Abstract
Methylation-controlled J protein (MCJ) is a newly identified member of the DnaJ family of cochaperones. Hypermethylation-mediated transcriptional silencing of the MCJ gene has been associated with increased chemotherapeutic resistance in ovarian cancer. However, the biology and function of MCJ remain unknown. Here we show that MCJ is a type II transmembrane cochaperone localized in the Golgi network and present only in vertebrates. MCJ is expressed in drug-sensitive breast cancer cells but not in multidrug-resistant cells. The inhibition of MCJ expression increases resistance to specific drugs by inducing expression of the ABCB1 drug transporter that prevents intracellular drug accumulation. The induction of ABCB1 gene expression is mediated by increased levels of c-Jun due to an impaired degradation of this transcription factor in the absence of MCJ. Thus, MCJ is required in these cells to prevent c-Jun-mediated expression of ABCB1 and maintain drug response.
Methylation-controlled J protein (MCJ) is a recently identified member of the DnaJ protein family of cochaperones, and its expression is controlled by methylation (56). DnaJ proteins are characterized by the presence of the DnaJ domain containing the His-Pro-Asp signature tripeptide. The DnaJ protein family is one of the largest cochaperone families that has members with diverse cellular localization patterns and functions (11). In addition to the DnaJ family, two other families of cochaperones have been identified based on the presence of the Bag domain (58) or the tetratricopeptide repeat clamp domain (53, 58). Cochaperones associate with the heat shock protein 70 (Hsp70) family of chaperones (Hsp90, Hsp70, and Hsc70) through these conserved domains and participate in protein folding and trafficking (64). Cochaperones have a modular architecture in which a chaperone binding domain (DnaJ, the tetratricopeptide, or Bag) is fused to other nonconserved sequences that can interact with specific proteins and mediate a variety of diverse activities including clathrin uncoating (63) and cytoskeletal functions (29). Some DnaJ cochaperones also participate in ubiquitin-dependent proteolysis either by tagging certain substrates for degradation or by facilitating the unfolding of folded proteins, thus allowing degradation by proteolysis (35).
MCJ has some unique features among the members of the DnaJ family. Comprising 150 amino acids (aa), it is a rather small protein (16 to 17 kDa) compared to other members (∼40 kDa). The DnaJ domain is located in the C terminus (56), while it is commonly present in the N terminus in other DnaJ proteins. In addition, a potential transmembrane domain distinguishes MCJ from most other DnaJ proteins that are present in the cytosol and interact with chaperones through the DnaJ domain. Thus, MCJ appears to be an atypical DnaJ family member.
The MCJ gene has been identified as a gene expressed in normal ovarian epithelial cells but absent or expressed at very low levels in a number of primary ovarian tumors and ovarian carcinoma cell lines (56). The loss of MCJ is correlated with increased drug resistance in ovarian cancer cell lines (56). The hypermethylation of a CpG island present within the first exon and first intron of the MCJ gene represses MCJ expression (60). The overexpression of MCJ in ovarian cancer cells increases sensitivity to antineoplastic drugs in vitro (56). A recent study of ovarian cancer patients demonstrates that the high levels of CpG island methylation correlate with the poor responses of the tumors to chemotherapy and overall poor survival rates (61). The methylation of the MCJ gene in some malignant pediatric brain tumors and in 90% of Wilms' tumors has also been reported, whereas very low levels of methylation in normal tissues have been found (17, 37). However, the relevance of MCJ gene hypermethylation for chemoresistance in these tumors has not yet been addressed.
While the regulation of MCJ gene expression has received a certain amount of interest, no information about the biology and function of MCJ, including its cellular localization pattern, is presently available. In addition, although the loss of MCJ gene expression by hypermethylation has been correlated with multidrug resistance in ovarian cancer, the mechanism by which this cochaperone regulates the drug response is completely unknown. We show here that MCJ is a Golgi compartment-associated, type II transmembrane DnaJ protein that arose in vertebrates as a result of gene duplication. We further demonstrate that MCJ is required to repress the expression of the ABCB1 drug transporter in breast cancer cells. The loss of MCJ expression leads to increased levels of c-Jun protein that trigger the expression of ABCB1 and thereby multidrug resistance.
MATERIALS AND METHODS
Cell culture.
MCF7 and MCF7/ADR cells were a kind gift from Ken Cowan (National Cancer Institute, Bethesda, MD). MCF7, MCF7/ADR, MCF7/siMCJ-1B, MCF7/siMCJ-3B, MDA-MB-321, MD22, MES-SA, and MES-Dx5 cells were maintained in RPMI 1640 (Life Technologies, Inc., Gaithersburg, MD) containing 5% fetal bovine serum (FBS; HyClone, Logan, UT), penicillin-streptomycin, and l-glutamine (Gibco). 293T cells were cultured in Dulbecco's modified Eagle's medium-F12 medium (Life Technologies, Inc.) supplemented with 10% FBS (HyClone, Logan, UT).
Plasmids.
A small interfering RNA (siRNA) construct targeting MCJ was generated using the human H1 RNA polymerase III promoter (6) cloned into the pCMV-EGFP-N1 vector (Invitrogen, Carlsbad, CA) upstream of the cytomegalovirus enhanced green fluorescent protein (EGFP) cassette to obtain the pSuper-EGFP vector. The siRNA for MCJ, 5′-gatccccGAAGATTTCAACTCCTAGCttcaagagaGCTAGGAGT TGAAATCTTCtttttggaag-3′ (Sigma Genosys, Woodlands, TX), was cloned into the BglII and HindIII sites downstream of the H1 promoter. The human MCJ target sequences are shown in capital letters. The pEGZ plasmid expressing hemagglutinin (HA)-tagged MCJ was generated by cloning the MCJ gene downstream of the HA tag sequence in the pEGZ plasmid containing the internal ribosome entry site-EGFP gene.
Transient and stable transfection.
For transient transfection, cells were transfected using Lipofectamine 2000 as recommended by the manufacturer (Invitrogen, Carlsbad, CA). For stable transfection, the above-described procedure was followed and clones were selected in medium containing 400 μg/ml of G418 (Life Technologies, Inc.) as described previously (10). 293T cells were transfected using calcium phosphate.
Luciferase assay.
MCF7, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were cotransfected with AP-1-luciferase reporter construct and pCMV-β-galactosidase (β-Gal) plasmids by using Lipofectamine 2000. After 24 h, the cells were washed, trypsinized, and lysed and the relative luciferase activity was determined as recommended by the manufacturer (Promega, Madison, WI) by using a TD-20/20 luminometer (Turner Biosystems, Inc., Sunnyvale, CA). Luciferase activity was normalized to β-galactosidase activity as a control for transfection efficiency.
Cell viability assay (MTT assay).
The responses of the cell lines to doses of different drugs were determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay as described previously (10, 57). Different concentrations of drugs were used, ranging from 0 to 100 μM for doxorubicin (adriamycin), 0 to 30 μM for paclitaxel, and 0 to 30 μM for 5-fluorouracil (5-FU). Fifty percent lethal doses (LD50) were calculated by nonlinear regression and plotted graphically as percentages of viable cells.
Analysis of drug internalization. (i) Flow cytometry.
MCF7, MCF7/ADR, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were treated with doxorubicin (0.3 or 3 μM) for 3 h at 37°C. Negative controls had no drug added. Cells were washed three times with phosphate-buffered saline (PBS) and trypsinized, and doxorubicin fluorescence was examined by flow cytometry using an LSRII flow cytometer (BD Biosciences, MA). A 695/40-nm-band-pass filter with a 685-nm long pass was used to measure doxorubicin fluorescence. For MCF7/siMCJ cells, EGFP was detected with a 530/30-nm-band-pass filter with a 505-nm long pass. No overlap of the two fluorescences could be detected.
(ii) Confocal microscopy.
MCF7, MCF7/ADR, and MCF7/siMCJ-1B and -3B cells (3 × 104) were seeded on BD BioCoat coverslips (BD Biosciences, Bedford, MA) and treated with doxorubicin in the presence or absence of verapamil (10 μM). After different periods of time (0, 2, and 3 h), cells were washed with PBS and fixed in 3.7% paraformaldehyde to be examined by confocal microscopy. Since both EGFP (present in MCF7/siMCJ cells) and doxorubicin are excited at 488 nm, the conventional multitrack type of analysis could not be used. Instead, we used the lambda mode analysis setting of the confocal microscope (LSM 510 META confocal laser scanning imaging system; Carl Zeiss Microimaging Inc., Thornwood, NY) that allows a precise analysis of the emission spectrum for each fluorochrome (7). The cells were excited at 488 nm, and emission peaks were readily separated by this methodology, with EGFP emission at 500 nm and doxorubicin emission at 600 nm (see Fig. S7 in the supplemental material).
Immunofluorescence staining by confocal microscopy.
MCF7, MCF7/ADR, MCF7/siMCJ-1B, MCF7/siMCJ-3B, and 293T cells were plated onto BD BioCoat poly-d-lysine coverslips. Cells were washed with PBS, fixed in 3.7% paraformaldehyde, and permeabilized in a blocking solution (Dulbecco's modified Eagle's medium-F12 with 5% FBS and 0.01% lysine) containing 0.1% Triton X-100 for 2 h at room temperature. Cells were then incubated with primary antibody in blocking solution for 1 h at room temperature, followed by incubation with the Alexa-568 anti-rabbit or Alexa-568 anti-mouse secondary antibody (Molecular Probes, Eugene, OR) for 30 min. For nuclear staining, either TOPRO-3 or Yoyo (Molecular Probes, Eugene, OR) was used, and Mitotracker-647 (Molecular Probes, Eugene, OR) was used to stain mitochondria. The primary antibodies used included the mouse anti-HA monoclonal antibody (mAb; Cell Signaling Technology, Inc.). The rabbit anti-MCJ polyclonal antibody was generated by using recombinant mouse MCJ (aa 62 to 150; Proteintech Group Inc., Chicago, IL). The rabbit antiserum was further purified over an antigen column to obtain the MCJ antibody. The mouse anti-MCJ mAb (WeNA12) was generated at the Green Mountain Antibodies Co. by immunizing mice with the N terminus peptide of MCJ. Screening of over 200 hybridomas was performed by immunostaining of MCJ-transfected and untransfected 293T cells and confocal microscopy searching for specificity and the expected subcellular distribution.
The semipermeabilization method of freeze-thaw was modified from a previously described method (39), and the staining was performed as mentioned above. Immunostained cells were examined by confocal microscopy using the LSM 510 META confocal laser scanning imaging system (Carl Zeiss Microimaging Inc., Thornwood, NY).
RNA isolation and RT-PCR.
Total RNA was isolated using Ultraspec RNA isolation systems as recommended by the manufacturer (Biotecx Laboratories, Inc., Houston, TX). The first strand of cDNA was obtained by reverse transcription as described previously (10). cDNA was used to detect human ABCB1, hypoxanthine phosphoribosyltransferase (HPRT), MCJ, ABCG2, and ABCC1 gene expression by conventional PCR or real-time reverse transcriptase PCR (RT-PCR). Previously described primers were used for the conventional RT-PCR amplification of ABCB1 (48), HPRT (10), ABCC1 (26), ABCG2 (16), and MCJ (56) genes. For the real-time RT-PCR analysis with the TaqMan system (Applied Biosystems), we used an assay-on-demand kit for human HPRT, ABCB1, and c-Jun genes (Sigma Genosys). For the detection of MCJ by real time RT-PCR, we used the following probe and primer set (Sigma Genosys): probe, 5′-CCTTGCCAGCAGATGGGCTTACACCTAAA-3′; sense primer, 5′-CAGAAAATGAGTAGGCGAGAAGC-3′; and antisense primer, 5′-TGAC TCTCCTATGAGCTGTTCTAATC-3′. The HPRT gene was used as an endogenous control to normalize the mRNA values in each sample. The relative values were determined by the comparative computed tomography analysis method.
Western blot analysis.
Whole-cell extracts were prepared in lysis buffer as previously described (51). Forty to 100 μg of protein was separated by electrophoresis and transferred onto a nitrocellulose membrane (10). Primary antibodies for Western blotting included anti-ABCB1 (JSB-1 clone; ALEXIS Corporation, Switzerland); rabbit polyclonal anti-MCJ, mouse anti-MCJ mAb, anti-HA, anti-Jun N-terminal protein kinase (anti-JNK), and phospho-JNK (Cell Signaling Technology, Inc.); and anti-c-Jun, antiactin, and the secondary antibodies goat anti-mouse horseradish peroxidase (HRP), goat anti-rabbit HRP, and donkey anti-goat HRP (Santa Cruz Biotechnology). The LumiGLO chemiluminescent substrate system (KPL, MD) was used to visualize the proteins. Levels of actin were determined as a loading control. For the coimmunoprecipitation of MCJ and c-Jun, whole-cell extracts from MG132-treated and untreated MCF7 cells were immunoprecipitated by using the anti-MCJ mAb as previously described (50). Immunoprecipitates were used to perform Western blot analysis using an anti-c-Jun antibody.
Immunoelectron microscopy.
293T cells were transfected with a plasmid containing HA-MCJ. After 24 h, transfected cells were fixed by immersion in 3% paraformaldehyde containing 0.1% glutaraldehyde, washed, and resuspended in 0.05 M ammonium chloride. Cells were embedded in agarose and refixed in paraformaldehyde-glutaraldehyde, washed, and dehydrated at a lower temperature in increasing concentrations of ethanol, followed by infiltration and embedding in the hydrophilic resin Lowicryl K4M at −35°C (30). Immunostaining was performed with anti-HA mAb (Cell Signaling) during overnight incubation at 4°C, followed by a secondary anti-mouse antibody and protein A-gold particles (10 nm). Contrasting was done using 3% aqueous uranyl acetate and lead citrate. Sections were examined using a JEOL 1210 transmission electron microscope (JEOL-USA, Inc., Peabody, MA).
Phylogenetic analysis.
BLASTp and PSI-BLAST (3) were used to search for homologous protein sequences in the GenBank (www.ncbi.nlm.nih.gov) nr protein database. A search with the human MCJ ortholog (accession number GI66472920) found 99 proteins (expect value, 0.001). A total of 47 sequences were used in the analysis, all of which were eukaryotic. The protein sequences were aligned using the T-Coffee program (49) with standard parameters. Five evolutionary clades (i.e., groups of sequences with a common ancestor) were identified: plant, yeast TIM14, fly-worm TIM14-like, vertebrate MCJ, and vertebrate TIM14-like clades. The level of confidence in each clade was determined by three parameters: (i) bootstrap support under maximum parsimony, (ii) bootstrap support using the neighbor-joining algorithm, and (iii) the posterior probability obtained from Markov Chain Monte Carlo simulation using the MrBayes program. The plant clade was set as the out-group. The confidence level for the yeast TIM14 group of proteins in fungi as a clade was 30/66/not applicable (NA) according to the above-listed parameters, and the confidence level for the ecdysozoan (fly and worm) MCJ and TIM-like sequences as a clade was 86/95/1.00. The confidence level for a gene duplication's leading to two vertebrate clades, the MCJ and the TIM14-like clades, was 86/95/1.00 (see Fig. 2B). A low confidence level (NA/54/NA) was obtained for a gene duplication event that occurred after the divergence of vertebrates from the fly-worm (ecdysozoan) lineage. Stronger support was found for MCJ's being the result of gene duplication prior to the divergence of ecdysozoa from vertebrates (50/NA/0.99). Phylogenetic analysis was performed using MrBayes 3.1 (28) for Bayesian methods and PROTPARS and PROTDIST/NEIGHBOR from the PHYLIP 3.6 package (21) for maximum parsimony and neighbor-joining methods, respectively.
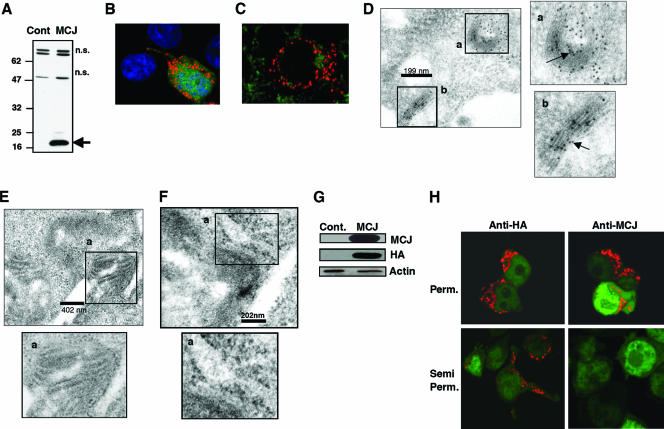
FIG. 2.
Localization of MCJ in the Golgi apparatus. (A) 293T cells were transfected with a plasmid expressing HA-tagged MCJ (MCJ) or an empty plasmid (Cont). Whole-cell extracts were examined for MCJ expression by Western blot analysis using an anti-HA mAb. MCJ is indicated with an arrow (n.s., nonspecific). Numbers on the left are molecular size markers. (B) 293T cells were transfected with an MCJ-expressing plasmid. Transfected cells were fixed, permeabilized, and stained. The MCJ intracellular distribution was examined by confocal microscopy. MCJ (red), EGFP (green), and TOPRO-3 nuclear stain (blue) are shown. (C) MCJ-transfected 293T cells were stained for MCJ (red) and Mitotracker (green; Molecular Probes). (D, E, and F) MCJ-transfected 293T cells were fixed and used to examine MCJ localization by immunoelectron microscopy. The presence of MCJ is shown by black immunodense particles (arrows). Scale bars are given for each set of panels. Areas in the micrographs that include the Golgi apparatus (D), mitochondria (E), and endoplasmic reticulum (F) are presented. The insets (a and b) show the specific organelles and have been magnified (side and bottom panels) for better resolution. (G) 293T cells were transfected with an MCJ-expressing plasmid (MCJ) or an empty plasmid (Cont). Whole-cell extracts were examined for MCJ expression by Western blotting using an anti-MCJ polyclonal antibody, an anti-HA antibody, and an antiactin antibody as a loading control. (H) 293T cells were transfected with a plasmid containing HA-tagged MCJ and EGFP. Either complete permeabilization with Triton X-100 (Perm.) or semipermeabilization with a rapid freeze-thaw procedure (Semi Perm.) was done, followed by fixation and staining with anti-HA or anti-MCJ antibody. The EGFP-positive (green) cells are the transfected cells.
Nuclear extracts and the electromobility shift assay (EMSA).
Mini nuclear extracts were made from cells as described previously (54). Binding reactions were done using 2 μg of nuclear protein in the presence of specific 32P-end-labeled double-stranded oligonucleotides as described previously (54). The oligonucleotides used in this study were as follows: AP-1 (4, 36), C/EBP (33), and NF-κB (31) gene sequences. The gel shift assay was performed using 1 μl of the anti-c-Jun, c-Fos, JunB, and Jun family antibodies (Santa Cruz).
RESULTS
MCJ is a unique transmembrane DnaJ protein, highly conserved in vertebrates.
Although some studies have examined the regulation of the MCJ gene by methylation, the biology and function of MCJ protein remain unknown. We therefore performed a PSI-BLAST search (2) by using the human MCJ sequence to examine the potential evolutionary association of MCJ with other proteins of known functions. The search revealed that MCJ is a member of a set of eukaryotic proteins that contain a conserved (66 to 100%) transmembrane domain and the C-terminal DnaJ domain (Fig. 1A; see Fig. S1 in the supplemental material). This set includes the previously described yeast TIM14, a component of a mitochondrial inner membrane translocase (42) (Fig. 1A). It also includes an uncharacterized human DnaJ protein that has been described as a “translocase of mitochondrion inner membrane” because of its high level of similarity (67%; P, 4 × 10−15) to the yeast TIM14. This TIM14 human ortholog, which will be referred to herein as a TIM14-like protein, is similar to human MCJ (74%; P, 6 × 10−32) but lacks the corresponding N terminus (Fig. 1A). The sequence similarity searches indicate no other human proteins within this eukaryotic phylogeny (see Fig. S1 in the supplemental material).
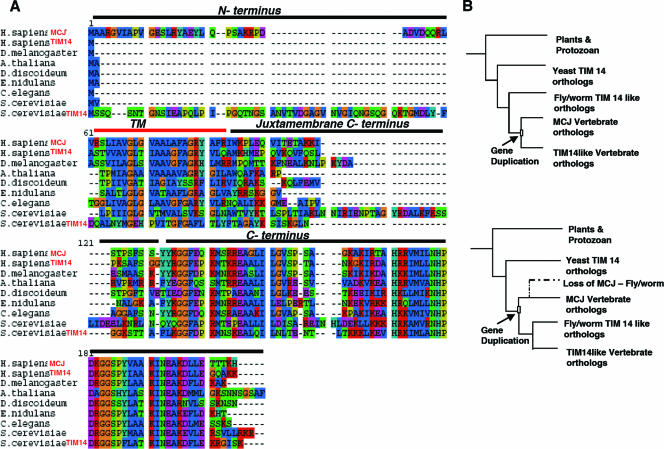
FIG. 1.
Phylogenetic analysis and sequence alignment of MCJ. (A) Homologous MCJ sequences from different species found by a BLASTp search. Alignments were made using the T-Coffee program. H. sapiens, Homo sapiens; D. melanogaster, Drosophila melanogaster; A. thaliana, Arabidopsis thaliana; D. discoideum, Dictyostelium discoideum; E. nidulans, Emericella nidulans; C. elegans, Caenorhabditis elegans; S. cerevisiae, Saccharomyces cerevisiae. (B) Possible scenarios for the gene duplication event that resulted in the evolution of MCJ in vertebrates. The models for gene duplication after the divergence of the fly-worm TIM14-like clade from the vertebrates (top panel) and gene duplication prior to the divergence of the fly-worm TIM14-like clade from the vertebrate clades with subsequent possible gene loss in the fly-worm lineage (bottom panel) are shown.
Five evolutionary clades (i.e., groups of sequences with a common ancestor) (Fig. 1B) were identified: plant, yeast TIM14, fly-worm TIM14-like, vertebrate TIM14-like, and vertebrate MCJ clades. The plant clade was set as the out-group, meaning that the other four clades share with one another a common ancestor that is not shared with the plant clade (Fig. 1B). The ecdysozoan (fly and worm) MCJ and TIM14-like sequences form a clade. We found with a high confidence level a gene duplication leading to two vertebrate clades, the MCJ and the TIM14-like clades (Fig. 1B). Although gene duplication may have occurred after the divergence of vertebrates from the fly-worm (ecdysozoan) lineage (Fig. 1B, upper panel), this possibility was not strongly supported by the results of our analysis (described in Materials and Methods). Stronger support was found for MCJ to be the result of gene duplication prior to the divergence of ecdysozoa from vertebrates (Fig. 1B, lower panel). This scenario would imply that MCJ must then have been lost in the fly-worm lineage(s).
Unlike the transmembrane and C-terminal DnaJ regions that are highly conserved within the five clades, the juxtamembrane C-terminal regions are distinct among clades, although they are conserved within each clade (Fig. 1A). Thus, vertebrate MCJ orthologs have a unique juxtamembrane C-terminal region that is not conserved in other clades. In addition, the N terminus region (35 aa) present in the MCJ clade is absent in the TIM14-like vertebrate and ecdysozoan clades and is highly variable in the yeast TIM14 clade (Fig. 1A). However, within the MCJ clade, seven sites of this N terminus are perfectly conserved and more than 90% of the sites exhibit some degree of conservation. The presence of the highly conserved N-terminal region specifically in the MCJ clade, but not in the TIM14-like vertebrate clade, suggests distinct functions of MCJ and TIM14 in vertebrates.
MCJ is a type II transmembrane protein localized in the Golgi compartment.
The above-described MCJ phylogenetic analysis revealed the evolutionary ancestor of MCJ as yeast TIM14, which is localized in the inner mitochondrial membrane (42). Since no previous studies have addressed the subcellular localization of MCJ, we generated a HA-tagged-MCJ-expressing construct that also contained an internal ribosome entry site-EGFP gene and transfected 293T cells with this construct. We first tested MCJ expression in transfected cells by Western blot analysis. In correlation with the predicted size of MCJ, a protein of approximately 16 to 18 kDa was detected only in MCJ-transfected 293T cells and not in cells transfected with an empty plasmid (Fig. 2A).
MCJ subcellular localization in transfected cells was examined by confocal microscopy analysis. MCJ was clearly a cytoplasmic protein with a distinct punctate distribution in well-defined areas of the cytosol that resembled the distribution of intracellular organelles (Fig. 2B). No MCJ staining in the untransfected cells was observed (Fig. 2B). To investigate whether MCJ was localized in the mitochondria, we costained MCJ-transfected cells with Mitotracker, a specific marker for mitochondria. However, MCJ did not colocalize with the Mitotracker (Fig. 2C). To test if MCJ was localized in the endoplasmic reticulum, cells were cotransfected with the MCJ-expressing plasmid and the pDsRed2-ER plasmid that expresses a red fluorescence protein targeted to the endoplasmic reticulum by the endoplasmic reticulum retention sequence (KDEL). No clear colocalization of MCJ with the pDsRed2-ER plasmid was observed by confocal microscopy (see Fig. S2 in the supplemental material). To precisely determine the organelle(s) where MCJ was localized, we performed immunoelectron microscopy. MCJ-transfected 293T cells were fixed, embedded, sectioned, and stained with antibody-coated gold particles (pAg10). A clear punctate distribution of MCJ specifically in the Golgi apparatus and other associated vesicles was visualized (Fig. 2D). No MCJ localization in the mitochondria (Fig. 2E), endoplasmic reticulum (Fig. 2F), or nuclear membrane (data not shown) was detected. Thus, MCJ is an intracellular transmembrane protein localized primarily in the Golgi apparatus and associated vesicles.
The superfamily Hidden Markov model protein topology prediction program predicted that MCJ was a type II transmembrane protein (i.e., a protein with an intracellular N terminus and an extracellular C terminus). Since our immunoelectron microscopy studies showed that MCJ was present in the Golgi apparatus, this prediction would suggest that the MCJ C terminus was in the Golgi lumen whereas the N terminus was cytoplasmic. To confirm this prediction and further characterize the orientation of MCJ, we used the permeabilization-semipermeabilization method previously described (39). This approach is based on comparative epitope accessibilities to the antibody for the detection of intracellular transmembrane protein topology. The classical method of immunostaining involves the permeation of fixed cells with a detergent (e.g., Triton X-100) that allows the antibodies to detect all intracellular proteins independently of their localization patterns. However, the semipermeabilization method uses a rapid freeze-thaw technique that selectively permeates the plasma membrane while the intracellular membranes remain impermeable. Since the HA tag was present at the N terminus of MCJ, we used the anti-HA antibody for the detection of the MCJ N-terminal region. For the detection of the C terminus, we generated a rabbit polyclonal antibody against the C terminus of MCJ. The specificity of this anti-MCJ antibody was examined by Western blot analysis using extracts from HA-MCJ-transfected and untransfected 293T cells. The anti-MCJ antibody was able to detect MCJ only in the MCJ-transfected 293T cells (Fig. 2G). The specificity of the detected band of MCJ was further demonstrated by reprobing the blot with anti-HA antibody (Fig. 2G).
We therefore used both anti-HA and anti-MCJ antibodies to detect MCJ by confocal microscopy analysis. 293T cells were transfected with an HA-MCJ-expressing plasmid, fixed, permeabilized, stained with either anti-HA or anti-MCJ antibody, and analyzed by confocal microscopy. Both anti-HA and anti-MCJ antibodies detected MCJ with similar patterns of expression in the transfected cells (Fig. 2H). In parallel, HA-MCJ-transfected cells were rapidly freeze-thawed for semipermeabilization, fixed, stained with anti-HA and anti-MCJ antibodies, and analyzed by confocal microscopy. MCJ was detected with the anti-HA antibody but not with the anti-MCJ antibody (Fig. 2H), indicating that the MCJ C terminus was not accessible to the antibody. These results confirm that the N terminus of MCJ is cytosolic whereas the C terminus resides within the Golgi lumen. Thus, MCJ is a type II cochaperone that resides within the Golgi compartment.
MCJ is expressed in drug-sensitive but not in drug-resistant breast cancer cells.
The loss of MCJ expression has been associated with increased resistance to chemotherapeutic drugs in ovarian cancer cells (56, 60, 61). Multidrug resistance is a common phenomenon observed in other cancer types such as breast cancer. To investigate whether the loss of MCJ expression in drug-resistant cells was extended to cancer types other than ovarian, we examined MCJ expression in breast cancer cells. We compared MCJ expression in drug-sensitive MCF7 breast cancer cells with MCJ expression in MCF7/ADR cells that are derived from MCF7 cells but are resistant to several drugs, including doxorubicin, paclitaxel, and vincristine (18). Total RNA was isolated from MCF7 and MCF7/ADR cells, and MCJ expression was examined by conventional RT-PCR using HPRT as an internal control. MCJ was expressed at high levels in MCF7 cells but was undetectable in MCF7/ADR cells (Fig. 3A). Similar results were obtained by quantitative real-time RT-PCR analysis of MCJ (Fig. 3B).
FIG. 3.
Loss of MCJ gene expression in multidrug-resistant breast cancer cells. (A) Total RNA was extracted from MCF7 and MCF7/ADR breast cancer cells and examined for the expression of MCJ and HPRT genes by RT-PCR. (B) The expression of the MCJ gene relative to that of the HPRT gene in MCF7 and MCF7/ADR cells was examined by quantitative real-time RT-PCR. (C) Total RNA was extracted from MCF7 and MCF7/IL-6 cells and examined for the expression of MCJ and HPRT genes by RT-PCR. (D) Total RNA was extracted from MCF7 cells cultured for 8 days in medium alone or in the presence of IL-6 (50 ng/ml). MCJ and HPRT gene expression was examined by RT-PCR. (E) MCF7 cells were cultured as described in the legend to panel D, and MCJ gene expression relative to that of HPRT was analyzed by real-time RT-PCR. (F and G) Total RNA was extracted from MDA-MB-321 (MDA) and MD22 breast cancer cells (F) and uterine cancer cells MES-SA (MES) and MES-SA/Dx5 (MES/Dx) (G) and examined for the expression of MCJ and HPRT genes by RT-PCR.
To confirm the loss of MCJ expression in multidrug-resistant cells, we used other MCF7-derived cells with a multidrug-resistant phenotype. We have previously shown that multidrug-resistant but not drug-sensitive breast cancer cells produce interleukin-6 (IL-6) and that the stable expression of IL-6 in MCF7 cells confers multidrug resistance (10). We therefore isolated total RNA from MCF7 cells and MCF7 cells stably expressing IL-6 (MCF7/IL-6 cells) and examined MCJ expression by RT-PCR. MCJ was expressed in MCF7 cells but not in the multidrug-resistant MCF7/IL-6 cells (Fig. 3C). We have also shown that the transient treatment of MCF7 cells with exogenous IL-6 increases the drug resistance of these cells (10). We tested whether this increased resistance was associated with decreased MCJ expression. MCF7 cells that were treated with exogenous IL-6 for 1 week contained reduced levels of MCJ mRNA compared with untreated MCF7 cells (Fig. 3D and E).
To rule out the possibility that the correlation between reduced MCJ expression and chemoresistance was restricted to this breast cancer cell line, we examined another breast cancer cell line, MDA-MB-321, and its doxorubicin-resistant derivative, the MD22 cell line (32). Unlike the multidrug-resistant MCF7/ADR cells, MD22 cells are resistant specifically to doxorubicin. We isolated RNA and performed RT-PCR to assess MCJ and HPRT expression. While MDA-MB-321 cells expressed high levels of MCJ, very low levels were detected in MD22 cells (Fig. 3F). To examine whether MCJ expression was also blocked in other multidrug-resistant cells, we examined the drug-sensitive MES-SA uterine cancer cell line and its multidrug-resistant derivative, the MES-SA/Dx5 cell line (27), by RT-PCR. As in MCF7 cells, MCJ was expressed in the MES-SA cells, but its expression was undetectable in MES-SA/Dx5 cells (Fig. 3G). Thus, the loss of MCJ gene expression in drug-resistant cells may be a common phenomenon in a number of solid tumors, suggesting that MCJ can be a multidrug resistance marker independent of the cancer type.
Since the sensitivity of the rabbit anti-MCJ polyclonal antibody was limited for the detection of endogenous MCJ (data not shown), we generated a monoclonal antibody against the N-terminal peptide (aa 1 to 35) of MCJ that has no homology with corresponding regions of other mammalian proteins, as described above (Fig. 1). To first test the specificity of the anti-MCJ mAb and its ability to recognize MCJ, we performed Western blot analysis using whole-cell extracts from MCJ-transfected and untransfected 293T cells. A unique band of the corresponding size was detected exclusively in MCJ-transfected cells (Fig. 4A). To further confirm the specificity of the anti-MCJ mAb, we performed confocal microscopy with MCJ-transfected and untransfected 293T cells. Anti-MCJ reactivity was detected only in the transfected and not in the untransfected cells (Fig. 4B), and the pattern of expression was similar to that observed with the antitag and the polyclonal anti-MCJ antibodies (Fig. 2B).
FIG. 4.
Loss of MCJ in multidrug-resistant breast cancer cells. (A) 293T cells were transfected with an MCJ-expressing plasmid (MCJ) or a control plasmid (Con) and lysed, and MCJ expression was determined by Western blot analysis using an anti-MCJ mAb (MCJ). Actin was examined as a loading control. (B) 293T cells were transfected with MCJ-expressing (MCJ) or control plasmids, fixed, stained using an anti-MCJ mAb (red), and examined by confocal microscopy. TOPRO (blue) was used as a nuclear dye. EGFP (green) denotes the transfected cells. The middle panel is a magnification (zoom) of a section of the upper panel. (C) MCJ expression in MCF7, MCF7/ADR (ADR), MCF7/IL-6 (M/IL-6), MDA-MB-321 (MDA), and MD22 cells was examined by Western blotting using whole-cell extracts and the anti-MCJ mAb (MCJ). Actin was used as a loading marker. (D) MCF7 and MCF7/ADR cells were fixed, stained with the anti-MCJ mAb (MCJ; red) or secondary antibody (control), and examined by confocal microscopy. TOPRO was used as a nuclear dye (blue). Magnified images of the anti-MCJ-stained MCF7 and MCF7/ADR cells are shown in the middle panels. (E) MCF7 cells were stained with a mouse anti-MCJ mAb and a rabbit anti-TGN46 antibody. TOPRO was used as a nuclear dye. Images show only MCJ staining (red), TGN46 staining (green), and the staining of both MCJ and TGN46 (MCJ+TGN) for the visualization of colocalization (orange).
We used the anti-MCJ mAb to further demonstrate the selective expression of MCJ in drug-sensitive breast cancer cells but not in drug-resistant cells by Western blot analysis. A distinct band of a size corresponding to MCJ was present in MCF7 cells but not in multidrug-resistant MCF7/ADR and MCF7/IL-6 cells (Fig. 4C). No other cross-reactive proteins in MCF7 cells could be detected with this mAb. In addition, MCJ was present in the drug-sensitive MDA-MB-231 breast cancer cells while only very low MCJ levels were detected in the doxorubicin-resistant MD22 cells (Fig. 4C). Thus, the expression of MCJ correlated with the MCJ mRNA levels detected in these cells, further demonstrating the loss of MCJ in drug-resistant breast cancer cells.
We also investigated the subcellular distribution of endogenous MCJ in MCF7 cells by immunostaining and confocal microscopy using the anti-MCJ mAb. Although lower than that in MCJ-overexpressing 293T cells, the level of expression of endogenous MCJ in MCF7 cells was clearly detectable (Fig. 4D). No MCJ could be detected in MCF7/ADR cells (Fig. 4D). Endogenous MCJ also showed a punctate distribution pattern in MCF7 cells, although it was more widely distributed through the cytoplasm. Several studies have shown that the Golgi compartments of MCF7 cells are diffusely localized throughout the cytoplasm (5, 40, 43, 59). We examined the colocalization of MCJ with the human trans-Golgi network protein TGN46 that has been previously used to detect the Golgi network in MCF7 cells (59). Confocal microscopy analysis showed that MCJ partially colocalized with TGN46 (Fig. 4E). Together, these results indicate that endogenous MCJ is localized in the Golgi compartment in drug-sensitive breast cancer cells but its expression is lost in multidrug-resistant cells.
MCJ is required for breast cancer cells to maintain the response to chemotherapeutic drugs.
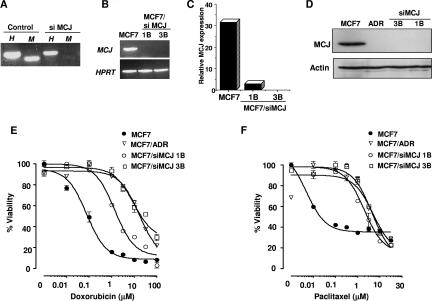
To address whether the absence of this Golgi compartment-associated protein by itself could induce multidrug resistance in breast cancer cells, we examined the effect of the inhibition of MCJ expression by RNA interference. An siRNA MCJ target sequence (siMCJ) was cloned downstream of the H1 RNA polymerase III promoter in pSuperEGFP, a modified version of the pSuper plasmid (6) that includes the EGFP gene the under control of the CMV promoter. MCF7 cells were transiently transfected with pSuperEGFP-siMCJ or an empty plasmid. Cells transfected with siMCJ had lower levels of MCJ mRNA than cells transfected with the empty plasmid, as determined by RT-PCR (Fig. 5A). We then performed the stable transfection of MCF7 cells with the pSuperEGFP-siMCJ construct, generating MCF7/siMCJ cells. Two clones, MCF7/siMCJ-1B and MCF7/siMCJ-3B, were selected for further expansion and characterization. Total RNA was isolated from MCF7, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells and examined for MCJ gene expression. No MCJ mRNA was detected in MCF7/siMCJ-3B and MCF7/siMCJ-1B cells by conventional RT-PCR (Fig. 5B), and very low levels were detected only in MCF7/siMCJ-1B cells by quantitative real-time RT-PCR (Fig. 5C).
FIG. 5.
MCJ is required for breast cancer cells to maintain a chemotherapy response. (A) MCF7 cells were transfected with the empty pSuperEGFP (control) or pSuperEGFP-siMCJ (siMCJ) plasmid. Total RNA was extracted 36 h after transfection, and MCJ (M) and HPRT (H) gene expression was examined by RT-PCR. (B) Total RNA from MCF7, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells was extracted and used to examine MCJ and HPRT gene expression by RT-PCR. (C) Total RNA isolated as described in the legend to panel B was used to examine MCJ gene expression relative to that of HPRT by real-time RT-PCR analysis. (D) Endogenous MCJ expression was examined by Western blot analysis. Whole-cell extracts from MCF7, MCF7/ADR (ADR), and MCF7/siMCJ (siMCJ)-1B and -3B cells were examined for MCJ with the anti-MCJ mAb. Actin expression was examined as a loading control. (E and F) Cell viability was determined by the MTT assay. LD50 were calculated by nonlinear regression. (E) LD50 of doxorubicin were 0.078 μM (MCF7), 16.60 μM (MCF7/ADR), 1.35 μM (MCF7/siMCJ-1B), and 9.29 μM (MCF7/siMCJ-3B). (F) LD50 of paclitaxel were 0.05 μM (MCF7), 4.5 μM (MCF7/ADR), 2.0 μM (MCF7/siMCJ-1B), and 4.99 μM (MCF7/siMCJ-3B). Results representative of those from four individual experiments are shown.
To confirm that the expression of siMCJ in MCF7/siMCJ cells abolishes not only MCJ mRNA expression but also protein expression, we examined endogenous MCJ levels by Western blotting using the anti-MCJ mAb. MCJ was clearly present in MCF7 cells, but it was almost undetectable in MCF7/siMCJ-1B and MCF7/siMCJ-3B cells, as well as in MCF7/ADR cells (Fig. 5D) These data indicate that MCJ mRNA and protein expression were abolished in MCF7/siMCJ cells.
The MCF7/siMCJ-1B and -3B cells had a rate of proliferation similar to that of MCF7 cells, and no difference in the viability of these cells in culture was observed (data not shown). We examined whether the inhibition of MCJ expression could increase resistance to doxorubicin (anthracycline), a commonly used chemotherapeutic drug for breast cancer treatment. MCF7, MCF7/ADR, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were cultured in the absence or presence of different concentrations of doxorubicin and the percent viability was measured by the MTT assay. In correlation with results from previous studies (1), MCF7 cells were highly sensitive to doxorubicin while MCF7/ADR cells were highly resistant (Fig. 5E). Interestingly, MCF7/siMCJ-1B and MCF7/siMCJ-3B cells were significantly more resistant to doxorubicin than MCF7 cells (Fig. 5E).
We also tested the response to paclitaxel (taxane), another commonly used chemotherapeutic drug for breast cancer. MCF7 cells were highly sensitive to paclitaxel (Fig. 5F). In contrast, MCF7/siMCJ-1B and MCF7/siMCJ-3B cells were highly resistant, similar to MCF7/ADR cells (Fig. 5F). Together, these results demonstrate that MCJ is required for breast cancer cells to respond to different drugs such as doxorubicin and paclitaxel and that the inhibition of MCJ expression causes multidrug resistance.
MCJ is required for the intracellular accumulation of chemotherapeutic drugs.
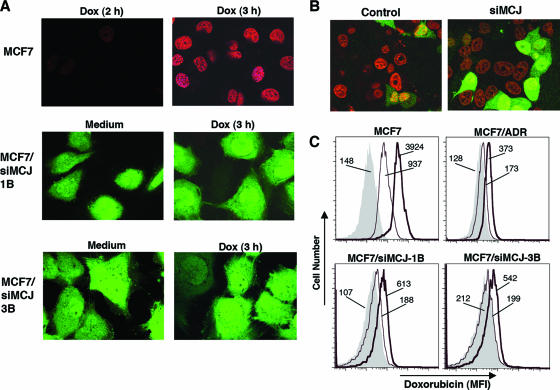
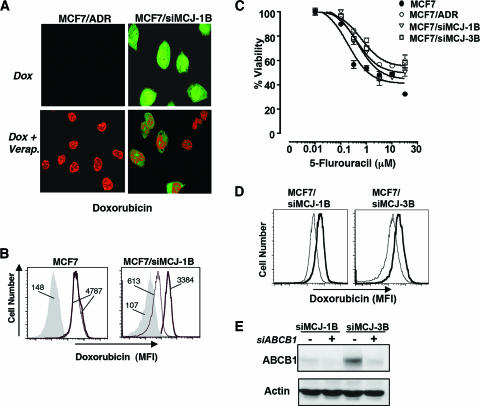
Impaired intracellular accumulation of chemotherapeutic drugs due to transport-mediated efflux is the best-characterized mechanism involved in multidrug resistance (38). We examined the effect of MCJ downregulation on the intracellular accumulation of doxorubicin by confocal microscopy. MCF7, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were treated with medium alone or with doxorubicin for 1, 2, or 3 h. Cells were then washed and fixed, and doxorubicin fluorescence was visualized by confocal microscopy. No doxorubicin fluorescence was detected in MCF7 cells treated with medium alone or with doxorubicin for only 1 h (data not shown). After 2 h of treatment, some doxorubicin fluorescence was detected in the MCF7 cells, but the maximum level of intracellular accumulation was reached after 3 h (Fig. 6A). Both MCF7/siMCJ-1B and MCF7/siMCJ-3B cells expressed EGFP, but no doxorubicin fluorescence was observed in these cells after 3 h of treatment (Fig. 6A). Similarly, no doxorubicin fluorescence in MCF7/siMCJ cells was detected after shorter (1- and 2-h) and longer (4-h) periods of treatment (data not shown).
FIG. 6.
MCJ is required for the intracellular accumulation of doxorubicin. (A) MCF7, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were incubated in the absence (medium) or the presence (Dox) of doxorubicin (3 μM) for the indicated periods of time. Doxorubicin (red) intracellular accumulation was detected by confocal microscopy. MCF7/siMCJ-1B and -3B cells expressed EGFP (green). (B) MCF7 cells were transiently transfected with the empty pSuperEGFP plasmid (control) or the pSuperEGFP-siMCJ plasmid (siMCJ). Thirty-six hours after the transfection, cells were treated with doxorubicin for 3 h and its intracellular accumulation (red) in untransfected (EGFP-negative) and transfected (EGFP-positive) cells was examined by confocal microscopy. (C) MCF7, MCF7/ADR, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were treated with medium alone (gray-filled profiles) or with doxorubicin at 0.3 μM (thin-line profiles) or 3 μM (thick-line profiles) for 3 h. Doxorubicin intracellular accumulation was examined by flow cytometry. Numbers represent the mean fluorescence intensities (MFI) of doxorubicin.
To demonstrate that this phenotype was due to the inhibition of MCJ expression rather than the selection of the siMCJ cell clones, MCF7 cells were transiently transfected with either an empty pSuperEGFP plasmid or the pSuperEGFP-siMCJ plasmid. Thirty-six hours after transfection, cells were treated with doxorubicin for 3 h and examined by confocal microscopy analysis. Transfected cells were identified by the presence of EGFP. Both EGFP-positive and EGFP-negative cells among the control plasmid-transfected MCF7 cells showed doxorubicin accumulation (Fig. 6B). In contrast, doxorubicin fluorescence could be detected only in EGFP-negative, not EGFP-positive, siMCJ-transfected MCF7 cells (Fig. 6B). Thus, the transient inhibition of MCJ expression interferes with the intracellular accumulation of the drug.
To confirm the confocal microscopy results, we examined the intracellular accumulation of doxorubicin by flow cytometry. MCF7, MCF7/ADR, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were treated with doxorubicin (0.3 and 3 μM) for 3 h, washed extensively, and examined by flow cytometry. High levels of doxorubicin were present in MCF7 cells even at the lower dose (Fig. 6C). In contrast, no intracellular accumulation of doxorubicin could be detected in MCF7/siMCJ-1B and -3B cells at the lower dose of doxorubicin (0.3 μM) and very low intracellular levels were detected at the higher dose (3 μM) (Fig. 6C). In addition, no doxorubicin in MCF7/ADR cells was observed (Fig. 6C). The intracellular accumulation of doxorubicin was also impaired in other cells that have lost MCJ, including MCF7/IL-6 and MES/DOX cells (Fig. S3 in the supplemental material) Together, these results demonstrate that the presence of MCJ is required to allow the intracellular accumulation of the drug.
MCJ suppresses ABCB1 gene expression.
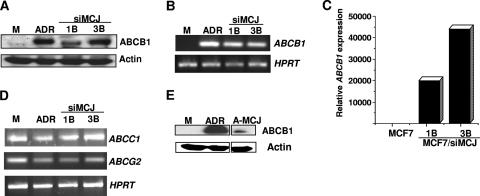
Specific ATP-binding cassette (ABC) transporters that promote drug efflux or drug retention in intracellular compartments of cancer cells provide one of the mechanisms to prevent drugs from reaching their specific intracellular targets. The ABC transporters constitute a large family, with 48 members in humans. Some ABC transporters that use specific drugs as substrates are overexpressed in the cancer cell lines and tumors that are multidrug resistant (24). The substrates of a large number of these transporters, however, remain unknown. The best-characterized member of this family is ABCB1 (also known as the mdr1 or P-glycoprotein). Since ABCB1 is known to be absent in drug-sensitive MCF7 cells (19), we examined its expression in MCF7/siMCJ cells by Western blot analysis. In contrast to the lack of ABCB1 in MCF7 cells, high levels of ABCB1 were present in MCF7/siMCJ-1B and -3B cells (Fig. 7A). As previously described, high levels of ABCB1 were also present in MCF7/ADR cells (Fig. 7A).
FIG. 7.
MCJ suppresses ABCB1 gene expression. (A) Whole-cell extracts from MCF7 (M), MCF7/ADR (ADR), and MCF7/siMCJ (siMCJ)-1B and -3B cells were used to examine ABCB1 expression by Western blot analysis. Actin was also examined as a loading control. (B) Total RNA extracted from MCF7 (M), MCF7/ADR (ADR), and MCF7/siMCJ-1B (siMCJ 1B) and -3B cells was used to examine ABCB1 and HPRT gene expression by RT-PCR. (C) Quantitative real-time RT-PCR analysis of ABCB1 gene expression relative to that of HPRT in MCF7 and MCF7/siMCJ-1B and -3B cells. (D) Total RNA was extracted from MCF7 (M), MCF7/ADR (ADR), and MCF7/siMCJ-1B (siMCJ 1B) and -3B cells and used to examine ABCC1, ABCG2, and HPRT gene expression by RT-PCR. (E) Whole-cell extracts from MCF7 (M), MCF7/ADR (ADR), and MCF7/ADR-MCJ (A-MCJ) cells were used to examine ABCB1 expression by Western blot analysis. Actin expression was examined as a loading control.
To determine whether the effect of MCJ on ABCB1 levels could be due to changes in the expression of the ABCB1 gene, we measured ABCB1 mRNA levels by conventional RT-PCR. The ABCB1 gene was not expressed in MCF7 cells (Fig. 7B), but it was highly expressed in MCF7/siMCJ-1B and -3B cells (Fig. 7B). Similar results were obtained by quantitative real-time RT-PCR (Fig. 7C). In addition, the inhibition of MCJ expression in MCF7 cells by transient transfection with the pSuperEGFP-siMCJ plasmid caused an upregulation of ABCB1 gene expression (data not shown). Unlike that of ABCB1, the expression of other multidrug ABC transporters like ABCC1 (for multidrug resistance-associated protein) (15) and ABCG2 (for breast cancer resistance protein) (16) that are expressed in MCF7 cells was not altered in MCF7/siMCJ cells (Fig. 7D). Thus, MCJ appears to selectively regulate ABCB1 gene expression.
To further demonstrate the negative role of MCJ in ABCB1 gene expression, we examined whether the expression of MCJ in MCF7/ADR cells downregulates ABCB1 expression. MCF7/ADR cells were transfected with an MCJ-expressing plasmid. Stably MCJ-transfected MCF7/ADR clones (MCF7/ADR-MCJ) were selected. The expression of MCJ did not affect the survival or the proliferation of these cells (data not shown). We examined the ABCB1 expression in these cells by Western blot analysis. Although not totally abolished, the ABCB1 levels in MCF7/ADR-MCJ were reduced compared with those in MCF7/ADR cells (Fig. 7E). Consistent with the reduced levels of ABCB1, MCF7/ADR-MCJ cells also showed increased intracellular doxorubicin accumulation compared with the parental MCF7/ADR cells (see Fig. S3 in the supplemental material) Together, these results indicate that MCJ is able to negatively regulate ABCB1 expression.
Multidrug resistance induced by the loss of MCJ expression is mediated by ABCB1.
To determine whether the presence of ABCB1 expression in MCF7/siMCJ cells was responsible for the inability of these cells to accumulate doxorubicin, we examined the effect of verapamil, a known pharmacological inhibitor of ABCB1 (8). MCF7/siMCJ-1B and MCF7/ADR cells were treated with doxorubicin for 3 h in the presence or absence of verapamil. Intracellular drug accumulation was examined by confocal microscopy. No doxorubicin fluorescence in MCF7/siMCJ-1B cells was detected, but clear intracellular accumulation after the addition of verapamil was observed (Fig. 8A). Similar results with MCF7/ADR cells were observed (Fig. 8A). To confirm these results, we measured doxorubicin fluorescence by flow cytometry analysis. Verapamil allowed the intracellular accumulation of doxorubicin in MCF7/siMCJ-1B cells and had no effect on MCF7 cells (Fig. 8B).
FIG. 8.
Multidrug resistance induced by the loss of MCJ expression is mediated by ABCB1. (A) MCF7/ADR and MCF7/siMCJ-1B cells (green) were treated with doxorubicin (3 μM) for 3 h in the absence (Dox) or the presence (Dox + Verap.) of the ABCB1 inhibitor verapamil (10 μM). The intracellular accumulation of doxorubicin fluorescence (red) was examined by confocal microscopy. EGFP expression (green) in MCF7/siMCJ-1B cells is also presented. (B) MCF7 and MCF7/siMCJ-1B cells were treated with medium alone (gray-filled profiles) or with 3 μM of doxorubicin (3 h) in the absence (thin-line profiles) or the presence (thick-line profiles) of verapamil (10 μM). The numbers represent mean fluorescence intensities (MFI) of doxorubicin. (C) MCF7, MCF7/ADR, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were plated and treated in the absence or presence of different concentrations of 5-FU. Cell viability was determined by the MTT assay. LD50s of 5-FU were 0.94 μM (MCF7), 2.34 μM (MCF7/ADR), 1.93 μM (MCF7/siMCJ-1B), and 1.71 μM (MCF7/siMCJ-3B). (D) MCF7/siMCJ-1B and -3B cells were transfected in the presence (thick line) or absence (thin line) of siRNA oligonucleotides corresponding to ABCB1. After 48 h, cells were treated with doxorubicin and the intracellular drug accumulation was examined by flow cytometry. (E) MCF7/siMCJ-1B (siMCJ 1B) and -3B cells were transfected as described in the legend to panel D, and after 48 h, cells were lysed to examine the levels of ABCB1 by Western blot analysis. siABCB1, siRNA oligonucleotides corresponding to ABCB1; −, absence; +, presence.
In contrast to anthracyclines and taxanes, 5-FU is not a substrate for ABCB1, and MCF7/ADR cells are therefore as sensitive to this drug as MCF7 cells (41). To further confirm the involvement of ABCB1 in the multidrug resistance of MCF7/siMCJ cells, we examined the response of these cells to 5-FU by using the MTT assay. The responses of MCF7, MCF7/ADR, and MCF7/siMCJ-1B and -3B cells to doses of 5-FU were comparable (Fig. 8C). Thus, the loss of MCJ confers resistance to specific drugs that have been associated with the ABCB1 transporter. To further confirm the contribution of ABCB1 to the multidrug resistance induced by the loss of MCJ, we examined the effect of the inhibition of ABCB1 expression by siRNA on intracellular doxorubicin accumulation. MCF7/siMCJ cells were transiently transfected with siRNA oligonucleotides targeting ABCB1, and after 48 h cells were treated with doxorubicin. The intracellular accumulation of doxorubicin was measured by flow cytometry. We observed increased intracellular levels of doxorubicin in MCF7/siMCJ cells transfected with ABCB1-targeting siRNA oligonucleotides (Fig. 8D). No effect on drug accumulation was detected when scrambled siRNA oligonucleotides or siRNA oligonucleotides corresponding to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were used as controls (Fig. S4 in the supplemental material). The partial effect of ABCB1 siRNA on drug accumulation correlated with an only partial decrease of ABCB1 levels as determined by Western blot analysis (Fig. 8E). Together, these results indicate that the inability of MCF7/siMCJ cells to accumulate doxorubicin is at least partially mediated by the upregulation of ABCB1, although we do not discard a potential contribution of other ABC transporters with unknown functions.
The absence of MCJ increases c-Jun levels and transcriptional activity.
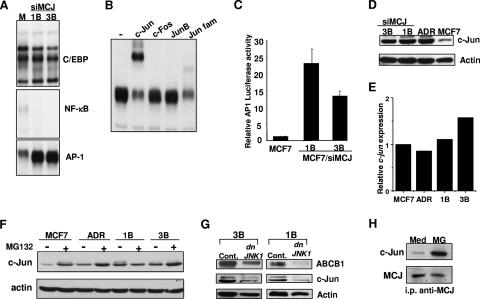
Several transcription factors have been shown to be involved in the regulation of ABCB1 gene expression, including AP-1, C/EBP, and NF-κB (55). To investigate the mechanism by which MCJ regulates ABCB1 expression, we examined AP-1, C/EBP, and NF-κB DNA binding by EMSA by using the nuclear extracts from MCF7 and MCF7/siMCJ cells. No difference in levels of C/EBP DNA binding between MCF7 and MCF7/siMCJ cells was observed (Fig. 9A). Low levels of NF-κB DNA binding in MCF7 cells could be detected, and the levels in MCF7/siMCJ cells were practically undetected (Fig. 9A). In contrast, the level of AP-1 DNA binding in MCF7/siMCJ cells was greatly increased compared with that in MCF7 cells (Fig. 9A).
FIG. 9.
MCJ downregulates ABCB1 expression by modulation of the AP-1 transcription factor. (A) Nuclear extracts from MCF7 (M) and MCF7/siMCJ (siMCJ)-1B and -3B cells were examined by EMSA using 32P-labeled double-stranded oligonucleotides specific for the C/EBP, NF-κB, and AP-1 genes. (B) AP-1 DNA binding was examined by EMSA using nuclear extracts from MCF7/siMCJ-1B cells and an AP-1 oligonucleotide. Binding reactions were performed in the absence (−) or presence of an anti-c-Jun, c-Fos, JunB, or Jun family (Jun fam.) antibody. (C) MCF7, MCF7/siMCJ-1B, and MCF7/siMCJ-3B cells were cotransfected with an AP-1-luciferase reporter construct and a β-Gal-expressing plasmid. After 24 h, luciferase values were measured and normalized to β-Gal activity as a control for the efficiency of the transfection. The error bars represent standard errors of the means (n = 3). (D) Whole-cell extracts from MCF7/siMCJ (siMCJ)-3B and -1B, MCF7/ADR (ADR), and MCF7 cells were analyzed for c-Jun expression by Western blot analysis. Actin was used as a loading control. (E) Total RNA isolated from MCF7/siMCJ-3B (3B) and -1B (1B), MCF7/ADR (ADR), and MCF7 cells was used to examine c-jun mRNA levels relative to levels of HPRT mRNA by real-time RT-PCR. (F) Whole-cell lysates from MCF7, MCF7/ADR (ADR), and MCF7/siMCJ-1B (1B) and -3B (3B) cells were treated in the absence (−) or presence (+) of proteasome inhibitor MG132 (5 μM) for 4 h and analyzed for c-Jun expression by Western blot analysis. (G) MCF7/siMCJ-3B (3B) and -1B (1B) cells were transfected with the empty control plasmid (Cont) or the dnJNK1 plasmid. Whole-cell lysates were analyzed for ABCB1, actin, and c-Jun by Western blotting. (H) MCF7 cells were treated with medium (Med) or MG132 (MG) for 4 h and lysed, and whole-cell extracts were used to immunoprecipitate MCJ (i.p. anti-MCJ). MCJ immunoprecipitates were examined for c-Jun by Western blot analysis using an anti-c-Jun antibody (c-Jun). The blot was further reprobed with the anti-MCJ mAb (MCJ).
AP-1 is composed of either heterodimers of Jun and Fos family members or homodimers of Jun family members (25). To identify the composition of the AP-1 complex in the MCF7/siMCJ cells, we performed supershift analysis with antibodies specific for AP-1 components by using nuclear extracts from these cells. Anti-JunB and anti-Fos antibodies did not substantially compete with AP-1 DNA binding, and no supershift complex could be detected, indicating that neither of these members was present in the complex (Fig. 9B). In contrast, anti-c-Jun antibody strongly inhibited the AP-1 binding and a supershift complex was present (Fig. 9B). Similarly, an antibody that does not supershift but competes with the DNA binding of the three Jun family members (c-Jun, JunB, and JunD) also inhibited the AP-1 complex present in the MCF7/siMCJ cells (Fig. 9B). These results indicate that the AP-1 complexes present in MCF7/siMCJ cells consist predominantly of c-Jun dimers.
To examine whether increased c-Jun DNA binding resulted in increased AP-1-mediated transcription, we transfected MCF7 and MCF7/siMCJ cells with an AP-1-luciferase reporter construct. In correlation with the high level of AP-1 DNA binding activity, increased AP-1 transcriptional activity was detected in MCF7/siMCJ cells (Fig. 9C). The loss of MCJ therefore induced AP-1-mediated transcription.
We determined whether the increased c-Jun DNA binding in MCF7/siMCJ cells could be due to an upregulation of c-Jun protein levels. We examined the levels of c-Jun by Western blot analysis using whole-cell lysates. Very low levels of c-Jun were detected in MCF7 cells, but high levels were present in MCF7/siMCJ cells (Fig. 9D), as well as in MCF7/ADR cells (Fig. 9D). In contrast, no significant difference in c-Jun mRNA levels was observed by quantitative real-time PCR (Fig. 9E), suggesting that MCJ may have an effect on c-Jun protein stability or synthesis. It has been previously reported that c-Jun levels can be regulated by ubiquitination and proteasome-mediated degradation (62). We examined whether the low levels of c-Jun in MCF7 cells were due to increased proteasomal degradation by treating these cells with the proteasome inhibitor MG132 and performing Western blot analysis. A remarkable increase in the levels of c-Jun in the MG132-treated MCF7 cells compared with those in untreated MCF7 cells was observed (Fig. 9F). MG132 treatment did not increase c-Jun protein levels in MCF7/siMCJ cells (Fig. 9F). Thus, the lower levels of c-Jun present in MCF7 cells than in MCF7/siMCJ cells are likely due to an increased rate of degradation of c-Jun in the former cells.
A functional AP-1 binding site within the ABCB1 promoter region that binds c-Jun dimers has been described, and a number of studies have shown the regulation of ABCB1 by c-Jun in multidrug-resistant cancer cells (12, 13, 34). To show that c-Jun is responsible for the induction of ABCB1 gene expression in MCF7/siMCJ cells, we inhibited c-Jun-mediated transcription. The phosphorylation of c-Jun at Ser-63 and Ser-73 by JNK leads to the activation of c-Jun (14). We transiently transfected MCF7/siMCJ cells with a dominant negative JNK1 (dnJNK1) mutant-expressing plasmid (14) to inhibit c-Jun activation. ABCB1 expression in untransfected and transfected cells was examined by Western blotting. The presence of dnJNK1 in MCF7/siMCJ-1B and -3B cells caused a substantial reduction of the ABCB1 levels (Fig. 9G). Thus, the induction of ABCB1 expression in the absence of MCJ required the c-Jun/JNK pathway. A reduction of the ABCB1 levels by the expression of dnJNK1 in MCF7/ADR cells was also observed (see Fig. S5A in the supplemental material).
In addition to the transcription activity, the phosphorylation of c-Jun by JNK has been shown to protect c-Jun from ubiquitination and degradation (22, 44). We therefore examined the levels of c-Jun in MCF7/siMCJ cells transfected with the dnJNK1 mutant. In correlation with the reduction of ABCB1 levels, the levels of c-Jun were substantially decreased in the presence of dnJNK1 (Fig. 9G). Similar results were obtained with MCF7/ADR cells (Fig. S5A in the supplemental material), but the expression of dnJNK did not affect the low c-Jun levels present in MCF7 cells (see Fig. S5B in the supplemental material). The amount of total JNK1/JNK2 as determined by Western blot analysis was not increased by the loss of MCJ in MCF7/siMCJ cells (see Fig. S5C in the supplemental material). Although the basal levels of phosphorylated JNK seemed to be slightly increased in both MCF7/siMCJ and MCF7/ADR cells compared with those in MCF7 cells, they were very low since a long film exposure was needed for detection by Western blot analysis (see Fig. S5C in the supplemental material).
Together, these results indicate that the presence of MCJ in MCF7 cells prevents the accumulation of c-Jun, probably by promoting its degradation, and that this process is impaired by the loss of MCJ, suggesting that MCJ may physically interact with c-Jun in MCF7 cells. To address this hypothesis, we performed coimmunoprecipitation analysis of MCJ with c-Jun. Since the levels of c-Jun in MCF7 cells are very low, we used whole-cell extracts from untreated MCF7 cells and from MCF7 cells treated for a short period of time with the proteosome inhibitor MG132 to allow the accumulation of c-Jun. MCJ was immunoprecipitated from whole-cell lysates generated from MCF7 cells or MG132-treated MCF7 cells. Immunoprecipitates were then analyzed for the presence of c-Jun by Western blot analysis. Low but detectable levels of c-Jun coimmunoprecipated with MCJ from MCF7 cells, and higher levels coimmunoprecipitated with MCJ from the MG132-treated cells (Fig. 9H). Thus, MCJ associates with c-Jun and this association likely favors c-Jun degradation.
DISCUSSION
Multidrug resistance is a complex and multifactorial phenomenon. It appears to be the major cause of chemotherapy failure in breast cancer since it is associated with the lack of response to a variety of drugs. The identification of tumor markers that can help to predetermine the response to a given type of chemotherapy is therefore an area of high priority in breast cancer research. The overexpression of markers (e.g., specific ABC transporters) exclusively in multidrug-resistant cancer cells and their absence in their drug-sensitive counterparts is the most frequent scenario (38). In contrast, the loss of expression of specific markers in association with multidrug resistance is less frequent. The loss of MCJ expression by methylation of the MCJ gene has been correlated with multidrug resistance in ovarian cancer cell lines. In addition, it has been recently shown that high levels of methylation of the MCJ gene correlate with a poor response of ovarian tumors to therapy and poor survival rates among patients (61). Here we show that the loss of MCJ gene expression also correlates with multidrug resistance in two independent breast cancer cell lines and in a uterine cancer cell line. Immunohistochemistry analysis of MCJ in breast tumor arrays indicated that MCJ expression is lost in a number of tumors (data not shown), but further studies are needed to show the correlation with the response to chemotherapy. Thus, MCJ may be a widely useable marker for chemoresistance among different types of cancer cells.
MCJ, however, is not just a marker for the response to chemotherapy. Here we show that the presence or absence of MCJ clearly modulates the response of breast cancer cells to specific chemotherapeutic drugs. MCJ has already been associated with the chemotherapeutic response in ovarian cancer cells, but no mechanism has yet been proposed. In this study, we show that the absence of MCJ prevents intracellular drug accumulation. This prevention is at least partially due to the upregulation of the ABCB1 transporter since verapamil reverses the intracellular accumulation of doxorubicin in the absence of MCJ and the expression of ABCC1 and ABCG2 is not affected. However, we cannot discard the possibility that MCJ may also regulate the expression of other uncharacterized ABC transporters of unknown functions that might be additional targets for verapamil.
We show here that the expression of the ABCB1 gene induced by the loss of MCJ is mediated by c-Jun. The intracellular concentration of c-Jun is normally tightly regulated through rapid turnover by ubiquitination and degradation, a process that is regulated in part by the phosphorylation of c-Jun. The phosphorylation of c-Jun by JNK has been shown to reduce c-Jun ubiquitination, leading to increased c-Jun stabilization (22, 44, 45). In contrast, the phosphorylation of c-Jun by COOH-terminal Src kinase at Y26 and Y170 appears to promote c-Jun degradation (65). Here we show that MCF7 cells did not contain significant amounts of c-Jun despite being tumor cells. However, the inhibition of the proteasome function greatly increased c-Jun levels, indicating that an active ubiquitination and degradation process prevents c-Jun from accumulating in these cells. In the absence of MCJ, however, c-Jun is able to accumulate in MCF7 cells. The accumulation of c-Jun and AP-1 activity have also been found in MCF/ADR cells (reference 13 and our unpublished data) in correlation with the loss of MCJ from these cells. Increased levels of c-Jun and ABCB1 in other drug-resistant cancer cell lines have also been observed (12, 34), but it remains to be examined whether or not these cells express MCJ. Thus, we propose that MCJ promotes the ubiquitination and/or the degradation of c-Jun. Although the mechanism needs to be further investigated in future studies, we have shown a physical association between MCJ and c-Jun in support of this model.
Recently, it has been shown that the ubiquitination of c-Jun is carried out by specific ubiquitin ligases such as Itch (23) and SCFFbw7 (47). In addition, c-Jun is also regulated by deubiquitination through the deubiquitinase POH1 (46). Both ubiquitination and deubiquitination have been subcellularly localized to lysosomes and endosomes (20, 46). Thus, MCJ may promote the ubiquitination or inhibit the deubiquitination of c-Jun and potentially other proteins. In this regard, we also found that the levels of hypoxia inducible factor 1α (HIF-1α), a transcription factor regulated primarily by ubiquitination and degradation (52), were higher in the MCF7/siMCJ cells than in the MCF7 cells (see Fig. S6 in the supplemental material). Although HIF-1α has also been involved in the transcription of the ABCB1 gene in hypoxia (9), the inhibition of HIF-1α expression in the MCF7/siMCJ cells did not interfere with ABCB1 gene expression (data not shown).
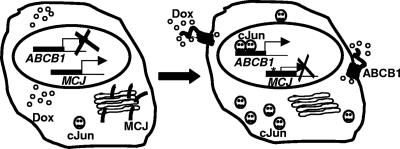
In summary, MCJ is a unique type II transmembrane DnaJ protein present in the Golgi compartment that acts as a repressor of the ABCB1 gene by promoting the degradation of c-Jun in breast cancer cells. The loss of MCJ expression causes an increased stabilization of c-Jun-mediated transcription, leading to the induction of ABCB1 expression. ABCB1 actively flushes the drug out of the cell and causes multidrug resistance as summarized in the model (Fig. 10).
FIG. 10.
Model representing MCJ in the Golgi network, associating with c-Jun and preventing its accumulation and inhibiting ABCB1 expression, allowing the intracellular accumulation of doxorubicin (Dox). In the absence of MCJ, increased levels of c-Jun upregulate ABCB1 gene expression. Doxorubicin is flushed out of the cell by the ABCB1 protein.
Supplementary Material
Acknowledgments
This research was supported by a Juckett Scholar Award (Lake Champlain Cancer Organization and Vermont Cancer Center).
We thank Robert Kerbel (Sunnybrook & Women's College Health Sciences Centre, Canada) for providing the MDA-MB-231 and MD22 cells. We thank Collete Charland for flow cytometry analysis, Marilyn Wadsworth for confocal microscopy and helpful discussion, Nicole DeLance for immunoelectron microscopy, Timothy Hunter for assistance with real-time PCR analysis, and Karen Fortner for critical reading of the manuscript.
Footnotes
Published ahead of print on 5 February 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alley, M., D. Scudiero, A. Monks, M. Hursey, M. Czerwinski, D. Fine, B. Abbott, J. Mayo, R. Shoemaker, and M. Boyd. 1988. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 48:589-601. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 5.Belhoussine, R., H. Morjani, S. Sharonov, D. Ploton, and M. Manfait. 1999. Characterization of intracellular pH gradients in human multidrug-resistant tumor cells by means of scanning microspectrofluorometry and dual-emission-ratio probes. Int. J. Cancer 81:81-89. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., and S. M. Simon. 2000. In situ biochemical demonstration that P-glycoprotein is a drug efflux pump with broad specificity. J. Cell Biol. 148:863-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. N., L. A. Mickley, A. M. Schwartz, E. M. Acton, J. L. Hwang, and A. T. Fojo. 1990. Characterization of adriamycin-resistant human breast cancer cells which display overexpression of a novel resistance-related membrane protein. J. Biol. Chem. 265:10073-10080. [PubMed] [Google Scholar]
- 9.Comerford, K. M., T. J. Wallace, J. Karhausen, N. A. Louis, M. C. Montalto, and S. P. Colgan. 2002. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 62:3387-3394. [PubMed] [Google Scholar]
- 10.Conze, D., L. Weiss, P. S. Regen, A. Bhushan, D. Weaver, P. Johnson, and M. Rincon. 2001. Autocrine production of interleukin 6 causes multidrug resistance in breast cancer cells. Cancer Res. 61:8851-8858. [PubMed] [Google Scholar]
- 11.Craig, E. A., P. Huang, R. Aron, and A. Andrew. 2006. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156:1-21. [DOI] [PubMed] [Google Scholar]
- 12.Cripe, L. D., V. M. Gelfanov, E. A. Smith, D. R. Spigel, C. A. Phillips, T. G. Gabig, S. H. Jung, J. Fyffe, A. D. Hartman, P. Kneebone, D. Mercola, G. S. Burgess, and H. S. Boswell. 2002. Role for c-jun N-terminal kinase in treatment-refractory acute myeloid leukemia (AML): signaling to multidrug-efflux and hyperproliferation. Leukemia 16:799-812. [DOI] [PubMed] [Google Scholar]
- 13.Daschner, P. J., H. P. Ciolino, C. A. Plouzek, and G. C. Yeh. 1999. Increased AP-1 activity in drug resistant human breast cancer MCF-7 cells. Breast Cancer Res. Treat. 53:229-240. [DOI] [PubMed] [Google Scholar]
- 14.Derijard, B., M. Hibi, I.-H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 15.Diah, S. K., P. K. Smitherman, J. Aldridge, E. L. Volk, E. Schneider, A. J. Townsend, and C. S. Morrow. 2001. Resistance to mitoxantrone in multidrug-resistant MCF7 breast cancer cells: evaluation of mitoxantrone transport and the role of multidrug resistance protein family proteins. Cancer Res. 61:5461-5467. [PubMed] [Google Scholar]
- 16.Doyle, L. A., and D. D. Ross. 2003. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene 22:7340-7358. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich, M., G. Jiang, E. Fiala, J. S. Dome, M. C. Yu, T. I. Long, B. Youn, O. S. Sohn, M. Widschwendter, G. E. Tomlinson, M. Chintagumpala, M. Champagne, D. Parham, G. Liang, K. Malik, and P. W. Laird. 2002. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene 21:6694-6702. [DOI] [PubMed] [Google Scholar]
- 18.Fairchild, C., S. Ivy, C. Kao-Shan, J. Whang-Peng, N. Rosen, M. Israel, P. Melera, K. Cowan, and M. Goldsmith. 1987. Isolation of amplified and overexpressed DNA sequences from adriamycin-resistant human breast cancer cells. Cancer Res. 47:5141-5148. [PubMed] [Google Scholar]
- 19.Fairchild, C. R., J. A. Moscow, E. E. O'Brien, and K. H. Cowan. 1990. Multidrug resistance in cells transfected with human genes encoding a variant P-glycoprotein and glutathione S-transferase-pi. Mol. Pharmacol. 37:801-809. [PubMed] [Google Scholar]
- 20.Fang, D., and T. K. Kerppola. 2004. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl. Acad. Sci. USA 101:14782-14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein, J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 266:418-427. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs, S. Y., L. Dolan, R. J. Davis, and Z. Ronai. 1996. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13:1531-1535. [PubMed] [Google Scholar]
- 23.Gao, M., T. Labuda, Y. Xia, E. Gallagher, D. Fang, Y. C. Liu, and M. Karin. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306:271-275. [DOI] [PubMed] [Google Scholar]
- 24.Gottesman, M. M., T. Fojo, and S. E. Bates. 2002. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2:48-58. [DOI] [PubMed] [Google Scholar]
- 25.Halazonetis, T. D., K. Georgopoulos, M. E. Greenberg, and P. Leder. 1988. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell 55:917-924. [DOI] [PubMed] [Google Scholar]
- 26.Harbottle, A., A. K. Daly, K. Atherton, and F. C. Campbell. 2001. Role of glutathione S-transferase P1, P-glycoprotein and multidrug resistance-associated protein 1 in acquired doxorubicin resistance. Int. J. Cancer 92:777-783. [DOI] [PubMed] [Google Scholar]
- 27.Harker, W. G., and B. I. Sikic. 1985. Multidrug (pleiotropic) resistance in doxorubicin-selected variants of the human sarcoma cell line MES-SA. Cancer Res. 45:4091-4096. [PubMed] [Google Scholar]
- 28.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 29.Izawa, I., M. Nishizawa, K. Ohtakara, K. Ohtsuka, H. Inada, and M. Inagaki. 2000. Identification of Mrj, a DnaJ/Hsp40 family protein, as a keratin 8/18 filament regulatory protein. J. Biol. Chem. 275:34521-34527. [DOI] [PubMed] [Google Scholar]
- 30.Jaskiewicz, E., G. Zhu, D. J. Taatjes, D. S. Darling, G. E. Zwanzig, Jr., and W. W. Young, Jr. 1996. Cloned beta 1,4N-acetylgalactosaminyltransferase: subcellular localization and formation of disulfide bonded species. Glycoconj. J. 13:213-223. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami, K., C. Scheidereit, and R. G. Roeder. 1988. Identification and purification of a human immunoglobulin-enhancer-binding protein (NF-kappa B) that activates transcription from a human immunodeficiency virus type 1 promoter in vitro. Proc. Natl. Acad. Sci. USA 85:4700-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klement, G., P. Huang, B. Mayer, S. K. Green, S. Man, P. Bohlen, D. Hicklin, and R. S. Kerbel. 2002. Differences in therapeutic indexes of combination metronomic chemotherapy and an anti-VEGFR-2 antibody in multidrug-resistant human breast cancer xenografts. Clin. Cancer Res. 8:221-232. [PubMed] [Google Scholar]
- 33.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1989. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 243:1681-1688. [DOI] [PubMed] [Google Scholar]
- 34.Ledoux, S., R. Yang, G. Friedlander, and D. Laouari. 2003. Glucose depletion enhances P-glycoprotein expression in hepatoma cells: role of endoplasmic reticulum stress response. Cancer Res. 63:7284-7290. [PubMed] [Google Scholar]
- 35.Lee, D., M. Sherman, and A. Goldberg. 1996. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, W., P. Mitchell, and R. Tjian. 1987. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell 49:741-752. [DOI] [PubMed] [Google Scholar]
- 37.Lindsey, J. C., M. E. Lusher, G. Strathdee, R. Brown, R. J. Gilbertson, S. Bailey, D. W. Ellison, and S. C. Clifford. 2006. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int. J. Cancer 118:346-352. [DOI] [PubMed] [Google Scholar]
- 38.Longley, D. B., and P. G. Johnston. 2005. Molecular mechanisms of drug resistance. J. Pathol. 205:275-292. [DOI] [PubMed] [Google Scholar]
- 39.Mardones, G., and A. Gonzalez. 2003. Selective plasma membrane permeabilization by freeze-thawing and immunofluorescence epitope access to determine the topology of intracellular membrane proteins. J. Immunol. Methods 275:169-177. [DOI] [PubMed] [Google Scholar]
- 40.Martersteck, C. M., N. L. Kedersha, D. A. Drapp, T. G. Tsui, and K. J. Colley. 1996. Unique alpha 2, 8-polysialylated glycoproteins in breast cancer and leukemia cells. Glycobiology 6:289-301. [DOI] [PubMed] [Google Scholar]
- 41.Mechetner, E., A. Kyshtoobayeva, S. Zonis, H. Kim, R. Stroup, R. Garcia, R. J. Parker, and J. P. Fruehauf. 1998. Levels of multidrug resistance (MDR1) P-glycoprotein expression by human breast cancer correlate with in vitro resistance to taxol and doxorubicin. Clin. Cancer Res. 4:389-398. [PubMed] [Google Scholar]
- 42.Mokranjac, D., M. Sichting, W. Neupert, and K. Hell. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22:4945-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morjani, H., N. Aouali, R. Belhoussine, R. J. Veldman, T. Levade, and M. Manfait. 2001. Elevation of glucosylceramide in multidrug-resistant cancer cells and accumulation in cytoplasmic droplets. Int. J. Cancer 94:157-165. [DOI] [PubMed] [Google Scholar]
- 44.Musti, A. M., M. Treier, and D. Bohmann. 1997. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275:400-402. [DOI] [PubMed] [Google Scholar]
- 45.Musti, A. M., M. Treier, F. A. Peverali, and D. Bohmann. 1996. Differential regulation of c-Jun and JunD by ubiquitin-dependent protein degradation. Biol. Chem. 377:619-624. [DOI] [PubMed] [Google Scholar]
- 46.Nabhan, J. F., and P. Ribeiro. 2006. The 19 S proteasomal subunit POH1 contributes to the regulation of c-Jun ubiquitination, stability, and subcellular localization. J. Biol. Chem. 281:16099-16107. [DOI] [PubMed] [Google Scholar]
- 47.Nateri, A. S., L. Riera-Sans, C. Da Costa, and A. Behrens. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303:1374-1378. [DOI] [PubMed] [Google Scholar]
- 48.Noonan, K. E., C. Beck, T. A. Holzmayer, J. E. Chin, J. S. Wunder, I. L. Andrulis, A. F. Gazdar, C. L. Willman, B. Griffith, D. D. Von Hoff, et al. 1990. Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumors by polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 50.Pedraza-Alva, G., M. Koulnis, C. Charland, T. Thornton, J. L. Clements, M. S. Schlissel, and M. Rincon. 2006. Activation of p38 MAP kinase by DNA double-strand breaks in V(D)J recombination induces a G2/M cell cycle checkpoint. EMBO J. 25:763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedraza-Alva, G., S. Sawasdikosol, Y. C. Liu, L. B. Merida, M. E. Cruz-Munoz, F. Oceguera-Yanez, S. J. Burakoff, and Y. Rosenstein. 2001. Regulation of Cbl molecular interactions by the co-receptor molecule CD43 in human T cells. J. Biol. Chem. 276:729-737. doi: 10.1074/jbc.M008494200. [DOI] [PubMed] [Google Scholar]
- 52.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1alpha (HIF- 1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 53.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scotto, K. W. 2003. Transcriptional regulation of ABC drug transporters. Oncogene 22:7496-7511. [DOI] [PubMed] [Google Scholar]
- 56.Shridhar, V., K. C. Bible, J. Staub, R. Avula, Y. K. Lee, K. Kalli, H. Huang, L. C. Hartmann, S. H. Kaufmann, and D. I. Smith. 2001. Loss of expression of a new member of the DNAJ protein family confers resistance to chemotherapeutic agents used in the treatment of ovarian cancer. Cancer Res. 61:4258-4265. [PubMed] [Google Scholar]
- 57.Sladowski, D., S. J. Steer, R. H. Clothier, and M. Balls. 1993. An improved MTT assay. J. Immunol. Methods 157:203-207. [DOI] [PubMed] [Google Scholar]
- 58.Sondermann, H., C. Scheufler, C. Schneider, J. Hohfeld, F. U. Hartl, and I. Moarefi. 2001. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291:1553-1557. [DOI] [PubMed] [Google Scholar]
- 59.Storey, H., A. Stewart, P. Vandenabeele, and J. P. Luzio. 2002. The p55 tumour necrosis factor receptor TNFR1 contains a trans-Golgi network localization signal in the C-terminal region of its cytoplasmic tail. Biochem. J. 366:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strathdee, G., B. R. Davies, J. K. Vass, N. Siddiqui, and R. Brown. 2004. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis 25:693-701. [DOI] [PubMed] [Google Scholar]
- 61.Strathdee, G., J. K. Vass, K. A. Oien, N. Siddiqui, J. Curto-Garcia, and R. Brown. 2005. Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol. Oncol. 97:898-903. [DOI] [PubMed] [Google Scholar]
- 62.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 63.Ungewickell, E., H. Ungewickell, S. E. Holstein, R. Lindner, K. Prasad, W. Barouch, B. Martin, L. E. Greene, and E. Eisenberg. 1995. Role of auxilin in uncoating clathrin-coated vesicles. Nature 378:632-635. [DOI] [PubMed] [Google Scholar]
- 64.Young, J. C., J. M. Barral, and F. Ulrich Hartl. 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28:541-547. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, F., B. Y. Choi, W. Y. Ma, Z. Zhao, Y. Zhang, Y. Y. Cho, H. S. Choi, A. Imamoto, A. M. Bode, and Z. Dong. 2006. COOH-terminal Src kinase-mediated c-Jun phosphorylation promotes c-Jun degradation and inhibits cell transformation. Cancer Res. 66:5729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.