Abstract
The six-subunit origin recognition complex (ORC) is a DNA replication initiator protein in eukaryotes that defines the localization of the origins of replication. We report here that the smallest Drosophila ORC subunit, Orc6, is a DNA binding protein that is necessary for the DNA binding and DNA replication functions of ORC. Orc6 binds DNA fragments containing Drosophila origins of DNA replication and prefers poly(dA) sequences. We have defined the core replication domain of the Orc6 protein which does not include the C-terminal domain. Further analysis of the core replication domain identified amino acids that are important for DNA binding by Orc6. Alterations of these amino acids render reconstituted Drosophila ORC inactive in DNA binding and DNA replication. We show that mutant Orc6 proteins do not associate with chromosomes in vivo and have dominant negative effects in Drosophila tissue culture cells. Our studies provide a molecular analysis for the functional requirement of Orc6 in replicative functions of ORC in Drosophila and suggest that Orc6 may contribute to the sequence preferences of ORC in targeting to the origins.
Eukaryotic cells duplicate their genomes with remarkable precision during the course of growth and division. This process depends on stringent regulatory molecular mechanisms that couple DNA replication and cell cycle progression. To efficiently duplicate large genomes, eukaryotes have evolved a mechanism for the initiation of DNA replication that involves multiple origins of replication (ori) along the chromosomal DNA. The utilization of such sites in multicellular organisms changes during development, and this process affects both gene expression and chromosome folding. The program of such spatial and temporal activation is not understood. Although not necessarily random, the origin site selection during early Drosophila and Xenopus development appears to be less dependent on specific DNA sequences (5, 25). In agreement with this idea, a number of studies suggest that specific replicator sequences might be dispensable (22, 38, 52, 53). Later in development origin usage becomes more specific (26, 49) and depends on many mechanisms for selection of the initiation events. Overall, with an exception of the budding yeast Saccharomyces cerevisiae, DNA sequences that define eukaryotic and especially metazoan replication origins are poorly characterized, mainly because of a lack of definitive biochemical or genetic assays (13, 17, 18).
The hexameric origin recognition complex (ORC) is an important component for eukaryotic DNA replication. It was originally discovered in the budding yeast S. cerevisiae, and subsequent studies both in yeast and higher eukaryotes laid the foundation for understanding the functions of this important key initiation factor. ORC binds to origin sites in an ATP-dependent manner and serves as a scaffold for the assembly of other initiation factors (3). ORC also directly participates in the loading of initiation factors (6, 45). Sequence rules for ORC DNA binding appear to vary widely. In budding yeast ORC recognizes specific ori elements; however, in higher eukaryotes origin site selection appears to be less dependent on the specific DNA sequence. Even though ORC is bound at specific chromosomal regions containing origins of replication in both differentiated insect and human somatic cells, little is known about how ORC finds these sequences (1, 2, 27). ORC localization and origin selection involve many elaborate pathways with many regulators intervening upstream and downstream of ORC chromatin association (13, 14, 15, 17, 18).
In addition to initiating DNA replication, ORC is involved in other functions (see references 3 and 9a for reviews). Some of these activities link cell cycle progression to DNA replication, whereas other functions seem distinct from replication. ORC assists in the establishment and maintenance of transcriptionally repressed domains in yeast and metazoans (see reference 51 for a review). The latheo gene product, Drosophila Orc3, is implicated in ion transport at neuromuscular junctions (43). Other core ORC subunits may regulate dendrite development in postmitotic neurons (23). The Orc6 subunit participates in cytokinesis in both Drosophila and human cells, probably through the interaction with septin proteins (9, 44).
The Orc6 protein is the least conserved of all ORC subunits, and amino acid alignments between budding yeast and metazoan proteins do not show statistically significant homologies. However, Schizosaccharomyces pombe and metazoan Orc6 proteins (7, 16, 29, 40) are homologous, similar in size, and considerably smaller than the S. cerevisiae Orc6. In S. cerevisiae, Orc6 is essential for viability but is not required for DNA binding in vitro (32, 34). In Xenopus and humans, Orc6 protein does not seem to be tightly associated with other core ORC subunits (16, 19, 56), but when human Orc6 is expressed in the baculovirus system with the other ORC genes, it does join a six-subunit ORC complex (55, 56). In contrast, Drosophila Orc6 is an integral part of the complex and is required for DNA binding by ORC. Moreover, the ORC consisting of subunits 1 to 5 [ORC(1-5)] and lacking the Orc6 subunit did not complement ORC-depleted extracts for DNA replication (8).
In our previous work we established two distinct functional domains in Drosophila Orc6. The C-terminal domain of the protein participates in cytokinesis through the interaction with the septin protein Pnut (9). In this study we show that the replication function of Drosophila Orc6 is associated with the N-terminal domain of the protein. Orc6 has a DNA binding ability, prefers poly(dA) sequences, and may contribute to the targeting of ORC to the origins of replication. Analysis of the N-terminal core replication domain allowed identification of amino acids that are essential for the DNA binding ability of Orc6. Alterations of these amino acids severely compromised the functions of reconstituted Drosophila ORC protein in DNA binding and in DNA replication in vitro. In vivo, mutant Orc6 proteins do not associate with chromosomes and have dominant negative effects when expressed in Drosophila tissue culture cells. Cells with overexpressed mutant proteins have a reduced replication activity, as judged by bromodeoxyuridine (BrdU) incorporation. Our studies further illustrate the biochemical mechanisms underlying the initiation site selection by ORC in metazoan species and provide evidence for the important role of Orc6 in DNA recognition and replication in Drosophila.
MATERIALS AND METHODS
Purification of recombinant Orc6 and Drosophila ORC.
His-tagged wild-type Orc6 (Orc6-wt) and mutants (Orc6 with a K76A mutation [Orc6-K76A], Orc6-S72A, and Orc6 incorporating a stop codon instead of amino acid residue 200 [Orc6-200]) were purified by using the QIAexpress Escherichia coli expression system. In brief, proteins were produced by expression from pET15b plasmid in E. coli strain BL21. Protein induction and purification using Ni-nitrilotriacetic acid-agarose beads were done according to QIAGEN recommendations. Peak fractions containing Orc6 proteins were further purified using Tricorn Superdex 75 (internal diameter, 10 mm; height, 300 mm) and HiTrap SP 5/5 (internal diameter, 5 mm; height, 50mm) columns. Recombinant baculoviruses were generated by using a Bac-to-Bac expression system (GIBCO/BRL). Viruses carrying Orc1 with a six-His N-terminal tag, Orc2, Orc3, Orc4, and Orc5 genes were mixed with either wt or mutant Orc6 baculovirus constructs. High Five cells were infected for 72 h, collected, and resuspended in buffer A (10 mM HEPES [pH 7.5], 15 mM KCl, 2 mM MgCl2, 2 mM β-mercaptoethanol, 1 tablet of protease inhibitor cocktail [Roche] per 50 ml of buffer A) and incubated on ice for 30 min. Nuclei were precipitated by centrifugation at 3,200 × g for 10 min. Nuclear extracts were prepared by incubation with buffer B (buffer A containing 450 mM NaCl) for 30 min on ice and centrifugation at 15,000 × g for 15 min. Supernatant was mixed with Ni-agarose (QIAGEN), incubated for 4 to 5 h at 4°C, and washed (buffer B with 20 mM imidazole), and proteins were eluted with buffer B, containing 250 mM imidazole, using a gravity flow column. Peak fractions were further purified as described previously (8).
Site-directed mutagenesis.
Orc6-wt was originally cloned into the pQE-30 QIAexpress vector. The single-amino-acid mutants Orc6-K76A and Orc6-S72A were generated by replacing amino acids K76 and S72 with alanines following Stratagene's site-directed mutagenesis protocol (http://www.stratagene.com/manuals/200516.pdf). The C-terminal mutant Orc6-200 was designed by using a PCR technique. All mutants were verified by sequencing to confirm that only the desired changes had been made.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (8, 47). Each reaction mixture contained ∼50 to 200 ng of purified Orc6 protein, 100 to 1,000 ng of competitor (see figure legends for details), and ∼1 ng of end-labeled specific DNA probes. ACE3 (320 bp), ori-β (810 bp), ori-β-R (310-bp fragment located to the right from ori-β), and the DNA fragment corresponding to the Orc6 cDNA (782 bp) were used as specific probes in EMSAs. Reaction mixtures were set up on ice and incubated at room temperature for 30 min. A total of 10 μl was loaded on a 4% native polyacrylamide gel. The size of synthetic competitor DNAs (Amersham Biosciences) used in competition EMSA experiments was from 500 to 3,000 bp.
EMSAs with purified recombinant Drosophila ORC protein were performed as described previously (8). See Results and figure legends for details.
In vitro replication in Drosophila egg extracts.
The preparation of egg extracts was based on a procedure described previously (12). In brief, embryos (0 to 2 h) were washed with extraction buffer, cold treated, and homogenized. The homogenate was centrifuged for 20 min at 13,000 rpm in a TLA100.3 Beckmann rotor. The middle layer was collected and recentrifuged. The supernatant was made 5% with respect to glycerol and 1 mM to ATP. The extract was frozen in 20-μl beads in liquid nitrogen.
Extract beads were thawed and supplemented with an ATP regeneration system (60 mM phosphocreatine and 150 μg/ml creatine phosphokinase) and immunodepleted with anti-Orc2 and anti-Orc6 antibodies. Xenopus sperm DNA was incubated for 1 h in extracts at a concentration of 2 to 5 ng/μl in the presence of [α32 P]dCTP. The replication rescue experiment was performed by the addition of increasing amounts of baculovirus-reconstituted ORC. DNA was ethanol precipitated, resuspended in Tris-EDTA buffer and subjected to electrophoresis in a 0.8% agarose gel. The gel was dried and autoradiographed.
BrdU incorporation and GFP-Orc6 expression in L2 cells.
Orc6-wt and mutant proteins were fused with green fluorescent protein (GFP) at the N terminus, subcloned into pMT/V5-B vector, and transfected into Drosophila L2 cells according to the manufacturer's (Invitrogen) recommendations. The metallothionein promoter was induced by 0.5 mM CuSO4, and cells were incubated with BrdU overnight at a final concentration of 10 μM. Cells were fixed with 2% formaldehyde, treated with DNase, incubated with anti-BrdU antibodies (Becton Dickinson), and subjected to immunofluorescent microscopy as described previously (9).
BrdU incorporation in salivary gland polytene chromosomes and immunostaining.
Salivary glands of third-instar larvae were incubated in phosphate-buffered saline (PBS) with 10 μM BrdU for 40 min, transferred to fixing solution (2% formaldehyde in PBS, 0.1% Triton X-100, 45% acetic acid), squashed in 45% acetic acid, and frozen in liquid nitrogen. Subsequently, an immunostaining procedure was performed, generally according to a previously described polytene chromosome immunostaining protocol (53a) using anti-BrdU (Becton Dickinson) and anti-Orc6 affinity-purified rabbit antibodies.
GFP-Orc6 expression in salivary glands.
Fused, wt GFP-Orc6 and GFP-Orc6 mutants were cloned into the pUAST vector and injected into w1118 fly embryos. Homozygous flies stocks were set up. To induce GFP-Orc expression, UAS-GFP-Orc flies were crossed to flies bearing GAL4 driven by Sgs-3 promoter (Bloomington stock w1118; P{w+mC = Sgs3-GAL4.PD}TP1). Sgs-3 promoter induces GAL4 in the salivary glands of third-instar larvae. Salivary glands of these larvae were dissected in PBS, stained with 4′,6′-diamidino-2-phenylindole (DAPI), and mounted in PBS containing 60% glycerol-4% N-propyl-gallate. Slides were sealed, and live images were taken within 2 h using a fluorescent microscope.
RESULTS
Drosophila Orc6 binds DNA on its own and prefers poly(dA) sequences.
We have shown before (8) that Drosophila ORC lacking the smallest Orc6 subunit was unable to bind DNA, suggesting that Orc6 is important for DNA recognition. To examine whether the Drosophila Orc6 subunit on its own was able to bind DNA, a series of gel shift experiments was carried out. ori-β and ACE3 DNA fragments with origins of DNA replication derived from the Drosophila chorion gene amplification locus were used as probes for EMSAs. Drosophila Orc6 expressed in E. coli as a His-tagged fusion protein was purified through several chromatographic steps (see Materials and Methods for details). A recombinant Orc6 formed a distinct complex with both ori-β and ACE3 fragments derived from the chorion locus (Fig. 1, lanes 1 to 12). Binding was very tight, with up to 90% of the labeled probe bound by the protein in the case of the ori-β fragment (Fig. 1A, lanes 2 to 5). The addition of more protein caused a stronger shift of the DNA-protein complex. The mobility of this DNA-protein complex changed when increasing amounts of the competitor DNA [poly(dI-dC)] were added to the reaction. This change in the mobility of the discrete Orc6-DNA complexes suggests the presence of multiple binding sites for Orc6 within the ACE3 and ori-β fragments. The complex migrated faster, probably due to a titration of excessive Orc6 protein away from the labeled DNA probe that resulted in reduced template occupancy. An Orc6-DNA complex formed even in the presence of a 1,000-fold excess of poly(dI-dC) competitor DNA. The same result was observed when baculovirus-expressed Orc6 was used in the reactions (data not shown). In contrast, non-origin fragments exhibited significantly reduced ability to bind with Orc6 under the same conditions. No discrete Orc6-DNA complexes were detected when the ori-β-R fragment, located between ori-β and the s15 chorion gene, was used in the EMSA (Fig. 1A, lanes 13 to 16). Only a minor shift was observed when Orc6 cDNA was used as a probe (Fig. 1A, lanes 17 to 19).
FIG. 1.
DNA-binding ability of Drosophila Orc6 protein. Binding to radiolabeled origin and non-origin DNA fragments was monitored in EMSAs. (A) Binding of Orc6 protein to ori-β (lanes 1 to 8), ACE3 (lanes 9 to 12), ori-β-R (lanes 13 to 16), and Orc6 cDNA (lanes 17 to 19) DNA fragments. Orc6-wt (lanes 2 to 5, 9 to 11, 13 to 15, and 18 to 19) or the Orc6-200 deletion mutant (lanes 7 and 8) protein (50 ng each) was incubated with origin (ACE3 and ori-β) and non-origin (ori-β-R and Orc6 cDNA) DNA fragments in the presence of increasing amounts of competitor poly(dI-dC) DNA. The amount of competitor was 100 ng, 200 ng, 500 ng, and 1,000 ng (lanes 2, 3, 4, and 5, respectively) for the ori-β probe; 100 ng, 200 ng, and 500 ng (lanes 9, 10, and 11, respectively) for ACE3 probe; 100 ng, 200 ng, and 500 ng (lanes 13, 14, and 15, respectively) for ori-β-R probe; and 100 ng and 200 ng (lanes 18 and 19, respectively) for Orc6 cDNA probe. A total of 200 ng of competitor DNA was used in lanes 7 and 8. Addition of affinity-purified polyclonal antibodies against Orc6 supershifted the Orc6-200-DNA complex (lane 8). Controls: lanes 1, 6, 12, 16, and 17 (no protein). (B) Binding preferences of Orc6-wt protein. Orc6-wt (100 ng) was incubated with the ori-β fragment in the presence of increasing amounts of various competitor DNAs (100 ng, 500 ng, and 1,000 ng). IC, poly(dI-dC) (lanes 2 to 4); I/C, poly(dI) · poly(dC) (lanes 5 to 7); AT, poly(dA-dT) (lanes 8 to 10); A/T, poly(dA) · poly(dT) (lanes 11 to 13); GC, poly(dG-dC) (lanes 14 to 16); G/C, poly(dG) · poly(dC) (lanes 17 to 19). Lane 1, no protein.
To identify the minimal domain of the protein that binds DNA, a deletion analysis was carried out. We have shown before (9) that the C-terminal region of Orc6 is important for the cytokinesis function of the protein. The deletion of that domain resulted in cytokinesis defects. However, DNA replication in cells overexpressing the Orc6 mutant Orc6-200, which lacks the C-terminal 57 amino acids, did not change (9). This suggests that the N-terminal domain is sufficient to support the replicative function of ORC. To explore this hypothesis further, the Orc6-200 mutant was analyzed by a gel shift assay. The C-terminal domain, required for cytokinesis function of Orc6, was dispensable for DNA binding (Fig. 1). Orc6-200 behaved similar to the wt protein in EMSA experiments (Fig. 1, lanes 7 and 8). The electrophoretic mobility of the complex decreased when affinity-purified polyclonal rabbit antibodies raised against Orc6 were added to the reaction mixture (Fig. 1, lane 8). Thus, the Drosophila Orc6 binds with DNA in vitro with a preference for ACE3 and ori-β fragments derived from the chorion gene amplification locus. Different fragments derived from the same locus had diminished affinity for Orc6, and small amounts of competitor DNA prevented the formation of DNA-protein complex (data not shown). We also tried to identify the high-affinity Orc6 binding site within the ori-β or ACE3 fragments by DNase I footprinting. However, we were unable to detect a discrete binding site using either E. coli- or baculovirus-expressed Orc6. Instead, we found that the addition of increasing amounts of Orc6 protein caused the entire DNA fragment to be protected (data not shown), as was observed in Drosophila ORC footprints (47).
ACE3 and ori-β fragments are AT rich, with the AT content at 60 to 70%. Therefore, to further define the Orc6-DNA interaction, Orc6 sequence preference was tested for several synthetic DNA substrates (Fig. 1B). Poly(dA) · poly(dT) was the most efficient competitor as a DNA-protein complex formed only when relatively small amounts (100 ng) of synthetic competitor were used (Fig. 1B, lanes 11 to 13). Higher amounts of poly(dA) · poly(dT) in the reaction completely abolished the formation of Orc6-DNA complex. Interestingly, poly(dA-dT) · poly(dA-dT), with the same AT content, was a much poorer competitor. Orc6-DNA complex was detected even in the presence of 1,000 ng of this competitor (Fig. 1B, lanes 8 to 10). Poly(dI) · poly(dC) synthetic DNA, which has a similar minor groove structure as poly(dA) · poly(dT) DNA, was also able to successfully compete for Orc6 in EMSAs (Fig. 1B, lanes 5 to 7). These results suggested a preference of Drosophila Orc6 for poly(dA) sequences and/or structural features associated with these sequences.
Next, all fragments derived from the chorion gene locus were analyzed for the presence of poly(dA) tracts of three and greater. The chorion gene locus was divided into 11 fragments of 300 to 350 bp as described before (47). The amount of poly(dA) tracts within the ACE3 or ori-β fragments was three to six times higher than in any other fragment derived from the chorion gene region or in the Orc6 cDNA (data not shown). This suggests that the repeats of A and T residues are significant for Orc6-DNA recognition.
Amino acids essential for DNA binding ability of Orc6.
Previous evidence indicated that the amino terminus of Drosophila and human Orc6 proteins (amino acids 1 to 203) might fold like human transcription factor TFIIB (9). A molecular modeling technique was used to dock the Orc6 to a DNA scaffold. Using the Swiss PDB program (21) (www.expasy.org/spdbv/) we replaced TFIIB with Orc6 to create a model of Orc6 protein bound to the DNA. Molecular graphics for structural representation of this model generated with the PyMol program (www.pymol.org) are shown in Fig. 2A and B. This model predicted a number of Orc6 amino acid residues in close proximity to DNA, which might contribute to DNA binding and recognition. Several blocks of these amino acid residues appear to be highly conserved in all metazoan species. Of particular interest was a consensus sequence between amino acids 69 and 79 (Fig. 2C) that in the Orc6 model forms a potential helix-turn-helix motif (Fig. 2A and B, highlighted in red). The corresponding motif of TFIIB interacts with DNA specifically with the assistance of the K189 residue (54). Two amino acids in this structural motif were selected for mutagenesis: lysine 76 (K76), which might contact the DNA, and serine 72 (S72), which might stabilize the helix-turn-helix motive (illustrated in Fig. 2B). Each amino acid was mutated into alanine by site-directed mutagenesis. Proteins, expressed in E. coli as His-tagged fusions, were purified. A silver-stained gel of the Orc6 proteins used in these studies is shown in Fig. 3A. Mutants Orc6-S72A and Orc6-K76A were analyzed for DNA binding in EMSAs. As shown in Fig. 3B (lanes 6 to 13), both alanine mutations abolished the Orc6 binding to the ori-β fragment, suggesting that DNA interactions were affected. This inability to bind with DNA was not due to the structural distortion of the mutant proteins. Mutants expressed and purified normally, indistinguishable from the Orc6-wt protein. When expressed together with other ORC subunits, point mutants readily entered the six-subunit ORC (see below).
FIG. 2.
Molecular modeling of the Orc6 structure and strategy for selecting point mutations. (A) Molecular model of the Orc6 protein in a complex with DNA. The structure is based on a predicted structural homology between Orc6 and TFIIB. The Swiss PDB program (21) (www.expasy.org/spdbv/) and PyMol program (Delano Scientific) (www.pymol.org) were used to dock Orc6 to the DNA scaffold. A putative helix-turn-helix motif of Orc6 that might be important for interaction with DNA is highlighted. (B) Amino acids serine 72 and lysine 76 are shown within a putative helix-turn-helix motif in the Orc6-DNA model. (C) Amino acid alignment of mouse, human, and Drosophila Orc6 proteins shows a conservative block between amino acids 69 and 79 (numbering is according to the Drosophila Orc6 sequence). Serine 72 and lysine 76 residues (indicated by stars) have been mutated to alanines.
FIG. 3.
(A) Silver-stained gel of purified wt and mutant Orc6 proteins. In each lane, 300 ng of protein was loaded. (B) DNA binding ability of wt and mutant Orc6 proteins. Proteins were incubated with ori-β fragment in the presence of increasing amounts of poly(dI-dC) competitor DNA (100 ng, 200 ng, 500 ng, and 1000 ng). A total of 100 ng of protein was used per reaction.
To visualize the Orc6 binding in vivo, GFP-fused wt and mutant Orc6 genes were subcloned into the P(UAST) vector, designed for P-element transformation. The UAS promoter in the construct allows for GAL4-induced expression using the GAL-4/UAS binary system. Fly stocks were set up for each fusion construct. GFP-Orc6 expression was induced in salivary glands of third-instar larvae to test chromosome binding of Orc6 mutants. Nuclei of Drosophila salivary glands contain polytene chromosomes that can be easily visualized with microscopy. Flies bearing GFP-fused Orc6 (wt or mutant) were crossed to flies carrying P{Sgs3-GAL4.PD}TP1. The Sgs-3 promoter drives GAL4-induced GFP fusion expression in salivary glands at a high level (data not shown). This makes picking up live animals for imaging an easy procedure. Salivary glands expressing GFP-fused Orc6 were dissected in PBS and stained with DAPI, and images were taken within 2 h. Figure 4 shows the localization of GFP-fused wt and mutant Orc6 expressed under the control of Sgs-3 promoter in the nucleus of Drosophila salivary glands. GFP-fused Orc6-wt protein and the Orc6-200 deletion mutant were tightly associated with polytene chromosomes (Fig. 4A and B). In contrast, the single-amino-acid mutants Orc6-S72A and Orc6-K76A failed to associate with chromosomes (Fig. 4C and D), in agreement with the in vitro DNA binding experiments illustrated in Fig. 3B.
FIG. 4.
GFP fused Orc6-wt (A), Orc6-200 (B), Orc6-S72A (C), and Orc6-K76A (D) mutants expressed in salivary glands. We induced the expression of various GFP-Orc6 proteins in salivary gland of third-instar larvae to test chromosome binding of Orc6 mutants. Flies bearing GFP-fused Orc6 genes were crossed to P{Sgs3-GAL.PD}TP1 flies, and progeny of third-instar larvae were analyzed for GFP expression under a UV dissecting microscope. The Sgs-3 promoter drives GAL4 expression in salivary glands of third-instar larvae at high levels, which makes picking up live larvae for imaging easy. Salivary glands expressing GFP-fused Orc6 were analyzed as described in Materials and Methods.
We conclude that the Drosophila Orc6 has DNA binding ability. This activity of the protein may be mediated by a potential helix-turn-helix motif between amino acids 70 and 79. Alterations of conserved amino acids within this motif disrupt the DNA binding activity of the protein both in vitro and in vivo.
Association of Drosophila Orc6 with chromosomes in vivo.
While experiments in vitro show that Orc6 prefers poly(dA) · poly(dT) tracts, in vivo binding preferences of the protein might be affected by nuclear DNA organization. To analyze the Orc6 localization along the chromosomes, salivary gland polytene chromosomes were used because of their giant size and well-determined structure which allows visualization of protein distribution on the chromosome. Polytene chromosomes of Drosophila melanogaster have a chromomeric structure. Generally, there are two major types of chromomers: bands and interbands. Bands contain more DNA and have a high package ratio and strong DAPI staining. In contrast, interbands contain less DNA and have a low package ratio, higher AT content, and weak DAPI staining. To detect Orc6 preferences on chromosomes, immunostained polytene chromosomes from the Drosophila wt stock Canton S were stained with anti-Orc6 antibodies. Orc6 was found at the sites of BrdU incorporation, as expected from its role in replication initiation (Fig. 5A, B, and C). At the next step, Orc6 was overexpressed in salivary glands using the GAL-4/UAS binary system. As Orc6 did not demonstrate high specificity in DNA binding in in vitro experiments, we expected Orc6 to be distributed evenly along chromosomes, proportional to the DNA content in chromomers. In other words, the Orc6 immunostaining pattern was expected to follow the DAPI staining. Surprisingly, Orc6 did not follow the DNA distribution along chromosomes but preferred less condensed, AT-rich interband regions (Fig. 5D, E, and F). For example, bands from regions 21C, 21D, 21E, 22A, 23A, 25A, and 31A of the 2L chromosome demonstrate strong DAPI staining (Fig. 5D) but lack Orc6 (Fig. 5E). In contrast, strong Orc6 staining is detected in regions with weak DAPI staining or interbands (Fig. 5F) which are associated with less condensed, early replicating chromatin (39).
FIG. 5.
Sites of active BrdU incorporation (marker for DNA replication) colocalize with the Orc6 protein in salivary gland polytene chromosomes. Immunostaining data are presented. Salivary glands were dissected in PBS, stained with antibody raised against Drosophila Orc6 protein (A) and anti-BrdU antibody (B) and counterstained with DAPI. A merged image is shown in panel C. White arrowheads indicate the examples of the colocalization. GFP-Orc6, overexpressed in Drosophila salivary glands using the GAL-4/UAS binary system, binds with chromosomes and prefers interband regions. DAPI staining is shown (D) as well as immunostaining with antibody raised against GFP protein (E). A merged image is shown in panel F. Bars and arrows indicate examples of heavily stained (DAPI) bands in regions 21C, 21D, 21E, 22A, 23A, 25A, and 31A (A) which do not contain Orc6 (E). The merged image confirms that the strong Orc6 signal comes from interband regions (F).
Orc6 is essential for DNA binding and DNA replication abilities of Drosophila ORC.
Drosophila ORC lacking the Orc6 protein did not bind to DNA (8). To test the DNA binding activity of Drosophila ORC containing mutant Orc6 subunits deficient in DNA binding, six-subunit ORCs were reconstituted using the baculovirus expression system as described previously (7). ORC-wt and ORC containing the mutant Orc6 subunits (ORC containing Orc6-200 [ORC(6-200)], ORC containing Orc6-S72A [ORC(6-S72A)], and ORC containing Orc6-K76A [ORC(6-K76A)]) were purified from baculovirus-infected High Five cells. The Orc1 subunit carried a His tag to facilitate purification using Ni-Sepharose. All ORCs assembled normally and behaved similarly during purification. Mutant Orc6 subunits readily assembled into the complexes with other ORC subunits, suggesting that structural integrity of Orc6 was not affected by these point mutations. A silver-stained gel showing purified wt and mutant reconstituted Drosophila ORCs is presented in Fig. 6A. These purified proteins were used in EMSAs with radioactively labeled ori-β probe. ORC-wt formed a complex with the ori-β fragment (Fig. 6B, lanes 2 to 5), as did ORC carrying the Orc6-200 deletion mutant (Fig. 6B, lanes 2 to 8). Thus, the deletion of 57 amino acids from the C terminus of Orc6 in this particular mutant had no noticeable effect on ATP-dependent DNA-protein complex formation. However, the DNA binding ability of ORCs carrying two Orc6 point mutations [ORC(6-S72A) and ORC(6-K76A)] was severely diminished in EMSAs (Fig. 6B, lanes 9 to 16). ATP-dependent ORC-DNA complexes were undetectable under the same conditions used for ORC-wt. Limited ATP-dependent ORC-DNA complex formation was observed when significantly larger amounts of ORC protein carrying Orc6 point mutants were added to the reaction (Fig. 6C).
FIG. 6.
Drosophila Orc6 protein is important for ORC-dependent DNA binding and DNA replication in vitro. (A) Silver-stained gel of recombinant purified wt and mutant Drosophila ORC proteins. In each lane 100 ng of protein was loaded. (B) DNA binding of ORC6-wt and ORC containing mutant Orc6 proteins. ORC amounts added (50 ng, 100 ng, and 150 ng) are shown above the lanes. (C) DNA binding of the ORC with the Orc6-K76A mutant [ORC(6-K76A)] and the Orc6-S72A mutant [ORC(6-S72A)]) was tested at higher concentrations (150 ng, 300 ng, and 450 ng) of the protein. (D) DNA replication in Drosophila extracts. Xenopus sperm DNA was incubated for 1 h in Drosophila extract (with membranes) at a concentration of 2 to 5 ng/μl in the presence of [32P]dCTP. Where indicated, extracts were depleted of ORC by using antibodies raised against Orc2 and Orc6. An add-back experiment was performed by the addition of 50, 100, or 150 ng of recombinant ORC proteins to depleted extracts. RE, nondepleted replication extract control (lane 1).
In vitro DNA replication assays were performed with mutant and wt reconstituted ORC in early Drosophila embryonic extracts. In this assay radioactive precursor ([32P]dCTP) incorporation into high-molecular-weight DNA was measured by autoradiography of gels after electrophoresis. For these experiments Drosophila preblastula embryo extracts were immunodepleted for ORC by using antibody against Orc2 and Orc6 subunits. This removes all ORC components from the replication extract (7). The effectiveness of immunodepletion was verified by immunoblotting (data not shown). Demembranated sperm chromatin was added to the depleted extracts, and the replication activities of mutant and wt recombinant ORCs were compared to the endogenous level of replication. As anticipated, ORCs containing mutant Orc6-S72A and Orc6-K76A were inactive in DNA replication. The mutant proteins were 10- to 20-fold below the activity of wt recombinant ORC in restoring replication to the extracts (Fig. 6D, lanes 6 to 11). The reconstituted Drosophila ORC containing Orc6-200 was effective in rescue of extract replication ability but showed between 50 and 100% of wt complex replication activity in multiple experiments (Fig. 6D, lanes 12 to 14). We conclude that Orc6 protein is necessary for ORC DNA binding and DNA replication functions.
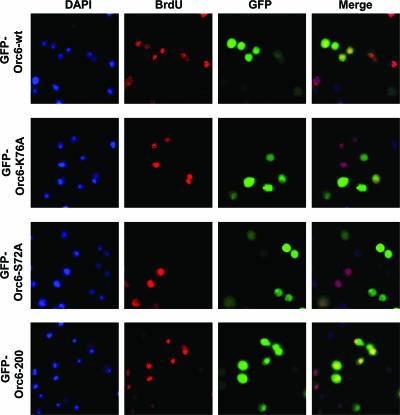
Orc6 is an integral part of the ORC complex in Drosophila, and it colocalizes with other core ORC subunits during fly embryo development (8). In Drosophila polytene chromosomes isolated from third-instar larvae, Orc6 colocalizes with the sites of active BrdU incorporation (Fig. 5A, B, and C). We and others have shown that ablation of Orc6 resulted in decreased BrdU incorporation and cell death in both Drosophila and human cells (9, 44). What effect would the Orc6 mutants described in this study have if expressed in Drosophila cultured cells? N-terminal GFP fusions of wt and mutant Orc6 proteins were transiently expressed in Drosophila L2 cells as described previously (9). Our goal was to compare the localization of Orc6 proteins in cells and DNA replication activity using BrdU incorporation. The results are quantified and summarized in Table 1 and shown in Fig. 7. Overexpression of Orc6-wt protein did not produce noticeable effects on either cell morphology or the ability of cells to replicate DNA in side-by-side comparison to nontransfected cells. Multiple experiments were performed in order to quantitate the level of BrdU incorporation as an indicator of DNA replication in cells transfected with wt and mutant Orc6 constructs. BrdU was incorporated in 32 to 43% of the total number of cells expressing GFP-Orc6-wt after 10 to 12 h of incubation. However, the overexpression of point mutants Orc6-S72A and Orc6-K76A resulted in a two- to fourfold decrease of BrdU incorporation in the cells expressing the corresponding GFP fusion proteins. Only 13 to 17% of GFP-positive cells were found to contain BrdU when the Orc6-K76A mutant was overexpressed in L2 cells. This number dropped even lower, to 9 to 12%, for the Orc6-S72A mutant (Table 1 and Fig. 7). Prolonged exposure of L2 cells to the Orc6 point mutants resulted in a further decrease of BrdU incorporation (data not shown). We did not observe cytokinesis defects in cells expressing Orc6 point mutants; therefore, it is unlikely that the observed dominant-negative effects were due to the inhibition of Orc6's cytokinetic role indirectly affecting BrdU incorporation, even though this possibility cannot be completely excluded. We would like to stress that all mutants tested here were able to incorporate effectively into recombinant ORC and thus could compete with the endogenous wt protein for complex formation when overexpressed in cultured cells. These dominant negative results suggest that a core replication domain of Orc6 was not affected by the deletion of 57 amino acids from the C terminus of the protein. Point mutations within Orc6, however, caused a significant reduction in the replication activity of transfected L2 cells as judged by BrdU incorporation. We conclude that our results provide the first evidence that DNA-binding activity of Orc6 is associated with the N-terminal domain of the protein. This activity is essential for the DNA binding and replication functions of the entire Drosophila ORC.
TABLE 1.
BrdU incorporation in Drosophila L2 cells expressing GFP-fused Orc6 wt and mutant proteinsa
| Protein | No. of cells incorporating:
|
BrdU/GFP (%)b | |
|---|---|---|---|
| GFP | BrdU | ||
| Orc6-wt | 342 | 119 | 34.7 |
| Orc6-200 | 296 | 112 | 38 |
| Orc6-K76A | 346 | 55 | 16 |
| Orc6-S72A | 301 | 33 | 11 |
Data from one representative experiment are shown. See text and Fig. 7 for more details.
No. of BrdU-positive cells/no. of GFP-positive cells.
FIG. 7.
BrdU incorporation in L2 cells expressing GFP-Orc6-wt and mutants GFP-Orc6-K76A, GFP-Orc6-S72A, and GFP-Orc6-200. GFP-tagged Drosophila Orc6 gene constructs under the control of the metallothionein promoter were transiently transfected into Drosophila L2 cells. The metallothionein promoter was induced by 0.5 mM CuSO4, and cells were incubated with BrdU overnight at a final concentration of 10 μM. Cells were fixed by using 2% paraformaldehyde, stained with anti-BrdU antibody, and subsequently subjected to immunofluorescent microscopy (magnification, ×40; Carl Zeiss Axioplan).
DISCUSSION
Replication origins in most eukaryotic species and especially in metazoans are poorly defined, making the identification of replication initiation sites difficult (13, 18). Development of a unified theory is even more complicated by the fact that initiation events appear to be specific at some loci and random at others. Whereas some experiments indicate that only particular DNA sequences possess replicator activity, other experiments indicate that any DNA sequence of sufficient length can support initiation. Our studies were undertaken to address the biochemical mechanism underlying initiation site selection in metazoans by ORC and define in more detail the DNA binding mechanisms of ORC to the origins.
The role of Orc6 in Drosophila is particularly interesting, as in budding yeast this subunit is dispensable for DNA binding (32). Drosophila ORC is also different from Xenopus and human ORC in the avidity of Orc6 association (19, 55, 56), suggesting that Orc6 may be involved in differential regulation of ORC in these organisms. Therefore, given the crucial and highly conserved role of ORC during replication initiation, it is very interesting that the DNA binding ability of ORC appears to be mediated by different subunits (or their combinations) in different species. Our previous work (8, 9) showed that Drosophila Orc6 is important for ORC-dependent DNA binding and DNA replication. Orc6 includes two distinct functional domains. The long N-terminal region is important for DNA replication, whereas the smaller 57-amino-acid C-terminal domain interacts with the septin protein Peanut and is involved in cytokinesis (9). These findings were confirmed by biochemical and cell-based genetic assays. We found that ORC(1-5) protein lacking the Orc6 subunit cannot bind DNA efficiently and does not support ORC-dependent DNA replication in Drosophila extracts (8), suggesting that Orc6 is strongly involved in both of these activities.
In this study we show that Orc6 by itself binds DNA with a preference for poly(dA) sequences. Synthetic DNAs with poly(dA) tracts competed successfully for binding of Orc6 to genomic DNA fragments containing origins of replication in Drosophila. Poly(dA) · poly(dT) was ∼10 to 100-fold better than any other synthetic DNA tested. Sequence analysis of the ACE3 and ori-β fragments of Drosophila origins shows that these sequences are enriched with poly(dA) and poly(dT) tracts compared to other fragments derived from the chorion gene amplification locus. On the other hand, the average AT content of these fragments does not differ significantly throughout the locus. This explains preferential binding of Orc6 and reconstituted Drosophila ORC to ACE3 and ori-β fragments. Austin and coworkers (1) have shown that ORC preferentially binds to the ACE3 fragment both in vitro and in vivo but did not quantitatively address the issue of ORC-DNA binding specificity. ORC associates with AT-rich Sciara coprophila origin II/9A but not with flanking fragments in vivo, and reconstituted Drosophila ORC binds the same sequence in vitro (4). Remus and others used EMSAs to test quantitatively the relative affinity of Drosophila ORC to various fragments derived from the third chromosome chorion gene cluster that included both ACE3 and ori-β regions (47). The authors concluded that the ORC DNA binding to the “nonspecific” sequences was up to sixfold lower than the “specific” (ACE3 and ori-β) fragments and that the topological state of the DNA significantly influences the affinity of ORC to DNA (47). Genome-wide analysis demonstrated that ORC in Drosophila localizes to AT-rich chromosomal sites, many of which coincide with early replication origins (36). Moreover, ORC was excluded from sequences with low AT content in these experiments, suggesting that increased AT content is necessary for ORC association (36). In agreement with these data, we found that Orc6 overexpressed in Drosophila salivary glands localizes at interband regions in polytene chromosomes (Fig. 5D, E, and F). Interband regions are extremely AT rich with a high concentration of poly(dA) stretches (50). The same regions replicated early during amplification of these giant chromosomes (39).
Thus, DNA-binding properties of the Orc6 protein and reconstituted Drosophila ORC might explain the connection between origins of DNA replication and poly(dA) blocks in vivo. The observed preference of Orc6 for poly(dA) tracts and interband regions of polytene chromosomes might be explained in part by the chromatin structure associated with these sequences. Interestingly, poly(dA) tracts form a rigid structure that is hard to deform and do not wrap easily around nucleosome cores (31), (41, 37). This feature of poly(dA) sequences might provide the basis for the assembly of a less condensed chromosome structure with a low package ratio, like interbands, making it more open and accessible to proteins like Orc6 and ORC.
The preferential binding of Drosophila Orc6 and ORC to synthetic AT-rich DNA is also seen in the fission yeast S. pombe ORC (10, 30, 33). DNA binding of the S. pombe ORC to AT-rich DNA is mediated by a unique N-terminal domain in the Orc4 subunit, which contains nine AT-hook motifs known to make minor groove contacts (11). S. pombe ORC binds more selectively than Drosophila ORC and human ORC to AT-rich DNA (10, 11, 33, 47, 55), and AT tracts have been shown to be important for the function of some origins in S. pombe (28, 42), whereas no such requirement has been demonstrated for metazoan origins. It is particularly interesting that S. pombe origins can be substituted by clustered poly(dA) stretches without noticeably affecting origin functions (42).
The helical structure of A-tract DNA is often referred to as the B* form. Characteristic features of the B*-form helix include an unusually narrow minor groove and a high base propeller twist (24, 41). The most peculiar and intensely studied feature of A-tract sequences is their propensity to cause helical axis bending when incorporated into otherwise non-A-tract DNA, which may promote DNA-protein interactions. A similar structure of a minor groove is found in poly(dI) · poly(dC) polymers, which might explain the ability of poly(dI) · poly(dC) to compete successfully for Orc6 in our EMSA experiments (Fig. 1B, lanes 5 to 7). However, Orc6 does not contain canonical AT-hook consensus sequences, so any minor groove interactions must be mediated by other structural motifs. To identify these motifs we used molecular modeling together with sequence comparison of metazoan Orc6 homologues. Aside from the hypothetical nature of this modeling, we would like to emphasize several points. First, the break in a predicted TFIIB homology domain occurs at amino acid 203 of the Drosophila Orc6 protein sequence, in good agreement with our biochemical and cell-based genetic assays. The C-terminal domain, which has been shown to be important for cytokinesis, does not fit into this fold. Second, the sequence comparison between Orc6 metazoan homologues revealed conserved amino acids that form a putative helix-turn-helix motif in our model. A corresponding structural motif is important for DNA recognition by TFIIB. Third, point mutations of two amino acids within this structural motif abolished DNA binding activities of Orc6 and, even more importantly, severely compromised DNA binding and DNA replication activities of reconstituted Drosophila ORC containing mutant Orc6 subunits. Moreover, mutant Orc6 proteins had an inhibitory effect on DNA replication in vivo, consistent with the requirement of Orc6 for ORC-dependent DNA binding and replication.
Of all metazoan ORCs, only Drosophila, Xenopus, and human proteins have been purified and biochemically characterized in detail (1, 7, 8, 19, 20, 46, 47, 48, 55, 56). Drosophila and human ORCs display similar DNA binding activities. Both Drosophila and human ORCs can bind DNA in the absence of ATP, although DNA binding is stimulated two- to fivefold by ATP. Although the Drosophila ORC, like the human ORC, can bind nonspecifically to many different DNA sequences in vitro, both proteins exhibit a preference for AT-rich sites. Drosophila ORC localizes preferentially to AT-rich ACE3 and ori-β sequences in vivo and binds the same fragments in vitro (1, 8, 47). Recombinant reconstituted human ORC shows a preference for poly(dA) sequences in vitro (55). It would be interesting to investigate if the human Orc6 protein, similar to the Drosophila homologue, displays an affinity for poly(dA) sequences.
All experimental data available to this date indicate that Orc6 is essential for ORC-dependent DNA binding and DNA replication in Drosophila. In Xenopus and human systems, published data suggest that Orc6 may not be important for these activities. This apparent inconsistency may reflect the difference in affinity of Orc6 for core ORC(1-5) complex in distant metazoan species. Drosophila ORC purifies as a tight six-subunit complex, even though a free pool of the Orc6 subunit is detected during purification from Drosophila egg extracts (7, 8, 20). In contrast, Xenopus and human Orc6 subunits are consistently underrepresented compared with the other subunits in preparations of Xenopus and human ORC, either recombinant or purified from extracts (19,46, 55, 56). It appears that in Xenopus and humans, Orc6 is less tightly associated with the core complex than other subunits and can be purified separately. During in vitro replication reactions in Xenopus egg extracts, recombinant human ORC was able to initiate DNA replication from essentially any DNA sequence (55). Human ORC(1-5) was able to restore DNA replication in Xenopus extracts depleted for ORC using antibody against the Orc2 subunit (55). ORC(1-5) was shown to be sufficient for licensing of replication origins in Xenopus (19). As Orc6 is less tightly associated with ORC in human and Xenopus, it is possible that this subunit was not completely removed from the extract when antibodies, raised against other ORC subunits, were used for immunodepletion. In Drosophila, ORC(1-5) was unable to support DNA replication when extracts were immunodepleted of both ORC(1-5) and Orc6 (8). The presence of a free pool of Orc6 in the early Drosophila egg extract, if not depleted, restored a functional six-subunit ORC that was active in both chromatin binding and DNA replication. It would be interesting to see whether immunodepletion of Orc6 from Xenopus extract has an effect on replication and licensing activities of human and/or Xenopus ORC(1-5). One recent result suggests that this indeed might be a case. The addition of either the Xenopus or human Orc6 subunit together with recombinant human ORC(1-5) to Xenopus extracts immunodepleted of all six subunits of ORC significantly increased the ability of human ORC to replicate DNA in Xenopus extracts (J. Blow and M. Gossen, personal communication), providing evidence that Orc6 might act as an assembly and/or activation factor for ORC(1-5).
In conclusion, our data strongly indicate that Orc6 in Drosophila is a DNA binding subunit of ORC, which is necessary for ORC replicative function. Orc6 is an integral part of the entire complex and functions in targeting ORC to the origins of DNA replication. Orc6 binds DNA directly, but it does not recognize a specific sequence. Rather it has an affinity for structural and topological features associated with poly(dA) stretches such as a minor groove structure. The importance of the topological state of DNA for the entire Drosophila ORC DNA binding activity has been shown recently (47). In vivo Orc6 associates with AT-rich, early replicating, interband regions. Genome-wide analysis of the entire ORC distribution in Drosophila revealed its preferential localization to AT-rich transcriptionally active chromosomal sites, many of which coincide with early replication origins (35, 36). According to our model the binding of Drosophila ORC to the origin DNA is mediated by the Orc6 subunit, which works as an anchor and targets ORC to the origins of DNA replication. This initial binding step is followed by ATP-dependent binding of the entire ORC. Interestingly, human and Xenopus Orc6 proteins are less tightly associated with core ORC subunits and may act, according to this model, as an assembly factor for ORC at the origins of DNA replication. In this case the Orc6 protein marks the origins and helps target the core ORC(1-5) complex to the DNA. As a result, six-subunit ORC is assembled at the origin at the end of mitosis and activated to ensure correct and timely origin licensing. This model may serve as a unifying mechanism for the initial stages of ori recognition in all metazoan species.
Acknowledgments
We thank Kirill Popov, Pat Higgins, and Manfred Gossen for helpful discussions and advice. We also thank Julian Blow and Manfred Gossen for communicating unpublished data.
This work is supported by a grant from NIH to I.C. (GM69681).
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Austin, R. J., T. L. Orr-Weaver, and S. P. Bell. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, E. L., J. R. Manak, S. Zhou, M. Bell, J. S. Lipsick, and M. R. Botchan. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833-837. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 4.Bielinsky, A. K., H. Blitzblau, E. L. Beall, M. Ezrokhi, H. S. Smith, M. R. Botchan, and S. A. Gerbi. 2001. Origin recognition complex binding to a metazoan replication origin. Curr. Biol. 11:1427-1431. [DOI] [PubMed] [Google Scholar]
- 5.Blow, J. J., P. J. Gillespie, D. Francis, and D. A. Jackson. 2001. Replication origins in Xenopus egg extract are 5-15 kilobases apart and are activated in clusters that fire at different times. J. Cell Biol. 152:15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers, J. L., J. C. Randell, S. Chen, and S. P. Bell. 2004. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell 16:967-978. [DOI] [PubMed] [Google Scholar]
- 7.Chesnokov, I., M. Gossen, D. Remus, and M. Botchan. 1999. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 13:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokov, I., D. Remus, and M. Botchan. 2001. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. USA 98:11997-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesnokov, I. N., O. N. Chesnokova, and M. Botchan. 2003. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc. Natl. Acad. Sci. USA 100:9150-9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Chesnokov, I. N. 2007. Multiple functions of the origin recognition complex. Int. Rev. Cytol. 256:69-109. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, R. Y., L. Chretien, J. Dai, and T. J. Kelly. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 15:15. [DOI] [PubMed] [Google Scholar]
- 11.Chuang, R. Y., and T. J. Kelly. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA 96:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crevel, G., and S. Cotterill. 1991. DNA replication in cell-free extracts from Drosophila melanogaster. EMBO J. 10:4361-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cvetic, C., and J. C. Walter. 2005. Eukaryotic origins of DNA replication: could you please be more specific? Semin. Cell Dev. Biol. 16:343-353. [DOI] [PubMed] [Google Scholar]
- 14.DePamphilis, M. L. 2005. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle 4:70-79. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis, M. L. 2003. The “ORC cycle”: a novel pathway for regulating eukaryotic DNA replication. Gene 310:1-15. [DOI] [PubMed] [Google Scholar]
- 16.Dhar, S. K., and A. Dutta. 2000. Identification and characterization of the human ORC6 homolog. J. Biol. Chem. 201:279. [DOI] [PubMed] [Google Scholar]
- 17.Diffley, J. F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778-R86. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, D. M. 2004. In search of the holy replicator. Nat. Rev. Mol. Cell. Biol. 5:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie, P. J., A. Li, and J. J. Blow. 2001. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gossen, M., D. T. Pak, S. K. Hansen, J. K. Acharya, and M. R. Botchan. 1995. A Drosophila homolog of the yeast origin recognition complex. Science 270:1674-1677. [DOI] [PubMed] [Google Scholar]
- 21.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 22.Harland, R. M., and R. A. Laskey. 1980. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell 21:761-771. [DOI] [PubMed] [Google Scholar]
- 23.Huang, Z., K. Zang, and L. F. Reichardt. 2005. The origin recognition core complex regulates dendrite and spine development in postmitotic neurons. J. Cell Biol. 170:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hud, N. V., and J. Plavec. 2003. A unified model for the origin of DNA sequence-directed curvature. Biopolymers 69:144-158. [DOI] [PubMed] [Google Scholar]
- 25.Hyrien, O., K. Marheineke, and A. Goldar. 2003. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25:116-125. [DOI] [PubMed] [Google Scholar]
- 26.Hyrien, O., C. Maric, and M. Mechali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 27.Keller, C., E. M. Ladenburger, M. Kremer, and R. Knippers. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 277:31430-31440. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S. M., and J. A. Huberman. 1998. Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol. Cell. Biol. 18:7294-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kneissl, M., V. Putter, A. A. Szalay, and F. Grummt. 2003. Interaction and assembly of murine pre-replicative complex proteins in yeast and mouse cells. J. Mol. Biol. 327:111-128. [DOI] [PubMed] [Google Scholar]
- 30.Kong, D., and M. L. DePamphilis. 2001. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol. 21:8095-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel, G. R., and H. G. Martinson. 1981. Nucleosomes will not form on double-stranded RNA or over poly(dA) · poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 9:6869-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, D. G., and S. P. Bell. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. K., K. Y. Moon, Y. Jiang, and J. Hurwitz. 2001. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl. Acad. Sci. USA 98:13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, J. J., and I. Herskowitz. 1993. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262:1870-1874. [DOI] [PubMed] [Google Scholar]
- 35.MacAlpine, D. M., and S. P. Bell. 2005. A genomic view of eukaryotic DNA replication. Chromosome Res. 13:309-326. [DOI] [PubMed] [Google Scholar]
- 36.MacAlpine, D. M., H. K. Rodriguez, and S. P. Bell. 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 18:3094-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marx, K. A., Y. Zhou, and I. Q. Kishawi. 2006. Evidence for long poly(dA) · poly(dT) tracts in D. discoideum DNA at high frequencies and their preferential avoidance of nucleosomal DNA core regions. J. Biomol. Struct. Dyn. 23:429-446. [DOI] [PubMed] [Google Scholar]
- 38.Mechali, M., and S. Kearsey. 1984. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 38:55-64. [DOI] [PubMed] [Google Scholar]
- 39.Mishra, A., and S. C. Lakhotia. 1982. Replication in Drosophila chromosomes. VII. Influence of prolonged larval life on patterns of replication in polytene chromosomes of Drosophila melanogaster. Chromosoma 85:221-236. [DOI] [PubMed] [Google Scholar]
- 40.Moon, K. Y., D. Kong, J. K. Lee, S. Raychaudhuri, and J. Hurwitz. 1999. Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 96:12367-12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, H. C., J. T. Finch, B. F. Luisi, and A. Klug. 1987. The structure of an oligo(dA) · oligo(dT) tract and its biological implications. Nature 330:221-226. [DOI] [PubMed] [Google Scholar]
- 42.Okuno, Y., H. Satoh, M. Sekiguchi, and H. Masukata. 1999. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol. Cell. Biol. 19:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto, S., D. G. Quintana, P. Smith, R. M. Mihalek, Z. H. Hou, S. Boynton, C. J. Jones, M. Hendricks, K. Velinzon, J. A. Wohlschlegel, R. J. Austin, W. S. Lane, T. Tully, and A. Dutta. 1999. latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron 23:45-54. [DOI] [PubMed] [Google Scholar]
- 44.Prasanth, S. G., K. V. Prasanth, and B. Stillman. 2002. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297:1026-1031. [DOI] [PubMed] [Google Scholar]
- 45.Randell, J. C., J. L. Bowers, H. K. Rodriguez, and S. P. Bell. 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell 21:29-39. [DOI] [PubMed] [Google Scholar]
- 46.Ranjan, A., and M. Gossen. 2006. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc. Natl. Acad. Sci. USA 103:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Remus, D., E. L. Beall, and M. R. Botchan. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remus, D., M. Blanchette, D. C. Rio, and M. R. Botchan. 2005. CDK phosphorylation inhibits the DNA-binding and ATP-hydrolysis activities of the Drosophila origin recognition complex. J. Biol. Chem. 280:39740-39751. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki, T., T. Sawado, M. Yamaguchi, and T. Shinomiya. 1999. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, Iu. V., S. A. Demakov, and I. F. Khimulev. 1998. Cloning and analysis of DNA from interband regions 85D9/D10 and 86B4/B6 of Drosophila melanogaster polytene chromosomes. Genetika 34:1081-1089. [In Russian.] [PubMed] [Google Scholar]
- 51.Shore, D. 2001. Transcriptional silencing: replication redux. Curr. Biol. 11:R816-R819. [DOI] [PubMed] [Google Scholar]
- 52.Smith, J. G., and M. P. Calos. 1995. Autonomous replication in Drosophila melanogaster tissue culture cells. Chromosoma 103:597-605. [DOI] [PubMed] [Google Scholar]
- 53.Spradling, A., and T. Orr-Weaver. 1987. Regulation of DNA replication during Drosophila development. Annu. Rev. Genet. 21:373-403. [DOI] [PubMed] [Google Scholar]
- 53a.Sullivan, W., M. Ashburner, and R. Scott Hawley. 2000. Drosophila protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Tsai, F. T., and P. B. Sigler. 2000. Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J. 19:25-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vashee, S., P. Simancek, M. D. Challberg, and T. J. Kelly. 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276:26666-26673. [DOI] [PubMed] [Google Scholar]