Abstract
UV-induced DNA damage stalls DNA replication forks and activates the intra-S checkpoint to inhibit replicon initiation. In response to stalled replication forks, ATR phosphorylates and activates the transducer kinase Chk1 through interactions with the mediator proteins TopBP1, Claspin, and Timeless (Tim). Murine Tim recently was shown to form a complex with Tim-interacting protein (Tipin), and a similar complex was shown to exist in human cells. Knockdown of Tipin using small interfering RNA reduced the expression of Tim and reversed the intra-S checkpoint response to UVC. Tipin interacted with replication protein A (RPA) and RPA-coated DNA, and RPA promoted the loading of Tipin onto RPA-free DNA. Immunofluorescence analysis of spread DNA fibers showed that treating HeLa cells with 2.5 J/m2 UVC not only inhibited the initiation of new replicons but also reduced the rate of chain elongation at active replication forks. The depletion of Tim and Tipin reversed the UV-induced inhibition of replicon initiation but affected the rate of DNA synthesis at replication forks in different ways. In undamaged cells depleted of Tim, the apparent rate of replication fork progression was 52% of the control. In contrast, Tipin depletion had little or no effect on fork progression in unirradiated cells but significantly attenuated the UV-induced inhibition of DNA chain elongation. Together, these findings indicate that the Tim-Tipin complex mediates the UV-induced intra-S checkpoint, Tim is needed to maintain DNA replication fork movement in the absence of damage, Tipin interacts with RPA on DNA and, in UV-damaged cells, Tipin slows DNA chain elongation in active replicons.
Cancer develops in tissue with active cell proliferation, and experimental studies demonstrate that S-phase cells are most susceptible to initiation of carcinogenesis by environmental carcinogens (19, 28). Mutations and chromosomal aberrations are induced during the replication of damaged DNA (30, 61). Sunlight is a ubiquitous environmental carcinogen, contributing to a million new cases of skin cancer in the United States yearly. Solar radiation that penetrates the Earth's atmosphere includes UVB wavelengths (290 to 320 nm) that damage DNA. The principal forms of DNA damage induced by UVB and UVC (240 to 290 nm) are cyclobutane pyrimidine dimers (CPDs) and 6,4-pyrimidine-pyrimidone photoproducts ([6-4]PPs). Both photoproducts are mutagenic and block DNA replication by the replicative DNA polymerases, alpha and delta. However, the great majority of mutations induced by UVB in mammalian cells appear to arise at CPDs, consistent with the more rapid repair of the [6-4]PPs (66). Patients with xeroderma pigmentosum (XP) display over 2,000-fold-increased incidence rates of skin cancer due to inherited defects in repair of UV-induced DNA damage (31, 53). Nucleotide excision repair of CPDs and [6-4]PPs protects against solar carcinogenesis by removal of DNA lesions before DNA replication. Error-free bypass of CPDs by DNA pol eta, a specialized translesion DNA polymerase that is mutated in XP-variant patients (26, 40), also suppresses UV carcinogenesis. The mechanisms that human cells use to replicate damaged DNA are, therefore, of considerable interest because, in the process, heritable genetic alterations are induced and carcinogenesis may be initiated.
Recent analyses of replication of UV-damaged DNA templates in various model systems suggest a coordination among cell cycle checkpoints, recombinational repair, and translesion synthesis machinery for preserving the integrity of replicating chromosomes. Irreparable DNA damage induced by UV in Saccharomyces cerevisiae causes an uncoupling of leading and lagging strands, producing extended regions of single-stranded DNA (ssDNA) (39). A similar uncoupling of leading and lagging strands was observed when human cell extracts replicated a circular DNA molecule containing a single CPD (9, 54), and evidence for such uncoupling has also been seen for UV-irradiated human cells (10). Genetic studies with yeast have revealed a large number of gene products that act to preserve replication fork structure and sustain DNA replication in the presence of DNA damage, including the checkpoint kinases, Mec1 and Rad53 (38, 55), factors that promote synthesis through DNA lesions by Rad6- and Rad18-dependent means that include translesion synthesis by DNA polymerases eta and zeta (20), and a nonrecombinational error-free repair pathway mediated by Rad5, Mms2, and Ubc13 (56). Additional components, such as the Rad9/Hus1/Rad1 and RFC/Rad17 complexes, Mrc1 (human Claspin), and Tof1 (human Timeless [Tim]) (57), may mediate responses to replicative stresses. Mrc1 and Tof1 promote replication fork progression and recovery from replicative stress in budding yeast (57), and their human counterparts, Claspin and Tim, have been shown to mediate the intra-S checkpoint response to UV (7, 58). Murine Hus1 has been shown to contribute to the intra-S checkpoint response to UV- and benzo[a]pyrene diolepoxide-induced DNA damage (62) and to the inhibition of DNA chain elongation in cells treated with high doses of ionizing radiation (IR) and camptothecin (60). Cells have evolved an elaborate and complex system to coordinate responses to DNA damage during DNA replication.
Human cells respond to various forms of DNA damage with a stereotypical inhibition of replicon initiation (29). The intra-S checkpoint response to UV differs from the response to IR in that ATM and the Mre11/Rad50/Nbs1 complex are not required for the response to UV (22). Instead, ATR signals through Claspin (7), Tim (58), and Chk1 (22) to inhibit replicon initiation. The intra-S checkpoint response reduces the numbers of active replicons in UV-damaged S-phase cells, thereby reducing the probability that replication forks encounter UV-induced [6-4]PPs and CPDs before these potentially mutagenic and clastogenic lesions can be repaired. The S. cerevisiae homologue of Tim, Tof1, is known to form a stable complex with Csm3, and the two proteins stabilize DNA replication forks when DNA is damaged (16, 41, 46). Swi1, the fission yeast homologue of Tof1, forms a complex with Swi3 (homologous to Csm3), which stabilizes arrested replication forks in a configuration that is recognized by the replication checkpoint sensors (48, 49). The Swi1/Swi3 complex mediates the activation of Cds1, the fission yeast homologue of budding yeast Rad53 and human Chk2 (also known as hCds1). The Tof1/Csm3 and Swi1/Swi3 complexes associate with stalled replication forks (5, 32, 47), the Swi1/Swi3 complex is associated with the Hsk-Dfp1/Him1 kinase that regulates the initiation of DNA synthesis (41), and the Tof1/Csm3 complex participates in the establishment of sister chromatid cohesion (42, 64). The mammalian orthologue of Csm3/Swi3 is Tim-interacting protein (Tipin) (17). Several recent studies show that Tim forms a complex with Tipin in human cells, and this complex mediates the intra-S checkpoint response to DNA damage (8, 18, 65). Here, we show that Tim and Tipin form a heterodimeric complex in human cells that coordinates the intra-S checkpoint response to UV-induced DNA damage. The intra-S checkpoint appears not only to inhibit the initiation of downstream replicons but also to slow DNA chain elongation in active replicons.
MATERIALS AND METHODS
Plasmids.
A OneStep reverse transcription-PCR kit from QIAGEN was used to clone a human Tipin cDNA from total RNA, which was isolated from HeLa cells. pcDNA3-Flag-Tipin was constructed using an N-terminal PCR primer that has a Flag sequence. The human Tipin cDNA was amplified from total RNA by using primers 5′-GATCGGATCCAGGATGGACTACAAGGACGACGATGACAAGCTAGAACCACAGGAGAATGGC-3′ and 5′-GATCCTCGAGTTATCTAGCTTCAGTAATATTTCTGG-3′. The PCR product was digested with BamHI and XhoI and inserted into a mammalian expression vector, pcDNA3 (Invitrogen).
A pFast-His-Flag-Tipin construct was generated by inserting the same PCR product that was described above into a pFastBacHTb expression vector to express Tipin in insect cells. The BamHI- and XhoI-digested insert was ligated to pFastBacHTb through the same sites.
pcDNA3-Flag-Tipin was used as a template for PCR amplification of Tipin to generate a pFast-His-Tipin construct. The PCR product was digested and ligated into the pFastBacHTb vector by using primers 5′-GATCTCTAGACTAGAACCACAGGAGAATGGC-3′ and 5′-GATCGGTACCTTATCTAGCTTCAGTAATATTTCTGG-3′. The PCR product was ligated into vector through the XbaI and KpnI sites, and the N-terminal His tag was obtained from the sequence within the pFastBacHTb vector (GIBCO/BRL) backbone.
The pcDNA4.1-Flag-Tim mammalian expression vector (58) was used as a template for amplification of Tim by PCR. The PCR product was digested and ligated into pFastBacHTb to generate pFast-His-Flag-Tim for the expression and purification of His-Flag-Tim from insect cells by using an N-terminal PCR primer that has a Flag sequence. The primers used were 5′-ACTTCTAGAAGGATGGACTACAAGGACGACGATGACAAGGACTTGCACATGATGAACTGTGAAC-3′ and 5′-ACTGGTACCTCAGTCATCCTCATCATCCTCAATCTGG-3′. The PCR product was amplified with Platinum Pfx DNA polymerase (Invitrogen) and then digested with restriction enzymes XbaI and KpnI.
p11d-tRPA Escherichia coli expression vector was used as a template for amplification of Flag-RPA34 by using primers 5′-GATCGGATCCAGGATGGACTACAAGGACGACGATGACAAGTGGAACAGTGGATTTCGAAAGC-3′ and 5′-GATCTCTAGATTATTCTGCATCTGTGGA-3′. The PCR product was digested with BamHI and XbaI restriction enzymes and ligated into the pcDNA3 (Invitrogen) mammalian expression vector through the same enzyme sites to generate N-terminal Flag epitope-tagged RPA34.
pcDNA4-Flag-XPA was constructed using an N-terminal PCR primer that has a Flag sequence. The XPA cDNA was amplified by using primers 5′-CTAGGAGGATCCACCATGGATTACAAAGACGATGACAAGGCGGCGGCCGACGGGGCTTTG-3′ and 5′-CGCGCTCGAGCATTTTTTCATATGTCAG-3′. The PCR product was digested with BamHI and XhoI and inserted into the pcDNA4a (Invitrogen) mammalian expression vector.
We received pBSChk1 from Stephen Elledge. Chk1 was amplified by using the following primers: 5′-TGTGGTGGAATTCCTCGAGATGGCAGTGCCCTTTGTGG-3′ and 5′-CGGGTTTAAACGTTAACTCACTTATCGTCATCGTCCTTGTAGTCCATACCCTCGAGTGTGGCAGGAAGCCAAAC-3′. The 3′ primer contains the XhoI site, a Flag epitope, and a PmeI site. The recipient vector, pcDNA4MycHis, was digested from EcoRI and PmeI sites to remove the Myc and His tags. The PCR product and vector were digested with EcoRI and PmeI, gel purified, and then ligated together to generate pcDNA4-Chk1-Flag.
Cell lines and antibodies.
Mammalian cell lines were maintained in Dulbecco's minimal essential medium (DMEM) with 10% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. A monolayer of Sf21 insect cells (Invitrogen) was grown in Grace's insect medium (GIBCO/BRL) supplemented with 10% fetal bovine serum. Rabbit polyclonal anti-Tim antibody was generated as described previously (63). Monoclonal anti-Flag antibody was purchased from Sigma. Phosphospecific anti-Chk1 antibody (P-S345) was purchased from Cell Signaling. Anti-actin antibody was purchased from Santa Cruz Biotechnology, Inc. Monoclonal anti-His antibody was purchased from Abgent.
Expression and purification of recombinant proteins.
Baculovirus for expression of Flag-Tim was generated with a Bac-to-Bac baculovirus expression system (GIBCO/BRL) and protocols suggested by the manufacturer. The optimal multiplicity of infection for recombinant virus was empirically determined. Monolayered Sf21 insect cells were infected with virus and harvested 48 h later. The cells were washed with phosphate-buffered saline (PBS) and lysed in 20 packed-cell volumes of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM β-glycerophosphate, 10% glycerol, 1% Tween 20, 0.1% NP-40, 1 mM Na3VO4, 1 mM NaF, and protease inhibitors [Roche Molecular Biochemicals]) for 30 min on ice. After centrifugation at 32,000 × g for 30 min, the supernatants were incubated with anti-FLAG agarose (Sigma) overnight at 4°C. The resin was washed three times with washing buffer that contained 50 mM Tris-HCl (pH 7.5) and 300 mM NaCl; the proteins were eluted in elution buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, protease inhibitors, 200 μg/ml of Flag peptide [Sigma]). For purification of the Tim-Tipin complex, cells were coinfected with viruses expressing Flag-Tim and His-Tipin. The complex was first bound to Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) for 2 h at 4°C and protein eluted with 50 mM Tris-HCl (pH 7.5), 150 mM NaCl buffer containing 200 mM imidazole. The eluate was then purified with anti-FLAG agarose, and protein was eluted with elution buffer as described above.
Purification of the Tim-Tipin-RPA complex.
The Tim-Tipin complex was first sequentially purified from insect cells as described above; before the Flag-peptide elution step, the Flag-beads were incubated for one hour on ice with recombinant replication protein A (RPA) that was purified from E. coli. The resin was then washed three times with washing buffer that contained 50 mM Tris-HCl (pH 7.5) and 300 mM NaCl, and the protein complex was eluted in elution buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, protease inhibitors, 10% glycerol, 200 μg/ml of Flag peptide).
Immunoprecipitation assays.
HEK293T cells were either singly transfected or cotransfected with the indicated plasmids by a calcium phosphate method. After 16 h of incubation at 37°C, the cells were washed twice in serum-free DMEM, fresh medium was added, and the cells were incubated for another 48 h. The cells were lysed for 30 min in ice-cold lysis buffer (described above). Lysates were clarified by centrifugation at 30,000 × g, and the supernatants were incubated with anti-FLAG M2 affinity gel (Sigma) at 4°C overnight. Protein-bound beads were then washed three times with washing buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl), and bound proteins were eluted with TBS buffer containing 200 μg/ml of Flag peptide (Sigma).
siRNA.
The small interfering RNA (siRNA) duplexes were 21 bp, including a 2-base deoxynucleotide overhang. The sequences of the Tim siRNA oligonucleotides were GUAGCUUAGUCCUUUCAAAdTdT and UUUGAAAGGACUAAGCUACdTdT, and the Tipin siRNA oligonucleotides were UGAUUGACCUACCAGAUUAdTdT and UAAUCUGGUAGGUCAAUCAdTdT (synthesized by Dharmacon Research, Inc.). Where transfections with control siRNA are indicated, Dharmacon's siCONTROL Non-Targeting siRNA #2 (catalog number D-001210-02) was used.
For transfections, HeLa cells were plated in 6-cm tissue culture plates and transfected at 40% confluence with the siRNA duplex, by using Oligofectamine (Invitrogen) transfection reagent and following the manufacturer's suggested protocol. Transfections were repeated 24 h later, and cells were analyzed 72 h after the first transfections.
Gel mobility shift assay.
RPA purified from E. coli or His-Tipin purified from insect cells at the indicated concentrations were incubated with 5 fmol of 50-bp duplex DNA for 30 min at 30°C in a 10-μl reaction mixture; the preparation and sequence of the DNA duplex and the composition of the incubation reactions were described previously (33). In supershift assays, antibodies were mixed with the proteins before the addition of the DNA to the reactions. Glycerol was added to a concentration of 8.5%, and samples were resolved by electrophoresis in a 5% nondenaturing polyacrylamide gel at room temperature and a constant current of 25 mA in 1× Tris-borate-EDTA buffer as described before (33). The gel was dried and exposed to PhosphorImager screening (Amersham Biosciences).
Generation of Tet-inducible stable cell lines.
To generate tetracycline (Tet)-inducible expression of Tipin, Flag-Tipin cDNA was cloned into BamHI-XhoI sites of the vector pcDNA5/FRT/TO (Invitrogen). This construct was then cotransfected with the Flp recombinase expression plasmid pOG44 into the Flp-In T-REx 293T cell line (Invitrogen). This cell line stably expresses the Tet repressor and contains a single integrated FRT (Flp recombination target) site. Flp recombinase expression from the pOG44 vector mediated insertion of Flag-Tipin into the genome at the integrated FRT site through site-specific DNA recombination. After 48 h, transfected cells were selected for resistance to 150 μg/ml hygromycin B and 15 μg/ml blasticidin. Cell clones appeared after 10 to 15 days. Isogenic pooled clones were expanded and used for induction of Flag-Tipin by Tet (Sigma). A similar strategy was used to generate stable transfectants with empty plasmid pcDNA5/FRT as control clones.
Radioresistant DNA synthesis assay.
HeLa cells were plated (2 × 105 per 6-cm tissue culture dish) in 4 ml of DMEM containing 10 nCi/ml of [14C]thymidine (ICN Radiochemicals)/ml to uniformly label DNA. The next day, cells were transfected with siRNA oligomers in the presence of [14C]thymidine. The medium containing [14C]thymidine was replaced the following day with fresh medium, and cells were transfected for the second time with siRNA oligonucleotides. Twenty-four hours later, cells were either unirradiated or irradiated with UVC (2.5 J/m2) and incubated at 37°C for 30 min before incubation with 25 μCi/ml of [3H]thymidine/ml for 15 min. Cells were then processed for determination of [3H]/[14C] ratios as previously described (22).
Immunofluorescence microscopy of DNA fibers.
Cells were double labeled by incubation, first in medium with 100 μM IdU for 10 min, and second after treatment with 2.5 J/m2 UVC, in 50 μM CldU for 20 min. DNA spreads were made as described previously (24), with certain modifications. Briefly, the cells were trypsinized and resuspended in ice-cold PBS at ∼200 to 400 cells/μl. Two microliters of cell suspension was spread on a Silane-Prep slide (S4651; Sigma-Aldrich), close and parallel to the label. The sample was allowed to evaporate until almost but not completely dry and was then overlaid with 10 μl of spreading buffer (0.5% sodium dodecyl sulfate [SDS] in 200 mM Tris-HCl [pH 7.4], 50 mM EDTA). After ∼10 min, the slide was tilted at ∼15° to allow the cell lysate to slowly move down the slide, and the resulting DNA spreads were air dried, fixed in 3:1 methanol/acetic acid for 2 min, and refrigerated overnight.
Modifications of published protocols were used for staining of DNA fibers (1, 51, 59). Briefly, each slide set included control samples from cells labeled with CldU alone or IdU alone, and the person doing the immunolabeling and scoring of fibers did not know the identity of the samples (i.e., a single-blind study). The slides were treated with 2.5 M HCl for 30 min, washed several times in PBS, and blocked in 2% bovine serum albumin in PBS for 60 min. The slides were incubated at room temperature with the antibodies indicated below, rinsed three times in PBS, and incubated for 30 min in blocking buffer between each of the following incubations: (i) 1 h in 1:250 rat antibromodeoxyuridine (detects CldU) (OBT0030; Accurate) plus 1:250 mouse antibromodeoxyuridine (detects IdU) (Becton Dickinson), (ii) 30 min in 1:300 Alexa Fluor 488-conjugated chicken anti-rat antibody (Molecular Probes) plus 1:400 Alexa Fluor 594-conjugated rabbit anti-mouse antibody, and (iii) 30 min in 1:250 Alexa Fluor 488-conjugated goat anti-chicken antibody plus Alexa Fluor 594-conjugated goat anti-rabbit antibody. In addition, prior to the blocking step between the first and second antibody incubations, the slides were placed for 10 min in a stringency buffer containing 10 mM Tris HCl (pH 7.4), 400 mM NaCl, 0.2% Tween 20, and 0.2% NP-40 to remove any nonspecifically bound primary antibodies. The slides were rinsed three times in PBS and mounted in antifade reagent (UNC Microscopy Core). Microscopy was carried out using an Olympus FV500 confocal microscope in sequential scanning mode.
When analyzing red and green track lengths, we tested whether two group means fell within a certain range in various binary comparisons. This range was taken to be 0.6 μm, as measurements of green track lengths in five different experiments under the same conditions used in this analysis yielded an average of 5.7 ± 0.6 μm. The null hypothesis was formulated as H0: μ1 − μ2 ≤ 0.6 μm, where μ1 and μ2 are the mean track lengths for groups 1 and 2, respectively. The alternative hypothesis was H1: μ1 − μ2 > 0.6 μm. The test can be considered an intersection-union test which involves two one-sided t tests, each at the 0.05 significance level. Each test returned a P value, and the overall P value was the minimum of the two.
RESULTS
Tipin forms a complex with Tim.
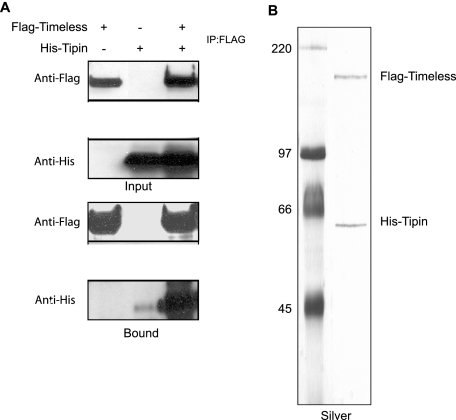
To investigate the interaction of human Tim with Tipin, we generated plasmids for expressing Flag-tagged Tim and His-tagged Tipin in mammalian cells and tested the interaction by cotransfection in HEK293T cells. Tim was immunoprecipitated with anti-Flag resin, and the immunoprecipitates were analyzed for His-tagged Tipin. As shown in Fig. 1A, Tim and Tipin interact specifically. To confirm and analyze the nature of the interaction, the Tim-Tipin complex was purified from insect cells by sequential immunoaffinity chromatography. Silver staining of the purified complex (Fig. 1B) showed that Tim and Tipin were in a stable protein complex at a 1:1 ratio following Flag-peptide elution. The sequential immunoaffinity chromatography procedure also yielded pure His-Tipin that was free of Tim in the flowthrough of the Flag-binding, and this His-Tipin was used in gel mobility shift assays.
FIG. 1.
Tim forms a complex with Tipin. (A) Tim-Tipin interaction in mammalian cells. HEK293T cells were transfected either with Flag-Tim or with His-Tipin or were cotransfected with Flag-Tim and His-Tipin and then immunoprecipitated (IP) with anti-Flag antibodies. Immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and then immunoblotted with antibodies to Flag and His as indicated. Input represents 1/30 of the cell lysates used for immunoprecipitation. (B) Purification of Tim-Tipin complex. Insect (Sf21) cells were coinfected with baculoviruses expressing His-Flag-Tim and His-Tipin, and proteins from extracts were first collected with Ni-NTA agarose and eluted with 200 mM imidazole. The eluates were then collected with anti-Flag agarose, and proteins were eluted by Flag peptide. The profile of the purified complex was visualized on a silver-stained SDS-polyacrylamide gel.
Tim and Tipin are required for the intra-S checkpoint response to UV.
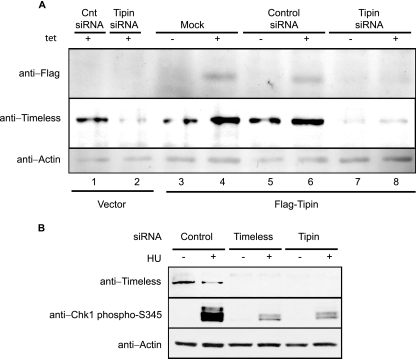
Knockdown of Tim in HeLa cells was shown previously to attenuate the replication checkpoint response to hydroxyurea (HU) and the intra-S checkpoint response to UV (58). siRNA-mediated knockdown was used to determine the role of the Tim binding partner Tipin in these checkpoint responses. To establish an effective siRNA for knockdown of Tipin, HEK293T cells were engineered to express Flag-Tipin under the control of a Tet-responsive promoter (Fig. 2A). The addition of Tet to culture medium induced expression of Flag-Tipin, and this induction was reversed by transfection of Tipin siRNA (Fig. 2A, lane 8) as measured by anti-Flag immunoblotting. Knockdown of Tipin by siRNA in cells expressing Flag-Tipin caused a significant downregulation of Tim as well (Fig. 2A, lanes 7 and 8). When 293 cells carrying the empty Flag expression vector were treated with Tipin siRNA, the Tim immunoblot showed that Tipin siRNA caused a significant downregulation of Tim relative to treatment with the scrambled-control siRNA (Fig. 2A, lanes 1 and 2). The downregulation of Tim seen in the vector-control cell line confirmed the successful knockdown of endogenous Tipin. Given the results described above, that Tipin and Tim form a stable complex in human cells, this downregulation suggests that endogenous Tim is destabilized in the absence of Tipin.
FIG. 2.
Loss of Tipin downregulates the expression level of Tim and attenuates Chk1 activation. (A) Tet-inducible expression of Flag-Tipin. Proteins were induced in the presence of Tet (1 mM) for 24 h and detected with anti-Flag antibodies (upper panel). HEK293T cells with inducible expression of Flag-Tipin and the control cell line expressing vector only were transfected with scrambled control or Tipin siRNA two times over a 3-day period. Forty-eight hours after the initial transfection, cells were treated with Tet or left untreated. A vector-expressing cell line (Mock) was used as a negative control for inducible protein expression and positive control for the siRNA treatment, where reduced expression of Tim in Tipin siRNA-treated cells can be seen as a result of successful knockdown of endogenous Tipin (lane 2). Cell lysate proteins (80 μg) were immunoblotted with anti-Tim, anti-Flag, and anti-actin antibodies (lower panel). Cnt, control. (B) Tipin is required for HU-induced Chk1 activation. HeLa cells were transfected with control, Tim, or Tipin siRNA two times over a 3-day period. Seventy-two hours after the initial transfection, cells were treated with 10 mM HU for 1.5 h or left untreated. Cell lysate proteins (150 μg) were immunoblotted with anti-Tim, anti-Chk1-phosphoS345, and antiactin antibodies.
To determine whether Tipin contributes to checkpoint responses, HeLa cells were transfected with either Tim or Tipin siRNA, and checkpoint signaling was monitored after treatment with HU. As previously reported (58) knockdown of Tim reduced the phosphorylation of Chk1 in response to replication stress (Fig. 2B). Similarly, transfection of Tipin siRNA attenuated phosphorylation of Chk1 in response to HU. Transfection of Tipin siRNA also reduced the expression of Tim in HeLa cells.
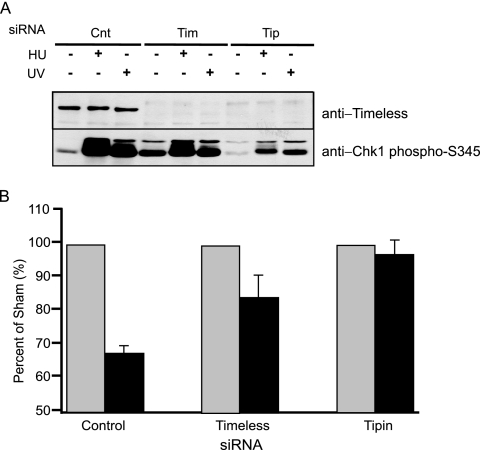
The intra-S checkpoint response to UV was monitored by immunoblot analysis of Chk1 phosphorylation and quantification of the incorporation of [3H]thymidine 30 to 45 min after irradiation of HeLa cells with 2.5 J/m2 (Fig. 3). Previous studies showed that knockdown of Tim in HeLa cells did not increase Chk1 phosphorylation (58), and so the variation in phospho-Chk1 in undamaged cells as demonstrated in Fig. 3A probably reflected biological or experimental variation. Knockdown of Tim and Tipin attenuated the phosphorylation of Chk1 in response to UVC (Fig. 3A). Cells transfected with a scrambled control siRNA responded to UV with 33% inhibition of DNA synthesis (Fig. 3B). As demonstrated previously (58), knockdown of Tim significantly attenuated this response. After knockdown of Tipin, DNA synthesis in UV-treated cells occurred at 95% of the rate measured in untreated control cells (Fig. 3B). Knockdown of Tipin reduced the overall rate of DNA synthesis by 59% (measured as the 3H/14C ratio; Fig. 3 legend) consistent with a similar value (45%) reported by Yoshizawa-Sugata and Masai (65). Knockdown of Tim also inhibited DNA synthesis by 33%, similar to the value of 14% reported by Gotter et al. (18). The results indicate that DNA replication and intra-S checkpoint response to UV were disturbed by the knockdown of Tipin and Tim.
FIG. 3.
Knockdown of Tipin attenuates the intra-S checkpoint response to UV. (A) Tipin is required for UV-induced Chk1 activation. HeLa cells were transfected with scrambled control (Cnt), Tim, or Tipin siRNA two times over a 3-day period. Seventy-two hours after the initial transfection, cells were treated with either 10 mM HU for 1.5 h or UV (6 J/m2) for 1 h or were left untreated. Cell lysate proteins (150 μg) were immunoblotted with anti-Tim and anti-Chk1-phosphoS345. (B) HeLa cells that were transfected with scrambled control, Tim, and Tipin siRNA were grown in the presence of [14C]thymidine for 40 h to label DNA uniformly until the second transfection and then grown in nonradioactive medium for an additional 24 h. Cells were exposed to UV (2.5 J/m2) or left untreated, incubated at 37°C for 30 min, and then labeled for 15 min in medium containing [3H]thymidine. Relative DNA synthesis was estimated from the incorporated [3H]thymidine normalized to total DNA by the 14C radioactivity. Data are expressed as percentages of the control samples (no UV irradiation) and plotted as means ± standard deviations (SD; n = 3). 3H/14C ratios in untreated cells were 67 ± 21% and 41 ± 21% (mean ± SD) of the scrambled control cells for Tim and Tipin knockdown cells, respectively.
Tipin interacts with RPA on DNA.
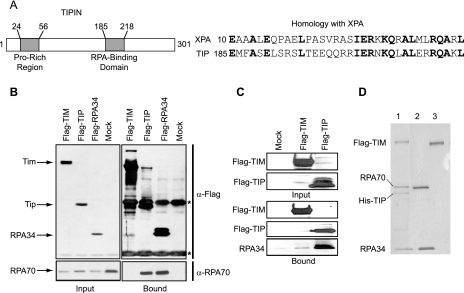
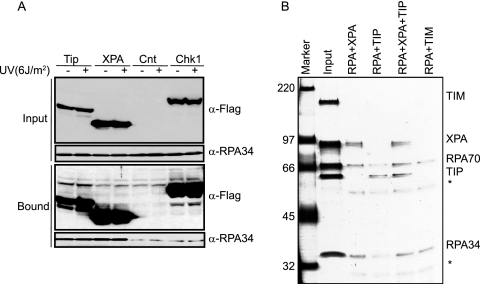
Searches for conserved domains within Tipin found a 34-amino-acid domain that resembles a sequence contained within XPA (16), a protein that is essential for nucleotide excision repair. The 53-amino-acid N-terminal region of XPA that shares homology to Tipin has been mapped to bind the 34-kDa subunit of RPA (23, 43) (Fig. 4A). Therefore, we tested for Tipin-RPA interaction in vivo and in vitro. HEK293T cells were transfected with empty plasmid or plasmids expressing Flag-Tim, Flag-Tipin, or Flag-RPA34, and anti-Flag immunoprecipitates were probed for the RPA70 subunit. As seen in Fig. 4B, a significant amount of RPA70, the large subunit of the RPA complex, was present in the anti-Flag immunoprecipitates only from cells that overexpressed Flag-Tipin and Flag-RPA34. These in vivo data show that Tip binds to RPA, a key protein for DNA replication and repair.
FIG. 4.
Interaction of Tipin with RPA. (A) Schematic representation of human Tipin protein with proline-rich and RPA-binding domains and protein sequence alignment of XPA and Tipin homology regions. (B) Tipin-RPA interaction in mammalian cells. HEK293T cells were transfected with vector (Mock; lane 4) or plasmids expressing Flag-Tim (Flag-TIM; lane1), Flag-Tipin (Flag-TIP; lane 2), and Flag-RPA34 (lane 3). Flag immunoprecipitates (Bound) as well as the whole-cell extract (Input) were separated by SDS-PAGE and blotted with anti-Flag and anti-RPA70 antibodies. Asterisks (*) indicate the heavy and light chains of the anti-Flag antibody. Input represents 1/30 of the cell lysates used for immunoprecipitation. (C) Tipin-RPA interaction in vitro. Insect cells (Sf21) were either uninfected (Mock; lane 1) or infected with viruses expressing Flag-Tim (lane 2) and Flag-Tipin (lane 3). Flag-purified proteins were incubated with 3.5 μg of RPA while still on the beads overnight at 4°C, and then the Flag agarose beads were washed three times. The bound proteins were eluted with Flag peptide and separated by SDS-PAGE and blotted with anti-Flag and anti-RPA34 antibodies. (D) Formation of Tim-Tipin-RPA complexes. Insect cells were either coinfected with viruses expressing Flag-Tim and His-Tipin (lane 1) or infected only with Flag-Tim virus (lane 3). The Tim-Tipin complex was purified by chromatograpy with Ni-NTA-agarose and anti-Flag agarose as described in Materials and Methods. Tim was purified by chromatography with anti-Flag agarose. Proteins bound to beads were incubated with RPA (lanes 1 and 3) on ice for one hour. After extensive washing, the proteins bound to beads were eluted with Flag peptide, and an aliquot of the first elution was analyzed by SDS-PAGE followed by silver staining. The second lane contains 1/15 of the RPA used in the binding assay.
To further test for direct Tipin-RPA interaction, we expressed Flag-Tim and Flag-Tipin in Sf21 insect cells, isolated protein by immunoprecipitation, and then incubated the protein with recombinant RPA (all three subunits) purified from E. coli. As seen in Fig. 4C, RPA specifically bound Tipin but not Tim in vitro, leading us to conclude that the Tipin-RPA interaction is a direct interaction and that Tipin may mediate the interaction of Tim with RPA. We tested the prediction that Tipin might mediate the interaction between Tim and RPA by reconstituting the tertiary complex with the purified Tim-Tipin complex (from Fig. 1B) and purified RPA (from E. coli). We observed that the Tim-Tipin complex indeed was able to bind to RPA, while Tim alone was not (Fig. 4D). Taken together, these data suggest that Tipin, the small binding partner of Tim, binds to RPA. The Tim-Tipin complex is stable upon interaction of Tipin, with RPA leading to the formation of a Tim-Tipin-RPA ternary complex.
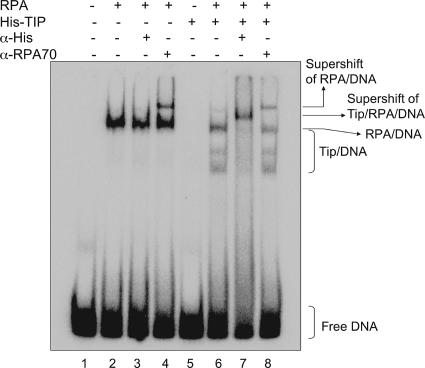
To determine whether the interaction between Tipin and RPA was influenced by DNA damage and whether the Tipin-RPA interaction might modulate excision repair by competing with XPA for binding to RPA, we compared the binding of RPA to Tipin and XPA before and after treating HEK293T cells with UV (Fig. 5A). Tipin had an affinity for RPA34 that was similar to that of XPA, but the interaction of RPA34 with Tipin and XPA was not altered by treatment with UV. This result suggests that binding of Tipin to RPA, at least in the soluble nucleoplasm, is not stimulated by posttranslational modifications in response to DNA damage. Flag-Chk1 served as a negative control, since its affinity for endogenous RPA34 was no more than that of the vector control. To assess binding stoichiometries and compare the affinities of XPA and Tipin to RPA, purified proteins were mixed in vitro and then immunoprecipitated with anti-RPA34 antibody. Assessments of complex formation after silver staining of the gel indicated that similar levels of XPA and Tipin were immunoprecipitated in what appeared to be 1:1 complexes with RPA34 and RPA70 (Fig. 5B), indicating that Tipin binds to RPA with the same avidity as XPA through a similar binding module. However, the significance of this finding for checkpoint-repair coordination is unclear at present. A competition assay indicated that the addition of increasing concentrations of Tipin to mixtures containing RPA and XPA produced decreasing amounts of XPA in anti-RPA immunoprecipitates (results not shown). Thus, Tipin appeared to compete with XPA for binding to RPA. DNA binding studies were performed to investigate the potential functional significance of the Tipin-RPA interaction. RPA and XPA are among the major damage recognition proteins involved in the early stage of nucleotide excision repair. RPA and XPA are able to bind damaged DNA independently, although the RPA interaction stimulates XPA binding to damaged DNA (21). Along this line, we tested the binding of Tipin to DNA in the presence or absence of RPA by gel mobility shift assays using 50-bp duplex DNA. Tipin alone did not bind to double-stranded DNA, but it bound to DNA in the presence of RPA (Fig. 6, lanes 5 and 6). Two additional retarded bands were observed when Tipin was included with RPA, perhaps representing DNA molecules with one or two molecules of bound Tipin. The formation of DNA-protein complexes was verified by supershifting the complexes with anti-His antibodies for Tipin binding and with anti-RPA70 antibodies for RPA binding (Fig. 6, lanes 7 and 8). The two higher-mobility DNA bands in lane 6 were shifted by anti-His antibody but not anti-RPA70 antibody, suggesting that both higher-mobility forms were free of RPA. Lane 6 also shows a DNA-protein band with mobility similar to that of the one formed with RPA in the absence of Tipin (lane 2). This slower-mobility band in lane 6 was also shifted with anti-His antibody, suggesting that Tipin associated with RPA-DNA complexes. We tested the binding of Tipin to naked and RPA-covered ssDNA; Tipin bound to naked ssDNA with low affinity, and binding was stimulated in the presence of RPA (data not shown). These data suggest that Tipin interacts with RPA on DNA and that RPA facilitates binding of Tipin to DNA but does not remain bound to Tipin-DNA complexes during gel electrophoresis. The binding of Tipin to DNA and to RPA-DNA complexes may enable the Tim-Tipin complex to carry out its fork-stabilizing function.
FIG. 5.
Tipin and XPA bind to RPA with comparable affinities. (A) UV damage does not stimulate the interactions between either XPA and RPA or Tipin and RPA. HEK293T cells transfected with vector (Cnt), Flag-Tipin (Tip), Flag-XPA (XPA), or Flag-Chk1 (Chk1) were either left untreated or treated with UV and then lysed 1 h later. An equal amount of cell lysate (2 mg) was immunoprecipitated with anti-Flag agarose and separated by SDS-PAGE. Flag immunoprecipitates (Bound) as well as the whole cell extract (Input) were blotted with anti-Flag and anti-RPA34 antibodies. Input represents 1/30 of the whole-cell lysate used for immunoprecipitation. (B) Tipin and XPA bind to RPA in vitro with comparable affinities. RPA protein (10 μg) was either mixed with 2 μg of XPA (RPA+XPA; lane 3), 2 μg of Tipin (RPA+TIP; lane 4), 2 μg of Tipin and 2 μg XPA (RPA+XPA+TIP; lane 5), or 2 μg of Tim (RPA+TIM; lane 6). Immunoprecipitation experiments were carried out with anti-RPA34 antibody. A fraction of bound proteins was separated by SDS-PAGE and visualized by silver staining. Input lane contains 1/10 of the amount of XPA, Tipin, and Tim and 1/40 of that of RPA used in the binding assay (lane 2). Asterisks (*) indicate the heavy and light chains of the anti-RPA34 antibody.
FIG. 6.
RPA recruits Tipin to DNA. Terminally labeled 50-bp duplex DNA was incubated with ∼200 ng of RPA, 150 ng of His-Tipin, or both proteins, and the DNA-protein complexes were separated on a 5% polyacrylamide gel. Anti-His and anti-RPA70 antibodies were added to the samples during the reaction where indicated. Lane 1 is DNA only.
Replication dynamics after knockdown of Tim and Tipin.
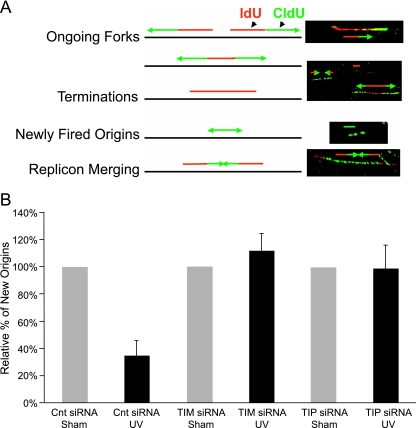
The yeast homologues of Tim and Tipin form a replication fork protection complex that stabilizes replication forks under stress. To further explore the roles for human Tim and Tipin in regulation of DNA replication, an immunofluorescence microscopy method used spread DNA fibers to visualize replication dynamics in UV-damaged cells. HeLa cells were incubated with IdU for 10 min before treatment with 2.5 J/m2 UV and with CldU for 20 min after treatment. Through the use of appropriate antibodies and fluorescent tags, tracks of IdU-labeled DNA were colored red, and tracks of CldU-labeled DNA were colored green (Fig. 7A). After cell lysis and the spreading of DNA fibers, it was possible to visualize the products of DNA synthesis at replication forks that were active before treatment with UV (IdU labeled only, colored red), after treatment (CldU labeled only, colored green), or before and after treatment (IdU and CldU labeled, colored red adjacent to green).
FIG. 7.
Knockdown of Tim and Tipin reverses the inhibition of replicon initiation in UV-treated HeLa cells. (A) Left, schematic illustration of results expected when a 10-min pulse of asynchronous cells with IdU is followed by a 20-min pulse with CldU and individual replication units are visualized by immunofluorescence detection of the incorporated halogenated nucleotides in DNA spreads. Right, the presence and relative positions of single and dual labeling in continuous replication tracks are used to interpret their representations of various stages of DNA synthesis. (B) Percentage (compared to the sham-treated control [Cnt]) of new origin initiation (green tracks). Tipin (Tip) or Tim protein levels were knocked down by siRNA prior to the exposure of HeLa cells to 2.5 J/m2 of UV (Table 1). The data are the means from three independent experiments (plus one SD). The results showed a significant UV-induced inhibition in green-only tracks in cells transfected with scrambled control siRNA (two-sided P value, <0.0001) and insignificant UV response in cells with knockdown of Tim (two-sided P value, 0.47) or Tipin (two-sided P value, 0.94).
A prior study showed that ionizing radiation reduced the fraction of green-only tracks in HeLa cells (44), consistent with a reduction in the rate of replicon initiation as a consequence of ATM-dependent intra-S checkpoint signaling (14, 50). We recently reported that a low fluence of UVC produced an ATR- and Chk1-dependent reduction in the frequency of green-only tracks (6), demonstrating that the method is capable of detecting the intra-S checkpoint response to UV. When HeLa cells were transfected with a nontargeting control siRNA, UV treatment caused a 64% reduction in the fraction of green-only tracks, indicative of an effective intra-S checkpoint response that inhibits replicon initiation (Fig. 7B; Table 1). Knockdown of Tim and Tipin reversed the UV-induced inhibition of replicon initiation; irradiation had no effect on the fractions of green-only tracks in Tim- or Tipin-depleted cells (Table 1).
TABLE 1.
Distribution of replication tracks with the IdU-only or CldU-only label or both labels
| siRNA condition | Treatmenta | No. of tracks (%)b
|
P valuec | ||
|---|---|---|---|---|---|
| Red only (IdU) | Red-green (IdU-CldU) | Green only (CldU) | |||
| Nontargeting | Sham | 200 (23) | 561 (66) | 95 (11) | <0.0001 |
| control | 2.5 J/m2 | 280 (38) | 422 (58) | 28 (4) | |
| Tim | Sham | 199 (21) | 659 (71) | 71 (8) | 0.4672 |
| 2.5 J/m2 | 229 (20) | 806 (71) | 97 (9) | ||
| Tipin | Sham | 189 (21) | 610 (69) | 89 (10) | 0.9365 |
| 2.5 J/m2 | 217 (25) | 557 (65) | 85 (10) | ||
HeLa cells were pulsed with 100 μM IdU for 10 min, treated with 2.5 J/m2 UV (or sham-treated), and then pulsed with 50 μM CldU for 20 min before preparation of the DNA fibers.
The numbers of tracks with IdU only (red only), CldU only (green only), or both IdU and CldU (red-green) were determined and are also expressed as percentages of the total. The values given are combinations of results from three independent experiments.
Fisher's exact test was performed for the three treatment conditions to examine whether there was a difference between the percentages of green-only tracks in UV- treated and sham-treated cells. The results showed a significant difference in the percentages of nontargeting control siRNA cells (two-sided P value, 4.705E10−8) and insignificant differences in those of Tim siRNA (two-sided P value, 0.4672) or Tipin siRNA (two-sided P value, 0.9365) cells.
Analysis of the lengths of red and green tracks provides a measure of replication fork displacement rates. Knockdown or knockout of Chk1 expression was recently shown to reduce the rate of fork displacement by about 50% in several vertebrate cell lines (52), and inactivation of Rad53 and Tof1 in budding yeast also reduced the fork displacement rate (57). While the analysis of inhibition of replicon initiation did not identify a difference between the Tim and Tipin knockdowns in response to UV (Fig. 3 and 7), analysis of fork displacement rates revealed a highly significant separation of function.
The lengths of red tracks that were adjacent to green tracks but labeled before irradiation were measured (Table 2); there were no differences between red track lengths in sham- and UV-treated cells, and so the results from both treatment conditions were combined. An analysis of red track lengths demonstrated that knockdown of Tim reduced fork displacement rates by 48% (P < 0.001; Table 2), while knockdown of Tipin produced a nonsignificant 11% reduction (P = 0.57; Table 2). Thus, the effect of Tipin and Tim knockdowns on the rate of replication fork displacement varied significantly, even before the cells were exposed to UV.
TABLE 2.
DNA replication fork progression in sham-treated and UV-irradiated HeLa cells
| siRNA condition(s) and treatment for indicated expta | Mean track length (μm) | SD (n) | B/Ad | P valuee |
|---|---|---|---|---|
| Red tracksb | ||||
| A. NTC | 5.3 | 2.9 (166) | ||
| B. Tim | 2.7 | 1.9 (225) | 0.52 | <0.0001 |
| A. NTC | 5.3 | 2.9 (166) | ||
| B. Tipin | 4.7 | 2.4 (201) | 0.89 | 0.57 |
| A. Tipin | 4.7 | 2.4 (201) | ||
| B. Tim | 2.7 | 1.9 (225) | 0.58 | <0.0001 |
| Green tracksc | ||||
| A. NTC, Sham | 5.8 | 3.0 (146) | ||
| B. NTC, 2.5 J/m2 | 2.9 | 2.0 (139) | 0.49 | <0.0001 |
| A. Tim, Sham | 2.6 | 1.5 (158) | ||
| B. Tim, 2.5 J/m2 | 2.2 | 1.1 (165) | 0.82 | 0.153 |
| A. Tipin, Sham | 3.8 | 2.4 (162) | ||
| B. Tipin, 2.5 J/m2 | 4.6 | 2.6 (161) | 1.20 | 0.721 |
NTC, nontargeting control. Other experimental details are as given in Table 1.
The lengths of red tracks that were adjacent to green tracks in sham- and UV-treated control cells from two independent experiments were combined for each siRNA condition. The total numbers of tracks that were analyzed are given in parentheses in the SD column. t tests were used to test the equality of mean red lengths in cells transfected with the indicated siRNAs.
The lengths of green tracks that were adjacent to red tracks in sham- and UV-treated cells from three independent experiments were combined for each sample. The total numbers of tracks that were analyzed are given in parentheses in the SD column. t tests were used to test the equality of mean green track lengths in cells with and without UV treatment.
B/A, ratio of mean track lengths in A and B samples.
Measurements of track lengths in five different samples (no gene knockdown or irradiation) that were processed under the same conditions used in this study yielded an average length of 5.7 ± 0.6 μm. Therefore, for tests of the equality of mean track lengths between sham-treated or irradiated cultures, or between cells with or without knockdown of specific gene products, the indicated P values reflect the statistical significance of changes in excess of 0.6 μm.
UV caused a significant inhibition of DNA synthesis in replicons that were active at the time of irradiation in the nontargeted control cells. When control siRNA-treated cells were irradiated with 2.5 J/m2 of UV, the average length of the green tracks that were adjacent to red tracks was reduced by 51%, from 5.8 μm to 2.9 μm (Table 2). Thus, UV-induced damage to DNA templates reduced the rate of replication fork progression. There was less inhibition of fork displacement rate after UV irradiation in the Tim-depleted cells. Because fork displacement in Tim-depleted cells was significantly reduced before UV irradiation, the lack of UV-induced inhibition of green track lengths may simply indicate that replication forks encountered fewer lesions during the 20-min postirradiation incubation. Cells with knockdown of Tipin appeared to be resistant to UV-induced inhibition of replication fork progression, and green track lengths in UV-treated, Tipin-depleted cells exceeded the track lengths in UV-irradiated, nontargeted control cells. In Tipin-depleted cells, replication forks moved 50% farther after UV irradiation than in nontargeted control cells and twice as far as in Tim-depleted cells. The UV-induced inhibition of DNA chain elongation in active replicons was significantly attenuated by knockdown of Tipin (Table 2).
DISCUSSION
The intra-S checkpoint response to UV in mammalian cells requires ATR, TopBP1, Chk1, Claspin, Hus1, and Tim to inhibit replicon initiation. The intra-S checkpoint response to UV differs from that provoked by IR-induced DNA damage in that ATM, MRE-11 and NBS1 are not required (22), Cdc25A is not degraded, and cyclin E/Cdk2 kinase is not inhibited (22a). A recent study reported that benzo[a]pyrene diolepoxide-induced DNA damage also provoked a Chk1-dependent intra-S checkpoint response that did not involve Cdc25A or inhibition of cyclin E/Cdk2 (36). In Xenopus egg extracts that generate intra-S checkpoint signaling in response to DNA damage (11, 12), activation of the checkpoint signal appears to derive from the uncoupling of helicase and DNA polymerase activities at replication forks, with extended regions of ssDNA stimulating activation of Chk1 by an ATR/ATRIP/TopBP1 complex (4, 34). Claspin acts downstream of TopBP1 to mediate phosphorylation of Chk1 by ATR (37). The intra-S checkpoint response to UV can produce significant inhibition of replicon initiation after doses with little or no toxicity. For example, a UV dose of 1 J/m2 produced a 50% reduction in the replicon initiation rate (22, 27) but no cytotoxicity (2) in diploid human fibroblasts. Thus, the intra-S checkpoint response to UV is an adaptive response associated with the survival of damaged cells. We show here that the mediator protein Tim forms a complex with Tipin in human cells, Tipin stabilizes the expression of Tim, Tipin binds to RPA in solution and in a complex with DNA, RPA facilitates binding of Tipin to DNA, and knockdown of Tipin attenuates the intra-S checkpoint response to UV. During the preparation of this report, three other papers were published confirming that the Tim/Tipin complex mediates an intra-S checkpoint response to DNA damage (8, 18, 65).
The Tipin-Tim complex appears to act like Claspin as a mediator that increases the efficiency whereby ATR phosphorylates and activates the signal transducer kinase Chk1 under conditions of replication stress provoked by the depletion of precursor pools or the presence of bulky lesions in DNA templates. However, the Tim-Tipin complex appears to have an additional function in regulating the rate of DNA chain elongation at active replication forks. This may be enforced through Tipin's ability to interact with RPA. Interestingly, Tipin contains a region of high homology to the domain in XPA that interacts with RPA (18). We demonstrated in this study that Tipin binds to RPA with the same avidity as XPA, a result which raised the possibility of Tipin serving as a link between excision repair and DNA damage checkpoints. Clearly, further studies are needed to determine whether Tipin influences the interaction between nucleotide excision repair and Chk1 activation (25).
Studies with budding and fission yeast have demonstrated that the Tim and Tipin homologues, Tof1/Csm3 and Sw1/Swi3, respectively, form stable complexes that remain associated with replication machinery and stabilize DNA replication forks when DNA is damaged (38, 39, 48, 49). In nonextracted human cells, Tim and Tipin appear to be evenly distributed throughout the nucleus, but after detergent extraction, a fraction of Tipin and Tim remains associated with PCNA on chromatin (65). Thus, the Tim/Tipin complex may interact with RPA at DNA replication forks or other sites of DNA metabolism. Mec1 and Rad53 were the first checkpoint genes shown to stabilize stalled replication forks (55); Tof1 and Mrc1 also perform this function (57). The human homologue of Rad53, Chk2, participates in the ATM-dependent intra-S checkpoint response to IR-induced DNA double-strand breaks (15). ATM, MRE-11, and NBS1 are not required for the intra-S checkpoint response to UV, which is enforced through ATR-dependent activation of Chk1 (22, 45). Human Chk1 thus appears to be the functional homologue of scRad53 in response to stalled replication forks. Knockdown of Chk1 in vertebrate cells reduced the rate of replication fork displacement (52), an effect similar to those elicited by the inactivation of Tof1 and Mrc1 in yeast (57) and the knockdown of Tim in HeLa cells (Table 2). As the Tim-Tipin complex appears to mediate the activation of Chk1 by ATR, we propose that a Tim-Tipin complex, analogous to the Swi1/Swi3 replication fork protection complex, continuously monitors the state of replication forks and promotes the interaction between ATR and Chk1 under conditions of replication stress when fork displacement rates slow, e.g., with depletion of DNA precursors or in the presence of replication-blocking lesions in template strands. Both of these conditions may uncouple helicase and polymerase activities at replication forks, generating extended regions of ssDNA that become coated with RPA. The depletion of Tim or Tipin interrupts the signaling circuit, causing continuous initiation of DNA replication even under conditions of replication stress. Stalled and uncoupled replication forks may be prone to breakage or collapse (39), leading to DNA double-strand breaks and the chromosomal aberrations detected cytogenetically (30). It should be noted that while ATM is not required for the intra-S checkpoint response to UV, it does protect against UV clastogenesis. Ataxia telangiectasia cells displayed 8- to 10-fold-increased frequencies of UV-induced chromosomal aberrations (13, 30), suggesting that after replication fork collapse, ATM signaling is required for efficient repair of DNA double-strand breaks.
While deficiencies of Chk1 (52) and Tim reduced the rate of fork displacement significantly, the knockdown of Tipin had little effect on this parameter. Knockdown of Tipin also reduced the levels of expression of Tim, but we could not ascertain whether knockdown of Tim had a reciprocal effect on Tipin. Recent studies of the Tim-Tipin complex in human cells demonstrated that knockdown of Tim reduced the expression of Tipin (8, 65), although the evidence presented did not exclude the possibility of a fraction of Tipin remaining after knockdown of Tim. In yeast, the inactivation of Swi1 led to reduced expression of Swi3 (49), supporting a conclusion that interaction between the two protein partners in the complex influences protein stability (the reciprocal experiment to determine whether inactivation of Swi3 affects stability of Swi1 was not done). However, Swi1- and Swi3-null yeast strains did not have identical phenotypes, with Swi3-null cells displaying greater sensitivity to HU (49). This result suggests that Swi3 has a function(s) that is independent of the binary complex. It is conceivable that with knockdown of Tim, Tipin still remains in sufficient quantity to exercise Tim-independent functions. Knockdown of Tipin removes both Tim-dependent and Tipin-dependent functions with additional biological effects. The observation that knockdown of Tipin did not significantly reduce fork displacement rates in undamaged cells, while knockdown of Tim did, suggests that Tipin may slow fork displacement when free of its binding partner, Tim. It cannot be determined from the available data whether such an effect is direct, through RPA-mediated loading of Tipin onto DNA, or indirect, through Tipin-mediated signaling.
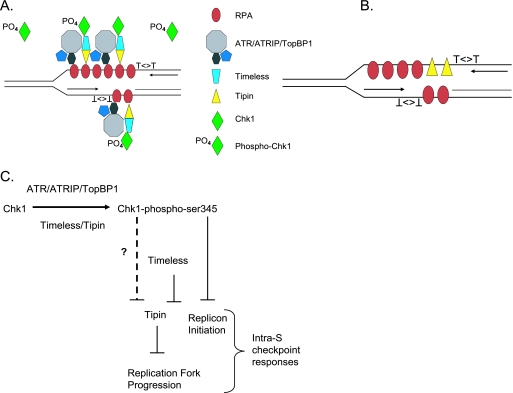
The data presented here suggest a model in which the Tim-Tipin complex mediates the inhibitions of replicon initiation and DNA chain elongation in UV-damaged cells. In undamaged cells, transient interactions of the ATR/ATRIP/TopBP1 and Tim/Tipin complexes with RPA may occur at DNA replication forks but without sufficient stability to activate Chk1. Tim was found to interact with Chk1, ATR, and ATRIP in mammalian cells, and the amount of Chk1 and ATRIP bound to Tim was increased by treatment with UV or HU (58). It remains to be determined whether posttranslational modifications of Tipin or Tim occur in response to DNA damage, although the damage-dependent interactions between Tim and Chk1, and between Tim and ATRIP, suggest that the interactions may be regulated. The uncoupling of DNA polymerase and helicase activities at sites of UV-induced template lesions generates long stretches of ssDNA that are coated with RPA. The Swi1/Swi3 complex is associated with replication forks in yeast (49), and Tipin binds to RPA-coated DNA. Thus, the Tim/Tipin complex appears to bring the highly diffusible Chk1 to ATR/ATRIP/TopBP1 at sites of stalled replication forks (Fig. 8A). Claspin, Hus1, and BRCA1, as other mediators of Chk1 phosphorylation by ATR (7, 35, 52), also contribute to the signaling complex.
FIG. 8.
Theoretical model depicting potential interactions in the intra-S checkpoint signaling pathway. (A) After UV-induced DNA damage, the Tim-Tipin complex brings Chk1 to sites of ssDNA coated with RPA, where the ATR/ATRIP/TopBP1 complex phosphorylates multiple molecules of Chk1 that diffuse away from the stalled fork to transduce signal throughout the nucleus (3). This model differs from that of Gotter et al. (18), as DNA damage did not destabilize the interaction between Tipin and RPA in checkpoint-competent HEK293T cells (Fig. 5A). Thus, with uncoupling of helicase and polymerase activities at sites of stalled replication forks, the increased amount of RPA-coated DNA increases the opportunity for Tim/Tipin/Chk1 complexes to interact with ATR/ATRIP/TopBP1 complexes to phosphorylate and activate Chk1. (B) Tipin molecules may be loaded onto DNA by RPA. When Tim is depleted by siRNA, loading of Tipin at replication forks may inhibit DNA chain elongation. (C) Tipin inhibits DNA chain elongation and replication fork progression, and this activity is inhibited by Tim. Chk1 also preserves replication fork progression (52), and this may be through an effect on Tipin or the Tim/Tipin complex (indicated by “?”).
In the absence of Tim, RPA may load free Tipin onto DNA (Fig. 8B), and it is conceivable that this loading of Tipin inhibits DNA chain elongation. Tipin may slow DNA synthesis by replacing RPA on ssDNA, and Tim suppresses this activity of Tipin in the absence of DNA damage (Fig. 8C). In the presence of damage, checkpoint signaling through Chk1 may alter the Tim/Tipin complex, allowing Tipin to inhibit chain elongation. As knockdown of Chk1 produces the same phenotype as knockdown of Tim, with inactivation of the inhibition of replicon initiation in response to damage and slowing of replication fork progression in the absence of damage, it is possible that Tim requires Chk1 to suppress the activity of Tipin.
Acknowledgments
This study was supported in part by PHS grants CA55065, CA81343, GM32833, ES11012, P30-CA16086, and P30-ES10126.
We are grateful for the skilled technical assistance of Yingchun Zhou.
Footnotes
Published ahead of print on 12 February 2007.
REFERENCES
- 1.Aten, J. A., P. J. Bakker, J. Stap, G. A. Boschman, and C. H. Veenhof. 1992. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem. J. 24:251-259. [DOI] [PubMed] [Google Scholar]
- 2.Bassett, E., N. M. King, M. F. Bryant, S. Hector, L. Pendyala, S. G. Chaney, and M. Cordeiro-Stone. 2004. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res 64:6469-6475. [DOI] [PubMed] [Google Scholar]
- 3.Bekker-Jensen, S., C. Lukas, R. Kitagawa, F. Melander, M. B. Kastan, J. Bartek, and J. Lukas. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173:195-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byun, T. S., M. Pacek, M. C. Yee, J. C. Walter, and K. A. Cimprich. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calzada, A., B. Hodgson, M. Kanemaki, A. Bueno, and K. Labib. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chastain, P. D., II, T. P. Heffernan, K. R. Nevis, L. Lin, W. K. Kaufmann, D. G. Kaufman, and M. Cordeiro-Stone. 2006. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle 5:2160-2167. [DOI] [PubMed] [Google Scholar]
- 7.Chini, C. C., and J. Chen. 2003. Human claspin is required for replication checkpoint control. J. Biol. Chem. 278:30057-30062. [DOI] [PubMed] [Google Scholar]
- 8.Chou, D. M., and S. J. Elledge. 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc. Natl. Acad. Sci. USA 103:18143-18147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordeiro-Stone, M., A. M. Makhov, L. S. Zaritskaya, and J. D. Griffith. 1999. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 289:1207-1218. [DOI] [PubMed] [Google Scholar]
- 10.Cordeiro-Stone, M., R. I. Schumacher, and R. Meneghini. 1979. Structure of the replication fork in ultraviolet light-irradiated human cells. Biophys. J. 27:287-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo, V., K. Robertson, C. Y. Ying, E. Kim, E. Avvedimento, M. Gottesman, D. Grieco, and J. Gautier. 2000. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol. Cell 6:649-659. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo, V., D. Shechter, P. J. Lupardus, K. A. Cimprich, M. Gottesman, and J. Gautier. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11:203-213. [DOI] [PubMed] [Google Scholar]
- 13.Ejima, Y., and M. S. Sasaki. 1986. Enhanced expression of X-ray- and UV-induced chromosome aberrations by cytosine arabinoside in ataxia telangiectasia cells. Mutat. Res. 159:117-123. [DOI] [PubMed] [Google Scholar]
- 14.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 15.Falck, J., J. H. Petrini, B. R. Williams, J. Lukas, and J. Bartek. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290-294. [DOI] [PubMed] [Google Scholar]
- 16.Foss, E. J. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotter, A. L. 2003. Tipin, a novel timeless-interacting protein, is developmentally co-expressed with Timeless and disrupts its self-association. J. Mol. Biol. 331:167-176. [DOI] [PubMed] [Google Scholar]
- 18.Gotter, A. L., C. Suppa, and B. S. Emanuel. 2007. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J. Mol. Biol. 366:36-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grisham, J., W. K. Kaufmann, and D. G. Kaufman. 1983. The cell cycle and chemical carcinogenesis. Surv. Synth. Pathol. Res. 1:49-66. [Google Scholar]
- 20.Haracska, L., I. Unk, L. Prakash, and S. Prakash. 2006. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 103:6477-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, Z., L. A. Henricksen, M. S. Wold, and C. J. Ingles. 1995. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature 374:566-569. [DOI] [PubMed] [Google Scholar]
- 22.Heffernan, T. P., D. A. Simpson, A. R. Frank, A. N. Heinloth, R. S. Paules, M. Cordeiro-Stone, and W. K. Kaufmann. 2002. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 22:8552-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Heffernan, T. P., K. Ünsal-Kaçmaz, D. A. Simpson, A. N. Heinloth, R. S. Paules, A. Sancar, M. Cordeiro-Stone, and W. K. Kaufmann. J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]
- 23.Hermanson, I. L., and J. J. Turchi. 2000. Overexpression and purification of human XPA using a baculovirus expression system. Protein Expr. Purif. 19:1-11. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, D. A., and A. Pombo. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, G., and A. Sancar. 2006. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol. Cell. Biol. 26:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann, W. K., and J. E. Cleaver. 1981. Mechanisms of inhibition of DNA replication by ultraviolet light in normal human and xeroderma pigmentosum fibroblasts. J. Mol. Biol. 149:171-187. [DOI] [PubMed] [Google Scholar]
- 28.Kaufmann, W. K., and D. G. Kaufman. 1993. Cell cycle control, DNA repair and initiation of carcinogenesis. FASEB J. 7:1188-1191. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann, W. K., and R. S. Paules. 1996. DNA damage and cell cycle checkpoints. FASEB J. 10:238-247. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann, W. K., and S. J. Wilson. 1994. G1 arrest and cell-cycle-dependent clastogenesis in UV-irradiated human fibroblasts. Mutat. Res. 314:67-76. [DOI] [PubMed] [Google Scholar]
- 31.Kraemer, K. H., M. M. Lee, A. D. Andrews, and W. C. Lambert. 1994. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch. Dermatol. 130:1018-1021. [PubMed] [Google Scholar]
- 32.Krings, G., and D. Bastia. 2004. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 101:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulaksýz, G., J. T. Reardon, and A. Sancar. 2005. Xeroderma pigmentosum complementation group E protein (XPE/DDB2): purification of various complexes of XPE and analyses of their damaged DNA binding and putative DNA repair properties. Mol. Cell. Biol. 25:9784-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumagai, A., J. Lee, H. Y. Yoo, and W. G. Dunphy. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124:943-955. [DOI] [PubMed] [Google Scholar]
- 35.Lin, S. Y., K. Li, G. S. Stewart, and S. J. Elledge. 2004. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. USA 101:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, P., L. R. Barkley, T. Day, X. Bi, D. M. Slater, M. G. Alexandrow, H. P. Nasheuer, and C. Vaziri. 2006. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25a/Cdk2-independent mechanism. J. Biol. Chem. 281:30631-30644. [DOI] [PubMed] [Google Scholar]
- 37.Liu, S., S. Bekker-Jensen, N. Mailand, C. Lukas, J. Bartek, and J. Lukas. 2006. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell. Biol. 26:6056-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 39.Lopes, M., M. Foiani, and J. M. Sogo. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21:15-27. [DOI] [PubMed] [Google Scholar]
- 40.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto, S., K. Ogino, E. Noguchi, P. Russell, and H. Masai. 2005. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J. Biol. Chem. 280:42536-42542. [DOI] [PubMed] [Google Scholar]
- 42.Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas, T. Kwok, R. Newitt, R. Aebersold, C. Boone, G. W. Brown, and P. Hieter. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15:1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mer, G., A. Bochkarev, R. Gupta, E. Bochkareva, L. Frappier, C. J. Ingles, A. M. Edwards, and W. J. Chazin. 2000. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103:449-456. [DOI] [PubMed] [Google Scholar]
- 44.Merrick, C. J., D. Jackson, and J. F. Diffley. 2004. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 279:20067-20075. [DOI] [PubMed] [Google Scholar]
- 45.Miao, H., J. A. Seiler, and W. C. Burhans. 2003. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J. Biol. Chem. 278:4295-4304. [DOI] [PubMed] [Google Scholar]
- 46.Mohanty, B. K., N. K. Bairwa, and D. Bastia. 2006. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103:897-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nedelcheva, M. N., A. Roguev, L. B. Dolapchiev, A. Shevchenko, H. B. Taskov, A. Shevchenko, A. F. Stewart, and S. S. Stoynov. 2005. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J. Mol. Biol. 347:509-521. [DOI] [PubMed] [Google Scholar]
- 48.Noguchi, E., C. Noguchi, L. L. Du, and P. Russell. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23:7861-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noguchi, E., C. Noguchi, W. H. McDonald, J. R. Yates III, and P. Russell. 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 24:8342-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Painter, R. B., and B. R. Young. 1980. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc. Natl. Acad. Sci. USA 77:7315-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panning, M. M., and D. M. Gilbert. 2005. Spatio-temporal organization of DNA replication in murine embryonic stem, primary, and immortalized cells. J. Cell. Biochem. 95:74-82. [DOI] [PubMed] [Google Scholar]
- 52.Petermann, E., A. Maya-Mendoza, G. Zachos, D. A. Gillespie, D. A. Jackson, and K. W. Caldecott. 2006. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell. Biol. 26:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sancar, A., L. A. Lindsey-Boltz, K. Ünsal-Kaçmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 54.Svoboda, D. L., and J. M. Vos. 1995. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc. Natl. Acad. Sci. USA 92:11975-11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 56.Torres-Ramos, C. A., S. Prakash, and L. Prakash. 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tourriere, H., G. Versini, V. Cordon-Preciado, C. Alabert, and P. Pasero. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell 19:699-706. [DOI] [PubMed] [Google Scholar]
- 58.Ünsal-Kaçmaz, K., T. E. Mullen, W. K. Kaufmann, and A. Sancar. 2005. Coupling of human circadian and cell cycles by the timeless protein. Mol. Cell. Biol. 25:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visser, A. E., R. Eils, A. Jauch, G. Little, P. J. Bakker, T. Cremer, and J. A. Aten. 1998. Spatial distributions of early and late replicating chromatin in interphase chromosome territories. Exp. Cell Res. 243:398-407. [DOI] [PubMed] [Google Scholar]
- 60.Wang, X., J. Guan, B. Hu, R. S. Weiss, G. Iliakis, and Y. Wang. 2004. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res. 32:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Watanabe, M., V. M. Maher, and J. J. McCormick. 1985. Excision repair of UV- or benzo[a]pyrene diol epoxide-induced lesions in xeroderma pigmentosum variant cells is ‘error free’. Mutat. Res. 146:285-294. [DOI] [PubMed] [Google Scholar]
- 62.Weiss, R. S., P. Leder, and C. Vaziri. 2003. Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Mol. Cell. Biol. 23:791-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao, J., C. Li, N. L. Zhu, Z. Borok, and P. Minoo. 2003. Timeless in lung morphogenesis. Dev. Dyn. 228:82-94. [DOI] [PubMed] [Google Scholar]
- 64.Xu, H., C. Boone, and H. L. Klein. 2004. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol. Cell. Biol. 24:7082-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshizawa-Sugata, N., and H. Masai. 2007. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J. Biol. Chem. 282:2729-2740. [DOI] [PubMed] [Google Scholar]
- 66.You, Y. H., D. H. Lee, J. H. Yoon, S. Nakajima, A. Yasui, and G. P. Pfeifer. 2001. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 276:44688-44694. [DOI] [PubMed] [Google Scholar]