Abstract
The A+U-rich elements (or AREs) are cis-acting sequences that activate rapid mRNA decay, yet the overall polarity of this process is unknown. The current study describes an unbiased approach to this using the Invader RNA assay (Third Wave Technologies, Inc.) to quantify the decay of each of the three exons of human β-globin mRNA without added instability elements or with the AREs from c-fos or granulocyte-macrophage colony-stimulating factor (GM-CSF) mRNA in the 3′ untranslated region. Each of these genes under tetracycline operator control was stably transfected into cells, and β-globin mRNA was quantified with exon-specific probes following transcription termination. There was little overall evidence for polarity in stable mRNA decay. Adding the c-fos ARE activated rapid and simultaneous decay from both ends of the mRNA. In contrast, the GM-CSF ARE activated decay primarily from the mRNA 5′ end. These data were supported by reciprocal RNA interference knockdowns, and we present evidence that the 5′-3′ and 3′-5′ decay pathways are functionally linked.
mRNA decay is one of the mechanisms controlling gene expression in both prokaryotes and eukaryotes, and change in the rate of mRNA decay is one factor that determines the overall expression of the protein product (reviewed in reference 25). The tractability of Saccharomyces cerevisiae for both genetic and biochemical investigation has provided a detailed picture of the molecular mechanisms of mRNA decay in this simple eukaryote. In yeast, the decay of both stable (e.g., phosphoglycerate kinase 1) and unstable (e.g., MFA2) mRNAs begins with poly(A) shortening (14), followed by removal of the 5′ cap and 5′-3′ degradation of the mRNA body (40), with decapping catalyzed by Dcp2p and 5′-3′ decay catalyzed by Xrn1p (11). The process of yeast mRNA decay is associated with a change in the composition of the substrate mRNP that includes loss of translation factors (eIF4E, Pab1p, and eIF4G) and association with the complex of Lsm1p-Lsm7p (50). The discovery that these proteins together with Dcp1p, Dcp2p, and Xrn1p are concentrated in discrete cytoplasmic foci (termed processing bodies, or P bodies) together with mRNA undergoing degradation created a paradigm shift in our understanding of the process of mRNA decay (47). More recent work further simplified this picture, essentially dividing the cytoplasmic mRNP pool into two categories: mRNPs that are actively engaged with translating ribosomes and those that are stored or degraded in P bodies (8, 12). P bodies are also present in mammalian cells (2, 5, 18, 19, 29, 31), and the growing list of component proteins (reviewed in reference 17) includes orthologs of the yeast P-body components plus eIF4E, eIF4E-T, the scaffold protein GW182, Hedls (39) (also called Ge-1 [56]), RNA-associated protein 55 (55), and components of the RNA-induced silencing complex together with mRNA targeted for degradation by microRNAs (37, 46).
The best-characterized mammalian mRNA instability elements are the AU-rich elements, or AREs. These can be divided into several classes depending on the number and positioning of AUUUA repeats and their impact on poly(A) shortening (reviewed in reference 4). AREs are targets for a number of RNA binding proteins, some of which exert a stabilizing effect (e.g., HuR [9]), some of which exert a destabilizing effect (e.g., tristetraprolin [TTP] and butyrate response factor 1 (BRF-1 [32, 48]) and at least one (Auf1 or hnRNP D) that can be stabilizing or destabilizing depending on the binding of different alternatively spliced isoforms (15, 54). In addition to progress made in identifying and characterizing proteins that bind to AREs (and other mRNA instability elements), we are beginning to understand the actual processes by which these mRNAs are degraded. Identification of 5′-3′ decay of unstable mRNAs in yeast was facilitated by the ability of a poly(G) tract to block processive exonucleolytic degradation by Xrn1p (40). Similar approaches in mammalian cells have met with limited success; however, decapped and deadenylated mRNAs have been identified (13), suggesting that the same 5′-3′ decay process also occurs in mammalian cells.
A number of studies used in vitro systems to study ARE-mediated mRNA decay, and in these the ARE accelerates poly(A) shortening and decay of the mRNA body (20-23). Several of these reports also show that AREs and/or associated ARE-binding proteins recruit the exosome to degrade these unstable mRNAs with 3′-5′ polarity (10, 26, 42), and one report described a decapping activity similar to that seen in yeast Dcp2 which is activated by an ARE (24). P bodies have also been identified as a major site for degradation of ARE-containing mRNA (17). As noted above, TTP and butyrate response factor 1 binding to the ARE activate decay, and Lykke-Andersen and Wagner (39) identified domains of these proteins responsible for this activation. In addition, they showed that increased expression of Dcp2 enhances the decay of mRNA carrying the granulocyte-macrophage colony-stimulating factor (GM-CSF) ARE. Ferraiuolo et al. (19) identified eIF4E-T, the binding partner of eIF4E, in P bodies, and showed both that eIF4E-T is essential for P-body integrity and that its knockdown by RNA interference (RNAi) stabilizes ARE-containing mRNAs. Using an RNAi screen for mRNA decay enzymes, Stoecklin et al. (49) showed that β-globin mRNA carrying the GM-CSF ARE is stabilized following knockdown of two key components of P bodies, Xrn1 and Lsm1. Comparable data supporting 5′-3′ decay for unstable mRNAs was obtained by introducing mRNAs with a number of novel cap analogs into cells (28).
In the course of our work on β-globin mRNA decay in erythroid cells, we sought a method to measure the decay rates of the 5′, middle, and 3′ portions of this mRNA that does not require primer binding to downstream sequences. This technique also had to be sufficiently sensitive to detect mRNA decay products without overexpressing the reporter mRNA. These criteria were met by the Invader RNA assay (Third Wave Technologies, Inc.) (16). The Invader RNA assay is a quantitative, isothermal assay in which a fluorescent signal accumulates linearly in proportion to the concentration of target RNA. Using this assay, we examined the polarity of decay of tetracycline-regulated β-globin mRNAs without the addition of any instability elements or with c-fos or GM-CSF AREs in the 3′ untranslated region (3′-UTR). A similar decay pattern for each exon was seen for stable β-globin mRNA, with little evidence for polarity in its decay. Both AREs activated mRNA decay but with distinctly different polarities. The c-fos ARE activated simultaneous decay from both ends of the mRNA, whereas the GM-CSF ARE activated decay primarily from the 5′ end. Unexpectedly, we observed that knockdown of Dcp2 reduces decay from the 3′ end of mRNA carrying the c-fos ARE, and knockdown of PM/Scl-100 or Rrp41 reduces decay from the 5′ end, indicating that the 5′ and 3′ decay pathways are functionally linked.
MATERIALS AND METHODS
Plasmid constructions and cell culture.
Tetracycline-regulated plasmids expressing the human β-globin gene without added instability elements (pTet-CMVmin-glo-SPA) or with the c-fos (pTet-CMVmin-glo-ARE[cfos]-SPA) or GM-CSF (pTet-CMVmin-glo-ARE[GMCSF]-SPA) A+U-rich instability elements in the 3′-UTR were described previously (19). For simplicity, the mRNAs expressed from these genes are referred to throughout this paper as control, c-fos, and GM-CSF mRNAs, respectively. LM(tk- tTA) cells are a line of murine fibroblasts that are stably transfected with a plasmid expressing the tetracycline repressor protein. Mixed cultures of stably transfected cells were created by cotransfecting each plasmid with pcDNA3 using Dreamfect (OZ Biosciences) followed by the addition of 500 μg/ml of G418 24 h later. Cells were maintained in G418 until confluent and grown in G418-containing medium each time they were passaged.
RNAi knockdown.
Small interfering RNA (siRNA) transfections were performed using Dreamfect transfection reagent (OZ Biosciences) according to the manufacturer's directions. The luciferase control GL2 siRNA and Smart Pool Dcp2 and Rrp41 siRNAs were purchased from Dharmacon. The siRNA to PM/Scl-100 consisted of 5′-GUUUCGAGAGAAGAUCGACAAdTdT (sense) and 5′-UUGUCGAUCUUCUCUCGAAACdTdT (antisense) and was shown previously to be effective in knocking down PM/Scl-100 and inactivating exosome activity (52). Cells were transfected with 100 nM of Dcp2, PM/Scl-100, or GL2 siRNAs or 50 nM Rrp41 siRNA 48 and 24 h before tetracycline was added to turn off reporter gene transcription (time zero). To determine the effectiveness of RNAi knockdown, Western blotting was performed on protein recovered at that time point using antibodies to Dcp2, PM/Scl-100 and Rrp41. These data are presented in Fig. 2C, 3C, and 4E.
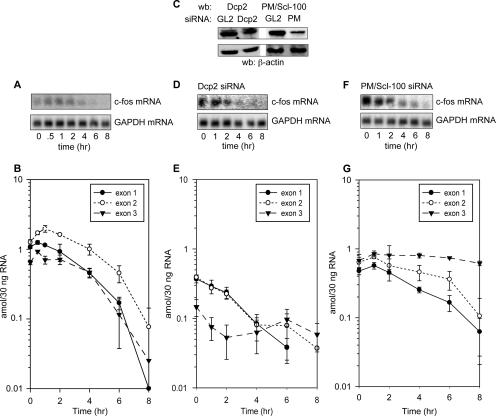
FIG. 2.
Analysis of stable mRNA decay by Northern blot and Invader RNA assay. (A) Log-phase cells expressing a tetracycline-regulated β-globin gene received tetracycline at time zero to turn off reporter gene transcription. RNA isolated at the indicated times over the next 8 h was assayed by Northern blotting for human β-globin mRNA and murine GAPDH mRNA. (B) The RNAs in panel A were analyzed by Invader assay using probe sets specific to exon 1, exon 2, and exon 3. Each sample was normalized to 30 ng of input RNA and quantified against a standard curve of in vitro-transcribed full-length β-globin mRNA. (C) The cells used in the preceding experiments were transfected 24 and 48 h prior to harvest with siRNAs to luciferase (GL2), Dcp2, or PM/Scl-100. Total protein isolated 24 h after the second round of transfection (time zero) was analyzed by Western blotting (wb) with antibodies to Dcp2, PM/Scl-100, or β-actin. (D) Tetracycline was added to cells 24 h after the second transfection with Dcp2 siRNA (time zero), and RNA isolated at the indicated times was analyzed by Northern blotting. (E) The RNA examined in panel D was analyzed by Invader assay as described above for panel B. (F) Tetracycline was added to cells 24 h after the second transfection with PM/Scl-100 siRNA (time zero), and RNA isolated at the indicated times was analyzed by Northern blotting. (G) The RNA examined in panel F was analyzed by Invader assay as described above for panel B. The points in each graph represent the means ± standard deviations (error bars) for triplicate determinations. To avoid bias introduced by curve-fitting programs, the actual decay lines are plotted.
FIG. 3.
Impact of the c-fos ARE on the polarity of mRNA decay. (A) Log-phase cells expressing a tetracycline-regulated β-globin gene with the c-fos ARE in the 3′-UTR (shown in Fig. 1) received tetracycline at time zero to turn off reporter gene transcription. RNA isolated at the indicated times over the next 8 h was assayed by Northern blotting for human β-globin mRNA and murine GAPDH mRNA. (B) The RNAs in panel A were analyzed by Invader assay as described in the legend to Fig. 2B using probe sets specific to each exon. (C) The cells used in the preceding experiments were transfected as described in the legend to Fig. 2C with GL2, Dcp2, or PM/Scl-100 siRNA. Total protein isolated 24 h after the second round of transfection (time zero) was analyzed by Western blotting (wb) with antibodies to Dcp2, PM/Scl-100, or β-actin. (D) Tetracycline was added to cells 24 h after the second transfection with Dcp2 siRNA (time zero), and RNA isolated at the indicated times was analyzed by Northern blotting. (E) The RNA examined in panel D was analyzed by Invader assay as described above for panel B. (F) Tetracycline was added to cells 24 h after the second transfection with PM/Scl-100 siRNA (time zero), and RNA isolated at the indicated times was analyzed by Northern blotting. (G) The RNA examined in panel F was analyzed by Invader assay as described above for panel B. The points in each graph represent the means ± standard deviations (error bars) for triplicate determinations.
FIG. 4.
Impact of the GM-CSF ARE on the polarity of mRNA decay. (A) Log-phase cells expressing a tetracycline-regulated β-globin gene with the GM-CSF ARE in the 3′-UTR received tetracycline at time zero to turn off reporter gene transcription. RNA isolated at the indicated times over the next 8 h was assayed by Northern blotting for human β-globin mRNA and murine GAPDH mRNA. (B) The RNAs in panel A were analyzed by Invader assay as described in the legend to Fig. 2B using probe sets specific to each exon. (C) Cells expressing β-globin mRNA with the GM-CSF ARE were transfected twice at 24-h intervals as described in the legend for Fig. 2C with Rrp41 siRNA. The corresponding Western blot for protein recovered 24 h after the second transfection is shown in panel E. Tetracycline was added 24 h after the second transfection with Rrp41 siRNA (time zero), and RNA isolated at the indicated times was analyzed by Northern blotting. (D) The RNA examined in panel C was analyzed by Invader assay as described in the legend to Fig. 2B. (F) Cells expressing β-globin mRNA with the GM-CSF ARE were transfected twice at 24-h intervals as described above with luciferase siRNA (GL2). The corresponding Western blot for protein recovered 24 h after the second transfection is shown in panel E. Tetracycline was added to these cells 24 h after the second transfection with GL2 siRNA (time zero), and RNA isolated at the indicated times was analyzed by Invader assay. The points in each graph represent the means ± standard deviations (error bars) for triplicate determinations.
RNA isolation and Northern blotting.
Adherent cells were washed twice with phosphate-buffered saline and harvested by scraping and centrifugation at 1,000 × gmax (maximum g)for 1 min at 4°C. The cell pellet from each 60-mm dish was resuspended in 200 μl of cytoplasmic extraction buffer (10 mM Tris-HCl, pH 8.0, 0.14 M NaCl, 1.5 mM MgCl2, 0.025% [vol/vol] NP-40, 10 mM dithiothreitol, 30 units RNase OUT/100 μl) and incubated on ice for 10 min. Nuclei were removed from the lysed cells by centrifuging at 12,000 × gmax for 5.5 min at 4°C in a refrigerated microcentrifuge. RNA was recovered from the remaining cytoplasmic fraction using TRIzol reagent (Invitrogen) or using the Absolutely RNA reverse transcription-PCR miniprep kit (Stratagene). Northern blotting was performed as described previously (7). DNA probes for both β-globin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were synthesized using the Random Primers DNA labeling system (Invitrogen). The template for synthesis of the β-globin probe was a 322-bp EcoRI fragment spanning a portion of exon 1, all of exon 2, and a portion of exon 3. The template for GAPDH probe synthesis was a 517-bp PCR product from CHOAA8 cDNA using primers CHOGAPDH5 (5′-AACTTTGGCATTGTGGAAGGAC) and CHOGAPDH3 (5′-TTCTTACTCCTTGGAGGCCATG).
Western blotting.
Cells were harvested by scraping with phosphate-buffered saline and centrifugation at 1,000 × gmax, and the resulting pellets were resuspended in MPER mammalian protein extraction reagent (Pierce) plus protease inhibitor cocktail (Sigma) for isolation of total cell protein. Debris was removed by centrifugation, and protein concentration was measured using the bicinchoninic acid assay (Pierce). Fifteen to 25 μg protein was separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane. Membranes were probed with rabbit antibodies to PM/Scl-100 and Rrp41 (provided by G. Pruijn), Dcp2 (provided by M. Kiledjian), or β-actin or GAPDH (Santa Cruz Biotechnology). Proteins were visualized using Super Signal West Femto or Pico maximum sensitivity substrate (Pierce).
Invader RNA assay.
The Invader RNA assay essentially followed the protocol described previously by Eis et al. (16), and detailed protocols describing the application of the probes developed here to study mRNA decay are described elsewhere (43). The sequences of the individual component oligonucleotides for each Invader probe set are listed in Table 1. Primary reaction mixtures were composed of 4 μl primary reaction buffer (supplied with the reagent kit), 0.25 μl β-globin primary oligonucleotides (40 μM probe oligonucleotide, 20 μM Invader oligonucleotide, 12 μM stacking oligonucleotide in Te buffer [10 mM Tris, 0.1 mM EDTA; Technologies, Inc.]), 0.25 μl of 1:2 diluted GAPDH primary oligonucleotides, 0.5 μl Cleavase enzyme (Third Wave Technologies, Inc.), and 5 μl sample RNA (30 to 35 ng cytoplasmic RNA in 20 ng/ml tRNA in H2O). Reaction mixtures were overlaid with 10 μl Chill-Out 14 liquid wax (MJ Research) and incubated in a hybridization oven at 60°C for 90 min at which time 5 μl secondary reaction mix was added. The secondary reaction mix contained 2 μl secondary reaction buffer, 0.75 μl 6-carboxyfluorescein (FAM) oligonucleotides containing 2 μM secondary reaction template and 13.4 μM FAM-labeled fluorescent oligonucleotide, 0.75 μl β-globin secondary oligonucleotides (53.4 μM arrestor oligonucleotides in Te buffer; Technologies, Inc.), 0.75 μl GAPDH secondary oligonucleotides (including a Redmond Red-labeled probe oligonucleotide), and 0.75 μl TE buffer. Primary and secondary reaction buffers, Cleavase enzyme, primary and secondary GAPDH oligonucleotides, and GAPDH in vitro transcript were purchased from Third Wave Molecular Therapeutics. β-Globin primary and secondary oligonucleotides were originally obtained from Third Wave Technologies, Inc. In later experiments, the primary β-globin probe oligonucleotides were obtained from IDT, and the secondary probe oligonucleotides were obtained from Eurogentec. In experiments where the secondary probe oligonucleotides and were obtained from Eurogentec, the amount of FAM-labeled oligonucleotides used in the second reaction was increased to 1.5 μl and no additional Te buffer (Technologies, Inc.) was added. Reaction mixtures were incubated at 60°C and read at 60 min or 90 min. All reactions were performed in triplicate in a Microseal 96 v-bottom polypropylene microplate (MJ Research), and each experiment was repeated at least twice. The amount of β-globin mRNA detected with each probe set was quantified against a standard curve of a gel-purified in vitro transcript of full-length human β-globin mRNA including a 14-nucleotide (nt) poly(A) tail (see below) and a full-length GAPDH transcript that was obtained from Third Wave Technologies, Inc. FAM fluorescence was read at 485 nm (excitation) and 535 nm (emission); Redman Red fluorescence was read at 560 nm (excitation) and 612 nm (emission) using a Tecan GENios fluorescence plate reader, and data reduction was performed using the associated Magellan software package.
TABLE 1.
Oligonucleotides used in the Invader RNA assay
| Probe set | Oligonucleotidea | Sequence (5′ to 3′)b |
|---|---|---|
| Exon 1 | Probe | aacgaggcgcacCTTCATCCAC-NH2 |
| Invader oligonucleotide | CAGGGCCTCACCACCAAA | |
| Stacking oligonucleotide | GUUCACCUUGCC | |
| Arrestor oligonucleotide | GUGGAUGAAGGUGCGC | |
| FRET oligonucleotide | (6-FAM)-CAC-(EQ)-TGCTTCGTGG | |
| SRT | CCAGGAAGCAAGTGGTGCGCCTCGUUAA | |
| Exon 2 | Probe | aacgaggcgcacCTAAAGGCACC-NH2 |
| Invader oligonucleotide | GGTGAGCCAGGCCATCAA | |
| Stacking oligonucleotide | GAGCACUUUCUUGC | |
| Arrestor oligonucleotide | GGUGCCUUUAGGUGCGC | |
| FRET oligonucleotide | (6-FAM)-CAC-(EQ)-TGCTTCGTGG | |
| SRT | CCAGGAAGCAAGTGGTGCGCCTCGUUAA | |
| Exon 3 | Probe | aacgaggcgcacACCAGCACG-NH2 |
| Invader oligonucleotide | GTGATGGGCCAGCACACAGC | |
| Stacking oligonucleotide | UUGCCCAGGAG | |
| Arrestor oligonucleotide | CGUGCUGGUGUGCGC | |
| FRET oligonucleotide | (6-FAM)-CAC-(EQ)-TGCTTCGTGG | |
| SRT | CCAGGAAGCAAGTGGTGCGCCTCGUUAA |
FRET, fluorescence resonance energy transfer; SRT, secondary reaction template.
Lowercase bases indicate 5′ flap sequences. The cleavage base is indicated by boldface type. Underlined bases indicate 2′ O-methylated nucleotides. A 3′ NH2 was included to enhance assay performance. EQ, Eclipse quencher (Epoch Biosciences).
To determine that signals from RNA samples fell within the linear range of the assay, standard curves were constructed for each Invader set every time the Invader RNA assay was performed. The net signal, mean, and standard deviation were calculated for each set of triplicate in vitro β-globin transcript standard samples. To determine which concentrations of β-globin transcript fell within the linear range of the assay, t tests were performed for each point on the standard curve and zero and for each point on the standard curve and the next lowest value and the signal amplification change greater than zero was calculated. Only those data points with P of <0.05 by t test for both tests and changes (n-fold) of >0 greater than 1.15 were used to generate the standard curve which was plotted as relative light units (RLUs) versus attomole of in vitro transcript on a linear graph. Each standard curve was fitted to a linear equation (y = mx + b where y is RLU, x is attomoles of target RNA, m is the slope, and b is the y intercept) and the R2 value was calculated. 95% confidence values were calculated for each standard set of triplicate samples to determine the precision of the assay.
Each time course of β-globin mRNA decay generated following transcription turnoff was analyzed in triplicate. For each data point, the net RLU was calculated and the attomole of target per sample was determined by inserting the RLU value into the equation x = (y − b)/x from the corresponding standard curve and then normalized to 30 ng of RNA per well. For each set of triplicate samples, the mean and standard deviation was calculated, and results were plotted as attomole/30-ng sample of input cytoplasmic RNA versus time on a semilogarithmic graph.
Preparation of β-globin transcript used for standard curves.
Plasmid pG-glo-PLEB-cDNA full-length-pA14 was linearized by digestion with SstII and transcribed with SP6 RNA polymerase using the Megascript kit (Ambion) according to the manufacturer's instructions. Resulting transcripts were treated with DNase I, extracted with phenol-chloroform/isoamyl alcohol, and isopropanol precipitated. Each preparation was gel purified and quantified spectrophotometrically before use.
RESULTS
Stable β-globin mRNA decays with little evidence of polarity.
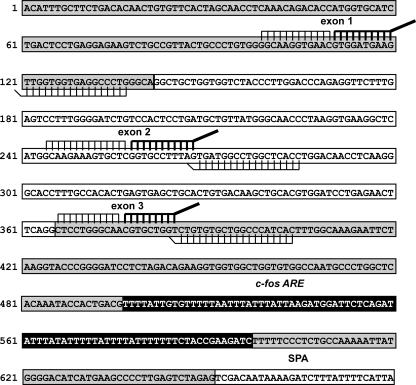
The Invader RNA assay is well suited to study the polarity of mRNA decay because the signal generated from each probe set is independent of its location on the mRNA. Before use, each Invader set must be tested for linearity, sensitivity, limit of detection, and background. Figure 1 shows the sequence of human β-globin mRNA with the c-fos ARE in the 3′-UTR that was used in a previous study characterizing the role of eIF4E-T in the structural integrity of P bodies and ARE-mediated mRNA decay (19). Ten Invader probe sets that spanned this mRNA up to the ARE were prepared, and the three indicated on the figure met these criteria. Set 2 is located in the middle of exon 1, set 5 is located in the middle of exon 2, and set 8 is located toward the 5′ end of exon 3. For simplicity throughout the text, these will be referred to as the exon 1, 2, and 3 probes. Each Invader set has three components: a stacking oligonucleotide, a probe oligonucleotide, and an Invader oligonucleotide, the sum of which span a region of ∼40 bases. The sequences of each probe set are presented in Table 1. The probe oligonucleotide has a sequence that hybridizes to the target RNA followed by a generic “flap” sequence. A stacker oligonucleotide binds immediately upstream of the probe to increase the melting temperature (Tm) of the probe, and the Invader oligonucleotide (shown beneath each sequence) hybridizes downstream of the probe with a 1-nt overlap at the junction of the “flap” sequence. This 1-nt overlap distorts the structure of the hybridized probe oligonucleotide, making it susceptible to cleavage by a modified form of Tth polymerase (16) termed Cleavase. The reaction is run at the Tm of the probe oligonucleotide so that flaps are generated in direct relationship to the concentration of the target mRNA. The flap serves as the Invader oligonucleotide in a second reaction that uses a common template and a probe bearing a quencher and fluorophores. Cleavage of the probe at the 1-nt overhang of this complex generates a fluorescent signal that is then read. Each experiment was done at least twice, and each Invader assay was performed on triplicate samples to obtain statistically significant results.
FIG. 1.
Locations of Invader probe sets on β-globin mRNA. The sequence of human β-globin mRNA is shown with exons 1 and 3 in gray, exon 2 in white, and the c-fos ARE in black. In the constructs used here, the β-globin polyadenylation signal has been replaced by a dominant synthetic poly(A) site (SPA [35]). Each of the Invader probe sets detailed in Table 1 is shown in its corresponding position on β-globin mRNA. In each set, the stacker oligonucleotide is on the left, the probe oligonucleotide with the unpaired flap is shown in thick black lines, and the Invader oligonucleotide is shown underneath the sequence with the overlapping base drawn at an angle.
The decay of each reporter β-globin mRNA was studied using murine fibroblasts [LM(tk- tTA) cells] that express the tetracycline repressor protein. These cells were stably transfected with tetracycline-regulated plasmids expressing control β-globin mRNA with no added instability elements or β-globin mRNA with the c-fos or GM-CSF ARE in the 3′-UTR as shown in Fig. 1. Technical limitations of the Invader assay limit the amount of RNA that can be analyzed in each reaction mixture. Because of this, we could not do transcription pulse-chase and instead examined decay from steady state following the addition of tetracycline to cells in log-phase growth.
The first experiments examined the decay after transcription turnoff of β-globin mRNA lacking any added instability element (Fig. 2). When assayed by Northern blotting, this mRNA decays with a half-life of ∼8 h (Fig. 2A). However, a more complex pattern emerged when individual portions of this mRNA were quantified using the Invader assay (Fig. 2B). By definition, this mRNA is at steady state at the time tetracycline is added to the medium (time zero). At this point, the exon 2 and 3 probes detected 3 amol of mRNA in a 30-ng sample of total RNA, but the exon 1 probe reproducibly detected only 2 amol. The difference in mRNA detected with each of the probe sets was maintained throughout the time course, suggesting that the Invader assay identified a population within this slowly decaying mRNA that is decapped and in the process of 5′-3′ decay.
Next we examined the impact of knocking down Dcp2 on the decay polarity of this stable mRNA. Western blots of each RNAi knockdown are presented in Fig. 2C. Transfecting cells with Dcp2 siRNA 48 and 24 h prior to transcription turnoff reduced this protein to 10% of control. When assayed by Northern blotting, this decreased level of Dcp2 had little impact on control β-globin mRNA during the first 4 h of decay; however, by 8 h it declined by 50% (Fig. 2D). A number of interesting differences became apparent when the decay of individual portions of this stable mRNA was studied by Invader assay (Fig. 2E). The first notable difference was that the steady-state levels of exons 1, 2, and 3 were half of those seen in Fig. 2B. Muhlrad and Parker (41) showed previously that loss of Dcp2 causes the steady-state level of reporter mRNA to decrease in a manner that is not linked to decay, and our results are consistent with their findings. Knockdown of Dcp2 also eliminated the quantitative difference seen at time zero between the amount of mRNA detected with probe to exon 1 versus to exons 2 and 3. In spite of this difference, knockdown of Dcp2 had no impact on the decay rates of exons 1, 2, and 3. Together, these data support the idea that stable mRNA decays incrementally faster from the 5′ end than from the 3′ end.
PM/Scl-100 is both nuclear and cytoplasmic (27), and the inhibition of 3′-5′ decay of nonsense-containing mRNA by knockdown of this protein (34) was the first in vivo demonstration of a role for the exosome in the decay process. An siRNA that was shown previously to effectively target PM/Scl-100 (52) reduced this protein by 90% in cells expressing each of the target mRNAs (Fig. 2C). When the decay of stable β-globin mRNA in cells treated with this siRNA was assayed by Northern blotting, there was a modest decrease in half-life from 8 h to 5.5 h (Fig. 2F). As one might anticipate, results with the Invader assay showed there was little impact of PM/Scl-100 knockdown on 5′ decay (Fig. 2G). As in Fig. 2B, there was less of exon 1 (0.9 amol) at time zero than exons 2 (1.5 amol) and 3 (1.4 amol), and this quantitative difference was maintained throughout the time course. We noted above that 3′ decay does not appear to be as important as 5′ decay for this mRNA, and the minor effect of PM/Scl-100 knockdown observed here supports this observation. Overall, these results indicate that a stable mRNA decays from both ends with 5′ decay proceeding incrementally faster than 3′ decay.
The c-fos ARE simultaneously activates 5′ and 3′ decay.
The AREs are classified by the number and nature of AUUUA repeats and the sequence context where they reside (3, 4). The class I AREs, which include c-fos, are characterized by multiple separate AUUUA elements in a U-rich context. These mRNAs undergo synchronous poly(A) shortening prior to decay of the mRNA body (53). Northern blot analysis showed that the c-fos ARE reduced the half-life of β-globin mRNA from 8 h to 3 h (Fig. 3A). However, a more complex picture emerged when the decay of each exon was quantified by Invader assay (Fig. 3B). After transcription turnoff, exons 1 and 3 decayed simultaneously, each with a half-life that matches the 3-h half-life determined by Northern blotting. This was followed by decay of exon 2, indicating that the c-fos ARE simultaneously activates decay from both the 5′ and 3′ ends of the mRNA.
The roles of the decapping and 5′-3′ decay pathways were examined as in Fig. 2C and D by knockdown of Dcp2. When decay was assayed by Northern blotting, the loss of Dcp2 had no impact on c-fos mRNA decay (Fig. 3D). Again, a more complex pattern emerged when each exon was quantified by Invader assay (Fig. 3E). As seen in Fig. 2E, knockdown of Dcp2 caused a twofold decrease in the steady-state level of reporter mRNA. At time zero, there was twice as much exon 1 and 2 than exon 3, indicating that Dcp2 knockdown inhibited 5′ decay of c-fos mRNA but had little impact on ARE-stimulated 3′ decay. Exon 3 decayed linearly until 2 h, at which time it reached the limit of detection for this experiment and plateaued. This was matched by decay of exons 2 and 1, with both disappearing with almost linear kinetics for 4 to 6 h after transcription turnoff. There are two possible interpretations for the similar decay of exons 1 and 2. In one, 5′ decay is inactivated and the delayed but simultaneous loss of exons 1 and 2 is due to processive 3′-5′ decay by the exosome. In the other, 5′ decay is slowed but not inactivated and the simultaneous decay of exons 1 and 2 results from a combination of this and exosome-mediated 3′-5′ decay. While we favor the former, at this time these cannot be formally distinguished.
PM/Scl-100 was again targeted by RNAi to characterize the 3′-5′ decay of c-fos mRNA. Knockdown of PM/Scl-100 increased the reporter mRNA half-life measured by Northern blotting from 3 h to 4 h (Fig. 3F). While this might be interpreted to be a minor effect, it masked major changes in the overall decay process. The most striking change was in the decay of exon 3, whose half-life changed from 3 h in untreated cells to essentially no decay (Fig. 3G). This is identical to that seen for control mRNA in Fig. 2G following knockdown of PM/Scl-100, indicating that loss of this exosome component effectively inactivated 3′ decay. In the absence of 3′ decay, the curves generated with the exon 1 and 2 probes show the contribution of 5′ decay to the turnover of c-fos mRNA. While exon 1 still decayed more rapidly than exon 2 and both decayed faster than the corresponding exons of control mRNA in Fig. 2, the overall rate of 5′ decay was slower than that seen in untreated cells (Fig. 3B). Notably, exon 1 decayed with the same half-life as that determined by Northern blotting, confirming this as the rate-limiting step in the absence of 3′ decay. Although P bodies are thought to be the sites where 5′ decay occurs, neither the transfection process itself nor knockdown of PM/Scl-100 reduced the size or number of these foci (data not shown).
The GM-CSF ARE preferentially activates 5′-3′ decay.
The class II AREs have multiple overlapping AUUUA repeats (3, 4), and unlike class I AREs, mRNAs with these elements undergo asynchronous poly(A) shortening (53). To determine whether the bidirectional decay observed with the c-fos ARE is a general property of ARE-stimulated decay, we examined the decay polarity of β-globin mRNA carrying the class II GM-CSF ARE. The Northern blot in Fig. 4A shows that GM-CSF mRNA decays with a half-life of 1.5 h. In contrast to the results seen with the c-fos ARE, mRNA with the GM-CSF ARE decays primarily with 5′-3′ polarity (Fig. 4B). Exons 1 and 2 decay rapidly and almost simultaneously with the 1.5-h half-life seen by Northern blotting, followed by exon 3.
The stabilization of c-fos mRNA exon 3 after knockdown of PM/Scl-100 confirmed the bidirectional nature of its decay process but also indicated that 5′ decay was not sufficiently processive to completely degrade the body of this mRNA. To determine whether there is a 3′ decay component to the loss of GM-CSF exon 3, we examined the impact of knocking down Rrp41, the catalytic subunit of the exosome (38). Results in Fig. 4E show that transfecting these cells twice at 24-h intervals with Rrp41 siRNA reduced its level to <10% of control. A control (GL2) siRNA had no impact on the amount of Rrp41 or on the preferential 5′-3′ decay of this mRNA (Fig. 4F). Knockdown of Rrp41 increased the half-life of GM-CSF mRNA from 1.5 to 2 h when assayed by Northern blotting (Fig. 4C), and results with the Invader assay show that this was matched by a similar decrease in the decay of exons 1 and 2 (Fig. 4D). This result is similar to the decreased rate of 5′ decay after PM/Scl-100 knockdown in Fig. 3G. As seen with c-fos mRNA following knockdown of PM/Scl-100, knockdown of Rrp41 significantly reduced the decay of exon 3, thus confirming a lesser but nonetheless significant role for the exosome in the decay of this mRNA.
DISCUSSION
In yeast, unstable mRNAs decay with 5′-3′ polarity and the identification of focal concentrations of enzymes involved in decapping and 5′-3′ decay (P bodies) in yeast and mammalian cells led to the general conclusion that these are the primary sites of mRNA decay. Most of the support for this concept in mammalian cells came from coprecipitation experiments that identified complexes containing decay-promoting proteins with proteins found in P bodies and from RNAi knockdown experiments where loss of P-body components stabilized ARE-containing mRNAs (19, 39, 49). The recent focus on P bodies contrasts with results from in vitro experiments that identified 3′-5′ decay catalyzed by the exosome as the primary mechanism that degrades ARE-containing mRNA (10, 42, 51). Here we describe a new approach using the Invader RNA assay that allowed us to quantify the decay of individual portions of a β-globin reporter mRNA independent of structural elements within the mRNA [e.g., poly(G) tracts] or RNAi knockdown of proteins involved in the decay process. By comparing these results with those obtained by Northern blotting, we were able to define the relative contribution of 5′ and 3′ pathways to the overall process of mRNA decay. All of the work described here used stable cell lines that express low levels of each reporter mRNA in order to avoid overexpression artifacts sometimes seen with transient transfection. This precluded the use of transcription pulse-chase experiments, since the amount of mRNA induced in the short intervals tested was at the limit of detection for the Invader assay. Instead we focused on decay from steady state after turning off transcription by adding tetracycline to the medium, and the first results showed that there is little evidence for differences in the decay rates of any of the exons of stable β-globin mRNA (Fig. 2). A different picture emerged with the Invader assay used to study the decay of two different ARE-containing mRNAs. In both cases, adding an ARE activated mRNA decay; however, c-fos mRNA decays simultaneously from both ends (Fig. 3B), whereas GM-CSF mRNA primarily undergoes 5′-3′ decay (Fig. 4B). Interestingly, evidence for simultaneous 5′ and 3′ decay of the same mRNA was seen previously in trypanosomes (30).
The individual contributions of the 5′ and 3′ decay pathways to the patterns observed with the Invader assay was supported by results using RNAi to reduce the level of Dcp2, PM/Scl-100, or Rrp41. Deleting Dcp2 from yeast causes a generalized decrease in reporter mRNA that is independent of mRNA decay (41), and we observed a similar phenomenon following knockdown of Dcp2. In spite of this, knockdown of Dcp2 also reduced 5′ decay without altering 3′ decay (Fig. 3E). PM/Scl-100 is the mammalian ortholog of Rrp6, a protein generally considered to be part of the nuclear exosome. When we began this work, it was the only exosomal protein whose knockdown had been shown to functionally impact mRNA decay (34), so this was targeted here. Others identified PM/Scl-100 in the cytoplasm (27, 34) and our data showing its knockdown stabilizes c-fos exon 3 points to a role for this protein in mRNA decay. Rrp41 was recently identified as the catalytic subunit of the mammalian exosome (38), and its knockdown caused a similar stabilization of exon 3 of GM-CSF mRNA (Fig. 4D).
A high-throughput approach is needed to more thoroughly characterize the contributions of the different classes of these elements to activating each of the major decay pathways. Nevertheless, we can draw a number of conclusions from our results. First, each of the major decay pathways participates to some extent in the turnover of unstable mRNAs. As noted above, in vitro decay experiments supported a predominant role for the exosome (10, 26, 42), whereas RNAi knockdown and coprecipitation experiments supported a predominant role for decapping and 5′-3′ decay (39, 49). The latter studies used a β-globin reporter mRNA carrying the GM-CSF ARE similar to the GM-CSF reporter used here, and our results demonstrating 5′-3′ polarity in the decay of this mRNA nicely complement their findings. Second, the two elements examined here represent the class I (c-fos) and class II (GM-CSF) AREs and while these have different effects on poly(A) shortening (53), our results show they also define the manner by which mRNA is degraded. An important area for future study will be to determine how the various ARE-binding proteins guide the relative contribution of each of the major decay pathways. The third conclusion is that the 5′-3′ and 3′-5′ decay pathways are functionally linked. While there was no discernible change in 3′ decay after knockdown of Dcp2, the 5′ decay of both c-fos and GM-CSF mRNAs was slowed by knockdown of PM/Scl-100 or Rrp41. This linkage is supported by results of a comprehensive yeast two-hybrid screen of human proteins involved in mRNA decay showing Ski2 interacts with both Xrn1 and PM/Scl-100 (33). Additional support for this is seen in the coprecipitation of Rrp4 with Dcp1a, Dcp2, and Xrn1 by TTP and BRF-1 (39) and the inhibition of GM-CSF ARE-stimulated mRNA decay by knockdown of PM/Scl-75 (49). TTP and BRF-1 possess specific activating domains, and it is tempting to speculate that these domains coordinate the 5′-3′ and 3′-5′ decay processes observed here.
Finally, the data presented here indicate it is an oversimplification to think that mRNA decay is restricted to P bodies. Several lines of evidence point to P bodies as sites where mRNAs accumulate during microRNA-mediated silencing (1, 36, 37, 44). In addition, there is evidence from yeast (8, 12) and mammalian cells (6) that mRNAs can reversibly transit into and out of P bodies. Each of the major exonuclease decay pathways participates in the decay of ARE-containing mRNA, and since the exosome is not present in P bodies (1), it is difficult to envision how, for example, c-fos mRNA can undergo simultaneous 5′ and 3′ decay if P bodies are the sole engine of mRNA decay. While knockdown of PM/Scl-100 inactivated 3′ decay, it had had no visible impact on the number and size of P bodies (data not shown). On the basis of our results and those of others linking the exosome to decapping (45, 51), we propose that the exosome and submicroscopic complexes of enzymes involved in decapping and 5′ decay are linked by a dynamic interaction that facilitates and coordinates the decay of unstable mRNAs.
Acknowledgments
This work was supported by PHS grant R01 GM38277 to D.R.S. E.L.M. was supported in part by PHS grant T32 CA09338, and support for core facilities was provided by PHS center grant P30 CA16058 from the National Cancer Institute to the Ohio State University Comprehensive Cancer Center.
We thank Yan Chen, Yong Peng, and Yuichi Otsuka for help with some of these experiments; Tsetka Takova for help with probe design, troubleshooting, and data analysis; and Peggy Eis and David Fritz for help with design of the primary and secondary oligonucleotide probe sets. We also thank Mike Kiledjian for antibody to Dcp2, Reinout Raijmakers and Ger Pruijn for antibodies to PM/Scl-100 and Rrp41, and Jim Dahlberg and members of the Schoenberg lab for helpful discussions.
Invader and Cleavase are registered trademarks of Third Wave Technologies, Inc.
Footnotes
Published ahead of print on 12 February 2007.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrei, M. A., D. Ingelfinger, R. Heintzman, T. Achesel, R. Rivera-Pomar, and R. Lührmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakheet, T., B. R. Williams, and K. S. Khabar. 2006. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 34:D111-D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreau, C., L. Paillard, and H. B. Osborne. 2006. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashkirov, V. I., H. Scherthan, J. A. Solinger, J. M. Buerstedde, and W. D. Heyer. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136:761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111-1124. [DOI] [PubMed] [Google Scholar]
- 7.Bremer, K. A., A. Stevens, and D. R. Schoenberg. 2003. An endonuclease activity similar to Xenopus PMR1 catalyzes the degradation of normal and nonsense-containing human β-globin mRNA in erythroid cells. RNA 9:1157-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life. Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 11.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861-890. [DOI] [PubMed] [Google Scholar]
- 12.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couttet, P., M. Fromont-Racine, D. M. Steel, R. Pictet, and T. Grange. 1997. Messenger RNA deadenylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. USA 94:5628-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable messenger RNAs in yeast—evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 15.DeMaria, C. T., and G. Brewer. 1996. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271:12179-12184. [DOI] [PubMed] [Google Scholar]
- 16.Eis, P. S., M. C. Olson, T. Takova, M. L. Curtis, S. M. Olson, T. I. Vener, H. S. Ip, K. L. Vedvik, C. T. Bartholomay, H. T. Allawi, W. P. Ma, J. G. Hall, M. D. Morin, T. H. Rushmore, V. I. Lyamichev, and R. W. Kwiatkowski. 2001. An invasive cleavage assay for direct quantitation of specific RNAs. Nat. Biotechnol. 19:673-676. [DOI] [PubMed] [Google Scholar]
- 17.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 18.Eystathioy, T., A. Jakymiw, E. K. L. Chan, B. Séraphin, N. Cougot, and M. J. Fritzler. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9:1171-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraiuolo, M. A., S. Basak, J. Dostie, E. L. Murray, D. R. Schoenberg, and N. Sonenberg. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford, L. P., P. S. Bagga, and J. Wilusz. 1997. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 17:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford, L. P., J. Watson, J. D. Keene, and J. Wilusz. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford, L. P., and J. Wilusz. 1999. 3′-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′→5′ exonuclease in vitro. Nucleic Acids Res. 27:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao, M., D. T. Fritz, L. P. Ford, and J. Wilusz. 2000. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell 5:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, M., C. J. Wilusz, S. W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20:1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garneau, N. L., J. Wilusz, and C. J. Wilusz. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 26.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571-583. [DOI] [PubMed] [Google Scholar]
- 27.Graham, A. C., D. L. Kiss, and E. D. Andrulis. 2006. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell 17:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grudzien, E., M. Kalek, J. Jemielity, E. Darzynkiewicz, and R. E. Rhoads. 2006. Differential inhibition of mRNA degradation pathways by novel cap analogs. J. Biol. Chem. 281:1857-1867. [DOI] [PubMed] [Google Scholar]
- 29.Ingelfinger, D., D. J. Arndt-Jovin, R. Lührmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489-1501. [PMC free article] [PubMed] [Google Scholar]
- 30.Irmer, H., and C. Clayton. 2001. Degradation of the unstable EP1 mRNA in Trypanosoma brucei involves initial destruction of the 3′-untranslated region. Nucleic Acids Res. 29:4707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fitzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai, W. S., and P. J. Blackshear. 2001. Interactions of CCCH zinc finger proteins with mRNA. Tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 276:23144-23154. [DOI] [PubMed] [Google Scholar]
- 33.Lehner, B., and C. M. Sanderson. 2004. A protein interaction framework for human mRNA degradation. Genome Res. 14:1315-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 35.Levitt, N., D. Briggs, A. Gil, and N. J. Proudfoot. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 36.Liu, J., F. V. Rivas, J. Wohlschlegel, J. R. Yates III, R. Parker, and G. J. Hannon. 2005. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7:1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, J., M. A. Valencia-Sanchez, G. J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Q., J. C. Greimann, and C. D. Lima. 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127:1223-1237. [DOI] [PubMed] [Google Scholar]
- 39.Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast mfa2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 41.Muhlrad, D., and R. Parker. 1999. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol. Biol. Cell 10:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray, E. L., and D. R. Schoenberg. Application of the Invader RNA assay to the polarity of vertebrate mRNA decay. Methods Mol. Biol., in press. [DOI] [PMC free article] [PubMed]
- 44.Rehwinkel, J., I. Behm-Ansmant, D. Gatfeld, and E. Izaurralde. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11:1640-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodgers, N. D., Z. Wang, and M. Kiledjian. 2002. Regulated alpha-globin mRNA decay is a cytoplasmic event proceeding through 3′-to-5′ exosome-dependent decapping. RNA 8:1526-1537. [PMC free article] [PubMed] [Google Scholar]
- 46.Sen, G. L., and H. M. Blau. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633-636. [DOI] [PubMed] [Google Scholar]
- 47.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoecklin, G., T. Mayo, and P. Anderson. 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tharun, S., and R. Parker. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8:1075-1083. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z., and M. Kiledjian. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107:751-762. [DOI] [PubMed] [Google Scholar]
- 52.West, S., N. Gromak, C. J. Norbury, and N. J. Proudfoot. 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell 21:437-443. [DOI] [PubMed] [Google Scholar]
- 53.Xu, N., C. Y. Chen, and A. B. Shyu. 1997. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 17:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, N., C.-Y. A. Chen, and A.-B. Shyu. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21:6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, W. H., J. H. Yu, T. Gulick, K. D. Bloch, and D. B. Bloch. 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, J. H., W. H. Yand, T. Gulick, K. D. Bloch, and D. B. Bloch. 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11:1795-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]