Wu, H. Piwnica-Worms, and S. J. Elledge. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497-1501. [DOI] [PubMed] [Google Scholar]
- 26.Sorensen, C. S., L. T. Hansen, J. Dziegielewski, R. G. Syljuasen, C. Lundin, J. Bartek, and T. Helleday. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7:195-201. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi, T., and A. D. D'Andrea. 2002. The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of FANCC. Blood 100:2457-2462. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414-2420. [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., P. R. Andreassen, and A. D. D'Andrea. 2004. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24:5850-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, B., S. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445-3450.11313470 [Google Scholar]
- 31.Yarden, R. I., S. Pardo-Reoyo, M. Sgagias, K. H. Cowan, and L. C. Brody. 2002. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30:285-289. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, H., and H. Piwnica-Worms. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21:4129-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.