Abstract
The mitogen-activated protein (MAP) kinases represent one of the most important classes of signaling cascades that are used by eukaryotic cells to sense extracellular signals. One of the major responses to these cascades is a change in cellular gene expression profiles mediated through the direct targeting of transcriptional regulators, such as the transcription factor Elk-1. Here we have identified human Rev7 (hRev7)/MAD2B/MAD2L2 as an interaction partner for Elk-1 and demonstrate that hRev7 acts to promote Elk-1 phosphorylation by the c-Jun N-terminal protein kinase (JNK) MAP kinases. As phosphorylation of Elk-1 potentiates the activity of its transcriptional activation domain, hRev7 therefore contributes to the upregulation of Elk-1 target genes, such as egr-1, following exposure of cells to stress conditions caused by DNA-damaging agents. Thus, given its previous roles in permitting DNA damage bypass during replication and regulating cell cycle progression, our data linking hRev7 to gene expression changes suggest that hRev7 has a widespread role in coordinating the cellular response to DNA damage.
The mitogen-activated protein (MAP) kinase pathways represent some of the most important pathways through which cells sense their external environment (reviewed in references 2 and 9). Proliferative signals are transduced by the extracellular signal-regulated kinase (ERK) pathway, whereas the c-Jun N-terminal protein kinase (JNK) and p38 pathways respond to a variety of stress signals. These stresses can vary from osmotic stress to DNA-damaging agents, such as UV irradiation. One of the major outcomes of MAP kinase signaling is a change in gene expression profiles, which is elicited to a large extent at the transcriptional level (reviewed in references 6, 16, and 27).
One of the best studied targets of these pathways is the ETS-domain transcription factor Elk-1 (reviewed in references 1, 17, and 18). Elk-1 is phosphorylated following activation of both the ERK pathway and the stress-activated p38 and JNK MAP kinase pathways. Phosphorylation takes place at multiple sites within the Elk-1 transcriptional activation domain (TAD), including Ser383. Phosphorylation leads to the potent activation of the TAD, and Ser383 plays a critical role in this process. MAP kinase-mediated Elk-1 regulation leads to the activation of a number of Elk-1 target genes, including c-fos and egr-1 (reviewed in references 17 and 18). The targeting of MAP kinases to Elk-1 occurs through docking domains that flank the TAD and determine the preferential recruitment of the ERK and JNK MAP kinases (8, 30, 31). Furthermore, additional mechanisms contribute to the responses of Elk-1 to the MAP kinase pathways. For example, we have recently shown that the Elk-1 coregulatory protein PIASxα can act to differentially determine the amplitude of the transcriptional response to the ERK and stress-activated p38 pathways (25, 26). Thus, although Elk-1 responds to all of the MAP kinase pathways, there appears to be additional layers of control which help determine the response of Elk-1 to individual MAP kinases.
Human Rev7 (hRev7) (also known as MAD2B and MAD2L2) is the homologue of Saccharomyces cerevisiae Rev7 and along with the catalytic subunit hRev3, represents a core subunit of DNA polymerase ζ (13; reviewed in reference 11). This DNA polymerase plays an important role in DNA translesion synthesis (TLS), permitting replication bypass of sites of DNA damage caused by agents such as UV or methyl methanesulfonate (MMS). However, hRev7 has also been shown to have a role independent from DNA polymerase ζ to inhibit the activity of the cell cycle regulator Cdh1-APC (anaphase-promoting complex), and to a lesser extent, Cdc20-APC (3, 14). Thus, hRev7 might act to coordinate cell cycle responses to DNA damage by acting at the level of DNA replication and on key cell cycle regulators.
In this study, we have identified hRev7 as an interaction partner for the transcription factor Elk-1. hRev7 potentiates the transcriptional activation activity of Elk-1 through promoting its phosphorylation by the JNK MAP kinase cascade. This enhancement of Elk-1 activity is particularly marked under DNA-damaging conditions. In addition to the previously defined roles of hRev7 in permitting DNA damage bypass during replication and regulating cell cycle progression, our data therefore demonstrate an additional important role for hRev7 in controlling gene expression. Thus, hRev7 appears to act as an integrator of cellular responses to DNA damage by acting on multiple processes.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were used in mammalian cell transfections. Plasmid pCH110 and plasmids pGL3-G5-E1B-Luc and pG5-TK-Luc (containing five GAL4 DNA binding sites cloned upstream of a minimal E1B and thymidine kinase [TK] promoter elements, respectively, and the firefly luciferase gene), were described previously (29). pEGR-1-Luc (kindly provided by Ian Stratford) contains the egr-1 promoter element and the firefly luciferase gene.
pAS2146 [pSG424 encoding GAL-c-Jun(1-223)], pAS2147 [pSG424 encoding GAL-c-Jun(1-223)(S63/73A)] (kindly provided by A. Whitmarsh), pAS383 [pCMV5-Flag-Elk-1] (25), pAS2238 (pCDNA3.1/his-hRev7; encoding hRev7; kindly provided by Y. Murakumo) (13), pAS2119 (pFlag-CMV2/hMAD2; encoding Flag-tagged human MAD2 (hMAD2) (kindly provided by K. Wassmann) (21), pAS1565 [pCMV-GAL-Elk(168-205)] (29), pAS900 [pCMV-GAL-Elk(310-428)] (30), pAS1553 [pCMV-GAL-Elk(223-428)] (24), and pAS883 [pCMV-GAL-MEF2A(266-413)] (28) were described previously.
pAS2114 [pEF1-GAL-p53(1-42)], pAS2115 [pEF1-GAL-VP16(1-94)], pAS2107 [pEF1-GAL-DBD], and pAS2108 [pEF1-GAL-Elk(310-428)] were constructed by taking either KpnI/XbaI (for pAS2114 and pAS2115) or BamHI/XbaI (for pAS2107 and pAS2108) fragments from pAS585 (kindly provided by S. Roberts), pAS287 (25), pAS571 (31), and pAS900 (30), respectively, and inserting them into the same sites in pEF1/Myc-HisB (Invitrogen). pAS2441 [pCDNA3-GAL-Elk(223-428)] and the mutant derivatives of this plasmid (mutations shown in parentheses after the plasmid) pAS2445 (S383A), pAS2446 (S389A), pAS2447 (T417A), pAS2448 (S422A), and pAS2449 (N/A [nine MAP kinase sites mutated to Ala]) were constructed by cloning BamHI/XbaI-cleaved PCR fragments from the respective pMluElk-1 plasmids described previously (4) (kindly provided by Richard Treisman) into the same sites of pCDNA3-GAL4.
pAS2145 (encoding hRev7 containing a silent mutation [hRev7_sil]) was constructed by QuikChange mutagenesis (Stratagene) using ADS1636/ADS1637 and p2238 as a template.
For bacterial expression, pAS2450 [encoding glutathione S-transferase (GST)-Elk-1(223-428)] was constructed by ligating a BamHI/XbaI fragment from pAS1553 into the same sites in pGEX-KG. pAS860 [encoding GST-MEF2A(266-413)] (28) and pAS2452 [encoding GST-c-Jun(1-79)[ (5) were used. Full-length Elk-1 was expressed in bacteria as a Flag-His-tagged protein from plasmid pAS278 (31). pAS2101 (pGEX4T-2/Rev7; encoding GST fused to full-length hRev7) (13) was described previously.
Primers.
The following primer pairs were used for reverse transcription-PCR (RT-PCR) experiments. c-fos and egr-1 have been described previously (25, 26). To detect 18S rRNA, primers ADS4005 (5′-CGGCTACCACATCCAAGGAA-3′) and ADS4006 (5′-GCTGGAATTACCGCGGCT-3′) were used.
The primer pairs used for chromatin immunoprecipitation (ChIP) experiments and detection of endogenous srf intronic sequences (25) were described previously. Primers ADS1644 (5′-GCTTCCCCAGCCTAGTTCAC-3′) and ADS1645 (5′-TGCCCAAATAAGGGTTGTTC-3′) were used to detect promoters from endogenous egr-1, and primers ADS1646 (5′-AGCGGATAGAATGGCG-3′) and ADS1647 (5′-CCAGTGCAAGTGCAG-3′) were used to detect promoters in the GAL-luciferase reporter.
Yeast two-hybrid screen.
A yeast two-hybrid screen was performed using GAL-Elk-1(223-428) as the bait in the pJ69-2A Saccharomyces cerevisiae strain and mating with a human fetal brain cDNA library as prey using the yeast strain Y187. Potential positives from the primary scheme were verified by retransforming either alone or together with the original bait plasmid, and positive clones were then sequenced.
Tissue culture, cell transfection, reporter gene assays, RT-PCR, and RNA interference.
293 and HeLa cells were grown, and transfection experiments were carried out as described previously (25). 293 cells containing short hairpin RNA (shRNA) constructs against hRev7 (293-sh-hRev7-1 and 293-sh-hRev7-3) or control cell lines containing empty shRNA vector (293-sh-control-2) were grown as described previously (12).
For phorbol myristate acetate (PMA) stimulation, cells were serum starved for 18 h and subsequently treated with 10 nM PMA for 35 min. For MMS stimulation, cells were serum starved for 22 h and subsequently treated with 0.01% MMS. For RT-PCR, ChIP, and Western blot analysis, cells were harvested immediately after stimulation for 0 to 2 h. However, for reporter gene analysis, cells were serum starved for 18 h and treated with MMS for 1 h, the MMS was then removed, and cells were washed and left in serum-free Dulbecco modified Eagle medium for a further 5 h before harvesting. For UV treatment, cells were serum starved for 18 h, the medium was removed, cells were treated with UV-C for 30 s, fresh serum-free medium was added, and the cells were left for a further 0 to 6 h. Where indicated, 10 μM of the inhibitors, SP600125 (for JNK) or SB203580 (for p38), was added to cells 1 h prior to MMS stimulation. For Gal4 fusion-driven luciferase reporter gene assays, 0 to 1.5 μg pCDNA-hRev7 (where used), 0.25 μg of reporter plasmid, and 0.25 μg of pCH110 were cotransfected with 0.1 μg of GAL4 fusion expression plasmids. Cell extracts were prepared, and equal amounts of protein were used in luciferase and β-galactosidase assays as described previously (31).
Real-time RT-PCR was carried out as described previously (25). Small interfering RNA (siRNA) against hRev7 and matched GAPDH control were constructed by the Silencer siRNA construction kit (Ambion). Human hRev7 target sequence was siRev7-1 (5′-AAGACCTCAACTTTGGCCAAG-3′) (ADS1462/3). The siRNAs against JNK1 and JNK2 (kindly provided by Alan Whitmarsh) and GAPDH (Dharmacon) controls were made synthetically. To carry out RNA interference, a two-step transfection protocol was carried out in 12-well plates as described previously (25).
Western blot and coimmunoprecipitation analysis.
Western blotting was carried out with the primary antibodies Flag (Sigma), Elk-1 (Santa Cruz Biotechnology and Epitomics), Gal4 DNA binding domain [Gal4(DBD)] (Santa Cruz Biotechnology), hemagglutinin antibody (Cancer Research UK), phospho-Elk (Cell Signaling), phospho-ERK, phospho-JNK, phospho-p38 (Cell Signaling), JNK1/2 (BD Biosciences), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam), hRev7 (BD Biosciences), and horseradish peroxidase-conjugated secondary antibodies (Transduction Laboratory) as described previously (25).
Coimmunoprecipitation analysis of overexpressed proteins was performed as described previously (29) whereas endogenous proteins were analyzed using exactaCruzC beads according to the manufacturer's instructions (Santa Cruz). GST pulldown analysis was performed essentially as described previously (19) with purified bacterially expressed recombinant GST-hRev7 and Flag-tagged full-length Elk-1.
ChIP assays.
Chromatin immunoprecipitation assays using antisera specific to hRev7 (BD Bioscience) and Gal4(DBD) (Santa Cruz Biotechnology) were performed as described previously (26) except that cross-linking was performed for 5 min. Bound promoters were detected by PCR as described previously (25).
Phosphorylation studies.
In vitro protein kinase assays were performed for 10 to 60 min at 30°C with 2 μg GST fusion proteins as substrates (purified from overexpressing bacterial strains), and kinases were pulled down from cell lysates by GST-hRev7 or endogenous hRev7 as described previously (31). Where indicated, 10 μM of the inhibitors, SP600125 (for JNK) or SB203580 (for p38), was added to kinase preparations 10 min prior to starting the kinase assay. hRev7-kinase complexes were isolated using either GST pulldown assays or by immunoprecipitating hRev7 from 293 cells. For GST pulldown assays, the standard procedure (19) was used, with a modified washing procedure in which phosphatase inhibitors were added into the pulldown buffer (10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerol phosphate, and 2 mM sodium pyrophosphate). A final wash in 1× kinase buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, 25 mM MgCl2, 0.5 mM dithiothreitol, 0.1 mM sodium orthovanadate) was added. To coimmunoprecipitate kinases with hRev7, cells were lysed in 600 μl of 50% lysis buffer (Tropix), 150 mM NaCl, 1× complete protease inhibitor cocktail (Roche), 10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerol phosphate, and 2 mM sodium pyrophosphate. After overnight incubation with antibodies, immunoprecipitations were carried out and precipitates were washed three times with 800 μl of the same buffer and then finally washed in 1× kinase buffer. In vivo phosphorylation was detected by using specific anti-phospho antibodies.
RESULTS
Rev7 binds to the transcription factor Elk-1.
A yeast two-hybrid screen was performed using the transcriptional activation domain of Elk-1 (amino acids 223 to 428) fused to the Gal4 DNA binding domain as the bait and a human fetal brain cDNA library. One of the positive clones isolated encodes hRev7.
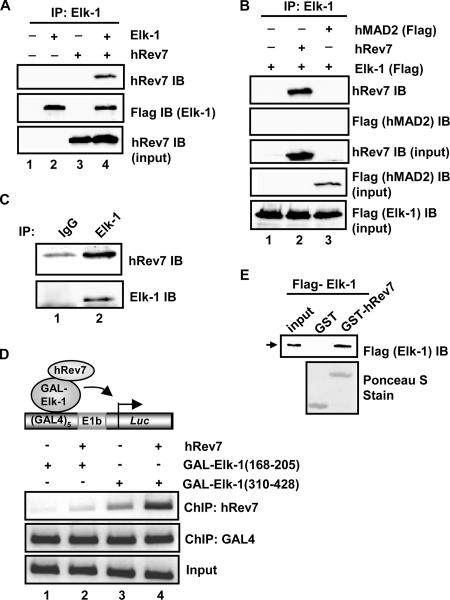
To establish whether hRev7 and Elk-1 could interact in mammalian cells, we coexpressed full-length Flag-tagged Elk-1 and hRev7 in human 293 cells and carried out a coimmunoprecipitation assay. When both proteins were coexpressed, anti-Elk-1 antibody was able to coprecipitate hRev7 (Fig. 1A). This interaction is specific, as the closely related protein human MAD2 (hMAD2) was not coprecipitated with Elk-1 (Fig. 1B). Importantly, endogenous Elk-1 and hRev7 in HeLa cells can be coprecipitated (Fig. 1C). To further demonstrate an interaction between Elk-1 and hRev7 in vivo, we asked whether GAL-Elk-1 fusion proteins could recruit hRev7 to a Gal4-driven reporter plasmid. ChIP analysis demonstrated that GAL-Elk-1(310-428) (encompassing the TAD) was able to recruit hRev7 to the promoter, whereas GAL-Elk-1(168-205) was unable to do so (Fig. 1D, compare lanes 2 and 4). In vitro GST pulldown assays using purified Elk-1 and GST-Rev7 demonstrated that the interaction between Elk-1 and hRev7 is direct (Fig. 1E). Further deletion analysis indicates that Elk-1 amino acids 349 to 399 (encompassing the TAD) are sufficient for interaction with hRev7 (data not shown).
FIG. 1.
Elk-1 interacts with hRev7 in vivo and in vitro. (A) Coimmunoprecipitation of hRev7 with Elk-1 from 293T cells. Cells were transfected (+) with constructs encoding Flag-Elk-1 (2 μg) and/or hRev7 (3 μg) where indicated. Elk-1 proteins were immunoprecipitated (IP) by Elk-1 antibody, and precipitated proteins were detected by immunoblotting (IB) using hRev7 or Flag (for Elk-1) antibodies. (B) Coimmunoprecipitation was performed as described above for panel A, but Flag-tagged hMAD2 was also transfected with Elk-1 where indicated. (C) Coimmunoprecipitation of endogenous Elk-1 and hRev7 proteins with Elk-1 antibody from HeLa cells. Immunoglobulin G (IgG) antibody was used as a control. Proteins were detected by the indicated antibodies (IB). (D) ChIP analysis of hRev7 on an Elk-1-driven promoter. A GAL4-driven E1b promoter-reporter construct and the indicated GAL4-Elk-1 constructs were transfected into 293T cells in the presence (+) and absence (−) of full-length hRev7. The indicated antibodies were used for precipitation of different proteins (ChIP), and coprecipitating DNA was detected by PCR. (E) GST pulldown analysis of Elk-1 interaction with hRev7. GST-hRev7 or GST alone was used in a GST pulldown assay with full-length Flag-tagged Elk-1 that had been expressed and purified from bacteria. Bound Elk-1 was detected by IB using an anti-Flag antibody.
These data therefore demonstrate that Elk-1 and hRev7 interact in mammalian cells through the Elk-1 TAD and that this interaction is direct.
hRev7 enhances the transactivation activity of Elk-1.
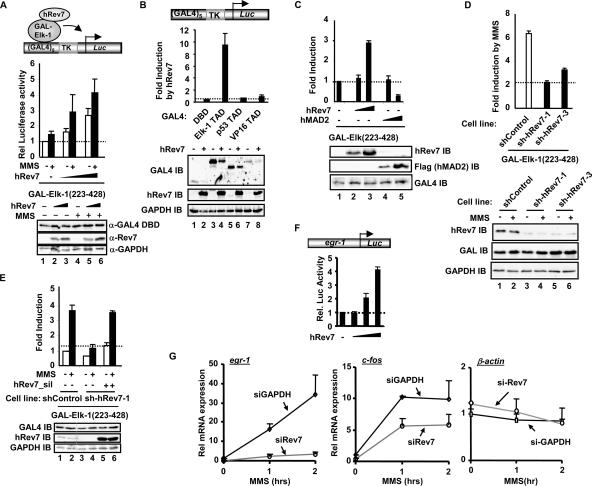
To establish the functional consequences of the hRev7-Elk-1 interaction, we first probed the role of hRev7 in controlling the transactivation capacity of Elk-1. The activity of Elk-1 was analyzed under basal conditions and in response to treatment with the DNA-damaging agent MMS which has previously been implicated in Rev7 function within the context of DNA polymerase ζ (20). Under basal conditions, increasing amounts of hRev7 caused an increase in the transactivation capacity of Elk-1 (Fig. 2A). This increase in transactivation was further potentiated upon stimulation with MMS. The ability of hRev7 to enhance the activity of TADs was specific to Elk-1, as it was unable to potentiate the transactivation capacity of other TADs, such as those found in p53 or VP16 (Fig. 2B). Moreover, the specificity of this effect was illustrated by the observation that the related protein, hMAD2, was unable to enhance the transactivation capacity of Elk-1 and actually reduced its activity (Fig. 2C). In all cases, the increase in Elk-1 activity was not due to increases in protein levels.
FIG. 2.
hRev7 activates Elk-1-dependent transcription. (A to E) Luciferase reporter assays of the indicated GAL4 fusion proteins in the presence of hRev7 on a GAL4-driven TK promoter-reporter plasmid (0.25 μg) (shown schematically at the top of panels A and B). (A) Dose-dependent Elk-1 activation. Constructs encoding GAL4-Elk-1(223-428) (0.2 μg) were transfected in the presence of increasing amounts of hRev7 constructs (0, 0.5 μg, 1.5 μg) in 293T cells. Where indicated, cells were treated with 0.01% MMS (+). Rel, relative; α-GAL4 DBD, anti-GAL4 DBD antibody. (B and C) Specificity of hRev7 effect. Each indicated GAL4-TAD fusion construct (0.1 μg) was transfected into 293T cells in the absence (−) and presence (+) of hRev7 (1 μg) (B) or increasing amounts (0.2 and 1 μg) of hRev7 or hMAD2 constructs (C). IB, immunoblotting. (D and E) Activity of Elk-1 following hRev7 knockdown. (D) GAL4-Elk-1(223-428) activity was monitored in control 293 cells or two different 293 cells stably expressing shRNA constructs against hRev7 in the presence (+) and absence (−) of MMS stimulation. Data are presented as changes in induction by MMS. (E) Rescue of Elk-1 activity by hRev7 reexpression. GAL4-Elk-1(223-428) activity was monitored in control and hRev7 knockdown cells in the presence and absence of MMS treatment and/or cotransfection of hRev7 containing a silent mutation (hRev7_sil). All luciferase reporter data were performed in duplicate, and the averages from three independent experiments are shown. Western blots are shown in the panels below each experiment with immunoblotting with the indicated antibodies to show the expression levels of transfected and endogenous proteins. (F) Luciferase reporter assays of the indicated egr-1 promoter-reporter plasmid (0.1 μg) (shown schematically at the top) in the presence of Elk-1 (25 ng) and increasing amounts of hRev7 constructs (0, 0.1 μg, 1 μg, and 2 μg) in 293T cells. Rel. Luc Activity, relative luciferase activity. (G) Quantitative RT-PCR analysis of egr-1, c-fos, and β-actin expression in MMS-treated HeLa cells. Endogenous hRev7 protein was reduced by siRNA treatment, and siRNA duplexes against GAPDH (siGAPDH) were used as a negative control. Experiments were performed in duplicate, and data were averaged from two independent experiments. Rel, relative.
Next we tested the effect of depleting hRev7 by using 293 cell lines that stably express shRNA constructs directed against hRev7 (12). The induction of Elk-1 transactivation capacity by MMS was reduced upon knockdown of hRev7, demonstrating the importance of hRev7 in this activation process (Fig. 2D). Furthermore, the reduction of Elk-1 transactivation capacity in the hRev7 knockdown cell lines could be restored by overexpression of a mutant form of hRev7 which harbors a silent mutation in the region covered by the shRNA hairpin (Fig. 2E).
To establish whether the hRev7-mediated changes in Elk-1 transactivation capacity had any effect on the induction of Elk-1 target genes, we first demonstrated that hRev7 activated the egr-1 promoter in a dose-dependent manner (Fig. 2F). Next, we analyzed the induction of endogenous egr-1 and c-fos in response to MMS treatment in 293 cells transiently transfected with siRNA constructs against either GAPDH or hRev7 (Fig. 2G). Both genes exhibited greatly reduced responsiveness to MMS treatment following hRev7 knockdown. However, β-actin expression, which is not an Elk-1 target gene, was not affected by hRev7 depletion, demonstrating the specificity of this effect.
These data therefore point to an important role for hRev7 in promoting maximal transcriptional activation responses from Elk-1 and its target genes following stimulation with DNA-damaging agents, such as MMS.
hRev7 enhances Elk-1 phosphorylation.
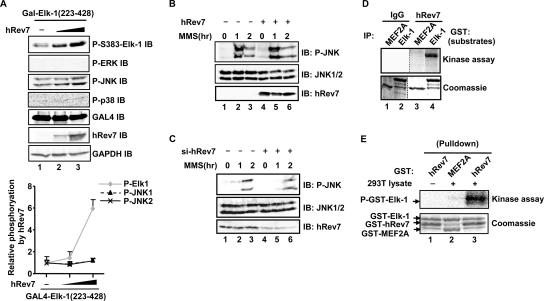
The Elk-1 TAD is activated through MAP kinase-mediated phosphorylation of a number of residues, the most important of which are Ser383 and Ser389 (reviewed in reference 17). As hRev7 binds to and potentiates the activity of the Elk-1 TAD, we tested whether it had any effect on Elk-1 phosphorylation status. First, we expressed increasing levels of hRev7 and monitored the phosphorylation of Ser383 in Elk-1 in vivo. A dose-dependent hRev7-mediated increase in Ser383 phosphorylation levels was observed (Fig. 3A, top blot). This increase in Elk-1 phosphorylation was not due to enhancement of activation of the ERK, JNK, and p38 MAP kinase pathways (Fig. 3A, P-ERK, P-JNK, and P-p38 blots). Indeed, while Elk-1 phosphorylation was enhanced more than sixfold by hRev7, only minimal increases in JNK phosphorylation (<0.3-fold) were observed (Fig. 3A, graph). Furthermore, overexpression of hRev7 did not perturb JNK activation by MMS (Fig. 3B). Similarly, depletion of hRev7 did not alter the activation of JNK by MMS, although treatment with both the control and hRev7 siRNAs caused a shift in the activation kinetics (Fig. 3C).
FIG. 3.
hRev7 enhances Elk-1 phosphorylation in vivo and in vitro. (A) hRev7 induces Elk-1 phosphorylation in vivo. 293T cells were transfected with 0.2 μg GAL4-Elk-1(223-428) and 0 (−), 0.2, or 1 μg hRev7 expression plasmid. Levels of phosphorylation and total protein were determined by immunoblotting (IB) using the appropriate standard or phospho-specific antibodies as indicated (P-S383-Elk-1, phospho-S383 Elk-1; P-ERK, phospho-ERK.). The graph shown below is the average level of Elk-1 and JNK phosphorylation (relative to the levels in the absence of hRev7, taken as 1) normalized for GAPDH levels, from three independent experiments (presented as mean ± standard deviation [error bar]). (B and C) hRev7 overexpression and depletion does not alter JNK activation. 293T cells were transfected (+) with 2.5 μg hRev7 expression plasmid (B) or siRNA duplexes against hRev7 (si-hRev7) (C) where indicated and treated with MMS for 0, 1, or 2 h. Levels of JNK phosphorylation and total protein were determined by immunoblotting (IB) using the appropriate standard or phospho-specific antibodies as indicated. (D) Immune complex in vitro kinase assays with kinases isolated by immunoprecipitation (IP) with either immunoglobulin G (IgG) or hRev7 antibodies from 293T cells transfected with hRev7 expression vectors. Purified GST-Elk-1 or GST-MEF2A were used as substrates. Phosphorylation levels were revealed by phosphorimaging (top blot) and total protein levels by staining with Coomassie blue (bottom blot). (E) GST pulldown and kinase assay. GST-hRev7 or GST-MEF2A beads were used to pull down kinases from 293T lysates where indicated, and the precipitated complexes were used to phosphorylate GST-Elk-1(223-428). Reactions were analyzed as described above for panel D.
To determine whether hRev7 or associated proteins could result in increased Elk-1 phosphorylation, we immunoprecipitated hRev7 and tested the activity of the precipitated complex towards Elk-1 in an in vitro kinase assay. Immunoprecipitated hRev7 could promote Elk-1 phosphorylation in vitro (Fig. 3D). However, purified recombinant hRev7 was unable to phosphorylate Elk-1 (data not shown), indicating that hRev7 most likely interacts with a coprecipitating kinase. Indeed, purified recombinant GST-Rev7 fusion protein was able to pull down a kinase from 293 cell lysates which could phosphorylate Elk-1 (Fig. 3E, lane 3).
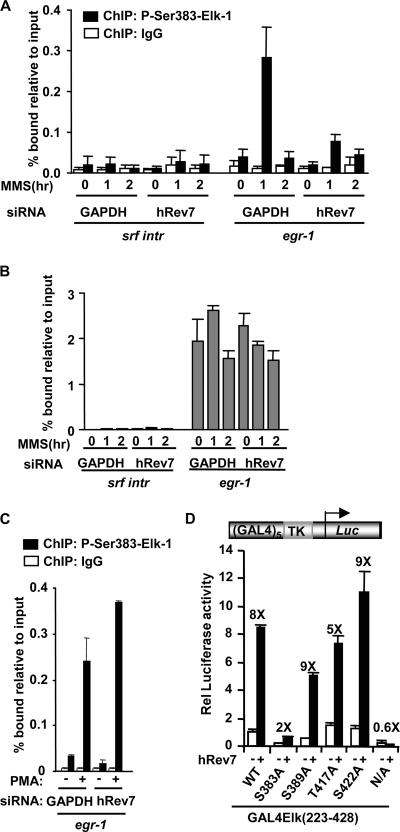
To establish a link between Elk-1 phosphorylation and target gene expression, we investigated whether hRev7 affected the levels of phosphorylated Elk-1 associated with the egr-1 promoter. Treatment of HeLa cells with MMS led to a large increase in the levels of Elk-1 phosphorylated at Ser383 on the egr-1 promoter (Fig. 4A). However, upon depletion of hRev7, this increase in phosphorylation was substantially reduced. Importantly, this was not due to alterations in Elk-1 binding to the promoter, as total Elk-1 levels on the egr-1 promoter are unaffected by either MMS treatment and/or hRev7 depletion (Fig. 4B). Moreover, hRev7 depletion did not reduce the levels of phosphorylated Elk-1 on the egr-1 promoter following PMA stimulation (Fig. 4C). Thus, hRev7 is specifically required for maximal induction of Ser383 phosphorylation upon treatment with the DNA-damaging agent MMS. This observation suggests that the presence of Ser383 is a critical determinant for hRev7-mediated transactivation. Indeed, Ser383 plays a vital role, as mutation of Ser383 virtually eliminated hRev7-mediated enhancement of Elk-1 activity (Fig. 4D). In comparison, individual mutations of other MAP kinase phosphorylation sites had little effect on the hRev7-mediated activation of Elk-1. Mutation of all nine MAP kinase sites abolished the stimulatory effect of hRev7.
FIG. 4.
hRev7 enhances Elk-1 phosphorylation in vivo. (A to C) ChIP assays of phosphorylated and total Elk-1 upon hRev7 depletion. (A) ChIP assays of MMS-induced phosphorylated Elk-1. ChIP was performed with immunoglobulin G (IgG) (white bars) or anti-phospho-Ser383 Elk-1 (black bars) antibodies on extracts from HeLa cells transfected with siRNA duplexes against GAPDH or hRev7 and stimulated with MMS for the indicated times. Promoter DNA corresponding to srf intron 3 (SRF intr) or the egr-1 promoter was detected by real-time PCR. Experiments were performed in duplicate, and data are the averages of two experiments. (B) ChIP assays of total Elk-1 following MMS stimulation as described above for panel A, except that ChIP was performed with an anti-Elk-1 antibody. (C) ChIP assays of phosphorylated Elk-1 following PMA stimulation as described above for panel A, except that cells were stimulated with PMA (+) rather than MMS for 35 min where indicated. (D) Luciferase reporter analysis of hRev7 activation of Elk-1 mutants in 293T cells. GAL4-driven TK promoter-reporter plasmids (0.25 μg) were transfected into cells with the indicated wild-type (WT) and mutant GAL4-Elk-1(223-428) constructs (0.2 μg). Plasmids encoding hRev7 (1.5 μg) were added where indicated (+). Experiments were performed in duplicate and averaged from three independent experiments. The change in stimulation by hRev7 is given above each set of bars. Rel, relative; N/A, nine MAP kinase sites mutated to alanine.
Together, these results demonstrate that hRev7 plays an important role in promoting phosphorylation of Elk-1 at Ser383 and hence potentiation of its transactivation capacity.
hRev7 promotes Elk-1 phosphorylation by JNK MAP kinase.
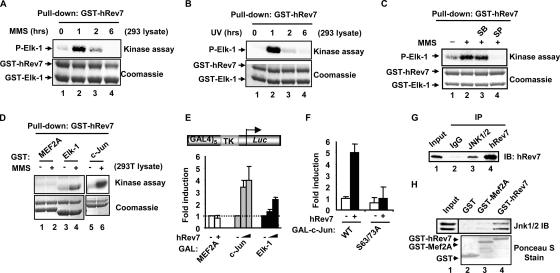
To determine the identity of the kinase that associates with hRev7, we first tested whether the activity of the kinase could be potentiated by different stress signals. Recombinant GST-hRev7 was used to pull down proteins from extracts of cells stimulated with MMS or UV irradiation, and coprecipitating kinases were tested using Elk-1 as a substrate. Large increases in kinase activity were observed upon either MMS (Fig. 5A) or UV (Fig. 5B) treatment of cells before making extracts. Under both conditions, the kinetics of kinase activation following MMS treatment was similar. Treatment of cells with a different stress signal, anisomycin, also elicited an increase in the activity of the hRev7-associated kinase, whereas the mitogen PMA caused no such enhancement (data not shown). MMS treatment also leads to enhanced levels of JNK activation within this time frame (10; data not shown). As Ser383 can be phosphorylated by MAP kinases, such as JNK, and the hRev7-associated kinase is stimulated by MMS and UV, we tested whether either a JNK inhibitor (SP600125) or a p38 inhibitor (SB203580) blocked the activity of the hRev7-associated kinase. Only the SP600125 inhibitor caused a reduction in the activity of the hRev7-associated kinase, suggesting that the kinase might be JNK (Fig. 5C). To further confirm the identity of the hRev7-associated kinase, we tested its activity against a p38 substrate (myocyte enhancer factor 2A [MEF2A]) and a JNK substrate (c-Jun). MEF2A was barely phosphorylated, but c-Jun was strongly phosphorylated by the hRev7-associated kinase (Fig. 5D). Furthermore, hRev7 was able to enhance the activity of the c-Jun TAD in vivo (Fig. 5E). This effect of hRev7 was dependent on the integrity of the JNK phosphorylation sites in c-Jun (Fig. 5F). These results therefore underscore the notion that hRev7 can associate with JNK MAP kinase. Indeed, hRev7 can be coprecipitated with endogenous JNK from 293T cells transfected with hRev7 (Fig. 5G). This interaction is likely to be direct, as purified hRev7 can bind to JNK MAP kinases from 293 cell lysates in GST pulldown assays (Fig. 5H).
FIG. 5.
hRev7 binds to an MMS-inducible kinase. (A to D) GST pulldown and kinase assays using GST-hRev7 to precipitate kinases from 293T cell lysates and GST-Elk-1(223-428) as a substrate. Lysates were from untreated cells (−) or cells treated with MMS (+) (A) and UV (B) for the indicated times or with MMS for 1 h (C and D). P-Elk-1, phospho-Elk-1. (C) The Rev7 precipitated proteins were preincubated with the inhibitor SB203580 (SB) or SP600125 (SP) for 10 min before starting the kinase assay. (D) The additional substrates GST-MEF2A and GST-c-Jun were tested. Phosphorylation levels were revealed by phosphorimaging (top blots) and total protein levels by staining with Coomassie blue (bottom blots). (E and F) Luciferase reporter analysis of hRev7 activation of c-Jun in 293T cells. GAL4-driven TK promoter-reporter plasmids (0.25 μg) were transfected into cells with the indicated GAL-fusion constructs (0.2 μg). (E) Increasing amounts of hRev7 (0, 0.2, and 1 μg) were added with GAL-c-Jun(1-223) and GAL-Elk-1(223-428) and either 0 or 1 μg hRev7 with GAL-MEF2A(266-413). (F) Wild-type (WT) or S63/73A GAL-c-Jun(1-223) constructs (0.2 μg) were transfected in the absence (−) and presence (+) of hRev7 (1 μg). Experiments were performed in duplicate and averaged from two (F) or three (E) independent experiments. (G) Coimmunoprecipitation of hRev7 with JNK from 293T cells. Cells were transfected with a construct encoding hRev7 (2.5 μg). hRev7 or JNK were immunoprecipitated (IP) with their respective antibodies (or immunoglobulin G [IgG] as a negative control), and precipitated proteins were detected by immunoblotting (IB) using a hRev7 antibody. (H) GST pulldown assay using the indicated GST fusion proteins and lysates from 293 cells. Precipitated JNK1/2 were detected by Western blotting. Input protein (5%) is shown in lane 1.
To definitively demonstrate that hRev7 promotes Elk-1 phosphorylation and subsequent potentiation of its transactivation capacity in vivo through enhancing its response to the JNK pathway, we depleted JNK1 and -2 using siRNA and examined the effect on hRev7 function. Knockdown of JNK caused a substantial reduction in the enhancement of Elk-1 phosphorylation at Ser383 (Fig. 6A) and transactivation capacity (Fig. 6B) in response to increasing amounts of hRev7. Similarly, pharmacological inhibition of JNK activity through treatment of cells with SP600125 severely attenuated the levels of Elk-1 phosphorylation (Fig. 6C) and Elk-1-mediated transactivation (Fig. 6D) caused by increased hRev7 expression.
FIG. 6.
hRev7-mediated Elk-1 activation is mediated through JNK. (A) Depletion of JNK1/2 diminishes Elk-1 phosphorylation induced by hRev7 in vivo. 293T cells were transfected with siRNA duplexes against JNK1/2 (si-JNK) (+) or control siRNA duplexes (−) and additionally Flag-tagged Elk-1 and 0, 0.2, or 1 μg hRev7 expression plasmids. Total protein levels and phosphorylated Elk-1 (P-S383 Elk-1) were determined by immunoblotting (IB). (B) Depletion of JNK1/2 diminishes Elk-1 activation induced by hRev7 in vivo. Luciferase reporter assay (0.25 μg of GAL4-driven TK promoter-reporter) in 293T cells transfected with siRNA duplexes against JNK1/2 (si-JNK) (+) (black bars) or control siRNA duplexes (−) (white bars) and additionally 0, 0.2, or 1 μg hRev7 expression plasmid. Experiments were performed in duplicate and averaged from two independent experiments. (C and D) Same as panels A and B except that the JNK inhibitor SP600125 was added in place of siRNA against JNK1/2 and the phosphorylation status of GAL-Elk-1(223-428) was analyzed instead of full-length Elk-1. (E) Quantitative RT-PCR analysis of egr-1 or c-fos in serum-starved HeLa cells as described in the legend to Fig. 2F. Experiments were performed in the presence of siRNA duplexes against JNK1/2 (siJNK) or control siRNA duplexes against GAPDH. (F) Model showing how hRev7 coordinates the cellular responses to DNA-damaging agents. hRev7 promotes TLS-mediated DNA repair in response to DNA damage induced by agents, such as MMS, and also promotes a transcriptional response through targeting the JNK pathway to transcription factors like Elk-1. The dotted line represents possible cross talk that might occur between the JNK pathway and hRev7 function in TLS. Pol, polymerase.
Finally, to establish that JNK is involved in the response of the Elk-1 targets genes egr-1 and c-fos to MMS treatment, we depleted JNK through siRNA treatment and examined their induction. JNK was important for the induction of both genes by MMS (Fig. 6E). Importantly, the same genes require Elk-1 (data not shown) and hRev7 (Fig. 2G) for their maximal induction in response to MMS treatment.
Collectively, these data therefore implicate JNK as the kinase through which hRev7 potentiates the activity of Elk-1 and its target genes in response to treatment with DNA-damaging agents, such as MMS.
DISCUSSION
Stress signals, including DNA-damaging agents, can lead to the upregulation of the p38 and JNK MAP kinase pathways (reviewed in reference 9). These MAP kinase pathways can then convert the signals into a cellular response, including a change in the cellular gene expression profile. One direct target of the MAP kinase pathways is the transcription factor Elk-1 which in turn activates target genes, such as egr-1 and c-fos (reviewed in references 1, 17, and 18). Here we have identified hRev7 as an important protein which is required for maximal JNK-mediated phosphorylation and activation of Elk-1 following treatment of cells with DNA-damaging agents.
Mechanistically, hRev7 binds to both JNKs and Elk-1, suggesting that it functions as an adapter molecule to permit efficient phosphorylation of Elk-1 by the JNKs. Elk-1 has previously been shown to be a JNK target (7, 23). Indeed, the JNKs themselves bind to a docking domain located N terminally to the Elk-1 TAD (30), while hRev7 binds to the TAD itself, meaning that JNK and hRev7 binding are not mutually exclusive. Attempts to map the binding surface on hRev7 for JNK and Elk-1 were unsuccessful, due to the nonmodular nature of hRev7 (data not shown). hRev7 having a role as an adapter molecule would be conceptually similar to the function of cytoplasmic scaffold proteins, like the JNK-interacting protein (JIP) family, which are thought to enhance the activity of the JNK pathway at least in part through colocalizing kinases and their downstream targets (22). It is however not known whether hRev7 recruits JNK to DNA-bound Elk-1 or before it engages with its target genes, as we were unable to detect hRev7 at Elk-1 target promoters in vivo. Current data suggest that hRev7 promotes the action of activated JNK on Elk-1 rather than participating in JNK activation itself, as changes in hRev7 levels have minimal effects of JNK phosphorylation and activation (Fig. 3). However, we cannot rule out the possibility that small changes in the activity of a subpopulation of JNK associated with transcription factors like Elk-1 might be physiologically significant.
We focused on the role of hRev7 in the response of Elk-1 to the DNA-damaging agent MMS, as hRev7 and its yeast homologue have previously been implicated in permitting TLS and subsequent repair of mutations generated by this agent. In TLS, hRev7 acts as an accessory subunit of DNA polymerase ζ (reviewed in reference 11). MMS also upregulates the activity of JNK (10; our unpublished data) and is also known to enhance the activity of Elk-1 target genes, such as egr-1 (15). hRev7 plays an important linker role in joining the JNK pathway to Elk-1 following MMS treatment. Importantly, another DNA-damaging agent, UV, is also implicated in JNK pathway upregulation (reviewed in reference 9), DNA polymerase ζ-mediated DNA repair (reviewed in reference 11), and hRev7-mediated activation of Elk-1 (our unpublished data). It is therefore tempting to speculate that hRev7 might provide a direct link in integrating these molecular events, not only through independently acting in TLS and in the upregulation of Elk-1 target genes but also through targeting the JNK pathway to DNA polymerase ζ (Fig. 6F). Further studies are needed to substantiate this hypothesis, but irrespective of their outcome, it is clear that hRev7 is an important coordinator of the cellular response to DNA damage. Indeed, hRev7 has also been implicated in control of the cell cycle through acting on Cdh1-APC (3, 14), further expanding its regulatory repertoire.
In summary, we have identified hRev7 as a novel interaction partner which promotes Elk-1 activation by promoting its phosphorylation by the JNK MAP kinases. This in turn leads to changes in gene expression profiles. This expands the regulatory roles attributed to hRev7 in coordinating the cellular responses to DNA-damaging agents.
Acknowledgments
We thank Anne Clancy for excellent technical assistance; Cathy Tournier and Alan Whitmarsh and members of our laboratory for comments on the manuscript and stimulating discussions; and Ian Stratford, Yoshiki Murakumo, Richard Treisman, Alan Whitmarsh, and Katja Wassmann for reagents.
This work was supported by grants from the Wellcome Trust.
Footnotes
Published ahead of print on 12 February 2007.
REFERENCES
- 1.Buchwalter, G., C. Gross, and B. Wasylyk. 2004. Ets ternary complex transcription factors. Gene 324:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J., and G. Fang. 2001. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 15:1765-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruzalegui, F. H., E. Cano, and R. Treisman. 1999. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene 18:7948-7957. [DOI] [PubMed] [Google Scholar]
- 5.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 6.Edmunds, J. W., and L. C. Mahadevan. 2004. MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J. Cell Sci. 117:3715-3723. [DOI] [PubMed] [Google Scholar]
- 7.Gille, H., T. Strahl, and P. E. Shaw. 1995. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr. Biol. 5:1191-1200. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs, D., D. Glossip, H. Xing, A. J. Muslin, and K. Kornfeld. 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13:163-175. [PMC free article] [PubMed] [Google Scholar]
- 9.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 10.Liu, Y., M. Gorospe, C. Yang, and N. J. Holbrook. 1995. Role of mitogen-activated protein kinase phosphatase during the cellular response to genotoxic stress. Inhibition of c-Jun N-terminal kinase activity and AP-1-dependent gene activation. J. Biol. Chem. 270:8377-8380. [DOI] [PubMed] [Google Scholar]
- 11.Murakumo, Y. 2002. The property of DNA polymerase zeta: REV7 is a putative protein involved in translesion DNA synthesis and cell cycle control. Mutat. Res. 510:37-44. [DOI] [PubMed] [Google Scholar]
- 12.Murakumo, Y., S. Mizutani, M. Yamaguchi, M. Ichihara, and M. Takahashi. 2006. Analyses of ultraviolet-induced focus formation of hREV1 protein. Genes Cells 11:193-205. [DOI] [PubMed] [Google Scholar]
- 13.Murakumo, Y., Y. Ogura, H. Ishii, S. Numata, M. Ichihara, C. M. Croce, R. Fishel, and M. Takahashi. 2001. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 276:35644-35651. [DOI] [PubMed] [Google Scholar]
- 14.Pfleger, C. M., E. Lee, and M. W. Kirschner. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 15:2396-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinones, A., K. U. Dobberstein, and N. G. Rainov. 2003. The egr-1 gene is induced by DNA-damaging agents and non-genotoxic drugs in both normal and neoplastic human cells. Life Sci. 72:2975-2992. [DOI] [PubMed] [Google Scholar]
- 16.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharrocks, A. D. 2002. Complexities in ETS-domain transcription factor function and regulation: lessons from the TCF subfamily. Biochem. Soc. Trans. 30:1-9. [DOI] [PubMed] [Google Scholar]
- 18.Shaw, P. E., and J. Saxton. 2003. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int. J. Biochem. Cell Biol. 35:1210-1226. [DOI] [PubMed] [Google Scholar]
- 19.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonoda, E., T. Okada, G. Y. Zhao, S. Tateishi, K. Araki, M. Yamaizumi, T. Yagi, N. S. Verkaik, D. C. van Gent, M. Takata, and S. Takeda. 2003. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 22:3188-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wassmann, K., V. Liberal, and R. Benezra. 2003. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J. 22:797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitmarsh, A. J. 2006. The JIP family of MAPK scaffold proteins. Biochem. Soc. Trans. 34:828-832. [DOI] [PubMed] [Google Scholar]
- 23.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403-407. [DOI] [PubMed] [Google Scholar]
- 24.Yang, S. H., D. C. Bumpass, N. D. Perkins, and A. D. Sharrocks. 2002. The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol. Cell. Biol. 22:5036-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, S. H., and A. D. Sharrocks. 2005. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 24:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, S. H., and A. D. Sharrocks. 2006. PIASxalpha differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol. Cell 22:477-487. [DOI] [PubMed] [Google Scholar]
- 27.Yang, S. H., A. D. Sharrocks, and A. J. Whitmarsh. 2003. Transcriptional regulation by the MAP kinase signaling cascades. Gene 320:3-21. [DOI] [PubMed] [Google Scholar]
- 28.Yang, S. H., P. Shore, N. Willingham, J. H. Lakey, and A. D. Sharrocks. 1999. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 18:5666-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, S. H., E. Vickers, A. Brehm, T. Kouzarides, and A. D. Sharrocks. 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21:2802-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, S. H., A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 17:1740-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, S. H., P. R. Yates, A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. The Elk-1 ETS-domain transcription factor contains a MAP kinase targeting motif. Mol. Cell. Biol. 18:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]