Abstract
Ribosome biogenesis is driven by a large number of preribosomal factors that associate with and dissociate from the preribosomal particles along the maturation pathway. We have previously shown that budding yeast Mak11, whose homologues in other eukaryotes were described as modulating a p21-activated protein kinase function, accumulates in Rlp24-associated pre-60S complexes when their maturation is impeded in Saccharomyces cerevisiae. The functional inactivation of WD40 repeat protein Mak11 interfered with the 60S rRNA maturation, led to a cell cycle delay in G1, and blocked green fluorescent protein-tagged Rpl25 in the nucleoli of yeast cells, indicating an early role of Mak11 in ribosome assembly. Surprisingly, Mak11 inactivation also led to a dramatic destabilization of Rlp24. The suppression of the thermosensitive phenotype of a mak11 mutant by RLP24 overexpression and a direct in vitro interaction between Rlp24 and Mak11 suggest that Mak11 acts as an Rlp24 cofactor during early steps of 60S ribosomal subunit assembly. Moreover, we found that Skb15, the Mak11 homologue in Schizosaccharomyces pombe, also associated with preribosomes and affected 60S biogenesis in fission yeast. It is thus likely that the previously observed phenotypes for MAK11 homologues in other eukaryotes are secondary to the main function of these proteins in ribosome formation.
Eukaryotic ribosome biogenesis begins in the nucleolus with the association of ribosomal and preribosomal proteins with a nascent rRNA precursor transcribed by RNA polymerase I. This precursor consists, in yeasts, of the sequences for mature 18S, 5.8S, and 25S rRNAs, separated by two internal transcribed spacers and flanked by additional 5′-end and 3′-end sequences. The rRNA precursor, as a large ribonucleoprotein particle, is next matured in well-ordered processing steps. Many auxiliary factors are required all along the pathway, but except for a few enzymatic functions, the molecular roles of these factors remain unclear. A cleavage in the region separating the precursor of the 18S rRNA and the 5.8S and 25S rRNAs leads to the generation of pre-60S and pre-40S intermediate particles. The third RNA component of the large ribosomal subunit, the 5S rRNA, associates in a precursor form with the pre-60S particles, and both pre-60S and pre-40S particles are further processed and exported to the cytoplasm, where final maturation events take place and generate the ribosomal subunits (56, 58).
Studies with budding yeast allowed the identification and functional characterization of a surprisingly large number of proteins and RNAs that participate in the maturation of eukaryotic ribosomes. Since 2001, generic purification methods such as tandem affinity purification (TAP), in association with developments in mass spectrometry, have allowed the identification of about 200 preribosomal factors (15, 16, 20, 29). In contrast to the ribosomal proteins, which associate with the preparticles and remain associated with the mature ribosomes, the preribosomal factors associate transiently with the precursors (for reviews see references 12 and 55). When ribosome biogenesis is blocked, the changes in purified preribosomal complex composition may indicate the order of protein association, dissociation, or subcomplex formation during the pathway (see for example references 19, 35, and 47). Such experiments can also establish the requirement of a given factor for subsequent association of other proteins with the precursors.
We identified Mak11 in complexes purified in association with the pre-60S essential protein Rlp24 and have shown that the amount of copurified Mak11 starkly increased when another essential pre-60S factor, the Nog1 GTPase, was depleted (47). Several known pre-60S factors (Nop7, Tif6, Erb1, and Nop2) were identified in association with Mak11 in a large-scale experiment (20). Recent purification of a large number of macromolecular complexes in yeast further confirmed the presence of Mak11 in predicted pre-60S complexes (15, 29). Moreover, the Mak11 fusion with green fluorescent protein (GFP) was shown to localize to the nucleus and concentrate in the nucleolus of yeast cells (22).
MAK11's name (maintenance of killer) comes from its identification in genetic screens looking for mutations that would affect the maintenance of the M1 toxin-encoding double-stranded RNA (dsRNA), a satellite of the L-A dsRNA yeast virus (54, 61, 62). The RNAs used for the synthesis of the viral proteins have neither a typical 5′ cap nor a poly(A) tail (60). Many of the isolated mak mutants showed polysome profiles typical for 60S ribosomal subunit biogenesis impairment (40). It is likely that the identification of 60S ribosomal subunit biogenesis factors among the mak genes was due to the fact that normal 60S ribosomal subunit levels, in contrast to 40S subunit levels, are required for the translation of uncapped and nonpolyadenylated RNAs like those encoding viral proteins (45, 50). Several mutations discovered during the mak screens affected genes like mak7, mak8, or mak18, later identified to be coding for ribosomal proteins (Rpl8a, Rpl3, and Rpl42b, respectively) (40). Other mutations affected genes coding for proteins recently shown to be physically associated with pre-60S particles by large-scale complex purification studies (15, 28). Recently, the requirements of Mak21/Noc1 (10, 36), Mak5 (64), and Mak16 (41) for 60S ribosomal subunit formation were described. One of the mutations described in the genetic screen and having effects on 60S levels affected MAK11, a gene shown to be essential for viability and suspected to be required for 60S subunit biogenesis (40).
Based on the available data, we predicted Mak11 to be an essential factor involved in nuclear maturation of 60S ribosomal subunits. Puzzlingly, while Mak11 putative orthologues exist in many eukaryotes, both the Schizosaccharomyces pombe homologue Skb15 (26, 27) and the human homologue hPip1 (63) have been previously described as direct binders and inhibitors of p21-activated protein kinases (PAKs), Shk1/Pak1 in fission yeast and Pak1 in Homo sapiens. PAKs link different receptors with modification of protein substrates by phosphorylation in the mitogen-activated protein kinase pathway and are directly activated by small GTPases of the Rho or Rac families (for a review see reference 21). PAK activation or inhibition affects major cellular pathways since PAK substrates play important roles in cell polarity and morphology, mitotic exit, and cytokinesis and mediate cellular responses to external stimuli. These signaling pathways are highly conserved as demonstrated by the heterologous complementation of the absence of yeast Ste20 by its human homologue Pak1 (8).
It was surprising that Mak11 homologues were described as modulators of PAK activity affecting central signaling pathways in other eukaryotes, while physical association and functional data strongly suggested that Mak11 is a novel factor involved in 60S ribosomal subunit formation in Saccharomyces cerevisiae. There were two explanations for this discrepancy: either Mak11 was not the homologue of Skb15 and hPip1 or the previously observed effects were secondary to the function of these factors in ribosome biogenesis. We thus tried to understand the role of Mak11 in ribosome biogenesis in S. cerevisiae and tested the putative ribosome biogenesis function for S. pombe Skb15.
We show here that the essential function of Mak11 is linked to an early, nucleolar step of 60S ribosomal subunit biogenesis, and we propose that it serves as an Rlp24 cofactor during the assembly of early pre-60S particles in S. cerevisiae. The ribosome biogenesis function of Mak11 was conserved during evolution, since we could show that Skb15 is required for 60S formation in S. pombe. We conclude that previously observed effects of Mak11 homologues are likely to be secondary to the primary effects of these factors on ribosome biogenesis.
MATERIALS AND METHODS
Strains and plasmids.
The strains used in this study are listed in Table 1. Chromosomal insertions were obtained by homologous recombination using PCR fragments (1).
TABLE 1.
S. cerevisiae and S. pombe strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| S. cerevisiae | ||
| MGD353-13D | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 | 44 |
| BY4741 | MATaura3Δ0 his3Δ1 leu2Δ0 met15Δ0 | 5 |
| BY4742 | MATα ura3Δ0 his3Δ1 leu2Δ0 lys2Δ0 | 5 |
| Y24870 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 Mak11::kanMX4/MAK11 | 17 |
| LMA160 | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 Rlp24-TAP-TRP1 | 47 |
| LMA260 | MATaura3Δ0 his3Δ1 leu2Δ0 lys2? met15? mak11::kanMX4 [pFL38-MAK11] | This study |
| LMA263-2 | MATaura3Δ0 his3Δ1 leu2Δ0 lys2? met15? mak11::kanMX4 [pFL36CII-mak11-2] | This study |
| LMA264 | MATaura3Δ0 his3Δ1 leu2Δ0 lys2? met15? mak11::kanMX4 [pFL36CII-MAK11] | This study |
| LMA326 | MATaura3Δ0 his3Δ1 leu2Δ0 lys2? met15? mak11::kanMX4 [pTG189-MAK11] | This study |
| LMA375 | MATaura3Δ0 his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 mak11-TAP-HIS3 | This study |
| LMA364 | MATaura3Δ0 his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 PGAL1::Nog1/KanMX6 mak11-TAP-HIS3 | This study |
| LMA371 | MATaura3Δ0 his3Δ1 leu2Δ0 met15Δ0 PGAL1::Rlp24/KanMX6 Mak11-TAP-HIS3 | This study |
| LMA431 | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 Ssf1-TAP-URA3 PGAL1::mak11/kanMX6 | This study |
| LMA437 | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 Nog1-TAP-TRP1 PGAL1::mak11/kanMX6 | This study |
| LMA515 | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 Rlp24-TAP-TRP1 PGAL1::mak11/kanMX6 | This study |
| S. pombe | ||
| LMA600 | h− (Msmt0) leu1-32 ura4-D18 adeM6-210 Skb15-TAP/kanMX6 | This study |
| Pb185 | h− (Msmt0) leu1-32 ura4-D18 adeM6-210 | B. Arcangioli |
| nmt-Skb15 | h90 ade6-M210 leu1-32 ura4-D18 skb15::ura4::nmt1-skb15-ADE2 | 27 |
| SP870 | h90 ade6-M210 leu1-32 ura4-D18 | 27 |
?, genotype not tested.
pCM190-RLP24 was obtained by subcloning the RLP24 sequence from the plasmid pGEX4-T-RLP24 (47) using BamHI and NotI into pCM190 (14). The MAK11 open reading frame was cloned after PCR amplification in the pDONR 201 vector using the Gateway system (Invitrogen). The resulting entry clone was used to generate the pTG189-MAK11 plasmid. pTG189, a Gateway-compatible TAP vector, was obtained from pCM189 (14) by cloning the N-terminal TAP cassette (43) downstream of the tetracycline-regulated promoter and upstream of an RfA Gateway cassette. A Gateway-compatible vector derived from pET32a (Novagen), a gift from E. Bertrand (IGM, Montpellier, France), was used to obtain a vector allowing (His)6-Mak11 expression in bacteria.
Nmt1-Skb15 and SP870 S. pombe strains were obtained from S. Marcus (University of Alabama). The Skb15-TAP strain was obtained by homologous recombination in the Pb185 strain (gift from B. Arcangioli, Institut Pasteur, Paris, France) using a method adapted from reference 57. Long recombination arms were generated by two successive PCRs from genomic DNA using oligonucleotides A-BamHI (ACG GGA TCC AGG CAA ATC TGT CTA CCC TGT TG), B-XmaI (TGA AAA GGA CGA AGC ATG CCC CCGGG), C-SalI (GGT CGA CCT TAA TAG GGA AAG GAC GGG), and D-BamHI (TGC TAG ATG AGC TAT TTG CCA CGG GAT CCA GG). The product of the second PCR was cloned in pCRBlunt (Invitrogen) and next subcloned in the pFA6a-CTAP-MX6 vector (52) using SalI and XmaI restriction enzyme sites. The resulting plasmid was linearized by digestion with BamHI and used to transform strain Pb185. Clones resistant to G418 were tested by immunoblotting for the presence of the fusion protein.
TAP and in vitro binding assay.
Complex purifications were performed as described in reference 44 with a few modifications, starting with 4 liters of yeast culture. Buffers contained 0.1 M NaCl. Eluted proteins were precipitated with methanol-chloroform, separated on a 5 to 20% polyacrylamide gradient-sodium dodecyl sulfate gel, and identified by either immunoblotting or mass spectrometry. Mass spectrometry protein identification was done using matrix-assisted laser desorption ionization-time of flight as described previously (32). For RNA association determination, only the first step of purification was performed, in the presence of vanadyl ribonucleoside complexes as RNase inhibitor. The associated RNAs were extracted twice with phenol-chloroform. To investigate the association of different proteins with purified complexes, tobacco etch virus (TEV) eluates were separated on denaturing polyacrylamide gels, transferred to nitrocellulose membranes (Bio-Rad), and probed with rabbit polyclonal antibodies to Nog2 (46), Nog1 and Rlp24 (47), Nsa2 (30), Arx1 (31), and Mak11 (this work) used at a 1:5,000 dilution. The peroxidase activity of secondary antibodies was detected using either the ECL+ (GE Healthcare) or the Immobilon Western (Millipore) chemiluminescence kit.
In vitro binding was tested as previously described (47) using plasmids expressing glutathione S-transferase (GST)-tagged Rlp24, Rpl3 and Rpl5 (controls) and (His)6-Mak11. The presence of Mak11 in the eluate was estimated by immunoblotting with anti-Mak11 antibodies.
Sucrose gradient sedimentation.
Polysomal extracts were obtained using glass bead vortexing. Polysomes were separated on a 10 to 50% sucrose gradient and centrifuged at 39,000 rpm for 2 h 45 min at 4°C in an SW41-Ti rotor. Fractions were recovered with an ISCO fractionator, and the 254-nm absorbance was measured. For protein identification by immunoblotting, the proteins from each fractions were precipitated with 10% trichloroacetic acid and separated on polyacrylamide gels. TAP-tagged proteins were revealed with a 1:10,000 dilution of peroxidase-antiperoxidase complex (Sigma).
RNA extraction, Northern blotting, and primer extension.
RNA extractions were performed using glass beads and phenol-chloroform. Large rRNAs were denatured with glyoxal and separated on 1% agarose gels, and small rRNAs were separated on 5% acrylamide-urea denaturing gels. Northern blot assays and primer extensions were performed using 32P-labeled oligonucleotides. The sequences of the oligonucleotides specific for S. cerevisiae were previously described (46). Those related to S. pombe were based on the detected pre-rRNA intermediates previously described (18) and were the following: CS151, TGT CGG AAA GCA TAG CAA GC, for U2 snRNA, used as a control; CS148, AAC AAA TTT TCG TTC AAC ACC TCA TC, used to detect 27S and 7S intermediates; and CS153, CGT TAA GGT TCA AAT ATA AAA GAG, specific for the 27S intermediates.
Screen for mak11 temperature-sensitive (ts) alleles.
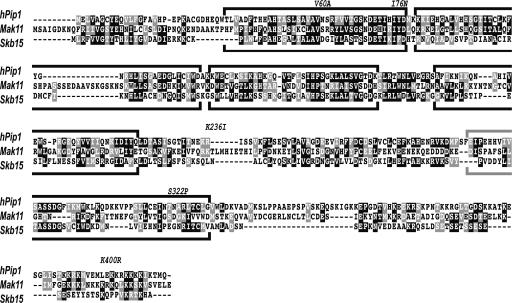
For the selection of thermosensitive MAK11 alleles, we used mutagenic PCR combined with gap repair (37). A region of genomic S. cerevisiae DNA encompassing the MAK11 open reading frame and 243 nucleotides upstream from the ATG sequence as well as 95 nucleotides downstream from the stop codon was amplified by PCR using oligonucleotides GCT CTA GAA GAC ATT TTT CTA GCT ACA TAA and AGG CGC GCC ATC ATC TTT AAC GAT TAA GATA. The resulting PCR product was cloned in the centromeric vector pFL38 (URA3 marker) using XbaI and AscI sites and verified by sequencing. The obtained pFL38-MAK11 plasmid was able to fully complement MAK11 deletion, and the resulting haploid strain was further used as LMA260. A PCR-based strategy was used for random mutagenesis of the MAK11 sequence. PCRs were performed with oligonucleotides AGA TGC GTA AGG AGA AAA TAC CGC ATC and CGA CTG GAA AGC GGG CAG TGA using pFL38-MAK11 as template. In one case the ratio of dATP to other nucleotides was 1:5 and we used 4 mM MgCl2 combined with 0.5 mM MnCl2, and in another case the ratio of dATP to other nucleotides was 1:10 and the reaction mixture contained 10% dimethyl sulfoxide. The strain LMA260 was transformed with PCR products and pFL36CII linearized by digestion with XbaI and HindIII. A total of 16,000 clones were obtained with around 5,000 of these being able to grow at 25°C on a selective medium containing 5-fluoroorotic acid. After replica plating on selective medium we obtained clones that were able to grow at 25°C but not at 35°C. Plasmids were extracted from nine candidates and tested by retransformation. The strongest ts phenotype was observed for the candidate mak11-2 strain, further used in this study. Sequencing of the mak11-2 allele showed several predicted amino acid changes depicted in Fig. 7.
FIG. 7.
Multiple sequence alignment between Mak11 and putative human (hPip1) and S. pombe (Skb15) homologues. Sequences of yeast Mak11, S. pombe Skb15, and human hPip1 were aligned using ClustalW (53), and similar or identical residues were shaded using BOXSHADE so that residues that have similar properties and are found in all three sequences have a black background while those with similar properties in two out of three sequences have a gray background. The residue changes indicated in the alignment correspond to the observed mutations in the mak11-2 strain. Boxed regions correspond to predicted WD40 motifs.
High-copy-number suppressor genetic screen.
The mak11-2 ts mutant strain (LMA263-2) was transformed with a yeast genomic high-copy-number library (generated in the pFL44L 2μm URA3 vector), a gift from F. Lacroute, CGM, Gif-sur-Yvette, France. The transformants were selected on minimal medium without uracil and leucine. After 24 h at 25°C the plates containing the transformants were incubated at 37°C and the clones growing at this restrictive temperature were selected. Plasmids were recovered and used to transform the LMA263-2 strain. The growth phenotype was compared with the growth phenotype of LMA264 transformed with an empty vector. After verification of the suppressor phenotype, the DNA inserts were amplified and the ends were sequenced. The complementation of the mak11-2 ts phenotype was verified using the pCM190-RLP24 vector.
Fluorescence microscopy.
Cells transformed with a centromeric plasmid expressing RPL25-enhanced GFP (eGFP) or cells expressing chromosomal TAP-tagged fusion proteins were cultured in minimal medium. The protein A part of the TAP tag was detected with anti-protein A antibodies and Cy3 secondary antibodies (42). Observation of the cells by epifluorescence was done as described previously (31).
Fluorescence-activated cell sorting (FACS) analysis.
To analyze the cell cycle distribution of the yeast cell population, we used exponentially growing cells in rich medium at 27°C or shifted for up to 6 h at 37°C. Aliquots of 2 ml were retrieved and fixed with 70% cold ethanol for 1 h at room temperature. RNA was digested with RNase A (1 mg/ml in 0.2 M Tris-HCl, pH 7.5, 20 mM EDTA for 1 h at 37°C), and DNA was stained with 50 μg/ml propidium iodide in 0.1 ml phosphate-buffered saline overnight at 4°C. The cell suspension was diluted to 1 ml with phosphate-buffered saline before analysis. Flow cytometry analysis was done using a FACSCalibur (BD Biosciences). One hundred thousand events were used to estimate the ratio between 1n and 2n DNA-containing cells.
Rlp24-TAP half-life estimation.
For the analysis of Rlp24-TAP half-life we used a procedure derived from reference 2. Addition of cycloheximide was done at the same time with a shift of the cultures from 27°C to 37°C. Immunoblotting was performed using peroxidase-antiperoxidase complexes (Sigma) and the Immobilon Western (Millipore) chemiluminescence kit. Images were obtained with a cooled digital camera (GeneGnome; Syngene) and quantitated using ImageJ (version 1.38a; W. S. Rasband, U.S. National Institutes of Health, Bethesda, MD [http://rsb.info.nih.gov/ij/]).
RESULTS
Mak11 is required for the maturation of 27SB pre-rRNA in the nucleolus.
MAK11 is essential in yeast (24), and a mutant allele of MAK11 leads to free 60S ribosomal subunit decrease and formation of halfmer polysomes (40). To investigate the involvement of Mak11 in ribosome biogenesis, and specifically in 60S ribosomal subunit formation, we tested the relative amounts of different mature and precursor rRNAs in yeast cells depleted of Mak11. We used a strain having the chromosomal copy of MAK11 deleted, where the expression of an N-terminal TAP fusion of MAK11 was placed under the control of a tetracycline-repressible promoter on a centromeric plasmid. In the absence of the repressor, the growth of the cells was comparable to the growth of a wild-type strain. Cell aliquots were collected at different time points after doxycycline addition, and total RNA was extracted and tested by primer extension or Northern blotting with specific oligonucleotides (Fig. 1). The 25S mature rRNA decreased whereas the 18S rRNA was only slightly affected (Fig. 1B), in agreement with the previously observed decrease of free 60S subunit levels. The only pre-rRNA species that showed a relative accumulation when Mak11 was depleted were the 27SB precursor and, as observed for other 60S mutants, the 35S precursor (Fig. 1B to D). The 27SB precursor did not accumulate compared with U2 or U5 snRNAs but was increased compared with the 27SA2 precursor. The apparent increase in U2 and U5 snRNA levels was due to the relative decrease of rRNA in the total RNA samples that were analyzed.
FIG. 1.
Mak11 depletion in yeast cells leads to a block in 27SB-to-7S conversion. A. Simplified drawing of different steps in rRNA maturation with the position of the oligonucleotides used for Northern blotting or primer extensions. (B to D) The amounts of large pre-rRNA and rRNA were estimated by Northern blotting (1% agarose gel with glyoxal denaturation) (B), by primer extension (C), or by Northern blotting after separation on denaturant urea-polyacrylamide (5%) gels (D). Equal amounts of total RNA were extracted at the indicated time points after addition of doxycycline from cells where endogenous MAK11 was deleted and where plasmidic TAP-MAK11 was under the control of a tetracycline-repressible promoter (strain LMA326). Oligonucleotides used to reveal the different RNAs are indicated in parentheses.
A decrease in the amounts of 7S pre-rRNA compared with earlier 27S intermediates could be detected after 6 h of Mak11 depletion, in correlation with the rapid decrease in the amount of Mak11 as judged by immunoblotting with antibodies binding to the N-terminal protein tag (not shown). The aberrant 23S rRNA processing intermediate, generated by cleavage at position A3 before A2, was detected at 10 h of depletion and strongly accumulated at later time points (21 h of shift to glucose) when 7S became undetectable. This aberrant cleavage was accompanied by a decrease of the 20S pre-rRNA levels (Fig. 1B).
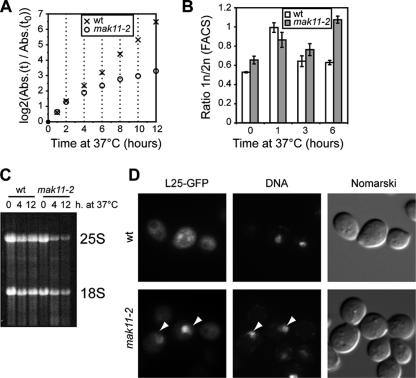
To perform additional functional studies, we searched for variants of Mak11 that, while supporting growth in yeast cells, were responsible for a thermosensitive phenotype. By mutagenic PCR coupled with plasmid gap repair, we obtained several alleles of mak11 that complemented the deletion of the gene at 25°C but supported only very slow growth at 37°C. The mak11-2 strain began to grow noticeably slower than a wild-type strain at 4 h after a shift of temperature from 25 to 37°C (Fig. 2A). The observed growth defect was associated with an increase in the G1 cell cycle phase length as determined by FACS analysis (Fig. 2B). Such a G1 cell cycle delay when ribosome biogenesis was impaired could be seen when other pre-60S factors like Nog1 were depleted (not shown) and was previously reported for small-subunit processome factor depletions (3). Total RNA extracted from the wild-type and mak11-2 strains after 4 and 12 h of growth at 37°C showed a decreased amount of the 25S rRNA compared with the 18S rRNA (Fig. 2C), in agreement with a role of MAK11 in 60S ribosomal subunit formation.
FIG. 2.
The mak11-2 ts mutant shows alterations in 25S synthesis, cell cycle, and preribosome export. (A) Growth curves at 37°C for the mak11-2 mutant (open circles) compared with a wild-type strain (crosses). The number of cells was estimated using the absorbance of the culture at 600 nm. (B) The cell cycle distribution of yeast cells was estimated by FACS analysis. Wild-type and mak11-2 haploid cells in rich medium were shifted from 27°C to 37°C for 0, 1, 3, and 6 h; aliquots were fixed with 70% ethanol and stained with propidium iodide. The ratios of cells having 1n DNA content to those having 2n DNA content were estimated in triplicate experiments (wild-type strain, white bars; mak11-2 strain, gray bars); error bars are standard deviations of the measured ratios. (C) Total RNA was extracted from a wild-type strain or the mak11-2 strain at time zero and 4 and 12 h after shift to the nonpermissive temperature, separated on a 1% agarose gel (glyoxal denaturation), and stained with ethidium bromide. (D) GFP fluorescence of the mak11-2 ts or isogenic wild-type cells, each type expressing Rpl25-eGFP, was detected after a shift to 37°C for 8 h. DNA was stained with Hoechst 33342. Arrowheads indicate the relative positions of the DNA-stained regions of the nuclei. wt, wild type.
Both protein depletion and mak11 mutation led to 60S formation defects. To look for the cellular localization of these blocked pre-60S particles, we used the Rpl25-GFP fusion previously described as a suitable marker for 60S ribosomal particle precursor export defects (23). When expressed in a mak11-2 strain at the nonpermissive temperature, Rpl25-GFP accumulated in the nuclei of yeast cells, in a region excluded from DNA staining (Fig. 2D). We concluded that Mak11 not only is required for the progression of rRNA maturation at the 27SB step but also plays a role in the exit of the 60S precursors from the nucleolus.
As predicted from our previous work, Mak11 acts thus early after the formation of the 60S precursors in the nucleolus. For a mechanistic analysis of Mak11 action we first defined the composition of preribosomal complexes that contained Mak11.
Complexes associated with Mak11 do not contain late preribosomal factors.
Mak11 was found associated with pre-60S complexes purified in association with Rlp24, and its levels increased in these particles when another pre-60S factor, Nog1, was depleted (47). To place Mak11 on the 60S assembly pathway, we purified the associated complexes and identified the proteins by mass spectrometry. Addition of the TAP tag (44) to Mak11 as an N-terminal (strain LMA326) or C-terminal (strain LMA375) fusion had no deleterious effects on Mak11 function since both versions supported growth to wild-type levels (not shown). Forty known and putative pre-60S factors, other than ribosomal proteins, were identified by mass spectrometry from TAPs using chromosomal C-terminal tagged Mak11 (Fig. 3A, second lane from left) as well as from TAPs using N-terminal tagged Mak11 expressed from a plasmid in a Δmak11 strain (Table 2). Notable absences in the list of the identified proteins are Arx1, Nog2, Nug1, and Nsa2, known to associate late in the nucleus with the preribosomes (30, 38, 46). We verified by immunoblotting with specific antibodies that Arx1, Nog2, and Nsa2 were present in complexes purified using Rlp24-TAP but absent when the complexes were purified using Mak11-TAP (Fig. 3B). The reverse experiment confirmed these results; neither Nog2-TAP nor Arx1-TAP was able to copurify Mak11 (Fig. 3C).
FIG. 3.
Mak11-associated complexes do not contain late pre-60S factors. (A) Proteins purified in association with Mak11-TAP from wild-type (wt) cells or cells depleted for Rlp24 or Nog1 (14 h in glucose medium) were separated on a 5 to 20% polyacrylamide gradient gel and stained with colloidal Coomassie blue. Proteins identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry, listed in Table 2, are indicated. (B) The presence or absence of proteins in TEV eluates from purifications using Mak11-TAP and Rlp24-TAP was determined by immunoblotting with specific antibodies. (C) The presence of Mak11 in complexes purified using Rlp24-TAP, Nog2-TAP, and Arx1-TAP was tested by immunoblotting, with Nog1 as a positive control. (D and E) RNAs associated with Mak11 complexes were enriched in the TEV protease eluate from a TAP-Mak11 (LMA326) purification. The recovered RNAs were extracted with phenol-chloroform and tested by primer extension (D) or Northern blotting (E) with the same oligonucleotides as those used for Fig. 1.
TABLE 2.
Nonribosomal proteins identified in complexes purified in association with TAP-Mak11 (N) and Mak11-TAP (C)
| Name | Open reading frame | pI | Size (kDa) | Band(s)a | Tagb |
|---|---|---|---|---|---|
| RRP5 | YMR229C | 6.1 | 193 | N | |
| UTP22 | YGR090W | 8.8 | 140 | 1 | N+C |
| ERB1 | YMR049C | 4.9 | 92 | 2, 3 | N+C |
| MAK5 | YBR142W | 8.7 | 87 | 5 | C |
| DRS1 | YLL008W | 5.5 | 85 | 4 | N+C |
| DBP7 | YKR024C | 10.0 | 83 | 8 | C |
| NOC2 | YOR206W | 8.9 | 82 | 6 | C |
| NOC3 | YLR002C | 5.8 | 76 | 8 | C |
| PUF6 | YDR496C | 6.9 | 75 | 8 | N+C |
| NOG1 | YPL093W | 9.4 | 74 | 10 | N |
| NOP7 | YGR103W | 5.4 | 70 | 9 | N+C |
| NOP2 | YNL061W | 4.8 | 70 | 7 | N+C |
| DBP9 | YLR276C | 10.0 | 68 | 11 | C |
| ROK1 | YGL171W | 9.9 | 64 | 14 | C |
| SIK1/NOP56 | YLR197W | 9.6 | 57 | N | |
| HAS1 | YMR290C | 10.1 | 57 | N | |
| CBF5 | YLR175W | 9.5 | 55 | N | |
| MAK11 | YKL021C | 8.6 | 54 | 16 | N+C |
| NOP12 | YOL041C | 10.2 | 52 | N | |
| NSA1 | YGL111W | 8.0 | 52 | 17 | N+C |
| SSF1 | YHR066W | 10.1 | 52 | 15 | N+C |
| YTM1 | YOR272W | 7.0 | 51 | 18 | C |
| RRP14 | YKL082C | 10.4 | 50 | 13 | C |
| EBP2 | YKL172W | 6.4 | 50 | 12 | C |
| FPR3/NPI46 | YML074C | 4.2 | 47 | 15 | N+C |
| FPR4 | YLR449W | 4.4 | 44 | N | |
| CIC1/NSA3 | YHR052W | 9.5 | 43 | 19 | C |
| RLP7 | YNL002C | 10.2 | 37 | 20 | N+C |
| MAK16 | YAL025C | 5.1 | 36 | 21 | C |
| RPF1 | YHR088W | 10.3 | 35 | 24 | N+C |
| NOP1 | YDL014W | 11.0 | 34 | 23 | N+C |
| BRX1 | YOL077C | 10.0 | 34 | 25 | N+C |
| RRP1 | YDR087C | 5.0 | 33 | 22 | N+C |
| RRP15 | YPR143W | 5.5 | 28 | 22 | C |
| MRT4 | YKL009W | 9.1 | 27 | 27 | C |
| NOP16 | YER002W | 10.1 | 27 | 26 | N+C |
| TIF6/CDC95 | YPR016C | 4.4 | 26 | 30 | C |
| NOP15 | YNL110C | 10.1 | 25 | 27 | C |
| RLP24 | YLR009W | 10.4 | 24 | 28 | C |
| LOC1 | YFR001W | 11.1 | 24 | 29 | C |
| NIP7 | YPL211W | 10.1 | 20 | 31 | C |
Band position is indicated for the proteins identified from the gel shown in Fig. 3.
Protein identified in purifications performed using N-terminally (N) or C-terminally (C) tagged Mak11 or both (N+C).
In view of the number of preribosomal proteins found in association with Mak11, we wondered what species of pre-rRNA were present in these particles. The analysis of the different pre-rRNA species enriched in the TEV protease eluate when TAP-Mak11 was purified revealed the presence of trace amounts of 27SA2, 7S, and 35S with a specific enrichment of the 27SB precursor (Fig. 3D and E).
Altogether, these results revealed Mak11's association with 27SB pre-rRNA in nucleolar complexes and its requirement for an essential maturation step of 27SB containing pre-60S particles. Mak11 seems to bind only transiently to nuclear 60S precursors, as it leaves these particles before the association of late pre-60S factors such as Nog2, Nsa2, or Arx1.
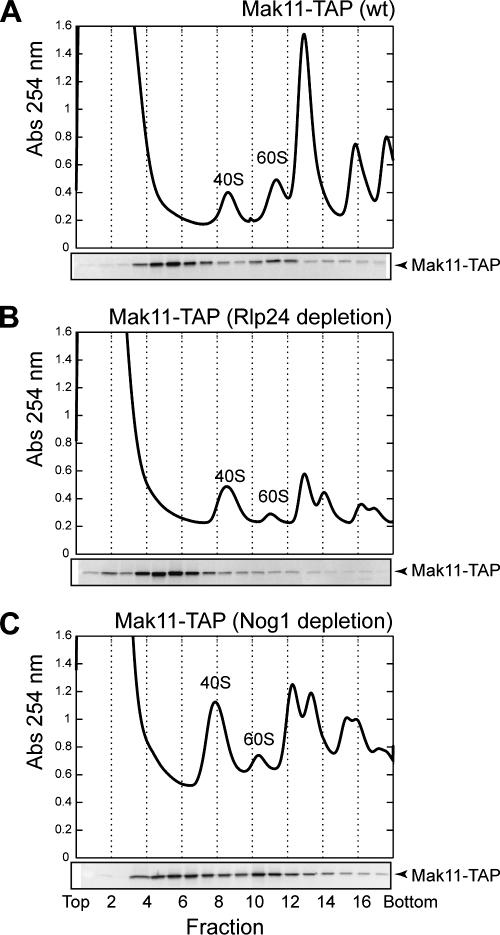
Mak11 is both associated with pre-60S particles and in a free form.
We observed that the amount of proteins purified in association with Mak11 was strongly decreased when the purification was performed in a strain depleted for Rlp24 and was increased when the purification was done after Nog1 depletion (Fig. 3A). To better understand what changed under these conditions, we separated total protein extracts from a wild-type strain or strains depleted for Rlp24 or Nog1, by ultracentrifugation on sucrose gradients. Mak11-TAP was detected in the different fractions by immunoblotting (Fig. 4). Surprisingly, even in a wild-type strain, the Mak11 sedimentation profile showed two peaks, a major one in fractions sedimenting in the upper part of the gradient and a minor one around the position of the 60S preribosomal particles (Fig. 4A). The ratio between these two fractions changed in opposite directions when either Rlp24 or Nog1 was depleted. Most of Mak11 was found in the smaller complexes, under conditions of low Rlp24 levels (Fig. 4B), whereas Mak11 was abundant in the pre-60S fractions when Nog1 was limiting for ribosome biogenesis (Fig. 4C). These changes were correlated with the small amount of preribosomal proteins purified with Mak11-TAP when Rlp24 was depleted while preribosomal proteins accumulated in the complexes associated with Mak11 under Nog1 depletion. In addition, our previous observation of Mak11 accumulating in Rlp24-TAP complexes under Nog1 depletion indicated that pre-60S complexes containing Mak11 were accumulating (47).
FIG. 4.
Dynamic association of Mak11 with pre-60S particles. Sucrose gradient ultracentrifugation analysis of Mak11-TAP in wild-type (wt) cells (A) and cells depleted for Rlp24 (B) or Nog1 (C) was followed by protein precipitation from the recovered fractions and immunoblotting to detect the TAP tag.
We tried to better characterize the form of Mak11 that sedimented in the upper part of the gradient by combining TAPs and sucrose gradients and by testing the putative oligomeric state of Mak11. No other proteins were found to be associated with the lower-sedimentation-rate Mak11-TAP when purified complexes were separated on a sucrose gradient. Moreover, other tagged forms of Mak11 (hemagglutinin and myc epitopes) have shown that the low-weight fraction of the protein sedimented in the first fractions of the gradients (not shown). The sedimentation of purified recombinant Mak11 from Escherichia coli matched the expected behavior for a monomeric protein (not shown). The relatively high sedimentation rate of the free TAP-tagged Mak11 form was thus probably due to interactions between Mak11 and the tag.
Mak11 seems to have a dynamic distribution between a free form and the pre-60S form, depending on the levels of different pre-60S intermediates. The free form might represent a “storage” of protein, readily available for variable 60S biogenesis demands.
Rlp24 cooperates with Mak11 for 60S ribosomal particle assembly.
To obtain further hints about Mak11 function, we performed a high-copy-number suppressor genetic screen with the mak11-2 thermosensitive strain. The large majority of the recovered plasmids (100) contained the MAK11 gene and were able to complement both the thermosensitivity of the test strain and the loss of a wild-type MAK11 plasmid. Nine other isolated plasmids allowed only partial complementation of the thermosensitive phenotype of the mak11-2 strain and contained sequences that had in common the RLP24 genomic region, from −619 upstream of the ATG to 1,074 nucleotides downstream of the stop codon. We further verified the complementation by using a 2μm plasmid derived from pCM190 (14) expressing RLP24 under the control of a doxycycline-repressible promoter. Only the pCM190-RLP24 plasmid could complement the mak11-2 ts phenotype, while the complementation failed if the expression of RLP24 was repressed (Fig. 5A).
FIG. 5.
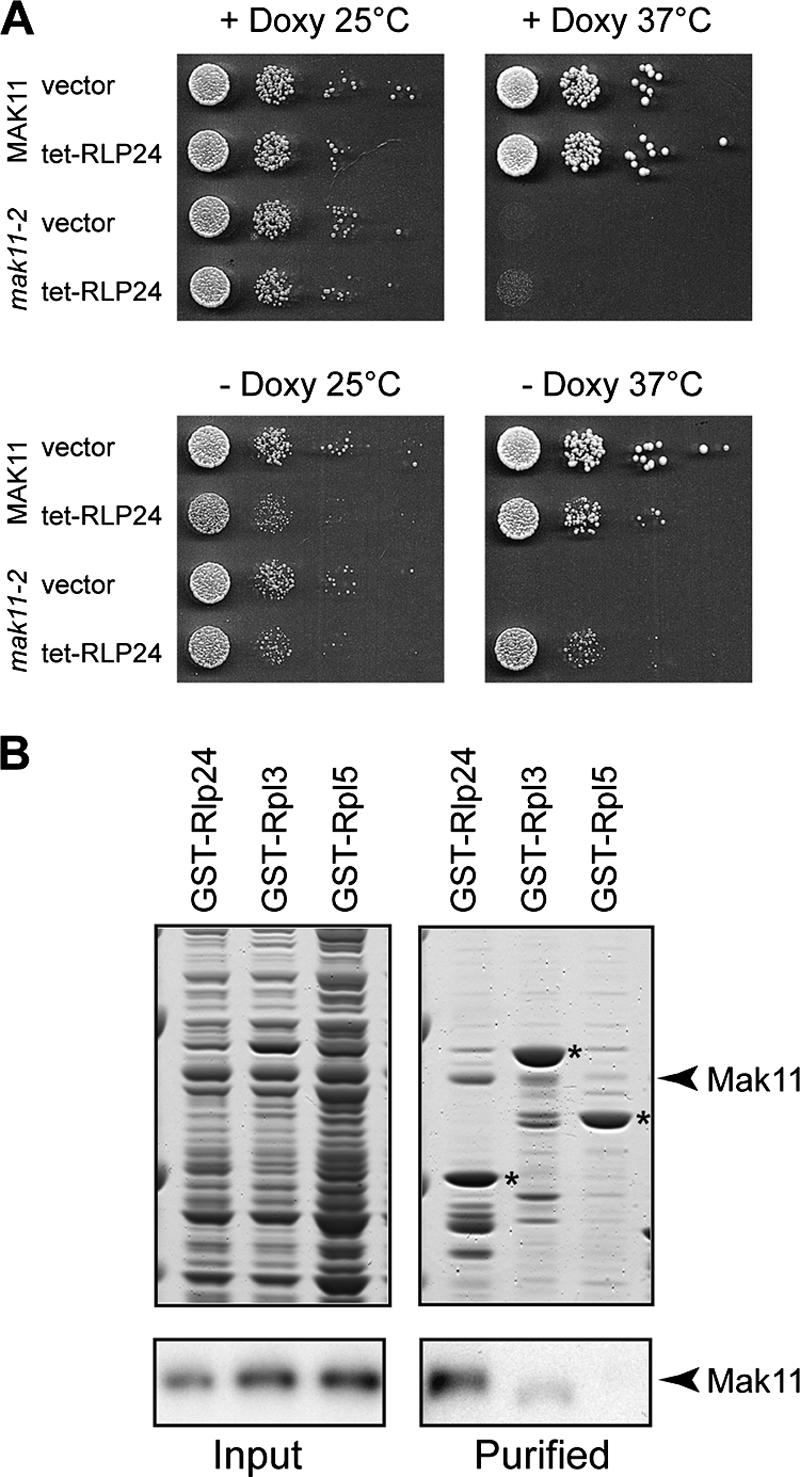
Mak11 and Rlp24 are functionally and physically linked. (A) RLP24 is a high-copy-number suppressor of the mak11-2 ts phenotype. Rlp24 was expressed from a high-copy-number vector (pCM190-RLP24) under the control of a tetracycline-repressible promoter in wild-type or mak11-2 cells at permissive and nonpermissive temperatures. Growth was estimated by 10-fold serial dilutions on solid yeast extract-peptone-dextrose medium with or without doxycycline. (B) Protein-protein interaction between (His)6-Mak11 and GST-Rlp24 was detected by mixing total extracts of E. coli overexpressing the proteins and pulling down the GST fusion proteins with glutathione-Sepharose. Eluates were separated on 4 to 12% Novex polyacrylamide gels and Coomassie blue stained. Immunoblot assays were performed in parallel with rabbit polyclonal antibodies raised against Mak11. Asterisks indicate GST-tagged proteins. Rpl3 and Rpl5 were used as controls.
We wondered if the complementation of the thermosensitive mak11-2 phenotype by RLP24 overexpression could be explained by a direct physical interaction between the two proteins. To this end, we tested in vitro the putative interaction between isolated Rlp24 and Mak11. After mixing of total bacterial extracts from E. coli expressing GST-Rlp24 or two ribosomal proteins, used as negative controls, with an extract from bacteria expressing (His)6-Mak11, GST-tagged proteins were purified on glutathione-Sepharose. The amount of copurified Mak11 was estimated by Western blotting. In contrast with the negative controls, GST-Rlp24 was able to copurify a substantial amount of Mak11 (Fig. 5B). This result suggests that functional complementation of Mak11 inactivation by Rlp24 correlates with a direct physical interaction, presumably on pre-60S particles in the nucleus.
A direct interaction between Mak11 and Rlp24 and the decrease of the amount of Mak11-associated preribosomal proteins when Rlp24 was depleted (Fig. 3A) suggested a possible role for Mak11 in Rlp24 association with pre-60S complexes. The amount of proteins purified in association with Rlp24 decreased dramatically during Mak11 depletion (not shown). The absence of a complex under these conditions was explained by a decrease in the total amount of the tagged Rlp24 (Fig. 6A). No change was observed when another tagged protein, Ssf1, was used in a similar experiment, and only a slight decrease in total protein concentration was observed with Nog1-TAP (Fig. 6B and C). No change was observed in the levels of the mRNA for Rlp24 when tested by reverse transcription and quantitative real-time PCR (not shown). These results strongly suggest that Mak11 is required at an early 60S assembly step either for Rlp24 association with precursor complexes or for the stability of these particles.
FIG. 6.
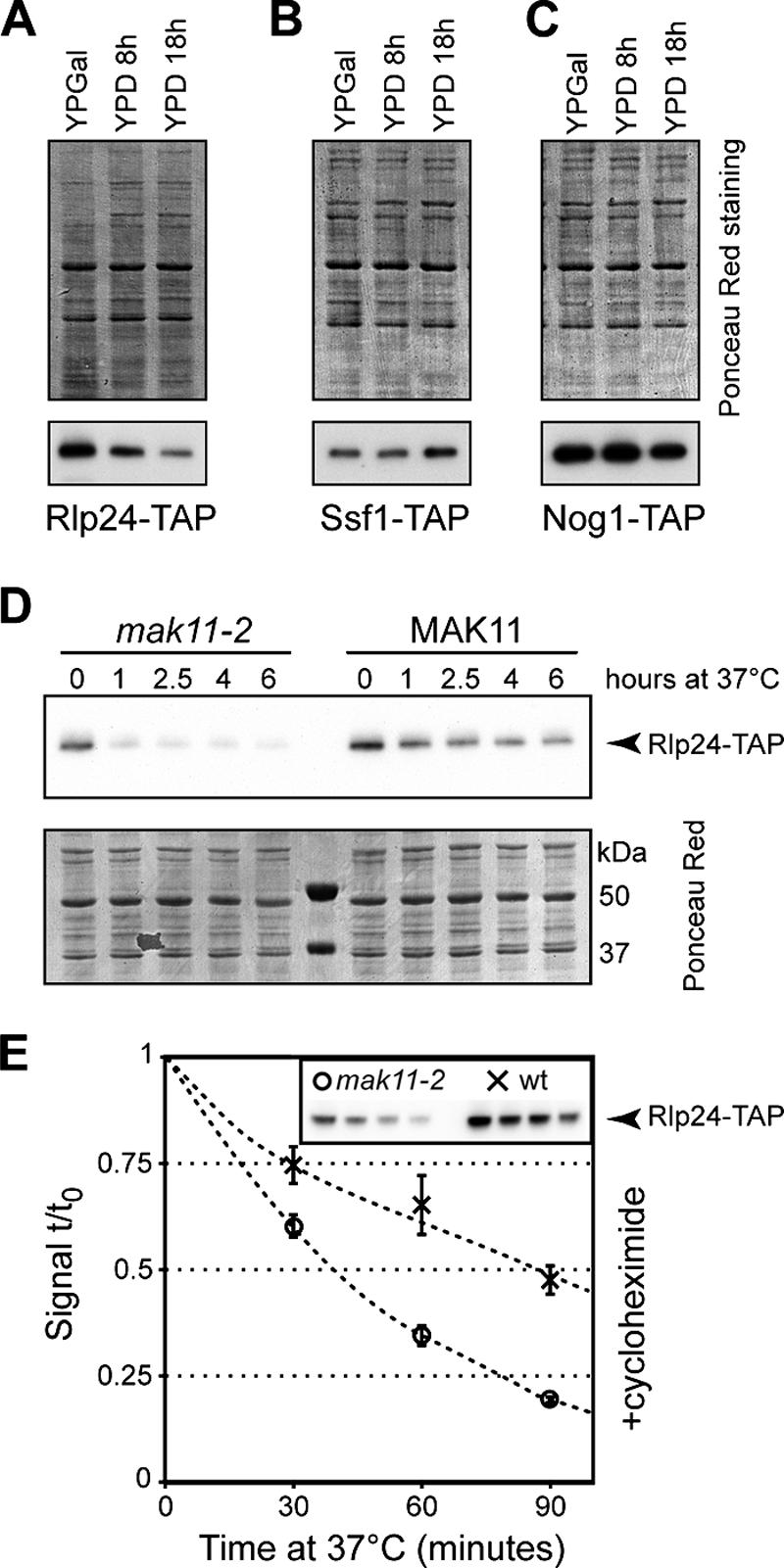
Mak11 stabilizes Rlp24. (A to C) Strains expressing Rlp24-TAP (A), Ssf1-TAP (B), and Nog1-TAP (C) with MAK11 under the control of a GAL1 promoter were grown on rich galactose medium and shifted to glucose for 8 and 18 h. The amounts of the tagged proteins were estimated by immunoblotting for the TAP tag; the total amounts of loaded proteins on each lane were estimated by Ponceau S staining of the nitrocellulose membranes. (D) Strains expressing Rlp24-TAP and either the mak11-2 allele or wild-type MAK11 were grown on yeast extract-peptone-dextrose at 27°C and shifted to 37°C for up to 6 h. The amount of Rlp24-TAP was estimated by immunoblotting (upper panel), and the amount of loaded total protein was visualized by Ponceau red staining (lower panel). (E) The stability of Rlp24-TAP in the mak11-2 (open circles) and wild-type (crosses) strains was measured by simultaneously shifting the culture from 27°C to 37°C and adding 35 μg/ml cycloheximide. The immunoblot signal was plotted as a ratio to the initial Rlp24-TAP amount. The inset shows a typical image used for quantification. The standard deviations of the results for three independent cultures are indicated as error bars.
To further investigate the destabilization of Rlp24 that follows the functional inactivation of MAK11, we transformed a strain expressing TAP-tagged Rlp24 and glucose-repressible MAK11 (LMA515) with a plasmid expressing mak11-2 or a wild-type allele. These yeast cells had a thermosensitive or a wild-type phenotype when grown on glucose-containing medium. The levels of Rlp24-TAP at 27°C in the mak11-2 strain were estimated by immunoblotting to be about 50% of the levels of Rlp24-TAP in the wild-type strain (Fig. 6D and E). Strikingly, when we performed a shift of the mak11-2 culture to the nonpermissive temperature, Rlp24-TAP levels dropped to barely detectable levels after only 1 hour of shift to 37°C compared with the wild-type background (Fig. 6D). For a quantitative estimation of the effects of Mak11 inactivation on Rlp24 stability, we looked for the decrease in the amount of Rlp24-TAP after a shift to 37°C, simultaneously with the addition of the translation inhibitor cycloheximide (Fig. 6E). Rlp24-TAP levels decreased at a higher rate in the mak11-2 background than in the wild-type strain with an estimated half-life of 38 min for the ts strain and 87 min for the wild-type strain. Mak11 is thus specifically required to maintain Rlp24 levels in yeast.
RLP24 overexpression complemented the phenotype of the mak11-2 strain, probably via a direct protein-protein interaction, and Rlp24 was destabilized in the absence of Mak11. These data strongly suggest that the essential function of MAK11 in S. cerevisiae is directly linked to 60S ribosomal subunit formation. Since another function has been described for putative orthologues of MAK11 in other organisms, we wondered whether these proteins were involved in the large subunit biogenesis in these organisms.
Skb15, the fission yeast Mak11 homologue, is involved in 60S ribosomal subunit biogenesis.
Mak11 fission yeast (Skb15) and human (hPip1) homologues were previously described as components of a mitogen-activated protein kinase signaling pathway, which delivers signals from the cell surface to different effectors (27, 63). Multiple alignments of the sequences for the human, fission yeast, and budding yeast proteins indicated a moderate level of similarity, with about 17% identity between any two aligned sequences (Fig. 7). Reciprocal BLAST searches against entire proteome sequences identified putative orthologues of Mak11 in many eukaryotes.
We wondered whether the fission yeast or the human protein complemented the lethal phenotype of S. cerevisiae cells deleted for MAK11. To test this hypothesis, we cloned the sequences coding for Mak11, Skb15, and hPip1 in vectors that would allow the expression of N-terminal protein A fusions in budding yeast. After transformation of the diploid heterozygote strain containing the deletion of MAK11, sporulation of the diploids was induced and the tetrads were dissected. Only the vectors bearing the MAK11 sequence were able to complement the absence of MAK11 (not shown). Either the sequence divergence is too important to allow functional complementation of MAK11 deletion by Skb15 or hPip1, or the Mak11 function in budding yeast is different from the Skb15 or hPip1 function in fission yeast or human cells.
Since we could not directly demonstrate the functional conservation of the Mak11 putative homologue Skb15 in budding yeast, we looked for potential implication of Skb15 in ribosome biogenesis in S. pombe. We took advantage of a previously described fission yeast strain where the expression of SKB15 is under the control of a repressible promoter (27) to assess 60S ribosomal subunit defects when Skb15 was depleted. We compared the polysome profile of extracts from the cells depleted for Skb15 with that of a corresponding wild-type strain. As expected for a Mak11 orthologue, depletion of Skb15 led to a polysome profile typical for 60S ribosomal subunit biogenesis defects with a large peak of free 40S, a decrease in the amount of free 60S, and a drastic decrease in the polysome levels (Fig. 8A and B). Skb15 is thus functionally linked to ribosome biogenesis in S. pombe.
FIG. 8.
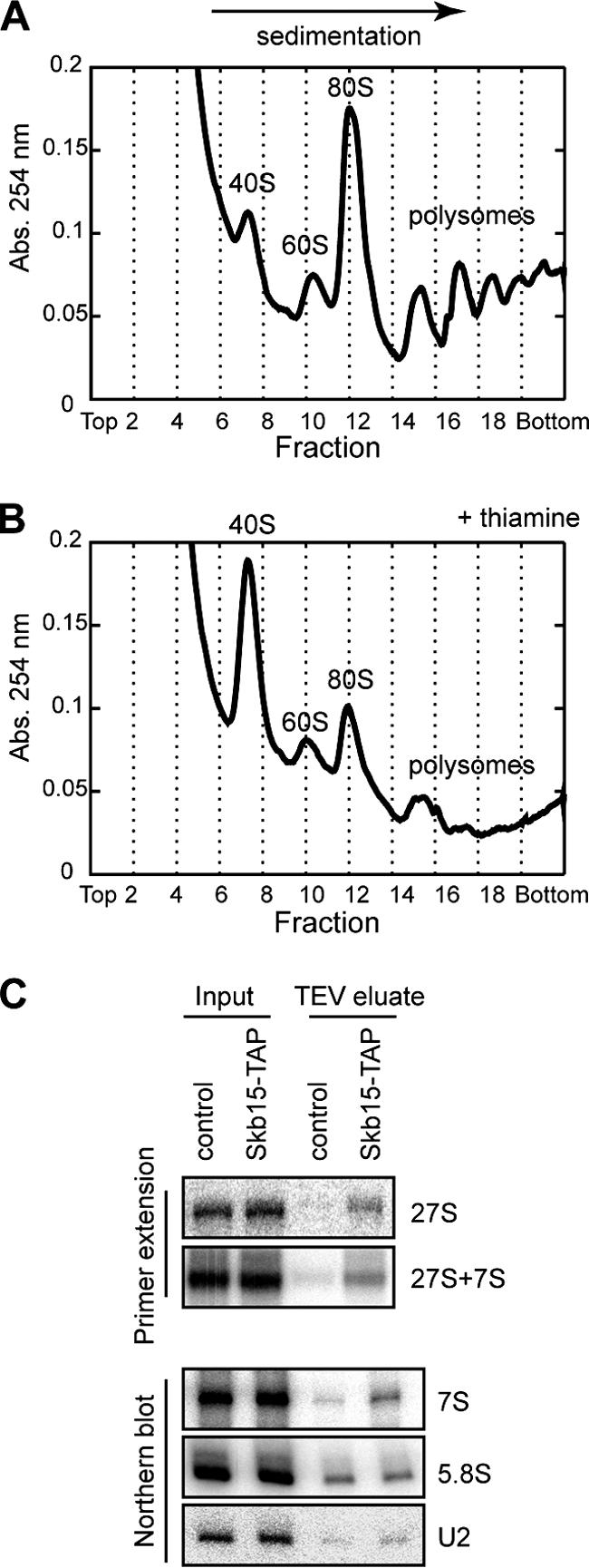
The Mak11 homologue in S. pombe, Skb15, is involved in 60S formation in fission yeast. (A and B) Cells expressing SKB15 under the control of the nmt1 thiamine-repressible promoter were grown in synthetic medium without thiamine (A) or with 5 μg/ml thiamine (B). Extracts were separated by sucrose gradient ultracentrifugation, and the amounts of free 40S, free 60S, and polysomes were estimated by the 254-nm absorbance profile. (C) A wild-type S. pombe strain and a strain expressing Skb15-TAP were used for the first step of TAP; RNAs were extracted with phenol-chloroform and tested by Northern blotting or primer extension with oligonucleotides specific for pre-rRNAs or U2 as control.
To get more mechanistic insights into Skb15 function in fission yeast, we generated an S. pombe strain producing the Skb15 protein fused to the TAP tag. We performed the first step of a TAP (TEV eluate) using extracts from this strain and from a corresponding wild-type strain as a control. The analysis of copurified rRNA precursors showed an enrichment of both 7S and 27S fission yeast equivalents of budding yeast pre-rRNA intermediates in the Skb15-associated complex (TEV eluate) in comparison with the wild-type control (Fig. 8C). These results indicate that Skb15 is a Mak11 orthologue in S. pombe that associates with pre-60S particles and is required for 60S ribosomal subunit biogenesis in fission yeast.
DISCUSSION
Building preribosomal complexes with WD40 repeats.
Based on genetic, copurification, and direct physical links, we show here that Mak11 is an essential, conserved preribosomal protein directly interacting with Rlp24 and affecting Rlp24 stability. Mak11 sequence contains several repeated motifs, with conserved tryptophan and aspartic acid residues and an average length of 40 amino acids (WD40). For the proteins with known structure that contain such motifs, it has been shown that the repeated sequences form a circularized beta-propeller structure mediating protein-protein interactions (for reviews see references 33 and 51).
There are more than 100 WD40 repeat proteins in yeast as listed by the Superfam database (34). Almost one-fifth of these proteins (22 out of 111) are factors annotated as being involved in ribosome biogenesis and assembly—a significant enrichment of this pathway (P < 4 × 10−11) over a random selection (GO Termfinder [4]). Many of the annotated ribosome biogenesis WD40 repeat proteins were shown to directly interact with, or regulate the levels of, other proteins (25) or to be components of multiprotein subcomplexes. A profusion of WD40 repeat proteins is found in discrete 80S processome subcomplexes. Four out of seven components of the tUTP complex (13) contain WD40 repeat proteins, and the UTP “B” complex (9, 29) is composed almost exclusively of WD40 repeat proteins (five of six proteins). One of the first pre-60S subcomplexes to be isolated contains two WD40 repeat proteins, Ytm1 and Erb1, in association with Nop7 (19, 29, 35). Another subcomplex, transiently associated with late, nuclear pre-60S precursors, is composed of four proteins and contains Ipi3 as a WD40 repeat member (29, 39).
Two examples of WD40 repeat ribosome biogenesis factors interacting directly with ribosomal proteins for 60S ribosomal subunit assembly are known. Rrb1 interacts with the ribosomal protein Rpl3 in the nucleus and regulates its levels (25, 48), and Sqt1 interacts with Rpl10 in the cytoplasm (11, 59). Both these proteins have a role in the association of the corresponding ribosomal protein with the nascent 60S ribosomal subunits and might regulate the levels of the corresponding ribosomal protein.
Only one ribosomal protein of the Rpl24e family exists in archaea, and its sequence is closer to the sequence of the preribosomal factor Rlp24 than to the sequence of the yeast ribosomal protein Rpl24 (discussed in reference 47). Yeast Rlp24 is essential to ribosome assembly, does not participate in translation, and thus may be considered an “assembly-only” version of a ribosomal protein. While it is assumed that most of the ribosomal proteins associate early with the precursors of the rRNA during ribosome formation, little is known about the timing and potential roles of preribosomal factors in coordinating this assembly process.
Our description of Mak11 as directly interacting with Rlp24 thus fits a pattern established by Sqt1 and Rrb1, factors that contain WD40 repeats and function as partners of ribosomal proteins. Both Sqt1 and Rrb1 were shown to exist mainly as free proteins or small complexes as demonstrated by their sedimentation on sucrose gradients (11, 48). However, the dynamics of the sedimentation pattern for these proteins under mutant conditions have not been evaluated in these previous studies. We show here that Mak11 is present in the cell both free and in association with pre-60S particles. This association is dynamic, and the relative ratios of the two forms vary under mutant conditions. An increase in the amount of the pre-60S-associated Mak11 was observed when Nog1 was depleted, in correlation with the change in Mak11 sedimentation pattern and the increase of the 60S-associated fraction. A similar effect was observed for the amount of Sqt1 copurified with preribosomal particles isolated in association with the cytoplasmic GTPase Lsg1. Blocked particles, containing a dominant-negative form of Lsg1, contained higher amounts of Sqt1 and Rpl10 than particles derived from wild-type cells (59).
In conclusion, the WD40 repeat structure is used by several proteins for binding specifically to ribosomal proteins during ribosome assembly. It would be interesting to test other preribosomal factors having WD40 repeats for direct physical and functional interactions with ribosomal proteins. It would be important to see whether transient and possibly regulated interactions between specific preribosomal factors and ribosomal proteins might play a role in the timing of ribosomal protein association with nascent ribosomes.
Functional conservation of Mak11 in eukaryotic ribosome biogenesis.
The sequence of Mak11 shows low levels of similarity with sequences of putative orthologues from other eukaryotes. Experiments with Skb15, the Mak11 homologue in S. pombe, allowed us to show that Skb15 associates with precursors of the 60S ribosomal subunits in fission yeast and that depletion of the protein leads to a decrease in free 60S levels. It is thus likely that the putative orthologues of Mak11 are involved in ribosome biogenesis in other eukaryotes as well. Our additional unpublished results suggest that even if SKB15 cannot complement MAK11 deletion, the protein and its mammalian homologues have features similar to those of Mak11. (i) Skb15, when expressed in S. cerevisiae, localized to the nucleus of budding yeast cells with a stronger signal in the nucleolus. With the use of a high-copy-number plasmid and a strong promoter, the signal for the human Mak11 homologue hPip1 was also detectable in the nucleus of yeast cells, including the nucleolus. The localization of Skb15 in fission yeast was previously described, and the published images of the GFP fusion protein localization (26) suggest the presence of the protein in the nucleolus of S. pombe cells. Moreover, hPip1 has been identified in highly purified nucleolar fractions of human cells (49). (ii) Total extracts of S. cerevisiae cells expressing tagged Skb15 were fractionated by sucrose gradient ultracentrifugation. While the tag alone was found exclusively in the upper part of the gradient, tagged Skb15 could also be detected in fractions around 60S—consistent with a weak but significant association of the protein with preribosomal particles (not shown). Skb15 thus contains a nuclear localization signal functional in S. cerevisiae and associates weakly with 60S size particles in budding yeast. (iii) In response to various environmental stresses, the changes in SKB15 mRNA levels are similar to the changes in the levels of mRNAs coding for factors predicted to be involved in ribosome biogenesis and assembly in S. pombe (7).
Both Skb15 and hPip1 have been previously described as PAK inhibitors (27, 63). Skb15 was identified by a two-hybrid screen performed using a truncated form of a PAK, Shk1. SKB15 deletion in S. pombe is lethal. Depletion of Skb15 led to cells with altered morphology and to an increase in the activity of protein kinase Shk1, estimated by the level of Shk1 autophosphorylation. Interestingly, expression of a mouse Skb15 homologue was able to complement the deletion of SKB15, indicating functional conservation between fission yeast and mammals (27).
While in vitro experiments were performed with the human Skb15 homologue hPip1 showing that hPip1 could inhibit the activity of human PAK1, no such experiments were reported for Skb15. We tried to reproduce these results with the S. cerevisiae pair of proteins. It has been previously shown that STE20 is functionally equivalent with human PAK1 since its deletion in yeast is complemented by the expression of human PAK1 (6). We tested in vitro the effect of adding purified Mak11 on the protein kinase activity of isolated Ste20 using myelin basic protein as a substrate. Ste20 activity was not affected by added purified Mak11, but we could observe that Mak11 itself was efficiently phosphorylated by Ste20 during these assays (not shown).
A simple hypothesis for the differences between our results and the published studies of Mak11 homologues is that these homologues acquired secondary functions in other eukaryotes, such as protein kinase inhibition. An alternative explanation is that Mak11 homologues function in 60S ribosome formation, as demonstrated here. The stimulation of Shk1 protein kinase activity by Skb15 depletion might thus be a secondary effect of decreasing 60S ribosomal subunit levels. This is an appealing hypothesis since it would suggest that the activation or inhibition of protein kinases might be important in the adaptation of cells to reduced ribosome biogenesis levels.
Acknowledgments
We thank Stevan Marcus (University of Alabama, Tuscaloosa) for the Skb15 strain and plasmid, Mingyao Liu (The University of Texas, Houston) for the hPip1 vector, Kathleen Gould (Vanderbilt University, Nashville, TN) for the pFA6a-CTAP-KanMX6 vector, Françoise Stutz (Centre Médicale Universitaire, Genève, Switzerland) for the pFA6a-TAP-Tag-His3MX6 vector, Edouard Bertrand (IGM-CNRS, Montpellier, France) for the Gateway-compatible pET32a vector, Ed Hurt (Biochemie-Zentrum, Heidelberg, Germany) for the centromeric plasmid expressing RLP25-eGFP, and François Lacroute (Centre de Génétique Moléculaire, Gif-sur-Yvette, France) for providing the high-copy-number vector genomic S. cerevisiae library. We are grateful to Benoit Arcangioli and Samia Ben Hassine (Institut Pasteur, Paris, France) for help with S. pombe manipulations.
This work was supported by the Ministère délégué à l'Enseignement Supérieur et à la Recherche (ACI-BCM0089-2003).
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belle, A., A. Tanay, L. Bitincka, R. Shamir, and E. K. O'Shea. 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 103:13004-13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, K. A., and S. J. Baserga. 2004. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell 15:5038-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, E. I., S. Weng, J. Gollub, H. Jin, D. Botstein, J. M. Cherry, and G. Sherlock. 2004. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. L., L. Stowers, M. Baer, J. Trejo, S. Coughlin, and J. Chant. 1996. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr. Biol. 6:598-605. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotteret, S., Z. M. Jaffer, A. Beeser, and J. Chernoff. 2003. p21-activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 23:5526-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosil, M., and X. R. Bustelo. 2004. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 279:37385-37397. [DOI] [PubMed] [Google Scholar]
- 10.Edskes, H. K., Y. Ohtake, and R. B. Wickner. 1998. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60 S ribosomal subunit biogenesis. J. Biol. Chem. 273:28912-28920. [DOI] [PubMed] [Google Scholar]
- 11.Eisinger, D. P., F. A. Dick, E. Denke, and B. L. Trumpower. 1997. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol. Cell. Biol. 17:5146-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 15.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 16.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 17.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 18.Good, L., R. V. Intine, and R. N. Nazar. 1997. The ribosomal-RNA-processing pathway in Schizosaccharomyces pombe. Eur. J. Biochem. 247:314-321. [DOI] [PubMed] [Google Scholar]
- 19.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 20.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, M. Tyers, J. S. Andersen, C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, and A. I. Lamond. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, C., M. Shepelev, and J. Chernoff. 2004. The genetics of Pak. J. Cell Sci. 117:4343-4354. [DOI] [PubMed] [Google Scholar]
- 22.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 23.Hurt, E., S. Hannus, B. Schmelzl, D. Lau, D. Tollervey, and G. Simos. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144:389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Icho, T., and R. B. Wickner. 1988. The MAK11 protein is essential for cell growth and replication of M double-stranded RNA and is apparently a membrane-associated protein. J. Biol. Chem. 263:1467-1475. [PubMed] [Google Scholar]
- 25.Iouk, T. L., J. D. Aitchison, S. Maguire, and R. W. Wozniak. 2001. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol. Cell. Biol. 21:1260-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, H., P. Yang, P. Catanuto, F. Verde, H. Lai, H. Du, F. Chang, and S. Marcus. 2003. The kelch repeat protein, Tea1, is a potential substrate target of the p21-activated kinase, Shk1, in the fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 278:30074-30082. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H. W., P. Yang, Y. Qyang, H. Lai, H. Du, J. S. Henkel, K. Kumar, S. Bao, M. Liu, and S. Marcus. 2001. Genetic and molecular characterization of Skb15, a highly conserved inhibitor of the fission yeast PAK, Shk1. Mol. Cell 7:1095-1101. [DOI] [PubMed] [Google Scholar]
- 28.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St. Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. [DOI] [PubMed] [Google Scholar]
- 29.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 30.Lebreton, A., C. Saveanu, L. Decourty, A. Jacquier, and M. Fromont-Racine. 2006. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 281:27099-27108. [DOI] [PubMed] [Google Scholar]
- 31.Lebreton, A., C. Saveanu, L. Decourty, J. C. Rain, A. Jacquier, and M. Fromont-Racine. 2006. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leger-Silvestre, I., P. Milkereit, S. Ferreira-Cerca, C. Saveanu, J. C. Rousselle, V. Choesmel, C. Guinefoleau, N. Gas, and P. E. Gleizes. 2004. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 23:2336-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, D., and R. Roberts. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58:2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madera, M., C. Vogel, S. K. Kummerfeld, C. Chothia, and J. Gough. 2004. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32:D235-D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miles, T. D., J. Jakovljevic, E. W. Horsey, P. Harnpicharnchai, L. Tang, and J. L. Woolford, Jr. 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 25:10419-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 37.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 38.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nissan, T. A., K. Galani, B. Maco, D. Tollervey, U. Aebi, and E. Hurt. 2004. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol. Cell 15:295-301. [DOI] [PubMed] [Google Scholar]
- 40.Ohtake, Y., and R. B. Wickner. 1995. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol. Cell. Biol. 15:2772-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellett, S., and J. W. Tracy. 2006. Mak16p is required for the maturation of 25S and 5.8S rRNAs in the yeast Saccharomyces cerevisiae. Yeast 23:495-506. [DOI] [PubMed] [Google Scholar]
- 42.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 43.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 44.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 45.Sachs, A. B., and R. W. Davis. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58:857-867. [DOI] [PubMed] [Google Scholar]
- 46.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaper, S., M. Fromont-Racine, P. Linder, J. de la Cruz, A. Namane, and M. Yaniv. 2001. A yeast homolog of chromatin assembly factor 1 is involved in early ribosome assembly. Curr. Biol. 11:1885-1890. [DOI] [PubMed] [Google Scholar]
- 49.Scherl, A., Y. Coute, C. Deon, A. Calle, K. Kindbeiter, J. C. Sanchez, A. Greco, D. Hochstrasser, and J. J. Diaz. 2002. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Searfoss, A., T. E. Dever, and R. Wickner. 2001. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 52.Tasto, J. J., R. H. Carnahan, W. H. McDonald, and K. L. Gould. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18:657-662. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toh-e, A., P. Guerry, and R. B. Wickner. 1978. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136:1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 56.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 57.Wang, L., R. Kao, F. D. Ivey, and C. S. Hoffman. 2004. Strategies for gene disruptions and plasmid constructions in fission yeast. Methods 33:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warner, J. R. 2001. Nascent ribosomes. Cell 107:133-136. [DOI] [PubMed] [Google Scholar]
- 59.West, M., J. B. Hedges, A. Chen, and A. W. Johnson. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 25:3802-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickner, R. B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickner, R. B. 1978. Twenty-six chromosomal genes needed to maintain the killer double-stranded RNA plasmid of Saccharomyces cerevisiae. Genetics 88:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickner, R. B., and M. J. Leibowitz. 1979. Mak mutants of yeast: mapping and characterization. J. Bacteriol. 140:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia, C., W. Ma, L. J. Stafford, S. Marcus, W. C. Xiong, and M. Liu. 2001. Regulation of the p21-activated kinase (PAK) by a human Gbeta-like WD-repeat protein, hPIP1. Proc. Natl. Acad. Sci. USA 98:6174-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zagulski, M., D. Kressler, A. M. Becam, J. Rytka, and C. J. Herbert. 2003. Mak5p, which is required for the maintenance of the M1 dsRNA virus, is encoded by the yeast ORF YBR142w and is involved in the biogenesis of the 60S subunit of the ribosome. Mol. Genet. Genomics 270:216-224. [DOI] [PubMed] [Google Scholar]